- 1Department of Psychological Sciences, University of Connecticut, Storrs, CT, United States
- 2Department of Physics, University of Connecticut, Storrs, CT, United States
A model for bacteriophage infections and bacteria defense is analyzed using the concepts of synergetics. The model order parameter is determined and the corresponding amplitude equations are derived. Within this framework it is shown how the order parameter defines a multi-species building block that captures the organization of infection outbreaks and the initial defense reaction and how the order parameter amplitude determines the corresponding temporal characteristics. Two approximative models with different domains of application are derived as well. In doing so, a supplementary perspective of bacteriophage infections that provides insights beyond the classical state space perspective is provided.
1 Introduction
In general, epidemiological systems are given by complex networks of interacting populations of different species (Pastor-Satorras et al., 1998). While the isolated populations typically exhibit a relatively simple dynamics, a challenge in the field of network physiology is to understand how the interactions between the different types of species shape the overall network dynamics. In this context, an important first step is to consider mean field approximations in terms of ODE and coupled differential equation models (Pastor-Satorras et al., 1998; Granger et al., 2024), which due to their relative simplicity frequently allow for analytical solution methods. For virus infections the ODE three-species TIV model captures the basic dynamics of interacting target cells, infected cells, and virus particles (Nowak and May 2000). Likewise, in the context of bacteriophage infections, we are dealing with target bacteria, infected bacteria, and bacteriophages–the latter act as viruses. Studying bacterial infections and the role of bacteriophages is an important task (Bloch and Wegrzyn, 2024; Geng et al., 2022; Zborowsky et al., 2025) and is an indispensable step when attempting to use phages in modern medicine to cure certain diseases in humans (Li et al., 2021; Zborowsky et al., 2025). To this end, both simplified and generalized three-species models have been studied in the literature (Li et al., 2021; Weitz et al., 2005; Zborowsky et al., 2025). In particular, as part of their comprehensive study, Skanata and Kussell (2021) considered bacteria that can exist as resistant and non-resistant phenotypes with respect to a given invading phage. Increasing the concentration of resistant phenotypes is a defense mechanism against phage attack because this mechanism decreases the effective contact rate between phage and non-resistant bacteria such that under appropriate conditions the infection dies out. While the dynamics systems perspective, in general, is an indispensable tool to analyze bacteriophage infection models, little attention has been paid to utilize the more specific dynamical systems concepts of synergetics (Haken, 2004; Hutt and Haken, 2020; Uhl, 1999; Wunner and Pelster, 2016) in this regard. However, in the wake of the COVID-19 pandemic it has been shown that synergetics can supplement existing dynamic systems approaches to understand epidemiological and virus dynamic models (Frank, 2022). In this Brief Report a parsimonious four-species model for bacteriophage infection and bacteria defense will be considered that involves resistant phenotypes as in Skanata and Kussell (2021). The relative simplicity of the model will allow for an analytical approach. The aim of the study is to derive explicitly the order parameter of the model and to show how it determines the initial organization of a phage attack and the corresponding defense reaction. Moreover, the aim is to derive the amplitude equations that determine the evolution of the system along the order parameter and the remaining directions. Two approximative models for the system dynamics in this context will be derived as well. An exemplary simulation will illustrate some of the analytical results.
2 Materials and methods
As reviewed above, our starting point is the three-species model that involves susceptible
with
where
where
Taking a synergetics perspective (Haken, 2004; Frank, 2022), for the model (1–3) the order parameter and the amplitude equations were derived. To this end, using the state vector
By constructing a bi-orthogonal basis (Haken, 2004; Frank, 2022) spanned by the vectors
where
were derived for
3 Results and discussions
3.1 Amplitude equation perspective
The fixed-point analysis showed that the model (1–3) exhibits the phage-free fixed points defined by
for
The linear stability analysis at
with
The linear stability analysis of the phage-free fixed point produced the eigenvectors
with
with
with
and
As indicated in Equation 11, the variables
3.2 Implications
3.2.1 Order parameter: essential building-block and initial organization
The model exhibits maximally one positive eigenvalue. Consequently, under the case II scenario with
with
Since
Equation 15 illustrates again that the order parameter describes the essential building-block that shapes an infection outbreak in the multi-species network under consideration including the defense reaction (see component
3.2.2 Stopping mechanisms
The exponential increase along
Note that
3.2.3 Linear predictor equations
Equation 15 implies that all species initially satisfy linear regression equations of the form as shown in Equation 17
Accordingly, any species of the network can be used to predict any other network species (assuming
where
3.2.4 Limited impact of bacterial growth term
Clearly, the bacterial growth term may affect the
3.3 2D infected/infectious species dynamics and double exponential dynamics approximation
The dynamics of the infected and infectious species
where
3.4 Scaled model
Using the variable transformations
with
and
3.5 Simulation
An Euler forward simulation scheme was used to solve Equations 21 and 22. In a first simulation, see Figure 1, only the passive defense mechanism was considered with
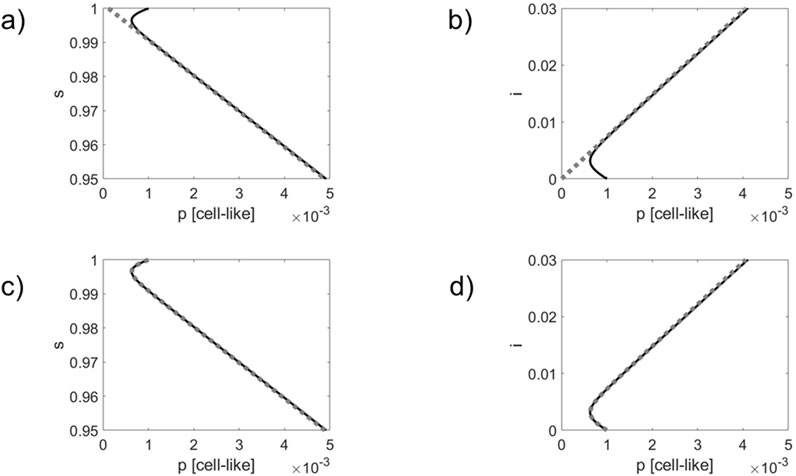
Figure 1. Comparison of simulated infection dynamics (solid black lines) with the order parameter dynamics (panels) (a,b) and the double exponential approximation (panels) (c,d) shown as gray dotted lines. Phase curves in 2D
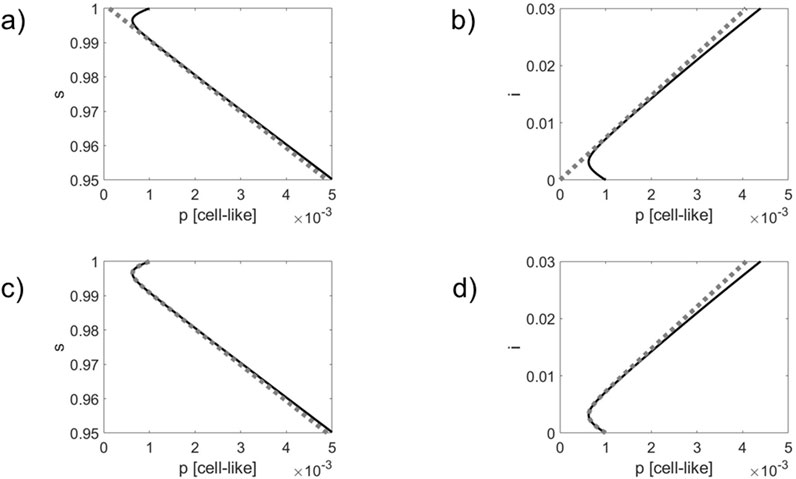
Figure 2. As for Figure 1 but for a simulation that takes the active defense mechanism via resistant bacteria into account. Comparison of simulated infection dynamics (solid black lines) with the order parameter dynamics (panels) (a,b) and the double exponential approximation (panels) (c,d) shown as gray dotted lines. Phase curves in 2D s – p state spaces (panels (a,c) and i – p state spaces (panels) (b,d) are shown.
As can be seen in panels (a) and (b) of Figures 1, 2, the phase curves quickly converge towards the order parameter
Comparing Figures 1, 2, it can be seen that due to the impact of the phage resistant bacteria the actual dynamics differs from the order parameter dynamics to a greater extent. Likewise, the actual dynamics departs earlier from the double exponential approximations. These observations do not come as a surprise since the active defense mechanism slows down and eventually stops the infection outbreak, see Equation 16. Therefore, the actual dynamics will deviate earlier from the order parameter dynamics, on the one hand, and the double exponential dynamics, on the other hand. Roughly speaking, the active mechanisms weakens the linear order parameter link between the network components.
4 Conclusions and limitations
We conclude that under appropriate conditions as specified in the Methods and Simulation sections the order parameter and its amplitude characterize the (self-)organization of a bacteriophage infection and the corresponding bacterial defense. In physics various experiments have been conducted to test specifically predictions of the theory of self-organization (and synergetics) as presented above. Therefore, just as in physics, the results presented above may serve as a basis for conducting laboratory experiments on bacteriophage infections to test the order parameter hypothesis. Moreover, we conclude that linear regression models as derived above may be used to estimate species that are difficult to observe on the basis of species that can be measured more conveniently. For sake of brevity, in the current study, properties of the endemic fixed point as studied, e.g., by Li et al. (2021) have not been examined in detail. Likewise, the current study was limited to consider one possible defense mechanism while alternative mechanisms (Skanata and Kussell, 2021; Weitz et al., 2005) were ignored. A more comprehensive study (which is beyond the scope of this Brief Report) may overcome such limitations by generalizing the results presented above.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
TF: Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedon, S. T. (2012). Bacterial immunity against bacteriophages. Bacteriophage 2, 50–54. doi:10.4161/bact.18609
Bloch, S., and Wegrzyn, A. (2024). Editorial: bacteriophage and host interactions. Front. Microbiol. 15, 1422076. doi:10.3389/fmicb.2024.1422076
Frank, T. D. (2022). COVID-19 epidemiology and virus dynamics: nonlinear physics and mathematical modeling. Berlin: Springer. doi:10.1007/978-3-030-97178-6
Geng, P., Flint, E., and Bernsmeier, C. (2022). Plasticity of monocytes and macrophages in cirrhosis of the liver. Front. Netw. Physiol. 2, 937739. doi:10.3389/fnetp.2022.937739
Granger, T., Michelitsch, T. M., Bestehorn, M., Riascos, A. P., and Collet, B. A. (2024). Stochastic compartment model with mortality and its application to epidemic spreading in complex networks. Entropy 26, 362. doi:10.3390/e26050362
Haken, H. (2004). Synergetics: introduction and advanced topics. Berlin: Springer. doi:10.1007/978-3-662-10184-1
Li, X., Huang, R., and He, M. (2021). Dynamics model analysis of bacteriophage infection of bacteria. Adv. Differ. Equations 2021, 488. doi:10.1186/s13662-021-03466-x
Nowak, M. A., and May, R. M. (2000). Viral dynamics: mathematical principles of immunology and virology. New York: Oxford University Press.
Pastor-Satorras, R., Castellano, C., Van Mieghem, P., and Vespignani, A. (1998). Epidemic processes in complex networks. Rev. Mod. Phys. 87, 925–979. doi:10.1103/RevModPhys.87.925
Skanata, A., and Kussell, E. (2021). Ecological memory preserves phage resistance mechanisms in bacteria. Nat. Commun. 12, 6817. doi:10.1038/s41467-021-26609-w
Uhl, C. (1999). Analysis of neurophysiological brain functioning. Berlin: Springer. doi:10.1007/978-3-642-60007-4
Weitz, J. S., Hartman, H., and Levin, S. A. (2005). Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 102, 9535–9540. doi:10.1073/pnas.0504062102
Wunner, G., and Pelster, A. (2016). Self-organization in complex systems: the past, present, and future of synergetics. Berlin: Springer. doi:10.1007/978-3-319-27635-9
Keywords: network physiology, bacteriophages, infection dynamics, order parameters, synergetics
Citation: Frank TD (2025) Analysis of a model for bacteriophage infections and bacteria defense: a synergetics perspective. Front. Netw. Physiol. 5:1657313. doi: 10.3389/fnetp.2025.1657313
Received: 01 July 2025; Accepted: 08 September 2025;
Published: 19 September 2025.
Edited by:
Eckehard Schöll, Technical University of Berlin, GermanyReviewed by:
Christian Uhl, Ansbach University of Applied Sciences, GermanyMarwa Ali, Purdue University, United States
Copyright © 2025 Frank. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: T. D. Frank, dGlsbC5mcmFua0B1Y29ubi5lZHU=
 T. D. Frank
T. D. Frank