- 1Department of Cardiology, The First Hospital of China Medical University, Shenyang, Liaoning Province, China
- 2Institute of Health Sciences, China Medical University, Shenyang, Liaoning Province, China
- 3Department of Pharmaceutical Toxicology, School of Pharmacy, China Medical University, Shenyang, China
Aging is a process in which organisms or cells undergo a decline in their functions. Epigenetic modification changes have been recognized as a senescence hallmark in both natural aging and stimulation-induced senescence. An acetylation modification is a dynamic process, which plays a crucial role in the senescence process through DNA stability, metabolism, and signaling pathways. We summarized the role and regulatory pathways of acetylation modifications in senescence. Various cell fate-determining proteins regulate multiple cellular processes through acetylation modifications. These processes interact and coordinate with each other, forming an integrated regulatory network framework that collectively drives cellular senescence via multiple systemic mechanisms. Based on these findings, we proposed the “acetylation-network regulation-cellular senescence” model, to elaborate how acetylation contributes to senescence. We believe this insight could provide new directions and intervention strategies for senescence and aging-related diseases.
1 Introduction
Aging is a complex process involving the decline of biological and metabolic activity, leading to the occurrence of various aging-related diseases, such as neurodegenerative diseases, cardiovascular diseases, metabolic diseases, immune system diseases, and cancer (Guo et al., 2023a). Over the past decade, our research has focused on age-related impairments in the cardiovascular system. We have observed that exogenous damaging stimuli frequently induces varying degrees of post-transcriptional modifications (PTMs) in functional and damage-associated molecules. These molecular alterations ultimately determine cellular fate and drive pathological changes in cardiac function. Our investigations have revealed that lactylation modification of α-myosin heavy chains (α-MHC) played a crucial role in maintaining myocardial fiber contractility. In angiotensin II (Ang II)-induced injury models, we observed significant reduction in α-MHC lactylation levels leading to heart failure pathogenesis (Zhang N. et al., 2023). Notably, acetylation modification of Septin4, a key apoptotic regulator, enhanced its pro-apoptotic capacity and exacerbated hypertensive nephropathy, whereas Sirt2-mediated deacetylation effectively ameliorated this pathological progression (Zhang et al., 2023b). Furthermore, during myocardial remodeling processes, ubiquitination-mediated degradation of poly (ADP-ribose) polymerase 1 (PARP-1) was found to attenuate apoptotic damage, thereby alleviating pathological cardiac hypertrophy and heart failure development (Zhang et al., 2020). These collective findings demonstrate that PTMs of critical cell fate regulators are actively involved in cellular decision-making processes under pathological stress conditions, ultimately determining tissue-specific pathological outcomes.
Cellular senescence, a state of irreversible cell cycle arrest, is a hallmark of aging and a key driver of age-related pathologies. Senescent cells accumulate with age due to persistent DNA damage, oxidative stress, or telomere shortening. Senescent cells contribute to the development of age-related diseases through the senescence-associated secretory phenotype (SASP) (Zhang L. et al., 2022). The SASP creates a toxic microenvironment that disrupts tissue homeostasis through multiple mechanisms. For instance, SASP factors like interleukin-6 (IL-6), matrix metalloproteinases (MMPs), and transforming growth factor-beta (TGF-β) promote chronic inflammation, extracellular matrix degradation, and fibroblast activation, contributing to conditions such as osteoarthritis, pulmonary fibrosis, and atherosclerosis (Li X. et al., 2023; Victorelli et al., 2023; Kandhaya-Pillai et al., 2022; Li et al., 2024a). And the acetylation plays critical roles in SASP-mediated senescence process (Li et al., 2024a). For example, the acetate-dependent acetyl-CoA synthetase 2 (ACSS2) drives the SASP and promotes cellular senescence by modulating the acetylation of PAICS (a key enzyme in purine biosynthesis) (Yang et al., 2025). The work by Giorgio et al. also revealed that the competitive interplay between acetyltransferases and deacetylases modulates the acetylation levels of histone (H3K27ac), thereby influencing cellular senescence processes (Di Giorgio et al., 2021). The loss of epigenetic information is recognized as a hallmark of cellular senescence and various age-related diseases, and thus we focus on the role and potential mechanism of acetylation modifications in senescence (Yang et al., 2023).
Increasing evidence confirms that PTM alterations are widespread in aging and related diseases. PTMs could bring unique changes that can lead to the generation of new phenotypes beyond those encoded by DNA sequences (Sen et al., 2016; Schroeder et al., 2013; Rando and Chang, 2012). Acetylation modification is fundamentally characterized by the covalent conjugation of acetyl groups (O=C-CH3), derived from acetyl-CoA as the biochemical donor, to either ε-amino groups of lysine residues or N-terminal α-amino groups, throughout the peptide chain. Mechanistically, this process is dynamically regulated by the opposing enzymatic activities of histone acetyltransferases and deacetylases (HATs/HDACs), which collectively maintain acetylation homeostasis through precise substrate recognition and catalytic modulation. Emerging research has fundamentally reshaped our understanding of acetylation enzymes. Contemporary studies have revealed that HATs/HDACs exhibit extensive substrate promiscuity, targeting not only histones but also non-histone cytoplasmic and nuclear proteins. Notably, these enzymes demonstrate specific affinity for pivotal cell fate regulators including p53, PARP-1, and signal transducer and activator of transcription 3 (STAT3) (Xia et al., 2022; Dai and Gu, 2010; Zhang N. et al., 2021). This paradigm shift has driven systematic reclassification of acetylation-related enzymes, with current nomenclature emphasizing their catalytic mechanism rather than substrate specificity. Given the exclusive modification of lysine residues, these enzymes are now more precisely designated as lysine acetyltransferases (KATs) and lysine deacetylases (KDACs), reflecting their biochemical specificity and functional diversity. Figure 1 summarized and clustered various enzymes which involved in the acetylation and deacetylation process (Semer et al., 2019; Agalioti et al., 2002; Ghosh et al., 2018; Fournier et al., 2016; Steffan et al., 2001; Lin et al., 2013; Murakami et al., 2005; Le Dour et al., 2022; Cazzalini et al., 2014; Masumi et al., 2006; Matsuzaki et al., 2005; Song et al., 2017; Brower-Toland et al., 2005; Kikuchi et al., 2023; Zhang L. et al., 2023; Jiang et al., 2011; Giaimo et al., 2018; Sun et al., 2009; Lin et al., 2012; Mishima et al., 2011; Rokudai et al., 2013; Ali et al., 2012; Yang et al., 2022; Izumikawa et al., 2019; Valerio et al., 2017; Liu et al., 2013; Kumar et al., 2012; Yang et al., 2011; Spencer et al., 1997; Zhang HL. et al., 2021; Saavedra et al., 2017; Chae and Kim, 2014; Donnarumma et al., 2022; Min et al., 2021; Wang et al., 2023; Wang et al., 2008; Chang et al., 2011; Aghdassi et al., 2012; Gao et al., 2020; Zhang et al., 2015; Chen et al., 2009; Lin and Wu, 2020; Gaddelapati et al., 2022; Grégoire et al., 2007; Meng et al., 2016; Deardorff et al., 2012; Qin et al., 2024; Luo et al., 2019; Han et al., 2022; Wang et al., 2021; Zhu et al., 2011; Ye et al., 2025; Liu et al., 2024; Sanguigno et al., 2023; Tao et al., 2007; Zhang et al., 2023d; Song et al., 2024; Zheng et al., 2015; Li et al., 2024b; Fan et al., 2020; Lee et al., 2015; Liu et al., 2025; Li et al., 2018; Radhakrishnan et al., 2015; Yamagata and Kitabayashi, 2009; Vaquero et al., 2004; Xie et al., 2024; Menssen et al., 2012; Chen et al., 2022; Wu et al., 2022; Xu et al., 2016; Jing et al., 2007; Seo et al., 2015; Cai et al., 2023; North et al., 2003; Xu et al., 2025; Papa et al., 2014; Xian et al., 2025; Novgorodov et al., 2016; Wang et al., 2022; Li C. et al., 2023; Zhang F. et al., 2022; Nakagawa et al., 2009; Shi et al., 2019; Guo Z. et al., 2023; Geng et al., 2020; Zhang et al., 2014; Barber et al., 2012; Blank et al., 2017; Mo et al., 2017; Vakhrusheva et al., 2008; Ryu et al., 2014; Liu et al., 2009; Núñez-Álvarez and Suelves, 2022). Current classification frameworks delineate that these enzymatic families are systematically partitioned based on their characteristic catalytic and structural domains, with distinct KAT and KDAC subgroups evolving to regulate spatially distinct molecular targets. Remarkably, the scope of acetylation substrates has expanded beyond chromatin-associated proteins to encompass pivotal molecular effectors governing cellular fate determination, including but not limited to metabolic enzymes, transcription factors, DNA repair machinery components, and inflammatory mediators (Núñez-Álvarez and Suelves, 2022; Hishikawa et al., 2019; Ma et al., 2019). This substrate diversification underscores the central regulatory role of lysine acetylation in integrating diverse cellular signaling pathways, with profound implications for developing precision therapies targeting acetylation-dependent pathological mechanisms.
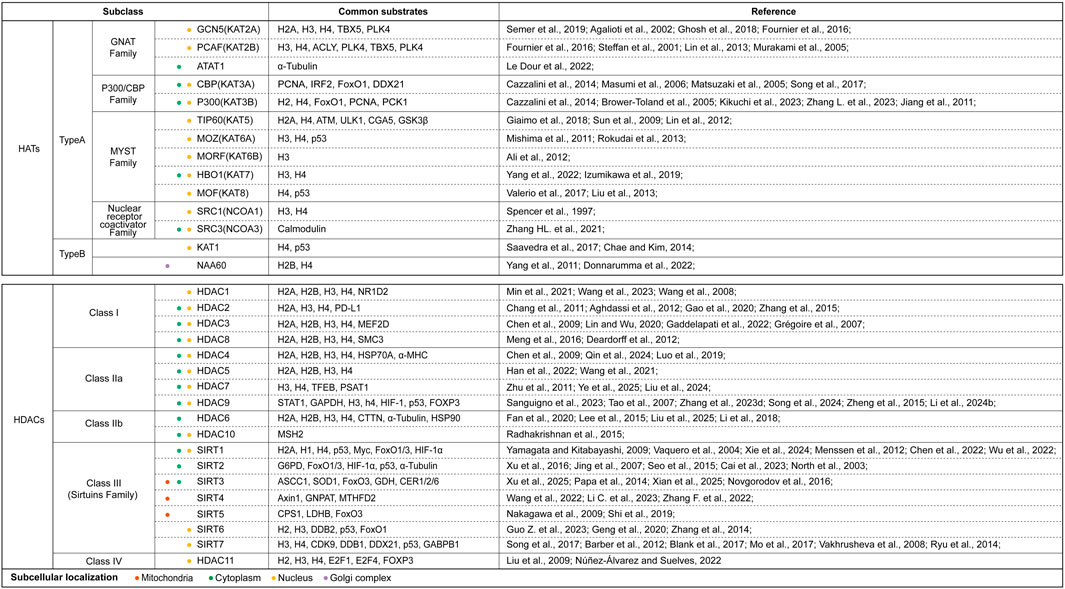
Figure 1. The summary of acetyltransferase and deacetylases. This figure systematically categorized and clustered various HATs and HDACs based on their structural families, subclasses. We also listed their subcellular localization (use different colors to represent different positioning) and main substrates which were reported in current studies. Abbreviations: ACLY, ATP-citrate lyase; ACSS1, acetyl coenzyme A synthetase 1; ATAT1, alpha-tubulin acetyltransferase 1; ATM, ataxia-telangiectasia mutated kinase; α-MHC, α-Myosin Heavy Chain; Axin1, Axis Inhibition Protein 1; CDK9, Cyclin-dependent kinase 9; CER1/2/6, eceriferum 1/2/6; CPS1, carbamoyl phosphate synthetase 1; CTTN, cortactin; CYCS, cytochrome c; DDB1/2, DNA damage-binding protein 1/2; DDX21, DEAD-Box RNA Helicase 21; E2F1/4, E2F Transcription factor 1/4; FoxO1/3, Forkhead box-O-1/3; FOXP3, Forkhead box P3; G6PD, glucose-6-phosphate dehydrogenase; GABPB1, GA-binding protein subunit beta-1; GCN5, general control non-depressible 5; GDH, glutamate dehydrogenase; GNAT, Gcn5-related acetyltransferase family; GNPAT, glyceronephosphate O-Acyltransferase; GSK3β, glycogen synthase kinase 3β; H, histone; HAT, histone acetyltransferase; HDAC, histone deacetylase; HIF-1α/2α, hypoxia inducible factor-1α/2α; HSP70A, heat shock protein 70A; IRF2, interferon regulatory factor 2; KAT, lysine acetyltransferase; LDHB, lactate dehydrogenase B; MEF2D, myocyte enhancer factor 2D; MORF, MOZ-related factor; MOZ, monocytic leukemic zinc finger; MSH2, MutS homolog 2; MTHFD2, methylenetetrahydrofolate dehydrogenase 2; NAA60, N-alpha-acetyltransferase 60; NCOA1/3, nuclear-receptor coactivator 1/3; NF-κB, nuclear factor-kappaB; NR1D2, nuclear receptor subfamily 1 group D member 2; P300/CBP, P300/CREB-binding protein; PCAF, P300/CBP-associated factor; PCNA, proliferating cell nuclear antigen; PCK1, cytosolic phosphoenolpyruvate carboxykinase 1; PLK4, polo-like kinase 4; PSAT1, phosphoserine aminotransferase 1; SIRT, Sirtuins family; SMC, structural maintenance of chromosomes; SOD1, superoxide dismutase 1; SRC1/3, steroid receptor coactivator 1/3; TBX5, T-box transcription factor 5; TFEB, transcription factor EB; TIP60, Tat-interacting protein 60; ULK1, UNC-52-like kinase 1.
2 Physiological functions of acetylation/deacetylation modifications
Acetylation and deacetylation maintain cellular homeostasis through a dynamic balance, regulating processes such as nucleic acid and protein dynamics, DNA replication/repair, protein synthesis/degradation, subcellular localization, and signaling pathways (Figure 2). Acetylation/deacetylation modification occurs throughout cellular processes.
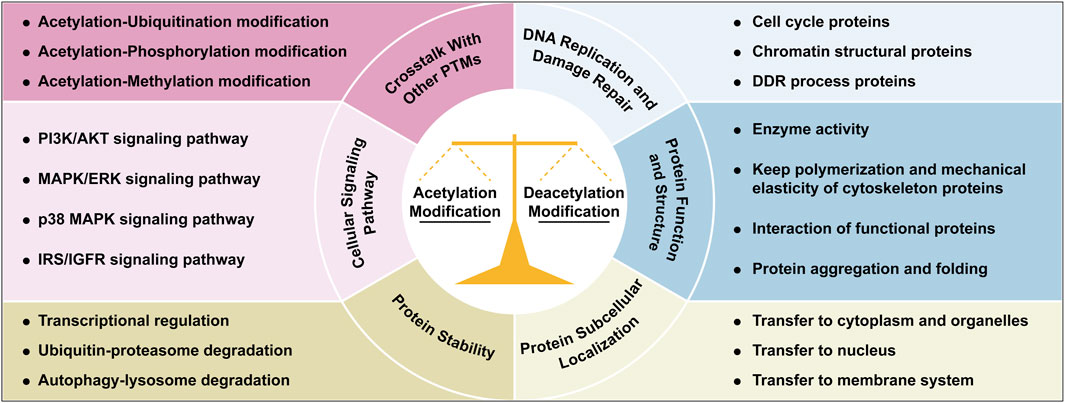
Figure 2. Integrated roles of acetylation/deacetylation in cellular regulation. This figure illustrates the multifaceted roles of acetylation and deacetylation modifications in coordinating cellular processes, through regulating genome homeostasis, protein function and activity, signaling pathways, protein dynamics and subcellular localization, and crosstalk with other PTMs. Each functional mechanism is illustrated with representative examples or key cellular processes. Abbreviations: AKT, protein kinase B; DDR, DNA damage repair; ERK, extracellular-signal-regulated kinases; IGFR, insulin like growth factor 1; IRS, insulin receptor substrate; MAPK, mitogen-activated protein kinases; PI3K, phosphatidylinositol 3-kinase; PTM, post-translational modification.
2.1 DNA replication and damage repair
Acetylation and deacetylation maintain cellular homeostasis by dynamically regulating processes such as DNA replication, protein function, and signaling. HDACs and HATs interact with cell cycle regulators like pRb and Mad/Max to control progression; for example, HDAC-pRb suppresses cyclin-dependent kinase 2 (Cdk2) expression, while p300 acetylates cyclin D1 to drive the progression of G1 phase (Knoepfler and Eisenman, 1999; Nomura et al., 1999; Albanese et al., 1999; Resnitzky et al., 1994). During mitosis, HAT5 acetylates histones (H2A/H3/H4), and HAT2B/HAT5 modify Aurora B and EB1 to ensure accurate chromosome segregation (Mo et al., 2016; Ward et al., 2013; Xia et al., 2012). Structural maintenance of chromosomes 3 (SMC3) acetylation by ESCO1/2 stabilizes sister chromatid cohesion, and its deficiency disrupts DNA repair (Zhang et al., 2008; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhao et al., 2010). In DSB repair, TIP60 activates ATM via acetylation, while ATP-citrate lyase (ACLY)-derived nuclear acetyl-CoA reduces TP53-binding protein 1 (53BP1) recruitment, favoring homology-directed repair (HDR) over non-homologous end-joining (NHEJ) (Sun et al., 2005; Sivanand et al., 2017).
2.2 Protein function and structure
Acetylation modulates enzyme activity by targeting catalytic lysine residues: mitochondrial ACSS2 is inhibited by acetylation but reactivated by Sirtuin deacetylase family 3 (SIRT3) mediating deacetylation, whereas PGAM1 acetylation enhances glycolysis but is suppressed by SIRT1 under glucose starvation (Schwer et al., 2006; Hallows et al., 2006; Fujino et al., 2001; Walter et al., 1999). Autoacetylation of HATs like P300/CREB-binding protein (CBP) and MYST1 activates their catalytic domains. Structurally, α-tubulin acetylation (alpha-tubulin acetyltransferase 1 (ATAT1)-mediated) strengthens microtubule elasticity, while HDAC6 deacetylation regulates dynamics (Akella et al., 2010; Hubbert et al., 2002; Zhang et al., 2003). Pathologically, Tau acetylation at Lys174 promotes neurotoxic aggregates in Alzheimer’s disease, while acetylation at Lys259/Lys353 inhibits hyperphosphorylation-driven aggregation (Cohen et al., 2011; Min et al., 2010; Min et al., 2015; Cook et al., 2014).
2.3 Protein subcellular localization
Acetylation alters protein charge or interactions to dictate localization. S-phase kinase-associated protein 2 (SKP2) acetylation promotes cytoplasmic retention by blocking nuclear import, whereas hepatocyte nuclear factor 4 (HNF4) acetylation enhances nuclear retention (Frescas and Pagano, 2008; Inuzuka et al., 2012; Soutoglou et al., 2000; Zhao LJ. et al., 2006). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) acetylated by P300/CBP-associated factor (PCAF) translocates to the nucleus for DNA repair, and kinase suppressor of Ras-1 (CNK1) acetylation directs it to the plasma membrane to amplify RAS/mitogen-activated protein kinases (MAPK) signaling (Ventura et al., 2010; Fischer et al., 2017). Mitochondrial p66Shc acetylation increases reactive oxygen species (ROS) production by facilitating its mitochondrial translocation (Kumar et al., 2017).
2.4 Protein stability
Histone acetylation loosens chromatin to activate transcription, exemplified by Spt-Ada-Gcn5 acetyltransferase (SAGA) complex recruitment of RNA Pol II and bromodomain-containing protein 4 (BRD4) binding to acetylated H3/H4 (Cazzalini et al., 2014; Fischer et al., 2017; Li et al., 2008). In Drosophila melanogaster, HAT8-mediated H4K16 acetylation preserves epigenetic memory across generations (Samata et al., 2020; Tadros and Lipshitz, 2009). Acetylation competes with ubiquitination to regulate degradation: small mothers against decapentaplegic 7 (Smad7) acetylation blocks Smad ubiquitination regulatory factor 1 (Smurf1)-mediated ubiquitination, stabilizing it, while DNA (cytosine-5)-methyltransferase 1 (DNMT1) acetylation triggers ubiquitin-like with PHD and RING finger domains 1 (UHRF1)-dependent degradation (Du et al., 2010). Autophagy targets acetylated pyruvate kinase (PKM2) and lactate dehydrogenase A (LDHA) for lysosomal degradation, and SIRT1 deficiency impairs basal autophagy (Lv et al., 2011; Zhao et al., 2013; Lee et al., 2008).
2.5 Cellular signaling pathways
Cellular signaling networks are information-processing networks that control almost all cellular functions, and acetylation of key proteins in signaling pathways enable cells to respond rapidly to internal and external signals. Phosphatase and tensin homolog (PTEN) acetylation by PCAF inhibits phosphatase activity, whereas protein kinase B (AKT)/3-phosphoinositide-dependent kinase 1 (PDK1) acetylation blocks phosphatidylinositol (3,4,5)-triphosphate (PIP3)-dependent membrane activation (Okumura et al., 2006). CNK1 acetylation recruits rapidly accelerated fibrosarcoma kinase (RAF) to the membrane, amplifying extracellular-signal-regulated kinases (ERK) signaling (Fischer et al., 2017). SIRT1 deacetylates insulin receptor substrate 2 (IRS2) to enhance insulin signaling, and p38 autoacetylation boosts ATP binding during stress (Li et al., 2008) (Pillai et al., 2011). Tat-interacting protein 60 (TIP60) acetylates ataxia-telangiectasia mutated kinase (ATM) to activate DNA damage responses, while mitogen-activated protein kinase phosphatase 1 (MKP1) acetylation enhances phosphatase activity to suppress MAPK signaling (Sun et al., 2005; Pillai et al., 2011).
2.6 Crosstalk with other PTMs
Acetylation interacts with ubiquitination, phosphorylation, and methylation. For example, p53 acetylation by CBP/P300 competes with murine double minute 2 (MDM2)-mediated ubiquitination, stabilizing p53 (Li et al., 2002). In IFNα signaling, STAT2 acetylation facilitates IFN-stimulated gene factor 3 (ISGF3) complex formation, amplifying antiviral gene transcription (Tang et al., 2007). TGF-β signaling involves Smad7 acetylation (by P300) to block ubiquitination, whereas HDAC1 deacetylation promotes Smurf1-mediated degradation (Grönroos et al., 2002; Simonsson et al., 2005).
3 Acetylation modifications are associated with senescence
Growing evidences suggest that acetylation is an important component of altering epigenetic marks during cellular senescence. Cellular senescence arises not from catastrophic loss of individual functional units or isolated molecular deficiency, but rather from dysregulation in multiscale biological processes. We believe the pathophysiological basis of aging manifests as dual-tiered coordination failures, which compromised homeostatic integration within cellular biochemical coordination and disrupted coordination across intercellular cross-talk. This hypothesis is substantiated by seminal work from Ana Carolina Leote et al., whose systematic analysis of aging transcriptomes revealed that while most gene-gene relationships in transcriptional regulatory networks remain preserved during aging, the deterioration of regulatory coupling predominantly stems from the loss of multiscale integration across cellular processes (Leote et al., 2024). These findings provide compelling validation for our hypothesis that senescence constitutes a systems-level failure of biological coordination rather than discrete component malfunction. Acetylation is a PTM that affects protein function regulating double strand break-DNA damage response (DSB-DDR), metabolic pathways and protein homeostasis. Hence, once acetylation modifications are disrupted, this balance is disrupted and eventually leads to cellular senescence (Kanfi et al., 2012; Levy et al., 2020). Figure 3 illustrates that an imbalance in acetylation-deacetylation homeostasis drives cellular senescence through three interconnected mechanisms, DNA damage accumulation, protein homeostasis disruption, and mitochondrial dysfunction.
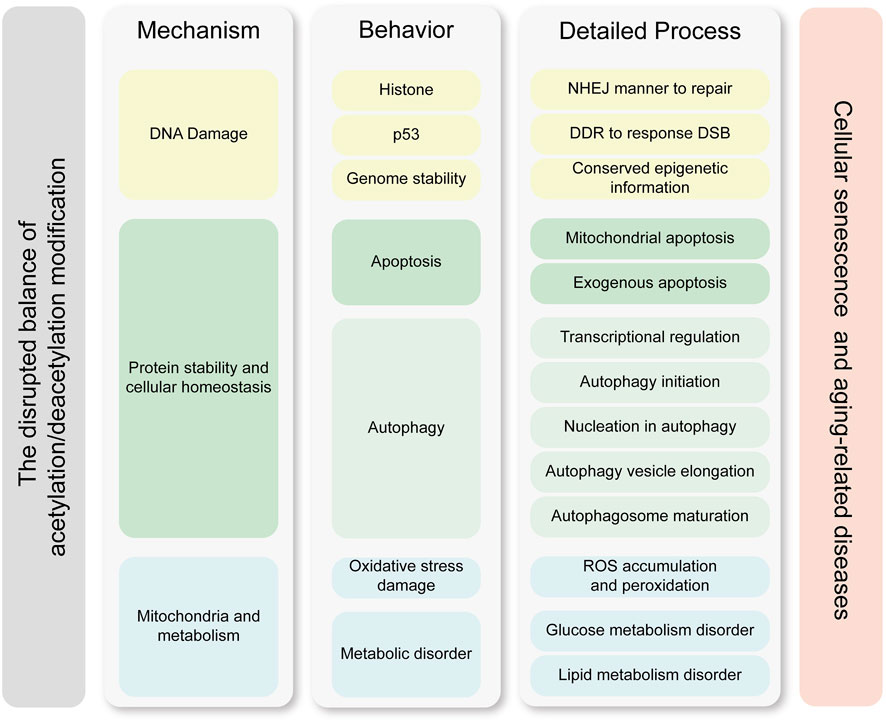
Figure 3. Molecular mechanisms linking acetylation dynamics to cellular senescence. This figure delineates the interplay between acetylation modifications and cellular senescence through three core mechanisms, which are DNA damage, protein stability and cellular homeostasis, and metabolic homeostasis. Key processes such as transcriptional regulation, genetic stability maintenance, and exogenous apoptosis further highlight how acetylation imbalances drive senescence-associated pathologies. The left column represents the core mechanisms by which acetylation modifications contribute to cellular senescence and age-related diseases. The middle column illustrates the key functional processes and effector molecules involved, while the right column details the specific molecular processes and pathways through which these effects are exerted. The mechanisms in the left column drive the process in the middle column, and the processes in the right column further refine the molecular pathways underlying these behaviors. The cellular processes and specific effective mechanisms are clustered by the similar color. Abbreviations: DDR, DNA damage repair; DSB, double strain break; NHEJ, non-homologous end-joining; ROS, reactive oxygen species.
3.1 DNA damage
Senescence is characterized by the loss of genomic integrity and stability, which involves chromatin remodeling, transcription dysregulation, and accumulation of DNA damage, as well as deficiencies in the ability to respond to DDR. HDAC1 plays a pivotal role in maintaining genomic integrity, supporting neuronal survival, and modulating synaptic plasticity, and collectively constituting a critical defense against neurodegenerative pathogenesis (Kim et al., 2008; Dobbin et al., 2013; Wang et al., 2013). Recruitment of HDAC1 by histones triggers hypoacetylation of H56K3 and H16K4, thereby promoting DSB repair through NHEJ, which is the major neuronal repair pathway (Miller et al., 2010; Madabhushi et al., 2014). p53 serves as a critical effector molecule in the DDR pathway following genotoxic stress (d'Adda di Fagagna, 2008). It governs cellular senescence and fate determination through mediating key processes including apoptosis and cell cycle arrest. Importantly, acetylation modifications have been established as a crucial regulatory mechanism linking p53 to senescence induction (Hong et al., 2010). Acetylated p53 attenuates the effect of Mdm2 on the ubiquitination degradation of p53 to enhance p53 stability, and further recruits HATs to the promoters of genes involved in the DDR process and cell cycle control mechanism, thereby mitigating DNA damage-induced senescence (Li et al., 2002; Ito et al., 2002). The SIRTs also participate in DDR processes; SIRT1, SIRT6, and SIRT7 have been shown to be directly involved in DSB repair. For example, SIRT7 was recruited to damage sites of DNA in a PARP-1 dependent manner and regulated H3K18 acetylation at the damaged sites. Acetylated H3K18 in turn affected the recruitment of the damage response factor 53BP1 to the damaged DNA sites, which ultimately enhanced the efficiency of NHEJ and promoted DNA damage repair. Correspondingly, the depletion of SIRT7 led to a decline in the repair capacity of DSBs (Vazquez et al., 2016).
In addition, the accumulation of mutant DNA during the genetic process has been found to be a major cause of senescence, while some studies indicated that mice or patients with higher mutation rates did not exhibit a premature aging phenotype. However, senescent mammalian cells still had the ability to clone new individuals with a normal life expectancy, suggesting there might be a mechanism beyond mutation-induced senescence that linked DNA stability and cellular senescence (Burgstaller and Brem, 2017; Robinson et al., 2021; De Majo et al., 2021). An increasing number of studies have shown that senescent cells have a deficiency of genetic information and a loss of epigenetic information. Some researchers believe that the epigenetic alterations caused by DNA damage that induce the relocation of chromatin modifying factors might be the major cause of senescence, and further proposed a hypothesis in which the loss of epigenetic information is the cause of senescence (Yang et al., 2023; Lu et al., 2020; Pan et al., 2023). DSBs are one of the major factors that cause time-varying epigenetic changes, and the epigenetic regulators SIRT2 and general control non-depressible 5 (GCN5) are required in the DDR process to repair DSBs, such as mediating acetylation of histones involved in aging. Senescent cells exhibit a higher acetylation level of H3K122 and lower acetylation level of H3K27. H3K27ac is usually enriched in accessible regions, including promoters and enhancers, but the highly accessible regions of senescent cells lose their original H3K27 acetylation, while less accessible regions have increased levels of H3K27 acetylation. This DSB-induced epigenomic change accelerates the production of age-related markers, and epigenetic remodeling can reverse these changes and restore the cellular epigenome to a youthful state (Yang et al., 2023).
ACLY is the important enzyme that catalyzes the reaction which products acetyl-CoA. ACLY and histone acetylation also play critical roles in regulating chromatin structure and gene expression. ACLY converts citrate derived from glucose metabolism into acetyl-CoA, the primary substrate for histone acetylation, thus directly influencing global histone acetylation levels (Wellen et al., 2009). During the DDR, ACLY is phosphorylated at Ser455 by ATM and AKT, leading to its nuclear localization (Sivanand et al., 2017). This promotes histone acetylation near DNA damage sites, such as H3K9ac and H4K16ac, which inhibits 53BP1 binding while enhancing BRCA1 recruitment, thereby driving homologous recombination (HR) repair (Sivanand et al., 2017). SIRT6, an NAD+ -dependent deacetylase, regulates chromatin stability and repair through deacetylation during DNA damage and aging, playing a central role in DDR and senescence (Onn et al., 2020). Age-related decline in NAD + levels may impair SIRT6 activity, resulting in accumulated histone acetylation and compromised repair capacity, exacerbating genomic instability. Upon DNA damage, studies demonstrate that SIRT6 directly recognizes and binds to DSBs via its core structural domain, acting as an independent damage sensor to activate downstream repair pathways, such as ATM recruitment and H2A histone family member X (H2AX) phosphorylation, and recruit repair proteins such as breast cancer type 1 susceptibility protein (BRCA1) and 53BP1 (Sivanand et al., 2017; Onn et al., 2020). The deacetylase activity of SIRT6 is essential for chromatin remodeling and DNA repair. Its deficiency leads to genomic instability, telomere dysfunction, and accelerated aging phenotypes (Onn et al., 2020). SIRT6 suppresses ACLY-mediated histone acetylation by removing acetyl groups from histone H3K9 and facilitates DNA-DNA dependent protein kinase catalytic subunit (PKcs) localization on chromatin to enhance DNA repair (McCord et al., 2009). The synergistic interaction between ACLY and SIRT6 (dynamic regulation of acetylation and deacetylation) likely holds significant implications for maintaining genomic integrity and delaying aging (Sivanand et al., 2017; Wellen et al., 2009; Onn et al., 2020; McCord et al., 2009).
3.2 Protein stability and cellular homeostasis
3.2.1 Apoptosis
Apoptosis is involved in all stages of development and senescence, and there is growing evidence that senescence and apoptosis are distinct cell fate outcomes that interact with each other (Basu, 2022; Childs et al., 2014; Schmitt, 2003). Aberrant apoptosis affects whether cells go into senescence or not, making apoptosis a potential mechanism to regulate senescence. The SIRTs affect various apoptotic regulation factors by regulating acetylation modifications. SIRT1, SIRT6, and SIRT7 mitigate p53-dependent apoptosis through deacetylation modifications. SIRT7, for example, was shown to enhance cardiomyocyte stress resistance and ultimately inhibit apoptosis by deacetylating p53, and SIRT7-deficient mice exhibited an elevated apoptotic response and severe cardiac inflammatory cardiomyopathy phenotype (Vakhrusheva et al., 2008; Zeng et al., 2022; Vaziri et al., 2001; Wood et al., 2018; Yao et al., 2018). SIRT1, SIRT2, and SIRT3 deacetylate and activate forkhead box-O-3a (FoxO3a) to promote the activation of the c-Jun N-terminal kinase (JNK) signaling pathway, which ultimately increased apoptosis (Yao et al., 2018; She et al., 2018; Peng et al., 2019). SIRTs also regulate the molecules that directly participate the apoptosis process. For example, SIRT5 deacetylated cytochrome C and reduced cytochrome C leakage to further alleviate endogenous apoptosis (Schlicker et al., 2008; Yang et al., 2021; Li et al., 2019).
3.2.2 Autophagy
Dysfunction of autophagy induces senescence and can promote aging phenotypes, and biochemical intervention or genetic manipulation to regulate autophagy can partially delay the aging process (Franco-Juárez et al., 2022; Goehe et al., 2012). Acetylation modifications are involved in the entire process of autophagy, including transcriptional regulation of genes in the initiation stage of autophagy to lysosomal degradation as the outcome of autophagy behavior (Sun et al., 2021). In terms of the transcriptional stage, activated transcription factor EB (TFEB) is an important factor in the transcriptional regulation of autophagy. Activated TFEB binds to CLEAR motifs to regulate the transcription of autophagy-lysosome related genes, promoting autophagic biogenesis and lysosomal fusion to increase autophagy, thereby effectively degrading molecular complex and preventing induced aging. ATAT1 mediated Lys91, Lys103, and Lys430 acetylation of TFEB to promote a transcriptional effect, while the HAT2/GCN5 mediated Lys116, Lys274, and Lys279 acetylation disrupted the dimerization of TFEB and decreased binding to the promoter of its target genes, indicating that the acetylation mode of TFEB directly affected transcriptional activity (Wang et al., 2020). The HAT8/SIRT1 system, which mediates H4K16 acetylation, suppresses the transcription of early-stage and end-stage autophagy related genes (Salminen and Kaarniranta, 2009; Füllgrabe et al., 2013). SIRT1 was also shown to indirectly regulate autophagy by upregulating the expression of the autophagy-related gene Bnip3 through the deacetylation of FoxO3 (Kume et al., 2010; Kroemer et al., 2010).
In the autophagy initiation process, starvation stimulation activates the glycogen synthase kinase 3β (GSK3β)/TIP60/UNC-52-like kinase 1 (ULK1) pathway to regulate autophagy behavior. GSK3β phosphorylates TIP60-Ser86 to an activated state, and the activated TIP60 further acetylates ULK1 on Lys162 and Lys606 sites to activate downstream protein kinases, subsequently initiating autophagy through the mechanistic target of rapamycin (mTOR) signaling pathways (Lin et al., 2012; Dossou and Basu, 2019; Nie et al., 2016). In the following autophagic lysosome formation process, P300-mediated acetylation controls the activity of the phosphatidylinositol 3-kinase catalytic subunit type 3/vacuolar protein sorting 34 (PIK3C3/VPS34) signaling pathway, which forms a complex with Beclin1 and further directly regulates autophagy nucleation (Su et al., 2017; Kary, 2018; Su and Liu, 2018). Furthermore, acetylated tripartite motif proteins (TRIMs) or heat shock protein 70 (Hsp70) indirectly affected autophagy by regulating other PTMs of key proteins in the autophagy nucleation process (Yang et al., 2013; Fusco et al., 2018). The P300/SIRT1 system is the key regulator of acetylation modifications during autophagic vesicle extension and the LC3 conjugation process (Liu and Klionsky, 2015; Sebti et al., 2014). In the last process, acetylated Beclin1 and TIP60 mediates the acetylation of downstream molecules to regulate autophagosome maturation through the RUN domain and Cysteine-rich domain containing, Beclin 1-interacting protein (RUBCN) bimolecular switch model (Matsunaga et al., 2009; Zhong et al., 2009; Cheng and Sun, 2019; Cheng et al., 2019). The reversible acetylation of α-microtubule proteins and HDAC6/SIRT1-mediated acetylation of cortactin protein is required for autophagosome–lysosome fusion (Lee et al., 2010; Zhang et al., 2009; Köchl et al., 2006).
3.3 Mitochondria and metabolism
Extensive experimental evidence demonstrates that mitochondrial dysfunction leads to aging processes, as mitochondria critically influence or regulate multiple key aspects of senescence (Sun et al., 2016). Notably, mitochondria-targeted interventions and strategies improving mitochondrial quality and function may exert profound beneficial effects, effectively mitigating the deleterious impact of senescence in aging tissues by restoring redox homeostasis and bioenergetic capacity (Sun et al., 2016; Correia-Melo et al., 2016).
3.3.1 Oxidative stress damage
In 1972, Harman synthesized the seminal proposal, which is defined as mitochondrial free radical theory of aging, that aging stems from cumulative free radical-induced damage to mitochondria. This hypothesis links the mitochondrial homeostasis and aging, which posits an age-associated elevation in mitochondrial ROS generation, wherein ROS accumulation inflicts oxidative lesions across critical biomolecules, such as mitochondrial DNA (mtDNA) mutations, lipid peroxidation, protein carbonyl formation and nucleic acid modifications (Harman, 1972). These molecular injuries propagate cellular dysfunction through compromised electron transport chain efficiency, impaired calcium homeostasis, and defective mitochondrial quality control mechanisms, ultimately manifesting as multi-tiered physiological decline across tissues and organ systems (Miwa et al., 2022; Guo et al., 2023a). Oxidative stress damage is one of the important factors of mitochondria-induced senescence, in which the oxidative stress-induced abnormal accumulation of ROS exceeds the cell’s scavenging capacity, leading to disrupted intracellular homeostasis and senescence. Increasing evidence has indicated that SIRTs regulate oxidative stress damage by deacetylation modification of various crucial factors involved in metabolism regulation, inflammation, and antioxidant responses. The most common SIRTs that participate in the antioxidant responses are SIRT1, SIRT3, and SIRT6.
AMP-activated protein kinase (AMPK) is a crucial kinase in the energy regulation process and is recognized as a core regulatory factor of eukaryotic cells. SIRT1 and SIRT3 were found to deacetylate live kinase B1 (LKB1) which is the upstream regulator of AMPK, and deacetylated LKB1 then activated AMPK to reduce ROS accumulation and lipid peroxidation (Li et al., 2020). SIRT6 directly interacted with AMPK and promoted the expression of antioxidant stress proteins, such as superoxide dismutase (MnSOD) and human catalase targeted to the mitochondria (mCAT), which inhibited oxidative stress damage (Wang et al., 2016). A similar mechanism was also observed in SIRT1-and SIRT3-mediated FoxO deacetylation that performed antioxidant function (Sundaresan et al., 2009; Kwon et al., 2015; Yang et al., 2016). SIRT1/2/6 regulated the transcriptional activity of nuclear factor erythroid 2-related factor 2 (Nrf2) by a deacetylation modification to protect cells from ROS damage (Chen et al., 2020; Zhao et al., 2021; Yu et al., 2019).
3.3.2 Glucose metabolism and lipid metabolism
Protein acetylation modifications have emerged as a major mechanism for regulating cellular metabolism. Phosphoenolpyruvate carboxy-kinase (PEPCK1) is the key enzyme and rate-limiting enzyme that catalyzes the synthesis of glucose from pyruvate. An acetylation modification regulates the stability of PEPCK1 to affect the balance between glucose metabolism and generation, thus achieving glucose homeostasis in vivo. When the glucose concentration increases, P300 mediates the acetylation of PEPCK1 and promotes interaction with the E3 ubiquitin ligase UBR5, facilitating further PEPCK1 ubiquitination and proteasome degradation, thereby limiting glucose production. SIRT1 also improves the glucose metabolism status and maintains energy homeostasis, such as by upregulating and activating AMPK (Peng et al., 2010; Silvestre et al., 2014). SIRT1 was also shown to downregulate the activity of protein tyrosine phosphatase 1B (PT1B), which is a key negative regulator of the insulin signaling pathway, thereby improving insulin sensitivity and reducing serum glucose (Gouranton et al., 2014). Another deacetylase with a substantial impact on metabolism is SIRT6. As the deacetylase of H3K9, SIRT6 inhibits the expression of several glycolytic genes (Zhong et al., 2010). SIRT6 also deacetylates FoxO1, which in turn increases the transcription and expression of glucose-dependent transporter 2 to maintain the glucose-sensitizing capacity of pancreatic islet β-cells and glucose tolerance, delaying high-glucose–induced chronic inflammation and senescence (Song et al., 2016).
Increasing evidence confirmed that Acetyl-CoA-induced acetylation modifications led to abnormal accumulation of fatty acids and eventually vascular aging (Lin et al., 2022). The SIRTs have been implicated as the major factors in the retardation of senescence in deacetylation modifications. The main target of SIRT1 are FoxOs, which act as important transcriptional regulators, and deacetylation of FoxOs promotes the transcription of fatty acid oxidation-related proteins (Mostoslavsky et al., 2006). SIRT3 plays a bridging role in lipid metabolic pathways and regulates mitochondrial acetylation levels. SIRT3 has been demonstrated to be involved in almost all processes of mitochondrial metabolism and homeostasis (Samant et al., 2014; Papa and Germain, 2017), such as deacetylating and enhancing isocitrate dehydrogenase 2 (IDH2) and long-chain acyl-CoA dehydrogenase (LCAD) activities to stimulate fatty acid β-oxidation and mitigate cellular senescence.
4 Acetylation modifications are associated with aging-related diseases
In addition to natural aging, cellular senescence is observed in various aging-related chronic diseases, such as tumors, hypertension, and neurodegenerative diseases. Cellular senescence is also an important factor accelerating disease progression, and is recognized as a common end-stage pathological condition of many diseases. Figure 4 illustrates that acetylation modifications orchestrate the pathogenesis of aging-related diseases through interconnected regulatory networks, dynamically coordinating DNA damage repair, protein homeostasis, and mitochondrial integrity.
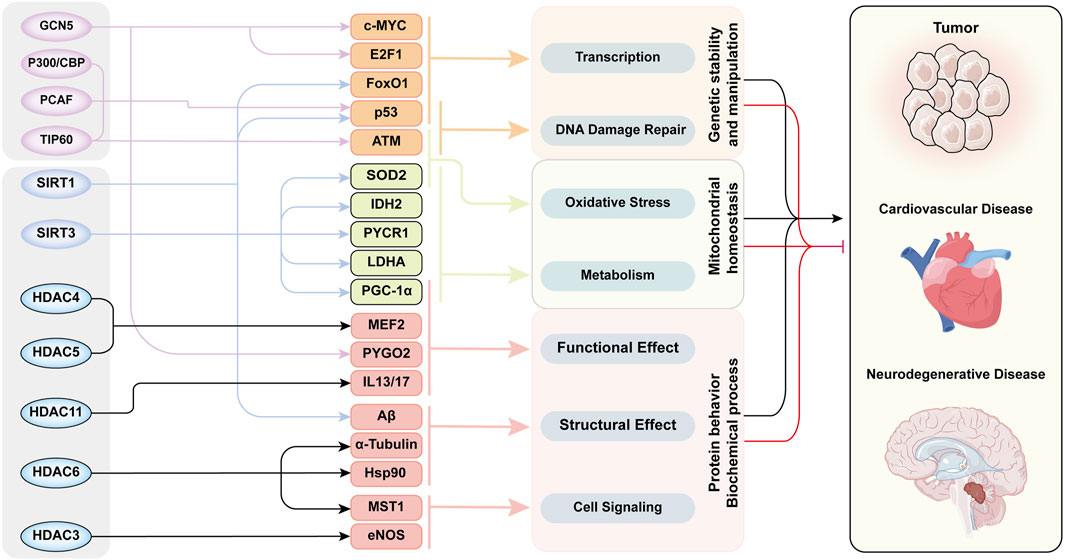
Figure 4. The Acetylation modification -Network mechanism -Senescence mode in aging-related diseases. This figure exhibited functional network of acetylation/deacetylation in cellular processes and disease pathogenesis. Multiple key proteins are acetylated by several HATs/HDACs to induce senescence through various cellular processes and biochemical reactions. The functions of these proteins intersected with each other and ultimately led to aging through three aspects of cellular behavior. The whole acetylation-related regulatory mechanism form a cross network to regulate the development of aging-related diseases. This diagram categorizes histone acetyltransferases and deacetylases in the leftmost column, their substrate targets in the second column, the associated cellular processes in the third column, and linked diseases in the rightmost column. Substrates are color-clustered by functional categories. Black lines between the third and fourth columns denote disease-promoting effects, while red lines indicate inhibitory roles. Abbreviations: ATM, ataxia-telangiectasia mutated kinase; E2F1, E2 promoter binding factor 1; eNOS, endothelial nitric oxide synthase; FoxO1, Forkhead box-O-1; GCN5, general control non-depressible 5; HDAC, histone deacetylase; Hsp90, heat shock protein 70; IDH2, isocitrate dehydrogenase 2; IL13/17, interleukin-13/17; LDHA, lactate dehydrogenase A; MEF2, myocyte enhancer factor 2; MST1, mammalian sterile 20-like kinase 1; P300/CBP, P300/CREB-binding protein; PCAF, P300/CBP-associated factor; PGC-1α, Peroxisome proliferator-activated receptor-gamma co-activator-1alpha; PYCR1, pyrroline-5-carboxylate reductase 1; PYGO2, Pygopus 2; SIRT, Sirtuins family; SOD2, superoxide dismutase 2; TIP60, Tat-interacting protein 60.
4.1 Tumors
Senescent cells represent a deficiency of genomic stability, which leads to an increasing probability of various DNA mutations and excessive accumulation of abnormal DNA, activating the expression of proto-oncogenes and ultimately promoting cellular carcinogenesis. Therefore, most tumors are considered to be aging-associated diseases. Acetylation modifications activate the enhancer of H4K16, further regulating chromatin structure and transcriptional regulation. whereas the loss of H4K16ac is considered to be a universal marker for the development and malignant transformation of tumors (Taylor et al., 2013; Fraga et al., 2005; Seligson et al., 2005). Changes in histone acetylation patterns can also be used to predict the prognosis and recurrence of cancer. For example, the insufficient acetylation of H3K9, H3K14, and H4K12 were strongly associated with an adverse prognosis of breast cancer, and low acetylated levels of H3K9, H3K18, and H4K16 were shown to be important markers for prostate cancer recurrence (Seligson et al., 2005). The acetylation of non-histone proteins is also involved in a variety of crucial life processes, such as DNA replication, metabolism, differentiation, and development, and abnormal acetylation modifications often disrupt the balance of these cellular processes, eventually leading to cancer (Glozak et al., 2005; Singh et al., 2010; Spange et al., 2009).
4.1.1 HATs in tumorigenesis
A large number of transcription factors are targeted substrates of HATs, including oncogenes and tumor suppressor genes associated with tumorigenesis, such as MYC and TP53. Myc protein, which is encoded by the gene c-MYC, is one of the most abundant oncogene proteins in cancer, and is stabilized by GCN5 with an acetylation modification at Lys323 (Patel et al., 2004). GCN5 can also be recruited by Myc to the template of Pol III, resulting in a significant increase in histone H3 acetylation, and ultimately promoting the transcription of c-MYC target genes (Kenneth et al., 2007). HAT2A regulated other transcription factors or tumor-promoting proteins, such as E2 promoter binding factor 1 (E2F1) and pygopus2 (PYGO2), to enhance the expression of target oncogenes in a similar manner, most of which stimulated abnormal cell proliferation and tumor growth (Chen et al., 2013; Chen et al., 2010). However, acetylation modifications also regulate the expression or function of tumor suppressor genes. Several HATs mediate tumor suppressor p53 acetylation modifications to increase the stability of p53 and enhance binding with DNA. CBP/P300 acetylates the carboxyl terminus of p53, preventing its proteasomal degradation (Li et al., 2002). TIP60 plays an important role in maintaining integrity of DNA and preventing the accumulation of tumor-inducing mutations. Under DSB conditions, acetylation and activation of the kinase ATM by TIP60 is the key step for subsequent recruitment of proteins involved in the DNA repair process that includes p53 (Sun et al., 2010).
4.1.2 HDACs in tumorigenesis
Different deacetylases cause various effects on the development of tumors by deacetylating different substrates, which drives tumor cells to different destinies. HDAC family-mediated deacetylation modifications are often observed in tumor progression and deterioration. HDAC6-mediated deacetylation of mammalian sterile 20-like kinase 1 (MST1) decreased protein stability and promoted the aggravation of breast cancer (Li et al., 2016). HDAC6 also promoted cancer development by deacetylating a-tubulin to enable cells to metastasize through malignant migration, chemotaxis, and angiogenesis (Valenzuela-Fernández et al., 2008; Kaluza et al., 2011). Hsp90 is another important substrate of HDAC6 which is activated after deacetylation modification. The activated Hsp90 further stabilized and promoted the transcription of the androgen receptor, eventually inducing the development of prostate cancer (Gao and Alumkal, 2010). We also noticed that HDACs negatively regulated the process of cancer and improved the prognosis. HDAC10 attenuated lymph node metastasis of cervical cancer by deacetylating the histones H3 and H4, which downregulated the expression of matrix metalloproteinase 2/9 (MMP2/9), thus mitigating migration and invasion (Song et al., 2013).
The SIRTs mainly mediate deacetylation of non-histone proteins and regulate cell behavior involving multiple tumorigenesis process, and cause different regulatory directions in tumor cells. SIRT1 is believed to be a factor in tumorigenesis and has been observed to be highly expressed in a variety of cancers, including prostate cancer and acute myeloid leukemia. SIRT1 was shown to bind and promote FoxO1 to translocate into the nucleus, enhancing the transcription of target genes involved in cancer development (Bosch-Presegué and Vaquero, 2011). SIRT1 also deacetylates c-Myc and further negatively regulates the transcription of tumor suppressor-related genes to promote the development of cancer (Yuan et al., 2009). Some studies observed downregulation of SIRT2 in glioma and gastric cancer, suggesting that SIRT2 might play a suppressive effect in cancer development (Inoue et al., 2007). SIRT3 regulates mitochondrial metabolism through deacetylation modifications and participates in cellular metabolism reprogramming, which plays a unique function by mediating the interaction between mitochondria and intracellular signal transduction, and is considered to be a hallmark molecule of cancer. SIRT3 plays different roles in various cancers and is specific to tumor type (Ouyang et al., 2022). SIRT3 regulated IDH2 dimerization by mediating Lys413 site acetylation of IDH2, which regulated the metabolism and progression of breast cancer (Zou et al., 2017). SIRT3 deacetylated the Lys228 site of pyrroline-5-carboxylate reductase 1 (PYCR1) to regulate the metabolism of proline, which promoted the development of breast cancer (Chen et al., 2019). In addition, a study revealed that SIRT3 participated in the mitochondrial metabolic balance of gastric cancer and promoted deacetylation and activation of LDHA, thereby enhancing glycolysis and proliferation of gastric cancer, and was considered a cancer promoting factor (Cui et al., 2015). Some studies focused on colon cancer have demonstrated that SIRT3 was highly expressed in colorectal cancer and was associated with its tumor stage and lymph node metastasis (Liu et al., 2014). SIRT3 has been confirmed to enhance chemotherapy resistance of colon cancer by modulating the acetylation of superoxide dismutase 2 (SOD2) and peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1α), which enabled SIRT3 to be an independent prognostic factor for colon cancer (Paku et al., 2021).
4.1.3 HDAC inhibitors in tumorigenesis
HDACs inhibitors induce cell cycle arrest, apoptosis, and necrosis in cancer cells. Some studies demonstrated that HDAC inhibitors enhanced the acetylation level and prolonged the half-life of p53, thereby increasing the interaction with the promoter of p21 (Gius et al., 2004; Zhao Y. et al., 2006; Richon et al., 2000). Additionally, HDAC competed with p53 to bind to p21, leading to decreased transcription of p21 (Ocker and Schneider-Stock, 2007; Mahyar-Roemer and Roemer, 2001). Therefore, HDAC inhibitors increase p21 expression by this mechanism, which blocks the dimerization of cell cycle proteins and arrests the cellular cycle, eventually inhibiting the proliferation of tumor cells (Richon et al., 2000; Sandor et al., 2000). HDAC inhibitors also induce endogenous apoptosis in tumor cells by regulating the transcription of pro-apoptotic or anti-apoptotic genes, inhibiting the development and progression of tumor cells (Kim and Bae, 2011; Minucci and Pelicci, 2006; Zhu et al., 2004). HDAC11 negatively regulated the expression of interleukin-13/17 (IL-13/17) and tumor necrosis factor-alpha (TNF-α) in Hodgkin lymphoma, suggesting that HDAC11 inhibitors killed tumor cells by activating inflammatory responses, immune system activity, and apoptosis processes (Buglio et al., 2011).
4.2 Cardiovascular diseases
Clinical and molecular studies have observed significant alterations in the expression levels of multiple HDAC and SIRT proteins in pulmonary arterial hypertension patients, which demonstrated the potential and strong association between acetylation modifications and pathological vascular remodeling processes, such as proliferation, inflammation, and fibrosis (Chelladurai et al., 2021).
4.2.1 Hypertension
Hypertension is a major factor in cardiovascular disease, and is mainly related to the activation of the renin-angiotensin-aldosterone system and sympathetic nervous system. Vascular injury is an important pathway for hypertension-induced target organ damage and multiple subsequent complications. The detailed pathological features include vascular smooth muscle cells (VSMCs) remodeling, mitochondrial dysfunction, and endothelial cell injury. It has been confirmed that HDAC4 and HDAC5 participate in Angiotensin II (Ang II)-induced hypertrophy of VSMCs, which led to vascular remodeling. Calmodulin kinase II promoted myocyte enhancer factor 2 (MEF2) activity through phosphorylation of HDAC4, thereby causing hypertrophy of VSMCs as well (Xu et al., 2007; Li et al., 2010). SIRT3-mediated deacetylation is essential for guaranteeing the activity of SOD2 to protect against vascular oxidative stress, and SIRT3 deficiency increased the production of mitochondrial superoxide free radicals and promoted the occurrence of hypertension (Dikalova et al., 2017). Endothelial nitric oxide synthase (eNOS) is the main producer of NO in vascular endothelial cells and is the core regulator of cardiovascular homeostasis; insufficient eNOS activity is a major pathogenic factor for endothelial dysfunction and hypertension. SIRT1 promotes endothelium-dependent vascular dilation by targeting eNOS for deacetylation and increasing its activity (Mattagajasingh et al., 2007). The vascular protective effect of aspirin may also be related to acetylation of eNOS (Jung et al., 2010), in which low-dose aspirin increased the vascular bioavailability of NO and promoted the binding of eNOS and calmodulin. In contrast, overexpression of HDAC3 inhibited aspirin-induced acetylation of eNOS, further antagonizing the effects of aspirin on NO production in endothelial cells (Jung et al., 2010).
4.2.2 Atherosclerosis
Atherosclerosis is recognized as the most common disease affecting the vascular system, especially the coronary artery. The pathological feature is lipid accumulation in the smooth muscle layer and vessel blockage, and the resulting narrowed arteries, atherosclerosis, and unstable plaques may lead to the occurrence of myocardial infarction (Libby et al., 2011). VSMCs are the main cellular component in the artery, and epigenetic modification of VSMCs has been shown to contribute to the formation of atherosclerotic lesions (Hai and Zuo, 2016). Research has demonstrated that SIRT1 deficiency caused over-acetylation of eNOS, resulting in functional inactivation of eNOS, which further promoted the development of atherosclerotic plaques in mice, while HDAC3-mediated Lys610 deacetylation of eNOS promoted the atherosclerosis process (Jung et al., 2010; Yang et al., 2017; Rössig et al., 2002; Schermuly et al., 2011). Cellular senescence is another cause of atherosclerosis, and causes cells to be more susceptible to damage and further develop to atherosclerosis. La-related protein 7 (LARP7) is an aging antagonist molecule, which was downregulated by DNA damage-induced activation of ATM, and thereby inhibited the deacetylase activity of SIRT1. The ATM-LARP7-SIRT1-p53/p65 aging axis, which is composed of these molecules, is actively involved in the process of vascular aging and atherosclerosis, and blocking its activation significantly alleviated atherosclerosis (Yan et al., 2021).
4.3 Neurodegenerative disease
The accumulation of senescent cells in the central nervous system (CNS) serves as a core driver of cognitive decline and neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (Melo Dos Santos et al., 2024). The senescent nervous cells face insufficient energy substrate supply due to impaired local microcirculation, which fundamentally disrupts glial cell metabolism and ultimately damages neural networks. Altered energy metabolism in glial cells and its impact on brain aging and related disorders are currently a major research focus (Mattson and Arumugam, 2018). For example, cellular senescence promotes amyloidogenic cleavage of amyloid precursor protein (APP) and Tau pathology, while the accumulation of Aβ and Tau further amplifies senescence marker expression. Concurrently, lipid peroxidation-modified proteins accumulate in the aging brain, enhancing γ-secretase-mediated APP cleavage and inducing the production of neurotoxic Aβ42. These vicious cycles exacerbate AD progression through reciprocal reinforcement of senescence and neurodegenerative pathways. Neurological degeneration is another important feature of aging, in which HAT- and HDAC-regulated acetylation modifications play an indispensable role. An imbalance of acetylation modifications can lead to progressive neuronal-specific loss, impaired neuronal function, and ultimately neuronal death. Increasing evidence has shown that abnormal acetylation and deacetylation were associated with the pathogenesis of various neurodegenerative diseases, especially AD (Saha and Pahan, 2006).
During the development of AD, P300-mediated acetylation of histone H3, located in the promoter region of presenilin-1 (PS1) and beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) aggravated neuronal injury, whereas targeted inhibition of P300 mitigated AD symptoms and abnormal protein aggregation. This suggested that P300 plays a key role in controlling the expression of AD-associated genes by regulating the acetylation of their promoter region (Lu et al., 2014). The stability of DNA replication guaranteed by HDAC1 has been found to play an important role in nervous system damage. The p25/Cdk5 complex involved in the AD process inactivated HDAC1, which led to aberrant cell cycle activity, double-stranded DNA breaks, and ultimately neurotoxicity, which was alleviated by the exogenous introduction of HDAC1 (Kim et al., 2008). In terms of molecular therapy for AD, SIRTs have been shown to regulate the progression of various neurodegenerative diseases by modulating transcription factor activity and protein toxicity (Gomes et al., 2019; Herskovits and Guarente, 2013). Abundant expression of SIRT1 inhibited Aβ polymerization and attenuated plaque formation, resulting in amelioration of behavioral deficits, suggesting a neuroprotective role for SIRT1 in AD (Jęśko et al., 2017). Small molecule SIRT activators, especially SIRT1 and SIRT2, have great potential in the treatment of age-related diseases, particularly neurodegenerative diseases (Han, 2009). They inhibit acetylation levels of various key process proteins and reduce toxic protein aggregates to restore protein homeostasis, and also enhance neuronal plasticity by promoting transcription of important genes responsible for memory and learning (Min et al., 2013). PD is defined as the most common neurological degeneration disease, especially in the geriatric population. α-Synuclein is an important effector of PD, which directly binds to histone H3 to mediate neurotoxicity. Various studies have confirmed that the inhibition of several HATs, such as P300/CBP and PCAF, reduced the level of H3 acetylation and alleviated α-synuclein-mediated nervous system damage, which has been verified in animal models. This provides support for histone deacetylase inhibitors as a potential treatment for PD (Kontopoulos et al., 2006).
5 Conclusions: “Acetylation-network regulation-cellular senescence” model
Based on the above evidence, we provide insight into the senescence mechanism and propose the concept that acetylation modifications play a pivotal effect in the senescence process. Macroscopically, acetylation modifications participate in the aging process through both direct and indirect pathways. Acetylation modifications play a pivotal role in orchestrating core cellular processes during senescence, encompassing the homeostasis and regulation of DNA maintenance and DDR process, protein stability and proteostasis, metabolic reprogramming and mitochondrial function. Acetylation modifications of histones ensure the stability of the DNA structure, and the acetylation of numerous DNA damage repair factors guarantees the integrity and accuracy of the DNA replication process. Acetylation modifications also occur on multiple transcription factors to regulate their transcriptional ability, altering the expression level of downstream target molecules. Acetylation modifications also directly regulate the activity, structure, or sub-location of various key proteins in senescent mechanisms, thereby affecting cellular fate through these senescence execution molecules. At the organelle level, mitochondria are the regulatory pivot of several cellular processes, and acetylation modifications regulate the activity of metabolism-related proteins to affect the metabolic function of mitochondria. At the organellar level, mitochondria act as a pivotal regulator of multifaceted cellular processes. Acetylation modifications orchestrate mitochondrial metabolic functions by dynamically modulating the activity of metabolism-associated proteins. Beyond metabolic regulation, acetylation modification critically coordinates non-metabolic pathways, including mitochondrial apoptosis and mitophagy. Such dual-axis regulation (metabolic/non-metabolic) ultimately governs cellular fate determination and senescence by integrating energy status with stress-responsive signaling. We believe that senescence likely operates through a networked model of interconnected and coordinated mechanisms. Upon exposure to endogenous damage or exogenous stressors, key nodes within this regulatory network simultaneously activate multiple pathways, including metabolic reprogramming, proteostasis regulation, genomic surveillance systems, and cell fate-determining signaling cascades, which collectively drive senescence through synergistic interactions. This model establishes an activation threshold requiring concurrent engagement of multiple pathways to reach critical signaling intensity, rather than relying on singular pathway activation. Crucially, this architecture prevents premature senescence commitment due to isolated pathway activation or single homeostatic imbalance. As a terminal cell fate, this multilayered control mechanism provides survival protective benefits by ensuring senescence occurs only when cumulative damage surpasses cellular adaptive capacity, thereby avoiding inappropriate cell cycle arrest under subcritical stress conditions.
Our proposed regulatory mode of acetylation modifications in senescence is a broad regulation of multiple key proteins in the senescence mechanism network. This mode regulates various downstream detailed senescence-related mechanisms at the same time, which is different from the classical mechanism of a single executive molecule affecting the terminal executors. We believe that the appropriate intervention of acetylation modifications is to move the intervention process forward, and intervene in multiple pathways at the same time through a mechanism network that does not rely on a single downstream senescence execution molecule. Acetylation modifications function as regulatory modalities rather than direct causative factors of senescence. These post-translational adjustments exert precise control over primary senescence-triggering processes, including metabolism, genomic homeostasis, mitochondrial function, and proteostasis networks, thereby indirectly modulating senescence-associated biological processes. This regulatory architecture explains the remarkable efficacy of acetylation-targeted interventions (e.g., chemical agents mimic HAT/HDAC activity) in ameliorating or even reversing senescent phenotypes, as they fundamentally recalibrate core aging mechanism through upstream pathway modulation. As a novel insight into the aging mechanism, the imbalance of cellular homeostasis caused by the disrupted acetylation modification balance systematically and comprehensively explains the changes of different cellular processes during senescence. For example, simple DNA damage might not lead to aging necessarily, while the reduced stability of the DNA-histone complex and the decreased activity of damage repair-related proteins caused by abnormal acetylation will cooperate to inevitably cause senescence. Therefore, the targeted intervention of the acetylation modifications of key nexus proteins or upstream regulators in the network mechanism is a new idea and direction for delaying or even reversing aging.
Author contributions
MX: Writing – original draft. WW: Writing – original draft. SL: Writing – original draft. MX: Writing – original draft. TZ: Writing – review and editing. YY: Writing – review and editing. CS: Writing – review and editing. JY: Writing – review and editing. NZ: Writing – review and editing. LC: Writing – review and editing. GS: Writing – review and editing. SC: Conceptualization, Supervision, Writing – review and editing. PW: Conceptualization, Project administration, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (NO. 82301778); The China Postdoctoral Science Foundation (NO.2023MD744274); The Science and Technology Program of Liaoning Province, China (NO.2022-MS-213).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agalioti T., Chen G., Thanos D. (2002). Deciphering the transcriptional histone acetylation code for a human gene. Cell 111 (3), 381–392. doi:10.1016/s0092-8674(02)01077-2
Aghdassi A., Sendler M., Guenther A., Mayerle J., Behn C. O., Heidecke C. D., et al. (2012). Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 61 (3), 439–448. doi:10.1136/gutjnl-2011-300060
Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., et al. (2010). MEC-17 is an alpha-tubulin acetyltransferase. Nature 467 (7312), 218–222. doi:10.1038/nature09324
Albanese C., D'Amico M., Reutens A. T., Fu M., Watanabe G., Lee R. J., et al. (1999). Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274 (48), 34186–34195. doi:10.1074/jbc.274.48.34186
Ali M., Yan K., Lalonde M. E., Degerny C., Rothbart S. B., Strahl B. D., et al. (2012). Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 424 (5), 328–338. doi:10.1016/j.jmb.2012.10.004
Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., et al. (2012). SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487 (7405), 114–118. doi:10.1038/nature11043
Basu A. (2022). The interplay between apoptosis and cellular senescence: Bcl-2 family proteins as targets for cancer therapy. Pharmacol. Ther. 230, 107943. doi:10.1016/j.pharmthera.2021.107943
Blank M. F., Chen S., Poetz F., Schnölzer M., Voit R., Grummt I. (2017). SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 45 (5), 2675–2686. doi:10.1093/nar/gkx053
Bosch-Presegué L., Vaquero A. (2011). The dual role of sirtuins in cancer. Genes Cancer 2 (6), 648–662. doi:10.1177/1947601911417862
Brower-Toland B., Wacker D. A., Fulbright R. M., Lis J. T., Kraus W. L., Wang M. D. (2005). Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346 (1), 135–146. doi:10.1016/j.jmb.2004.11.056
Buglio D., Khaskhely N. M., Voo K. S., Martinez-Valdez H., Liu Y. J., Younes A. (2011). HDAC11 plays an essential role in regulating OX40 ligand expression in Hodgkin lymphoma. Blood 117 (10), 2910–2917. doi:10.1182/blood-2010-08-303701
Burgstaller J. P., Brem G. (2017). Aging of cloned animals: a mini-review. Gerontology 63 (5), 417–425. doi:10.1159/000452444
Cai H., Wang Y., Zhang J., Wei Z., Yan T., Feng C., et al. (2023). Discovery of novel SIRT1/2 inhibitors with effective cytotoxicity against human leukemia cells. J. Chem. Inf. Model 63 (15), 4780–4790. doi:10.1021/acs.jcim.3c00556
Cazzalini O., Sommatis S., Tillhon M., Dutto I., Bachi A., Rapp A., et al. (2014). CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res. 42 (13), 8433–8448. doi:10.1093/nar/gku533
Chae Y. B., Kim M. M. (2014). Activation of p53 by spermine mediates induction of autophagy in HT1080 cells. Int. J. Biol. Macromol. 63, 56–63. doi:10.1016/j.ijbiomac.2013.10.041
Chang S., Wang R. H., Akagi K., Kim K. A., Martin B. K., Cavallone L., et al. (2011). Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 17 (10), 1275–1282. doi:10.1038/nm.2459
Chelladurai P., Boucherat O., Stenmark K., Kracht M., Seeger W., Bauer U. M., et al. (2021). Targeting histone acetylation in pulmonary hypertension and right ventricular hypertrophy. Br. J. Pharmacol. 178 (1), 54–71. doi:10.1111/bph.14932
Chen F. Q., Schacht J., Sha S. H. (2009). Aminoglycoside-induced histone deacetylation and hair cell death in the mouse cochlea. J. Neurochem. 108 (5), 1226–1236. doi:10.1111/j.1471-4159.2009.05871.x
Chen J., Luo Q., Yuan Y., Huang X., Cai W., Li C., et al. (2010). Pygo2 associates with MLL2 histone methyltransferase and GCN5 histone acetyltransferase complexes to augment Wnt target gene expression and breast cancer stem-like cell expansion. Mol. Cell Biol. 30 (24), 5621–5635. doi:10.1128/MCB.00465-10
Chen L., Wei T., Si X., Wang Q., Li Y., Leng Y., et al. (2013). Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, cyclin D1, and cyclin E1 expression. J. Biol. Chem. 288 (20), 14510–14521. doi:10.1074/jbc.M113.458737
Chen S., Yang X., Yu M., Wang Z., Liu B., Liu M., et al. (2019). SIRT3 regulates cancer cell proliferation through deacetylation of PYCR1 in proline metabolism. Neoplasia 21 (7), 665–675. doi:10.1016/j.neo.2019.04.008
Chen Y., An N., Zhou X., Mei L., Sui Y., Chen G., et al. (2022). Fibroblast growth factor 20 attenuates pathological cardiac hypertrophy by activating the SIRT1 signaling pathway. Cell Death Dis. 13 (3), 276. doi:10.1038/s41419-022-04724-w
Chen Z., Yu J., Fu M., Dong R., Yang Y., Luo J., et al. (2020). Dipeptidyl peptidase-4 inhibition improves endothelial senescence by activating AMPK/SIRT1/Nrf2 signaling pathway. Biochem. Pharmacol. 177, 113951. doi:10.1016/j.bcp.2020.113951
Cheng X., Ma X., Zhu Q., Song D., Ding X., Li L., et al. (2019). Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol. Cell 73 (4), 788–802.e7. doi:10.1016/j.molcel.2018.12.017
Cheng X., Sun Q. (2019). RUBCNL/Pacer and RUBCN/Rubicon in regulation of autolysosome formation and lipid metabolism. Autophagy 15 (6), 1120–1121. doi:10.1080/15548627.2019.1596500
Childs B. G., Baker D. J., Kirkland J. L., Campisi J., van Deursen J. M. (2014). Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 15 (11), 1139–1153. doi:10.15252/embr.201439245
Cohen T. J., Guo J. L., Hurtado D. E., Kwong L. K., Mills I. P., Trojanowski J. Q., et al. (2011). The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2, 252. doi:10.1038/ncomms1255
Cook C., Carlomagno Y., Gendron T. F., Dunmore J., Scheffel K., Stetler C., et al. (2014). Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 23 (1), 104–116. doi:10.1093/hmg/ddt402
Correia-Melo C., Marques F. D., Anderson R., Hewitt G., Hewitt R., Cole J., et al. (2016). Mitochondria are required for pro-ageing features of the senescent phenotype. Embo J. 35 (7), 724–742. doi:10.15252/embj.201592862
Cui Y., Qin L., Wu J., Qu X., Hou C., Sun W., et al. (2015). SIRT3 enhances glycolysis and proliferation in SIRT3-expressing gastric cancer cells. PLoS One 10 (6), e0129834. doi:10.1371/journal.pone.0129834
d'Adda di Fagagna F. (2008). Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8 (7), 512–522. doi:10.1038/nrc2440
Dai C., Gu W. (2010). p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 16 (11), 528–536. doi:10.1016/j.molmed.2010.09.002
Deardorff M. A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., et al. (2012). HDAC8 mutations in Cornelia de Lange syndrome affect the Cohesin acetylation cycle. Nature 489 (7415), 313–317. doi:10.1038/nature11316
De Majo F., Martens L., Hegenbarth J. C., Rühle F., Hamczyk M. R., Nevado R. M., et al. (2021). Genomic instability in the naturally and prematurely aged myocardium. Proc. Natl. Acad. Sci. U. S. A. 118 (36), e2022974118. doi:10.1073/pnas.2022974118
Di Giorgio E., Paluvai H., Dalla E., Ranzino L., Renzini A., Moresi V., et al. (2021). HDAC4 degradation during senescence unleashes an epigenetic program driven by AP-1/p300 at selected enhancers and super-enhancers. Genome Biol. 22 (1), 129. doi:10.1186/s13059-021-02340-z
Dikalova A. E., Itani H. A., Nazarewicz R. R., McMaster W. G., Flynn C. R., Uzhachenko R., et al. (2017). Sirt3 impairment and SOD2 hyperacetylation in vascular oxidative stress and hypertension. Circ. Res. 121 (5), 564–574. doi:10.1161/CIRCRESAHA.117.310933
Dobbin M. M., Madabhushi R., Pan L., Chen Y., Kim D., Gao J., et al. (2013). SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 16 (8), 1008–1015. doi:10.1038/nn.3460
Donnarumma F., Tucci V., Ambrosino C., Altucci L., Carafa V. (2022). NAA60 (HAT4): the newly discovered bi-functional golgi member of the acetyltransferase family. Clin. Epigenetics 14 (1), 182. doi:10.1186/s13148-022-01402-8
Dossou A. S., Basu A. (2019). The emerging roles of mTORC1 in macromanaging autophagy. Cancers (Basel) 11 (10), 1422. doi:10.3390/cancers11101422
Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., et al. (2010). DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal 3 (146), ra80. doi:10.1126/scisignal.2001462
Fan B., Wang J., Zha D., Qiu J., Chen F. (2020). ATP depletion induced cochlear hair cells death through histone deacetylation in vitro. Neurosci. Lett. 727, 134918. doi:10.1016/j.neulet.2020.134918
Fischer A., Mühlhäuser W. W. D., Warscheid B., Radziwill G. (2017). Membrane localization of acetylated CNK1 mediates a positive feedback on RAF/ERK signaling. Sci. Adv. 3 (8), e1700475. doi:10.1126/sciadv.1700475
Fournier M., Orpinell M., Grauffel C., Scheer E., Garnier J. M., Ye T., et al. (2016). KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun. 7, 13227. doi:10.1038/ncomms13227
Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., et al. (2005). Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37 (4), 391–400. doi:10.1038/ng1531
Franco-Juárez B., Coronel-Cruz C., Hernández-Ochoa B., Gómez-Manzo S., Cárdenas-Rodríguez N., Arreguin-Espinosa R., et al. (2022). TFEB; beyond its role as an autophagy and lysosomes regulator. Cells 11 (19), 3153. doi:10.3390/cells11193153
Frescas D., Pagano M. (2008). Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8 (6), 438–449. doi:10.1038/nrc2396
Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. (2001). Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 276 (14), 11420–11426. doi:10.1074/jbc.M008782200
Füllgrabe J., Lynch-Day M. A., Heldring N., Li W., Struijk R. B., Ma Q., et al. (2013). The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500 (7463), 468–471. doi:10.1038/nature12313
Fusco C., Mandriani B., Di Rienzo M., Micale L., Malerba N., Cocciadiferro D., et al. (2018). TRIM50 regulates Beclin 1 proautophagic activity. Biochim. Biophys. Acta Mol. Cell Res. 1865 (6), 908–919. doi:10.1016/j.bbamcr.2018.03.011
Gaddelapati S. C., Albishi N. M., Dhandapani R. K., Palli S. R. (2022). Juvenile hormone-induced histone deacetylase 3 suppresses apoptosis to maintain larval midgut in the yellow fever mosquito. Proc. Natl. Acad. Sci. U. S. A. 119 (11), e2118871119. doi:10.1073/pnas.2118871119
Gao L., Alumkal J. (2010). Epigenetic regulation of androgen receptor signaling in prostate cancer. Epigenetics 5 (2), 100–104. doi:10.4161/epi.5.2.10778
Gao Y., Nihira N. T., Bu X., Chu C., Zhang J., Kolodziejczyk A., et al. (2020). Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 22 (9), 1064–1075. doi:10.1038/s41556-020-0562-4
Geng A., Tang H., Huang J., Qian Z., Qin N., Yao Y., et al. (2020). The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res. 48 (16), 9181–9194. doi:10.1093/nar/gkaa661
Ghosh T. K., Aparicio-Sánchez J. J., Buxton S., Ketley A., Mohamed T., Rutland C. S., et al. (2018). Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb development. J. Mol. Cell Cardiol. 114, 185–198. doi:10.1016/j.yjmcc.2017.11.013
Giaimo B. D., Ferrante F., Vallejo D. M., Hein K., Gutierrez-Perez I., Nist A., et al. (2018). Histone variant H2A.Z deposition and acetylation directs the canonical Notch signaling response. Nucleic Acids Res. 46 (16), 8197–8215. doi:10.1093/nar/gky551
Gius D., Cui H., Bradbury C. M., Cook J., Smart D. K., Zhao S., et al. (2004). Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 6 (4), 361–371. doi:10.1016/j.ccr.2004.08.029
Glozak M. A., Sengupta N., Zhang X., Seto E. (2005). Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23. doi:10.1016/j.gene.2005.09.010
Goehe R. W., Di X., Sharma K., Bristol M. L., Henderson S. C., Valerie K., et al. (2012). The autophagy-senescence connection in chemotherapy: must tumor cells (self) eat before they sleep? J. Pharmacol. Exp. Ther. 343 (3), 763–778. doi:10.1124/jpet.112.197590
Gomes P., Leal H., Mendes A. F., Reis F., Cavadas C. (2019). Dichotomous sirtuins: implications for drug discovery in neurodegenerative and cardiometabolic diseases. Trends Pharmacol. Sci. 40 (12), 1021–1039. doi:10.1016/j.tips.2019.09.003
Gouranton E., Romier B., Marcotorchino J., Tourniaire F., Astier J., Peiretti F., et al. (2014). Visfatin is involved in TNFα-mediated insulin resistance via an NAD(+)/Sirt1/PTP1B pathway in 3T3-L1 adipocytes. Adipocyte 3 (3), 180–189. doi:10.4161/adip.28729
Grégoire S., Xiao L., Nie J., Zhang X., Xu M., Li J., et al. (2007). Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol. Cell Biol. 27 (4), 1280–1295. doi:10.1128/MCB.00882-06
Grönroos E., Hellman U., Heldin C. H., Ericsson J. (2002). Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10 (3), 483–493. doi:10.1016/s1097-2765(02)00639-1
Guo Y., Guan T., Shafiq K., Yu Q., Jiao X., Na D., et al. (2023a). Mitochondrial dysfunction in aging. Ageing Res. Rev. 88, 101955. doi:10.1016/j.arr.2023.101955
Guo Z., Yu X., Zhao S., Zhong X., Huang D., Feng R., et al. (2023b). SIRT6 deficiency in endothelial cells exacerbates oxidative stress by enhancing HIF1α accumulation and H3K9 acetylation at the Ero1α promoter. Clin. Transl. Med. 13 (8), e1377. doi:10.1002/ctm2.1377
Hai Z., Zuo W. (2016). Aberrant DNA methylation in the pathogenesis of atherosclerosis. Clin. Chim. Acta 456, 69–74. doi:10.1016/j.cca.2016.02.026
Hallows W. C., Lee S., Denu J. M. (2006). Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 103 (27), 10230–10235. doi:10.1073/pnas.0604392103
Han S. H. (2009). Potential role of sirtuin as a therapeutic target for neurodegenerative diseases. J. Clin. Neurol. 5 (3), 120–125. doi:10.3988/jcn.2009.5.3.120
Han X., Peng C., Huang L., Luo X., Mao Q., Wu S., et al. (2022). EGCG prevents pressure overload-induced myocardial remodeling by downregulating overexpression of HDAC5 in mice. Int. J. Mol. Med. 49 (1), 11. doi:10.3892/ijmm.2021.5066
Harman D. (1972). The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 20 (4), 145–147. doi:10.1111/j.1532-5415.1972.tb00787.x
Herskovits A. Z., Guarente L. (2013). Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res. 23 (6), 746–758. doi:10.1038/cr.2013.70
Hishikawa A., Hayashi K., Abe T., Kaneko M., Yokoi H., Azegami T., et al. (2019). Decreased KAT5 expression impairs DNA repair and induces altered DNA methylation in kidney podocytes. Cell Rep. 26 (5), 1318–1332.e4. doi:10.1016/j.celrep.2019.01.005
Hong L. Z., Zhao X. Y., Zhang H. L. (2010). p53-mediated neuronal cell death in ischemic brain injury. Neurosci. Bull. 26 (3), 232–240. doi:10.1007/s12264-010-1111-0
Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., et al. (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417 (6887), 455–458. doi:10.1038/417455a
Inoue T., Hiratsuka M., Osaki M., Oshimura M. (2007). The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle 6 (9), 1011–1018. doi:10.4161/cc.6.9.4219
Inuzuka H., Gao D., Finley L. W., Yang W., Wan L., Fukushima H., et al. (2012). Acetylation-dependent regulation of Skp2 function. Cell 150 (1), 179–193. doi:10.1016/j.cell.2012.05.038
Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., et al. (2002). MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. Embo J. 21 (22), 6236–6245. doi:10.1093/emboj/cdf616
Izumikawa K., Ishikawa H., Yoshikawa H., Fujiyama S., Watanabe A., Aburatani H., et al. (2019). LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST-type acetyltransferase KAT7 to rDNA. Nucleic Acids Res. 47 (19), 10357–10372. doi:10.1093/nar/gkz747
Jęśko H., Wencel P., Strosznajder R. P., Strosznajder J. B. (2017). Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem. Res. 42 (3), 876–890. doi:10.1007/s11064-016-2110-y
Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., et al. (2011). Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43 (1), 33–44. doi:10.1016/j.molcel.2011.04.028
Jing E., Gesta S., Kahn C. R. (2007). SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 6 (2), 105–114. doi:10.1016/j.cmet.2007.07.003
Jung S. B., Kim C. S., Naqvi A., Yamamori T., Mattagajasingh I., Hoffman T. A., et al. (2010). Histone deacetylase 3 antagonizes aspirin-stimulated endothelial nitric oxide production by reversing aspirin-induced lysine acetylation of endothelial nitric oxide synthase. Circ. Res. 107 (7), 877–887. doi:10.1161/CIRCRESAHA.110.222968
Kaluza D., Kroll J., Gesierich S., Yao T. P., Boon R. A., Hergenreider E., et al. (2011). Class IIb HDAC6 regulates endothelial cell migration and angiogenesis by deacetylation of cortactin. Embo J. 30 (20), 4142–4156. doi:10.1038/emboj.2011.298
Kandhaya-Pillai R., Yang X., Tchkonia T., Martin G. M., Kirkland J. L., Oshima J. (2022). TNF-α/IFN-γ synergy amplifies senescence-associated inflammation and SARS-CoV-2 receptor expression via hyper-activated JAK/STAT1. Aging Cell 21 (6), e13646. doi:10.1111/acel.13646
Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483 (7388), 218–221. doi:10.1038/nature10815
Kenneth N. S., Ramsbottom B. A., Gomez-Roman N., Marshall L., Cole P. A., White R. J. (2007). TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. U. S. A. 104 (38), 14917–14922. doi:10.1073/pnas.0702909104
Kikuchi M., Morita S., Wakamori M., Sato S., Uchikubo-Kamo T., Suzuki T., et al. (2023). Epigenetic mechanisms to propagate histone acetylation by p300/CBP. Nat. Commun. 14 (1), 4103. doi:10.1038/s41467-023-39735-4
Kim D., Frank C. L., Dobbin M. M., Tsunemoto R. K., Tu W., Peng P. L., et al. (2008). Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron 60 (5), 803–817. doi:10.1016/j.neuron.2008.10.015
Kim H. J., Bae S. C. (2011). Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 3 (2), 166–179.
Knoepfler P. S., Eisenman R. N. (1999). Sin meets NuRD and other tails of repression. Cell 99 (5), 447–450. doi:10.1016/s0092-8674(00)81531-7
Köchl R., Hu X. W., Chan E. Y., Tooze S. A. (2006). Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 7 (2), 129–145. doi:10.1111/j.1600-0854.2005.00368.x
Kontopoulos E., Parvin J. D., Feany M. B. (2006). Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 15 (20), 3012–3023. doi:10.1093/hmg/ddl243
Kroemer G., Mariño G., Levine B. (2010). Autophagy and the integrated stress response. Mol. Cell 40 (2), 280–293. doi:10.1016/j.molcel.2010.09.023
Kumar D., Rajanala K., Minocha N., Saha S. (2012). Histone H4 lysine 14 acetylation in Leishmania donovani is mediated by the MYST-family protein HAT4. Microbiol. Read. 158 (Pt 2), 328–337. doi:10.1099/mic.0.050211-0
Kumar S., Kim Y. R., Vikram A., Naqvi A., Li Q., Kassan M., et al. (2017). Sirtuin1-regulated lysine acetylation of p66Shc governs diabetes-induced vascular oxidative stress and endothelial dysfunction. Proc. Natl. Acad. Sci. U. S. A. 114 (7), 1714–1719. doi:10.1073/pnas.1614112114
Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., et al. (2010). Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Invest 120 (4), 1043–1055. doi:10.1172/JCI41376
Kwon D. N., Park W. J., Choi Y. J., Gurunathan S., Kim J. H. (2015). Oxidative stress and ROS metabolism via down-regulation of sirtuin 3 expression in Cmah-null mice affect hearing loss. Aging (Albany NY) 7 (8), 579–594. doi:10.18632/aging.100800
Le Dour C., Chatzifrangkeskou M., Macquart C., Magiera M. M., Peccate C., Jouve C., et al. (2022). Actin-microtubule cytoskeletal interplay mediated by MRTF-A/SRF signaling promotes dilated cardiomyopathy caused by LMNA mutations. Nat. Commun. 13 (1), 7886. doi:10.1038/s41467-022-35639-x
Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., et al. (2008). A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U. S. A. 105 (9), 3374–3379. doi:10.1073/pnas.0712145105
Lee J. Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y. S., et al. (2010). HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 29 (5), 969–980. doi:10.1038/emboj.2009.405
Lee S. W., Yang J., Kim S. Y., Jeong H. K., Lee J., Kim W. J., et al. (2015). MicroRNA-26a induced by hypoxia targets HDAC6 in myogenic differentiation of embryonic stem cells. Nucleic Acids Res. 43 (4), 2057–2073. doi:10.1093/nar/gkv088
Leote A. C., Lopes F., Beyer A. (2024). Loss of coordination between basic cellular processes in human aging. Nat. Aging 4 (10), 1432–1445. doi:10.1038/s43587-024-00696-y
Levy O., Amit G., Vaknin D., Snir T., Efroni S., Castaldi P., et al. (2020). Age-related loss of gene-to-gene transcriptional coordination among single cells. Nat. Metab. 2 (11), 1305–1315. doi:10.1038/s42255-020-00304-4
Li C., Chen F., Lin L., Li J., Zheng Y., Chen Q. (2023b). CSE triggers ferroptosis via SIRT4-mediated GNPAT deacetylation in the pathogenesis of COPD. Respir. Res. 24 (1), 301. doi:10.1186/s12931-023-02613-0
Li H., Li W., Gupta A. K., Mohler P. J., Anderson M. E., Grumbach I. M. (2010). Calmodulin kinase II is required for angiotensin II-mediated vascular smooth muscle hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 298 (2), H688–H698. doi:10.1152/ajpheart.01014.2009
Li L., Fang R., Liu B., Shi H., Wang Y., Zhang W., et al. (2016). Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene 35 (31), 4048–4057. doi:10.1038/onc.2015.476
Li M., Li C. M., Ye Z. C., Huang J., Li Y., Lai W., et al. (2020). Sirt3 modulates fatty acid oxidation and attenuates cisplatin-induced AKI in mice. J. Cell Mol. Med. 24 (9), 5109–5121. doi:10.1111/jcmm.15148
Li M., Luo J., Brooks C. L., Gu W. (2002). Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277 (52), 50607–50611. doi:10.1074/jbc.C200578200
Li T., Zhang C., Hassan S., Liu X., Song F., Chen K., et al. (2018). Histone deacetylase 6 in cancer. J. Hematol. Oncol. 11 (1), 111. doi:10.1186/s13045-018-0654-9
Li W., Yang Y., Li Y., Zhao Y., Jiang H. (2019). Sirt5 attenuates cisplatin-induced acute kidney injury through regulation of Nrf2/HO-1 and Bcl-2. Biomed. Res. Int. 2019, 4745132. doi:10.1155/2019/4745132
Li X., Chen M., Chen X., He X., Li X., Wei H., et al. (2024a). TRAP1 drives smooth muscle cell senescence and promotes atherosclerosis via HDAC3-primed histone H4 lysine 12 lactylation. Eur. Heart J. 45 (39), 4219–4235. doi:10.1093/eurheartj/ehae379
Li X., Li C., Zhang W., Wang Y., Qian P., Huang H. (2023a). Inflammation and aging: signaling pathways and intervention therapies. Signal Transduct. Target Ther. 8 (1), 239. doi:10.1038/s41392-023-01502-8
Li X., Yu Y., Zhang Q., Luo X., Yu L., Zhao Z. (2024b). Effect of HDAC9 on the differentiation of chicken embryonic stem cells into male germ cells. Anim. Reprod. 21 (2), e20240011. doi:10.1590/1984-3143-AR2024-0011
Li Y., Xu W., McBurney M. W., Longo V. D. (2008). SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 8 (1), 38–48. doi:10.1016/j.cmet.2008.05.004
Libby P., Ridker P. M., Hansson G. K. (2011). Progress and challenges in translating the biology of atherosclerosis. Nature 473 (7347), 317–325. doi:10.1038/nature10146
Lin R., Tao R., Gao X., Li T., Zhou X., Guan K. L., et al. (2013). Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol. Cell 51 (4), 506–518. doi:10.1016/j.molcel.2013.07.002
Lin S. Y., Li T. Y., Liu Q., Zhang C., Li X., Chen Y., et al. (2012). GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336 (6080), 477–481. doi:10.1126/science.1217032
Lin T., Yang W. Q., Luo W. W., Zhang L. L., Mai Y. Q., Li Z. Q., et al. (2022). Disturbance of fatty acid metabolism promoted vascular endothelial cell senescence via acetyl-CoA-induced protein acetylation modification. Oxid. Med. Cell Longev. 2022, 1198607. doi:10.1155/2022/1198607
Lin Y. T., Wu K. J. (2020). Epigenetic regulation of epithelial-mesenchymal transition: focusing on hypoxia and TGF-β signaling. J. Biomed. Sci. 27 (1), 39. doi:10.1186/s12929-020-00632-3
Liu C., Huang Z., Jiang H., Shi F. (2014). The sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed. Res. Int. 2014, 871263. doi:10.1155/2014/871263
Liu H., Hu Q., D'Ercole A. J., Ye P. (2009). Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia 57 (1), 1–12. doi:10.1002/glia.20729
Liu N., Wang J., Wang J., Wang R., Liu Z., Yu Y., et al. (2013). ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 73 (12), 3749–3760. doi:10.1158/0008-5472.CAN-12-3684
Liu Q., Wang X., Fang Z. T., Zhao J. N., Rui X. X., Zhang B. G., et al. (2025). Upregulation of ISG15 induced by MAPT/tau accumulation represses autophagic flux by inhibiting HDAC6 activity: a vicious cycle in Alzheimer disease. Autophagy 21 (4), 807–826. doi:10.1080/15548627.2024.2431472
Liu X., Klionsky D. J. (2015). TP53INP2/DOR protein chaperones deacetylated nuclear LC3 to the cytoplasm to promote macroautophagy. Autophagy 11 (9), 1441–1442. doi:10.1080/15548627.2015.1074373
Liu Y., Xun W., Zhao T., Huang M., Sun L., Wen G., et al. (2024). Interplay between acetylation and ubiquitination controls PSAT1 protein stability in lung adenocarcinoma. Commun. Biol. 7 (1), 1365. doi:10.1038/s42003-024-07051-2
Lu X., Deng Y., Yu D., Cao H., Wang L., Liu L., et al. (2014). Histone acetyltransferase p300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer’s disease. PLoS One 9 (7), e103067. doi:10.1371/journal.pone.0103067
Lu Y., Brommer B., Tian X., Krishnan A., Meer M., Wang C., et al. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature 588 (7836), 124–129. doi:10.1038/s41586-020-2975-4
Luo L., Martin S. C., Parkington J., Cadena S. M., Zhu J., Ibebunjo C., et al. (2019). HDAC4 controls muscle homeostasis through deacetylation of myosin heavy chain, PGC-1α, and Hsc70. Cell Rep. 29 (3), 749–763.e12. doi:10.1016/j.celrep.2019.09.023
Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., et al. (2011). Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42 (6), 719–730. doi:10.1016/j.molcel.2011.04.025
Ma Z., Liu D., Di S., Zhang Z., Li W., Zhang J., et al. (2019). Histone deacetylase 9 downregulation decreases tumor growth and promotes apoptosis in non-small cell lung cancer after melatonin treatment. J. Pineal Res. 67 (2), e12587. doi:10.1111/jpi.12587
Madabhushi R., Pan L., Tsai L. H. (2014). DNA damage and its links to neurodegeneration. Neuron 83 (2), 266–282. doi:10.1016/j.neuron.2014.06.034
Mahyar-Roemer M., Roemer K. (2001). p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene 20 (26), 3387–3398. doi:10.1038/sj.onc.1204440
Masumi A., Fukazawa H., Shimazu T., Yoshida M., Ozato K., Komuro K., et al. (2006). Nucleolin is involved in interferon regulatory factor-2-dependent transcriptional activation. Oncogene 25 (37), 5113–5124. doi:10.1038/sj.onc.1209522
Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11 (4), 385–396. doi:10.1038/ncb1846
Matsuzaki H., Daitoku H., Hatta M., Aoyama H., Yoshimochi K., Fukamizu A. (2005). Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 102 (32), 11278–11283. doi:10.1073/pnas.0502738102
Mattagajasingh I., Kim C. S., Naqvi A., Yamamori T., Hoffman T. A., Jung S. B., et al. (2007). SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 104 (37), 14855–14860. doi:10.1073/pnas.0704329104
Mattson M. P., Arumugam T. V. (2018). Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. 27 (6), 1176–1199. doi:10.1016/j.cmet.2018.05.011
McCord R. A., Michishita E., Hong T., Berber E., Boxer L. D., Kusumoto R., et al. (2009). SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 1 (1), 109–121. doi:10.18632/aging.100011
Melo Dos Santos L. S., Trombetta-Lima M., Eggen B., Demaria M. (2024). Cellular senescence in brain aging and neurodegeneration. Ageing Res. Rev. 93, 102141. doi:10.1016/j.arr.2023.102141
Meng J., Liu X., Zhang P., Li D., Xu S., Zhou Q., et al. (2016). Rb selectively inhibits innate IFN-β production by enhancing deacetylation of IFN-β promoter through HDAC1 and HDAC8. J. Autoimmun. 73, 42–53. doi:10.1016/j.jaut.2016.05.012
Menssen A., Hydbring P., Kapelle K., Vervoorts J., Diebold J., Lüscher B., et al. (2012). The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. U. S. A. 109 (4), E187–E196. doi:10.1073/pnas.1105304109
Miller K. M., Tjeertes J. V., Coates J., Legube G., Polo S. E., Britton S., et al. (2010). Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17 (9), 1144–1151. doi:10.1038/nsmb.1899
Min S., Lee H. S., Ji J. H., Heo Y., Kim Y., Chae S., et al. (2021). The chromatin remodeler RSF1 coordinates epigenetic marks for transcriptional repression and DSB repair. Nucleic Acids Res. 49 (21), 12268–12283. doi:10.1093/nar/gkab1093
Min S. W., Chen X., Tracy T. E., Li Y., Zhou Y., Wang C., et al. (2015). Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med. 21 (10), 1154–1162. doi:10.1038/nm.3951
Min S. W., Cho S. H., Zhou Y., Schroeder S., Haroutunian V., Seeley W. W., et al. (2010). Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67 (6), 953–966. doi:10.1016/j.neuron.2010.08.044
Min S. W., Sohn P. D., Cho S. H., Swanson R. A., Gan L. (2013). Sirtuins in neurodegenerative diseases: an update on potential mechanisms. Front. Aging Neurosci. 5, 53. doi:10.3389/fnagi.2013.00053
Minucci S., Pelicci P. G. (2006). Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 6 (1), 38–51. doi:10.1038/nrc1779
Mishima Y., Miyagi S., Saraya A., Negishi M., Endoh M., Endo T. A., et al. (2011). The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118 (9), 2443–2453. doi:10.1182/blood-2011-01-331892
Miwa S., Kashyap S., Chini E., von Zglinicki T. (2022). Mitochondrial dysfunction in cell senescence and aging. J. Clin. Invest 132 (13), e158447. doi:10.1172/JCI158447
Mo F., Zhuang X., Liu X., Yao P. Y., Qin B., Su Z., et al. (2016). Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12 (4), 226–232. doi:10.1038/nchembio.2017
Mo Y., Lin R., Liu P., Tan M., Xiong Y., Guan K. L., et al. (2017). SIRT7 deacetylates DDB1 and suppresses the activity of the CRL4 E3 ligase complexes. FEBS J. 284 (21), 3619–3636. doi:10.1111/febs.14259
Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124 (2), 315–329. doi:10.1016/j.cell.2005.11.044
Murakami M., Nakagawa M., Olson E. N., Nakagawa O. (2005). A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. U. S. A. 102 (50), 18034–18039. doi:10.1073/pnas.0509109102
Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009). SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137 (3), 560–570. doi:10.1016/j.cell.2009.02.026
Nie T., Yang S., Ma H., Zhang L., Lu F., Tao K., et al. (2016). Regulation of ER stress-induced autophagy by GSK3β-TIP60-ULK1 pathway. Cell Death Dis. 7 (12), e2563. doi:10.1038/cddis.2016.423
Nomura T., Khan M. M., Kaul S. C., Dong H. D., Wadhwa R., Colmenares C., et al. (1999). Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13 (4), 412–423. doi:10.1101/gad.13.4.412
North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11 (2), 437–444. doi:10.1016/s1097-2765(03)00038-8
Novgorodov S. A., Riley C. L., Keffler J. A., Yu J., Kindy M. S., Macklin W. B., et al. (2016). SIRT3 deacetylates ceramide synthases: implications for mitochondrial dysfunction and brain injury. J. Biol. Chem. 291 (4), 1957–1973. doi:10.1074/jbc.M115.668228
Núñez-Álvarez Y., Suelves M. (2022). HDAC11: a multifaceted histone deacetylase with proficient fatty deacylase activity and its roles in physiological processes. Febs J. 289 (10), 2771–2792. doi:10.1111/febs.15895
Ocker M., Schneider-Stock R. (2007). Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int. J. Biochem. Cell Biol. 39 (7-8), 1367–1374. doi:10.1016/j.biocel.2007.03.001
Okumura K., Mendoza M., Bachoo R. M., DePinho R. A., Cavenee W. K., Furnari F. B. (2006). PCAF modulates PTEN activity. J. Biol. Chem. 281 (36), 26562–26568. doi:10.1074/jbc.M605391200
Onn L., Portillo M., Ilic S., Cleitman G., Stein D., Kaluski S., et al. (2020). SIRT6 is a DNA double-strand break sensor. Elife 9, e51636. doi:10.7554/eLife.51636
Ouyang S., Zhang Q., Lou L., Zhu K., Li Z., Liu P., et al. (2022). The double-edged sword of SIRT3 in cancer and its therapeutic applications. Front. Pharmacol. 13, 871560. doi:10.3389/fphar.2022.871560
Paku M., Haraguchi N., Takeda M., Fujino S., Ogino T., Takahashi H., et al. (2021). SIRT3-Mediated SOD2 and PGC-1α contribute to chemoresistance in colorectal cancer cells. Ann. Surg. Oncol. 28 (8), 4720–4732. doi:10.1245/s10434-020-09373-x
Pan C., Zhou F., Zhang L. (2023). The loss of epigenetic information: not only consequences but a cause of mammalian aging. Signal Transduct. Target Ther. 8 (1), 140. doi:10.1038/s41392-023-01412-9
Papa L., Germain D. (2017). Correction for Papa and Germain, “SirT3” regulates a novel arm of the mitochondrial unfolded protein response. Mol. Cell Biol. 37 (13), e00191-1. doi:10.1128/MCB.00191-17
Papa L., Hahn M., Marsh E. L., Evans B. S., Germain D. (2014). SOD2 to SOD1 switch in breast cancer. J. Biol. Chem. 289 (9), 5412–5416. doi:10.1074/jbc.C113.526475
Patel J. H., Du Y., Ard P. G., Phillips C., Carella B., Chen C. J., et al. (2004). The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell Biol. 24 (24), 10826–10834. doi:10.1128/MCB.24.24.10826-10834.2004
Peng Y., Rideout D. A., Rakita S. S., Gower W. R., You M. (2010). Murr MM: does LKB1 mediate activation of hepatic AMP-protein kinase (AMPK) and sirtuin1 (SIRT1) after Roux-en-Y gastric bypass in obese rats? J. Gastrointest. Surg. 14 (2), 221–228. doi:10.1007/s11605-009-1102-5
Peng Y., Yang C., Shi X., Li L., Dong H., Liu C., et al. (2019). Sirt3 suppresses calcium oxalate-induced renal tubular epithelial cell injury via modification of FoxO3a-mediated autophagy. Cell Death Dis. 10 (2), 34. doi:10.1038/s41419-018-1169-6
Pillai V. B., Sundaresan N. R., Samant S. A., Wolfgeher D., Trivedi C. M., Gupta M. P. (2011). Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol. Cell Biol. 31 (11), 2349–2363. doi:10.1128/MCB.01205-10
Qin Y., Huang Y., Lin W., Huang R., Li K., Han X., et al. (2024). Neurotoxic effects induced by flunitrazepam and its metabolites in zebrafish: oxidative stress, apoptosis, and histone hypoacetylation. Sci. Total Environ. 917, 170521. doi:10.1016/j.scitotenv.2024.170521
Radhakrishnan R., Li Y., Xiang S., Yuan F., Yuan Z., Telles E., et al. (2015). Histone deacetylase 10 regulates DNA mismatch repair and may involve the deacetylation of MutS homolog 2. J. Biol. Chem. 290 (37), 22795–22804. doi:10.1074/jbc.M114.612945
Rando T. A., Chang H. Y. (2012). Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148 (1-2), 46–57. doi:10.1016/j.cell.2012.01.003
Resnitzky D., Gossen M., Bujard H., Reed S. I. (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell Biol. 14 (3), 1669–1679. doi:10.1128/mcb.14.3.1669-1679.1994
Richon V. M., Sandhoff T. W., Rifkind R. A., Marks P. A. (2000). Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. U. S. A. 97 (18), 10014–10019. doi:10.1073/pnas.180316197
Robinson P. S., Coorens T. H. H., Palles C., Mitchell E., Abascal F., Olafsson S., et al. (2021). Increased somatic mutation burdens in normal human cells due to defective DNA polymerases. Nat. Genet. 53 (10), 1434–1442. doi:10.1038/s41588-021-00930-y
Rokudai S., Laptenko O., Arnal S. M., Taya Y., Kitabayashi I., Prives C. (2013). MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. U. S. A. 110 (10), 3895–3900. doi:10.1073/pnas.1300490110
Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M., et al. (2008). Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321 (5888), 563–566. doi:10.1126/science.1157774
Rössig L., Li H., Fisslthaler B., Urbich C., Fleming I., Förstermann U., et al. (2002). Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ. Res. 91 (9), 837–844. doi:10.1161/01.res.0000037983.07158.b1
Ryu D., Jo Y. S., Lo S. G., Stein S., Zhang H., Perino A., et al. (2014). A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab. 20 (5), 856–869. doi:10.1016/j.cmet.2014.08.001
Saavedra F., Rivera C., Rivas E., Merino P., Garrido D., Hernández S., et al. (2017). PP32 and SET/TAF-Iβ proteins regulate the acetylation of newly synthesized histone H4. Nucleic Acids Res. 45 (20), 11700–11710. doi:10.1093/nar/gkx775
Saha R. N., Pahan K. (2006). HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 13 (4), 539–550. doi:10.1038/sj.cdd.4401769
Salminen A., Kaarniranta K. (2009). SIRT1: regulation of longevity via autophagy. Cell Signal 21 (9), 1356–1360. doi:10.1016/j.cellsig.2009.02.014
Samant S. A., Zhang H. J., Hong Z., Pillai V. B., Sundaresan N. R., Wolfgeher D., et al. (2014). SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol. Cell Biol. 34 (5), 807–819. doi:10.1128/MCB.01483-13
Samata M., Alexiadis A., Richard G., Georgiev P., Nuebler J., Kulkarni T., et al. (2020). Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 182 (1), 127–144.e23. doi:10.1016/j.cell.2020.05.026
Sandor V., Senderowicz A., Mertins S., Sackett D., Sausville E., Blagosklonny M. V., et al. (2000). P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br. J. Cancer 83 (6), 817–825. doi:10.1054/bjoc.2000.1327
Sanguigno L., Guida N., Anzilotti S., Cuomo O., Mascolo L., Serani A., et al. (2023). Stroke by inducing HDAC9-dependent deacetylation of HIF-1 and Sp1, promotes TfR1 transcription and GPX4 reduction, thus determining ferroptotic neuronal death. Int. J. Biol. Sci. 19 (9), 2695–2710. doi:10.7150/ijbs.80735
Schermuly R. T., Ghofrani H. A., Wilkins M. R., Grimminger F. (2011). Mechanisms of disease: pulmonary arterial hypertension. Nat. Rev. Cardiol. 8 (8), 443–455. doi:10.1038/nrcardio.2011.87
Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008). Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 382 (3), 790–801. doi:10.1016/j.jmb.2008.07.048
Schmitt C. A. (2003). Senescence, apoptosis and therapy--cutting the lifelines of cancer. Nat. Rev. Cancer 3 (4), 286–295. doi:10.1038/nrc1044
Schroeder E. A., Raimundo N., Shadel G. S. (2013). Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 17 (6), 954–964. doi:10.1016/j.cmet.2013.04.003
Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006). Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U. S. A. 103 (27), 10224–10229. doi:10.1073/pnas.0603968103
Sebti S., Prébois C., Pérez-Gracia E., Bauvy C., Desmots F., Pirot N., et al. (2014). BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc. Natl. Acad. Sci. U. S. A. 111 (11), 4115–4120. doi:10.1073/pnas.1313618111
Seligson D. B., Horvath S., Shi T., Yu H., Tze S., Grunstein M., et al. (2005). Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435 (7046), 1262–1266. doi:10.1038/nature03672
Semer M., Bidon B., Larnicol A., Caliskan G., Catez P., Egly J. M., et al. (2019). DNA repair complex licenses acetylation of H2A.Z.1 by KAT2A during transcription. Nat. Chem. Biol. 15 (10), 992–1000. doi:10.1038/s41589-019-0354-y
Sen P., Shah P. P., Nativio R., Berger S. L. (2016). Epigenetic mechanisms of longevity and aging. Cell 166 (4), 822–839. doi:10.1016/j.cell.2016.07.050
Seo K. S., Park J. H., Heo J. Y., Jing K., Han J., Min K. N., et al. (2015). SIRT2 regulates tumour hypoxia response by promoting HIF-1α hydroxylation. Oncogene 34 (11), 1354–1362. doi:10.1038/onc.2014.76
She D. T., Wong L. J., Baik S. H., Arumugam T. V. (2018). SIRT2 inhibition confers neuroprotection by downregulation of FOXO3a and MAPK signaling pathways in ischemic stroke. Mol. Neurobiol. 55 (12), 9188–9203. doi:10.1007/s12035-018-1058-0
Shi L., Yan H., An S., Shen M., Jia W., Zhang R., et al. (2019). SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol. Oncol. 13 (2), 358–375. doi:10.1002/1878-0261.12408
Silvestre M. F., Viollet B., Caton P. W., Leclerc J., Sakakibara I., Foretz M., et al. (2014). The AMPK-SIRT signaling network regulates glucose tolerance under calorie restriction conditions. Life Sci. 100 (1), 55–60. doi:10.1016/j.lfs.2014.01.080
Simonsson M., Heldin C. H., Ericsson J., Grönroos E. (2005). The balance between acetylation and deacetylation controls Smad7 stability. J. Biol. Chem. 280 (23), 21797–21803. doi:10.1074/jbc.M503134200
Singh B. N., Zhang G., Hwa Y. L., Li J., Dowdy S. C., Jiang S. W. (2010). Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer Ther. 10 (6), 935–954. doi:10.1586/era.10.62
Sivanand S., Rhoades S., Jiang Q., Lee J. V., Benci J., Zhang J., et al. (2017). Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67 (2), 252–265.e6. doi:10.1016/j.molcel.2017.06.008
Song C., Hotz-Wagenblatt A., Voit R., Grummt I. (2017). SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 31 (13), 1370–1381. doi:10.1101/gad.300624.117
Song C., Zhu S., Wu C., Kang J. (2013). Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J. Biol. Chem. 288 (39), 28021–28033. doi:10.1074/jbc.M113.498758
Song L., Zhong P., Yu R., Yuan Y., Zhou Y., Qian Y., et al. (2024). Effect of HDAC9-induced deacetylation of glycolysis-related GAPDH lysine 219 on rotavirus replication in rotavirus-infected Caco-2 cells. Virus Genes 60 (6), 621–634. doi:10.1007/s11262-024-02104-4
Song M. Y., Wang J., Ka S. O., Bae E. J., Park B. H. (2016). Insulin secretion impairment in Sirt6 knockout pancreatic β cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci. Rep. 6, 30321. doi:10.1038/srep30321
Soutoglou E., Katrakili N., Talianidis I. (2000). Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5 (4), 745–751. doi:10.1016/s1097-2765(00)80253-1
Spange S., Wagner T., Heinzel T., Krämer O. H. (2009). Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41 (1), 185–198. doi:10.1016/j.biocel.2008.08.027
Spencer T. E., Jenster G., Burcin M. M., Allis C. D., Zhou J., Mizzen C. A., et al. (1997). O’Malley BW: steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389 (6647), 194–198. doi:10.1038/38304
Steffan J. S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B. L., et al. (2001). Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413 (6857), 739–743. doi:10.1038/35099568
Su H., Liu W. (2018). PIK3C3/VPS34 control by acetylation. Autophagy 14 (6), 1086–1087. doi:10.1080/15548627.2017.1385676
Su H., Yang F., Wang Q., Shen Q., Huang J., Peng C., et al. (2017). VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell 67 (6), 907–921.e7. doi:10.1016/j.molcel.2017.07.024
Sun J., Tai S., Tang L., Yang H., Chen M., Xiao Y., et al. (2021). Acetylation modification during autophagy and vascular aging. Front. Physiol. 12, 598267. doi:10.3389/fphys.2021.598267
Sun N., Youle R. J., Finkel T. (2016). The mitochondrial basis of aging. Mol. Cell 61 (5), 654–666. doi:10.1016/j.molcel.2016.01.028
Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005). A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U. S. A. 102 (37), 13182–13187. doi:10.1073/pnas.0504211102
Sun Y., Jiang X., Price B. D. (2010). Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9 (5), 930–936. doi:10.4161/cc.9.5.10931
Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., et al. (2009). Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11 (11), 1376–1382. doi:10.1038/ncb1982
Sundaresan N. R., Gupta M., Kim G., Rajamohan S. B., Isbatan A., Gupta M. P. (2009). Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest 119 (9), 2758–2771. doi:10.1172/JCI39162
Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136 (18), 3033–3042. doi:10.1242/dev.033183
Tang X., Gao J. S., Guan Y. J., McLane K. E., Yuan Z. L., Ramratnam B., et al. (2007). Acetylation-dependent signal transduction for type I interferon receptor. Cell 131 (1), 93–105. doi:10.1016/j.cell.2007.07.034
Tao R., de Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., et al. (2007). Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13 (11), 1299–1307. doi:10.1038/nm1652
Taylor G. C., Eskeland R., Hekimoglu-Balkan B., Pradeepa M. M., Bickmore W. A. (2013). H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23 (12), 2053–2065. doi:10.1101/gr.155028.113
Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., et al. (2008). A molecular determinant for the establishment of sister chromatid cohesion. Science 321 (5888), 566–569. doi:10.1126/science.1157880
Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., et al. (2008). Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102 (6), 703–710. doi:10.1161/CIRCRESAHA.107.164558
Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., Sánchez-Madrid F. (2008). HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 18 (6), 291–297. doi:10.1016/j.tcb.2008.04.003
Valerio D. G., Xu H., Eisold M. E., Woolthuis C. M., Pandita T. K., Armstrong S. A. (2017). Histone acetyltransferase activity of MOF is required for adult but not early fetal hematopoiesis in mice. Blood 129 (1), 48–59. doi:10.1182/blood-2016-05-714568
Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004). Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16 (1), 93–105. doi:10.1016/j.molcel.2004.08.031
Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., et al. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107 (2), 149–159. doi:10.1016/s0092-8674(01)00527-x
Vazquez B. N., Thackray J. K., Simonet N. G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., et al. (2016). SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. Embo J. 35 (14), 1488–1503. doi:10.15252/embj.201593499
Ventura M., Mateo F., Serratosa J., Salaet I., Carujo S., Bachs O., et al. (2010). Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int. J. Biochem. Cell Biol. 42 (10), 1672–1680. doi:10.1016/j.biocel.2010.06.014
Victorelli S., Salmonowicz H., Chapman J., Martini H., Vizioli M. G., Riley J. S., et al. (2023). Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature 622 (7983), 627–636. doi:10.1038/s41586-023-06621-4
Walter R. A., Nairn J., Duncan D., Price N. C., Kelly S. M., Rigden D. J., et al. (1999). The role of the C-terminal region in phosphoglycerate mutase. Biochem. J. 337 (Pt 1), 89–95. doi:10.1042/bj3370089
Wang J., Liu N., Liu Z., Li Y., Song C., Yuan H., et al. (2008). The orphan nuclear receptor Rev-erbbeta recruits Tip60 and HDAC1 to regulate apolipoprotein CIII promoter. Biochim. Biophys. Acta 1783 (2), 224–236. doi:10.1016/j.bbamcr.2007.09.004
Wang S., Fairall L., Pham T. K., Ragan T. J., Vashi D., Collins M. O., et al. (2023). A potential histone-chaperone activity for the MIER1 histone deacetylase complex. Nucleic Acids Res. 51 (12), 6006–6019. doi:10.1093/nar/gkad294
Wang W. Y., Pan L., Su S. C., Quinn E. J., Sasaki M., Jimenez J. C., et al. (2013). Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 16 (10), 1383–1391. doi:10.1038/nn.3514
Wang X. X., Wang X. L., Tong M. M., Gan L., Chen H., Wu S. S., et al. (2016). SIRT6 protects cardiomyocytes against ischemia/reperfusion injury by augmenting FoxO3α-dependent antioxidant defense mechanisms. Basic Res. Cardiol. 111 (2), 13. doi:10.1007/s00395-016-0531-z
Wang Y., Huang Y., Liu J., Zhang J., Xu M., You Z., et al. (2020). Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 21 (1), e48335. doi:10.15252/embr.201948335
Wang Y., Li W., Schulz V. P., Zhao H., Qu X., Qi Q., et al. (2021). Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood 138 (17), 1615–1627. doi:10.1182/blood.2020007401
Wang Y., Yue J., Xiao M., Lu X., Chin Y. E. (2022). SIRT4-Catalyzed deacetylation of Axin1 modulates the wnt/β-catenin signaling pathway. Front. Oncol. 12, 872444. doi:10.3389/fonc.2022.872444
Ward T., Wang M., Liu X., Wang Z., Xia P., Chu Y., et al. (2013). Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288 (22), 15771–15785. doi:10.1074/jbc.M112.448886
Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324 (5930), 1076–1080. doi:10.1126/science.1164097
Wood M., Rymarchyk S., Zheng S., Cen Y. (2018). Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch. Biochem. Biophys. 638, 8–17. doi:10.1016/j.abb.2017.12.009
Wu H., Ma H., Wang L., Zhang H., Lu L., Xiao T., et al. (2022). Regulation of lung epithelial cell senescence in smoking-induced COPD/emphysema by microR-125a-5p via Sp1 mediation of SIRT1/HIF-1a. Int. J. Biol. Sci. 18 (2), 661–674. doi:10.7150/ijbs.65861
Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., et al. (2012). EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U. S. A. 109 (41), 16564–16569. doi:10.1073/pnas.1202639109
Xia Z., Kon N., Gu A. P., Tavana O., Gu W. (2022). Deciphering the acetylation code of p53 in transcription regulation and tumor suppression. Oncogene 41 (22), 3039–3050. doi:10.1038/s41388-022-02331-9
Xian Y., Liu B., Shen T., Yang L., Peng R., Shen H., et al. (2025). Enhanced SIRT3 expression restores mitochondrial quality control mechanism to reverse osteogenic impairment in type 2 diabetes mellitus. Bone Res. 13 (1), 30. doi:10.1038/s41413-024-00399-5
Xie L., Li C., Wang C., Wu Z., Wang C., Chen C., et al. (2024). Aspirin-mediated acetylation of SIRT1 maintains intestinal immune homeostasis. Adv. Sci. (Weinh) 11 (19), e2306378. doi:10.1002/advs.202306378
Xu G., Schell J., Quan S., Gao Y., Wei S. J., Pan M., et al. (2025). Mitochondrial ACSS1-K635 acetylation knock-in mice exhibit altered liver lipid metabolism on a ketogenic diet. Free Radic. Biol. Med. 232, 260–268. doi:10.1016/j.freeradbiomed.2025.03.009
Xu S. N., Wang T. S., Li X., Wang Y. P. (2016). SIRT2 activates G6PD to enhance NADPH production and promote leukaemia cell proliferation. Sci. Rep. 6, 32734. doi:10.1038/srep32734
Xu X., Ha C. H., Wong C., Wang W., Hausser A., Pfizenmaier K., et al. (2007). Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler. Thromb. Vasc. Biol. 27 (11), 2355–2362. doi:10.1161/ATVBAHA.107.151704
Yamagata K., Kitabayashi I. (2009). Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem. Biophys. Res. Commun. 390 (4), 1355–1360. doi:10.1016/j.bbrc.2009.10.156
Yan P., Li Z., Xiong J., Geng Z., Wei W., Zhang Y., et al. (2021). LARP7 ameliorates cellular senescence and aging by allosterically enhancing SIRT1 deacetylase activity. Cell Rep. 37 (8), 110038. doi:10.1016/j.celrep.2021.110038
Yang C. N., Lin S. K., Kok S. H., Wang H. W., Lee Y. L., Shun C. T., et al. (2021). The possible role of sirtuin 5 in the pathogenesis of apical periodontitis. Oral Dis. 27 (7), 1766–1774. doi:10.1111/odi.13723
Yang J. H., Hayano M., Griffin P. T., Amorim J. A., Bonkowski M. S., Apostolides J. K., et al. (2023). Loss of epigenetic information as a cause of mammalian aging. Cell 186 (2), 305–326.e27. doi:10.1016/j.cell.2022.12.027
Yang L., You J., Yang X., Jiao R., Xu J., Zhang Y., et al. (2025). ACSS2 drives senescence-associated secretory phenotype by limiting purine biosynthesis through PAICS acetylation. Nat. Commun. 16 (1), 2071. doi:10.1038/s41467-025-57334-3
Yang X., Wei J., He Y., Jing T., Li Y., Xiao Y., et al. (2017). SIRT1 inhibition promotes atherosclerosis through impaired autophagy. Oncotarget 8 (31), 51447–51461. doi:10.18632/oncotarget.17691
Yang X., Yu W., Shi L., Sun L., Liang J., Yi X., et al. (2011). HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44 (1), 39–50. doi:10.1016/j.molcel.2011.07.032
Yang Y., Fiskus W., Yong B., Atadja P., Takahashi Y., Pandita T. K., et al. (2013). Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. U. S. A. 110 (17), 6841–6846. doi:10.1073/pnas.1217692110
Yang Y., Kueh A. J., Grant Z. L., Abeysekera W., Garnham A. L., Wilcox S., et al. (2022). The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood 139 (6), 845–858. doi:10.1182/blood.2021013954
Yang Y., Wang W., Xiong Z., Kong J., Qiu Y., Shen F., et al. (2016). Activation of SIRT3 attenuates triptolide-induced toxicity through closing mitochondrial permeability transition pore in cardiomyocytes. Toxicol In Vitro 34, 128–137. doi:10.1016/j.tiv.2016.03.020
Yao H., Yao Z., Zhang S., Zhang W., Zhou W. (2018). Upregulation of SIRT1 inhibits H2O2-induced osteoblast apoptosis via FoxO1/β-catenin pathway. Mol. Med. Rep. 17 (5), 6681–6690. doi:10.3892/mmr.2018.8657
Ye J., Zhong S., Wan H., Guo X., Yao X., Liu Q., et al. (2025). Upregulated astrocyte HDAC7 induces Alzheimer-like tau pathologies via deacetylating transcription factor-EB and inhibiting lysosome biogenesis. Mol. Neurodegener. 20 (1), 5. doi:10.1186/s13024-025-00796-2
Yu J., Sun W., Song Y., Liu J., Xue F., Gong K., et al. (2019). SIRT6 protects retinal ganglion cells against hydrogen peroxide-induced apoptosis and oxidative stress by promoting Nrf2/ARE signaling via inhibition of Bach1. Chem. Biol. Interact. 300, 151–158. doi:10.1016/j.cbi.2019.01.018
Yuan J., Minter-Dykhouse K., Lou Z. (2009). A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 185 (2), 203–211. doi:10.1083/jcb.200809167
Zeng Z., Chen C., SiTu Y., Shen Z., Chen Y., Zhang Z., et al. (2022). Anoectochilus roxburghii flavonoids extract ameliorated the memory decline and reduced neuron apoptosis via modulating SIRT1 signaling pathway in senescent mice. J. Ethnopharmacol. 296, 115361. doi:10.1016/j.jep.2022.115361
Zhang F., Wang D., Li J., Su Y., Liu S., Lei Q. Y., et al. (2022b). Deacetylation of MTHFD2 by SIRT4 senses stress signal to inhibit cancer cell growth by remodeling folate metabolism. J. Mol. Cell Biol. 14 (4), mjac020. doi:10.1093/jmcb/mjac020
Zhang H. L., Han W., Du Y. Q., Zhao B., Yang P., Yin D. M. (2021b). SRC3 acetylates calmodulin in the mouse brain to regulate synaptic plasticity and fear learning. J. Biol. Chem. 297 (3), 101044. doi:10.1016/j.jbc.2021.101044
Zhang J., Shi X., Li Y., Kim B. J., Jia J., Huang Z., et al. (2008). Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31 (1), 143–151. doi:10.1016/j.molcel.2008.06.006
Zhang L., Pitcher L. E., Yousefzadeh M. J., Niedernhofer L. J., Robbins P. D., Zhu Y. (2022a). Cellular senescence: a key therapeutic target in aging and diseases. J. Clin. Invest 132 (15), e158450. doi:10.1172/JCI158450
Zhang L., Zhu K., Xu J., Chen X., Sheng C., Zhang D., et al. (2023c). Acetyltransferases CBP/p300 control transcriptional switch of β-catenin and Stat1 promoting osteoblast differentiation. J. Bone Min. Res. 38 (12), 1885–1899. doi:10.1002/jbmr.4925
Zhang N., Zhang Y., Qian H., Wu S., Cao L., Sun Y. (2020). Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 27 (9), 2605–2619. doi:10.1038/s41418-020-0523-2
Zhang N., Zhang Y., Wu B., Wu S., You S., Lu S., et al. (2021a). Deacetylation-dependent regulation of PARP1 by SIRT2 dictates ubiquitination of PARP1 in oxidative stress-induced vascular injury. Redox Biol. 47, 102141. doi:10.1016/j.redox.2021.102141
Zhang N., Zhang Y., Xu J., Wang P., Wu B., Lu S., et al. (2023a). α-myosin heavy chain lactylation maintains sarcomeric structure and function and alleviates the development of heart failure. Cell Res. 33 (9), 679–698. doi:10.1038/s41422-023-00844-w
Zhang P., Tu B., Wang H., Cao Z., Tang M., Zhang C., et al. (2014). Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. U. S. A. 111 (29), 10684–10689. doi:10.1073/pnas.1411026111
Zhang Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., et al. (2015). Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525 (7569), 389–393. doi:10.1038/nature15252
Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., et al. (2003). HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. Embo J. 22 (5), 1168–1179. doi:10.1093/emboj/cdg115
Zhang Y., Yang Y., Yang F., Liu X., Zhan P., Wu J., et al. (2023d). HDAC9-mediated epithelial cell cycle arrest in G2/M contributes to kidney fibrosis in male mice. Nat. Commun. 14 (1), 3007. doi:10.1038/s41467-023-38771-4
Zhang Y., Zhang M., Dong H., Yong S., Li X., Olashaw N., et al. (2009). Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28 (3), 445–460. doi:10.1038/onc.2008.388
Zhang Y., Zhang N., Zou Y., Song C., Cao K., Wu B., et al. (2023b). Deacetylation of Septin4 by SIRT2 (silent mating type information regulation 2 homolog-2) mitigates damaging of hypertensive nephropathy. Circ. Res. 132 (5), 601–624. doi:10.1161/CIRCRESAHA.122.321591
Zhao D., Zou S. W., Liu Y., Zhou X., Mo Y., Wang P., et al. (2013). Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell 23 (4), 464–476. doi:10.1016/j.ccr.2013.02.005
Zhao L. J., Subramanian T., Zhou Y., Chinnadurai G. (2006a). Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J. Biol. Chem. 281 (7), 4183–4189. doi:10.1074/jbc.M509051200
Zhao M., Zhang X., Tao X., Zhang B., Sun C., Wang P., et al. (2021). Sirt2 in the spinal cord regulates chronic neuropathic pain through nrf2-mediated oxidative stress pathway in rats. Front. Pharmacol. 12, 646477. doi:10.3389/fphar.2021.646477
Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., et al. (2010). Regulation of cellular metabolism by protein lysine acetylation. Science 327 (5968), 1000–1004. doi:10.1126/science.1179689
Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., et al. (2006b). Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol. Cell Biol. 26 (7), 2782–2790. doi:10.1128/MCB.26.7.2782-2790.2006
Zheng Y., Chen H., Yin M., Ye X., Chen G., Zhou X., et al. (2015). MiR-376a and histone deacetylation 9 form a regulatory circuitry in hepatocellular carcinoma. Cell Physiol. Biochem. 35 (2), 729–739. doi:10.1159/000369733
Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140 (2), 280–293. doi:10.1016/j.cell.2009.12.041
Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., et al. (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11 (4), 468–476. doi:10.1038/ncb1854
Zhu C., Chen Q., Xie Z., Ai J., Tong L., Ding J., et al. (2011). The role of histone deacetylase 7 (HDAC7) in cancer cell proliferation: regulation on c-Myc. J. Mol. Med. Berl. 89 (3), 279–289. doi:10.1007/s00109-010-0701-7
Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K. P., Göttlicher M. (2004). Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 5 (5), 455–463. doi:10.1016/s1535-6108(04)00114-x
Zou X., Zhu Y., Park S. H., Liu G., O'Brien J., Jiang H., et al. (2017). SIRT3-Mediated dimerization of IDH2 directs cancer cell metabolism and tumor growth. Cancer Res. 77 (15), 3990–3999. doi:10.1158/0008-5472.CAN-16-2393
Glossary
ACLY ATP-citrate lyase
ACSS2 acetate-dependent acetyl-CoA synthetase 2
AMPK AMP-activated protein kinase
APP amyloid precursor protein
ATM ataxia-telangiectasia mutated kinase
ATAT1 alpha-tubulin acetyltransferase 1
BACE1 beta-site amyloid precursor protein cleaving enzyme 1
BRCA1 breast cancer type 1 susceptibility protein
BRD4 bromodomain-containing protein 4
CBP CREB-binding protein
Cdk2 cyclin-dependent kinase 2
CNS central nervous system
CNK1 kinase suppressor of Ras-1
DNMT1 DNA (cytosine-5)-Methyltransferase 1
DDR DNA damage response
DSB double strand break
E2F1 E2 promoter binding factor 1
eNOS Endothelial nitric oxide synthase
ERK extracellular-signal-regulated kinases
FoxO3a forkhead box-O-3a
GAPDH Glyceraldehyde-3-phosphate dehydrogenase
GCN5 general control non-depressible 5
GSK3β glycogen synthase kinase 3β
H2AX H2A histone family member X
HATs acetyltransferases
HDACs deacetylases
HDR homology-directed repair
HNF4 hepatocyte nuclear factor 4
HR homologous recombination
Hsp70 heat shock protein 70
IDH2 isocitrate dehydrogenase 2
IFN-13/17 interleukin-13/17
IL-6 interleukin-6
IRS2 insulin receptor substrate 2
ISGF3 IFN-stimulated gene factor 3
JNK c-Jun N-terminal kinase
KATs lysine acetyltransferases
KDACs lysine deacetylases
LARP7 La-related protein 7
LCAD long-chain acyl-CoA dehydrogenase
LDHA lactate dehydrogenase A
LKB1 live kinase B1
MAPK mitogen-activated protein kinases
MDM2 murine double minute 2
MEF2 myocyte enhancer factor 2
MKP1 mitogen-activated protein kinase phosphatase 1
MMPs matrix metalloproteinases
MnSOD superoxide dismutase
mCAT catalase targeted to the mitochondria
mTOR mechanistic target of rapamycin
NHEJ non-homologous end-joining
Nrf2 nuclear factor erythroid 2-related factor 2
PARP-1 poly (ADP-ribose) polymerase 1
PCAF P300/CBP-associated factor
PD Parkinson’s disease
PDK1 3-phosphoinositide-dependent kinase 1
PEPCK1 Phosphoenolpyruvate carboxy-kinase
PGC-1α peroxisome proliferator-activated receptor-gamma co-activator-alpha
PIK3C3 phosphatidylinositol 3-kinase catalytic subunit type 3
PIP3 phosphatidylinositol (3,4,5)-triphosphate
PKcs DNA-dependent protein kinase catalytic subunit
PS1 presenilin-1
PT1B protein tyrosine phosphatase 1B
PTEN phosphatase and tensin homolog
PTMs post-transcriptional modifications
PYCR1 pyrroline-5-carboxylate reductase 1
PYGO2 pygopus2
RAF rapidly accelerated fibrosarcoma kinase
ROS reactive oxygen species
RUBCN RUN domain and Cysteine-rich domain containingBeclin 1-interacting protein
SAGA Spt-Ada-Gen5 acetyltransferase
SASP senescence-associated secretory phenotype
SIRT3 Sirtuins deacetylase family 3
SKP2 S-phase kinase-associated protein 2
Smad7 small mothers against decapentaplegic 7
SMC3 Structural maintenance of chromosomes 3
Smurf1 Sad ubiquitination regulatory factor 1
SOD2 superoxide dismutase 2
STAT3 signal transducer and activator of transcription 3
TFEB transcription factor EB
TGF-β transforming growth factor-beta
TIP60 Tat-interacting protein 60
TNF-α tumor necrosis factor-alpha
TRIMs tripartite motif proteins
UHRF1 ubiquitin-like with PHD and RING Finger domains 1
ULK1 UNC-52-like kinase 1
VSMCs vascular smooth muscle cells
VPS34 vacuolar protein sorting 34
53BP1 tp53-binding protein 1
Keywords: acetylation modification, aging, DNA stability, mitochondrial function, network mechanism, protein homeostasis
Citation: Xu M, Wang W, Lu S, Xiong M, Zhao T, Yu Y, Song C, Yang J, Zhang N, Cao L, Sun G, Chen S and Wang P (2025) The advances in acetylation modification in senescence and aging-related diseases. Front. Physiol. 16:1553646. doi: 10.3389/fphys.2025.1553646
Received: 31 December 2024; Accepted: 28 April 2025;
Published: 12 May 2025.
Edited by:
Dong Han, People’s Liberation Army General Hospital, ChinaReviewed by:
Celia Lía Quijano, Universidad de la República, UruguayShravan Kumar Paswan, University of Texas Health Science Center at Houston, United States
Copyright © 2025 Xu, Wang, Lu, Xiong, Zhao, Yu, Song, Yang, Zhang, Cao, Sun, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengbo Wang, cGJ3YW5nQGNtdS5lZHUuY24=; Sichong Chen, c2NjaGVuQGNtdS5lZHUuY24=
†These authors have contributed equally to this work
 Maiqi Xu1†
Maiqi Xu1† Wenbin Wang
Wenbin Wang Naijin Zhang
Naijin Zhang Guozhe Sun
Guozhe Sun Pengbo Wang
Pengbo Wang