- Department of Biological Sciences, University of Cincinnati, Cincinnati, OH, United States
Adaptive responses to hypoxia are likely accompanied by highly diverse changes in gene expression. Here, we examined the transcriptomic regulation in blood samples derived from independently-derived captive cave-dwelling fish. These fish are members of the species Astyanax mexicanus, which comprises two morphs: an obligate subterranean form, and a “surface-dwelling” form that lives in rivers and streams located near cave localities. These morphs diverged ∼20,000–200,000 years ago, and cavefish derived from multiple, distinct cave localities have adapted to life in hypoxic waters. Here, we focused on captive-reared Astyanax morphs since elevated hemoglobin levels persist in cavefish despite rearing in the normoxic conditions of a laboratory. A GO enrichment analysis revealed several instances of convergent gene regulation between some, but not all, cavefish populations. This finding suggests that different gene expression patterns have evolved in response to hypoxia across geologically-distinct cave localities. Additionally, we identified differential regulation of numerous genes of the canonical hypoxic response pathway. Interestingly, some genes activating this pathway were expressed lower in captive-reared cavefish. These patterns of gene expression may have evolved in cavefish as a consequence of negative pleiotropic consequences associated with prolonged hif gene expression. At present, it is unknown whether this finding is a function of captivity, or whether these expression patterns are also present in wild populations. Collectively, this work provides new insights to the transcriptomic regulation of hypoxia tolerance using a cavefish model evolving in distinct oxygenated environments.
Introduction
A number of transcriptomic studies in teleosts reveal that adaptation to low oxygen is accompanied by diverse changes in gene regulation. These gene expression alterations impact diverse processes such as metabolic suppression, intracardiac cooperation, increase in gill surface area, vasculature growth, and red blood cell overproduction. Hemoglobin family members are common targets of hypoxic stress, including the preferential expression of hemoglobin (hb) isoforms with unusually high oxygen affinity and sensitivity to allosteric regulators [reviewed in Nikinmaa and Rees (2005), Xiao (2015), Fago and Jensen (2015)]. Many of these traits are controlled by changes in expression of the hypoxia inducible factor (Hif) (Mandic et al., 2021).
Hypoxia is present in a variety of environments including frozen ponds, reef platforms at low tide, high altitude, deep-sea, aquatic environments with algal blooms, and caves (Storz, 2018). Here, we examined adaptation to hypoxia in cavefish with ancestors that evolved in a limestone cave complex in the Sierra de El Abra region of northeastern Mexico (Figure 1). Over 30 caves populated by cavefish populations are found in this region (Miranda-Gamboa et al., 2023). EL Abra caves are characterized by limited or absent light, minimal nutrition, and lower dissolved oxygen compared to the terrestrial environment. Each cave, however, is unique with respect to formation process, elevation, size, inhabitant fauna, volume of terrestrial input, and other factors (Elliott, 2018). Despite these differences, cavefish derived from these habitats evolve a number of convergent phenotypic features.
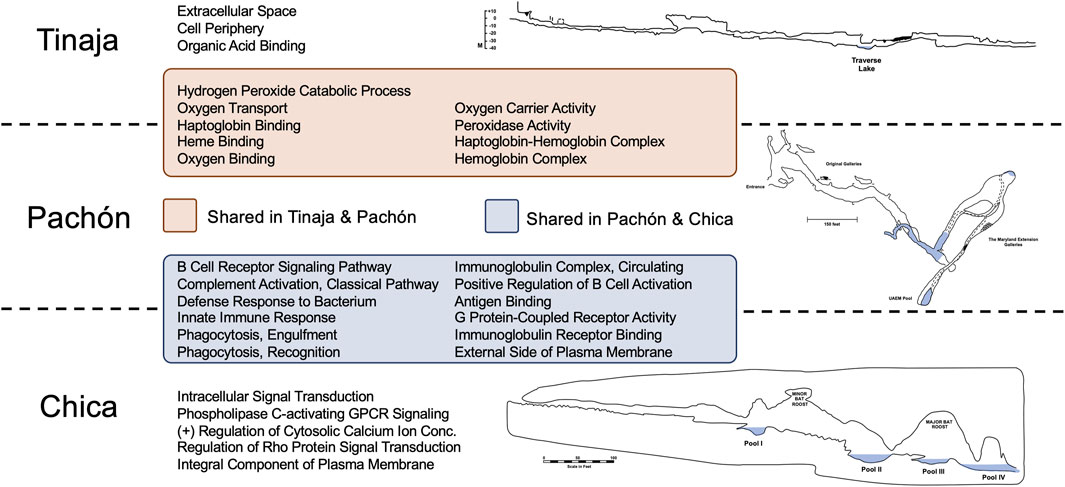
Figure 1. GO Enrichment revealed many shared and unique terms across cave populations relating to oxygen transport and immune function. Statistically overrepresented terms in Tinaja and Pachón cavefish compared to surface fish largely represented oxygen binding and transport. The majority of overrepresented terms in Pachón and Chica cavefish related to immune function. Interestingly, there were no overlapping terms between Tinaja and Chica compared to surface fish. The distinct geography and environmental histories of these caves have likely impacted global gene expression patterns differently in Astyanax cavefish.
These fish are members of the species Astyanax mexicanus, which comprises two morphotypes: an obligate cave-dwelling fish lacking eyes and pigmentation, and a terrestrial “surface” fish with stereotypical teleost features. It is estimated that ∼20,000–200,000 years ago these cave environments were colonized by surface-dwelling lineages (Herman et al., 2018; Fumey et al., 2018; Moran et al., 2023; Garduño-Sánchez et al., 2023). Extant cave and surface morphs inhabit starkly contrasting environments, providing the opportunity to examine evolutionary changes in closely-related morphotypes inhabiting environments marked by different levels of oxygen.
van der Weele and Jeffery (2022) discovered juvenile cavefish from the Pachón cave locality grow normally in hypoxic conditions, but surface fish do not. By 36 h post fertilization (hpf), cavefish produce more red blood cells than surface fish. This red blood cell expansion is accompanied by increased expression of certain hemoglobin genes, expanded hematopoietic domains, and elevated expression of several hif gene family members. Interestingly, prior work has shown that adult Astyanax cavefish, reared in captivity, show increased hemoglobin protein concentration in three different populations (Pachón, Tinaja, and Chica) compared to surface fish (Boggs et al., 2022). This concentration of adult hemoglobin is underpinned, in part, by larger red blood cells. However, elevated hemoglobin protein levels are mediated by the expression of different hemoglobin gene family members (Boggs and Gross, 2025).
Here, we examined how hypoxic adaptation impacts gene regulation by measuring broad scale transcriptomic regulation. We focused our attention to captive-reared Astyanax morphs since hemoglobin elevation persists in cave morphs, despite being reared in normoxic conditions. The results of a gene enrichment analysis of the blood transcriptomes of different morphs revealed shared enrichment patterns between Pachón and Chica cavefish, and Pachón and Tinaja cavefish (however Chica and Tinaja cavefish showed no overlap). Notably, many instances of overlap likely reflect convergent increases in expression of hemoglobin genes. However, we also identified numerous genes associated with canonical hypoxia response pathways. Many genes normally activating these pathways were expressed lower in cavefish compared to surface fish, and certain genes typically suppressing these pathways were expressed higher in cavefish. These surprising patterns may reflect the negative consequences that can arise as a function of prolonged hif expression. At present, it is unclear if these findings are a function of the captive conditions in which cavefish are reared in the lab, and whether these observations translate to natural populations as well. In any respect, this work provides news insight to the transcriptomic architecture of hypoxia tolerance, through use of a unique model that permits intraspecific comparison of morphs evolving in different oxygenated environments.
Results and discussion
Convergent and divergent regulation suggests cavefish suppress canonical hypoxia response pathways in normoxic captivity
We examined transcriptional gene regulation of blood by performing statistical overrepresentation analyses of Gene Ontology (GO) terms using PANTHERdb (Mi et al., 2019; Thomas et al., 2022). Accordingly, we scored annotated genes demonstrating significant two-fold (or higher) differences in gene expression in each cave population relative to surface fish. This resulted in six analyses, i.e., three pair-wise comparisons performed for both over- and under-expression. GO terms enrichments (FDR adjusted p-value <0.05) from each analysis were compared to identify convergent/divergent expression patterns between cave populations. A prior study using the same dataset provided expression validation through analysis of five genes subjected to quantitative real-time PCR (qPCR) and calculated delta Cq using ssr3 as the reference gene [see Boggs and Gross (2025)]. This study revealed an average correlation coefficient of 0.89, indicating a strong relationship (Cohen et al., 2009) and validation of our RNA-seq dataset.
Interestingly, we discovered substantial overlap between Tinaja and Pachón, and Pachón and Chica (Figure 1). These overlapping sets included every identified GO term for Pachón, however no overlap was observed for Chica and Tinaja cavefish. Many GO terms shared between Tinaja and Pachón cavefish were significant due to hemoglobin genes [see Boggs and Gross (2025)] including: oxygen transport, heme binding, oxygen binding, and hemoglobin complex (Figure 1). Overlapping terms between Pachón and Chica mostly reflected terms associated with immune system function, including: defense response to bacterium, innate immune response, phagocytosis recognition, and antigen binding.
These results were not entirely surprising given that prior GO enrichment studies in Astyanax identified convergent mechanisms of cave adaptation, including broad development processes (Riddle et al., 2020), metabolism (Krishnan et al., 2020), and immunity (Krishnan et al., 2022). Additionally, the number and diversity of genes within a test list affects the outcome of overrepresentation studies (Wijesooriya et al., 2022). Given that whole blood is a highly complex tissue (capable of predicting an estimated 60% of gene expression for dozens of tissues) (Basu et al., 2021), this likely impacted the statistical outcomes of the analysis.
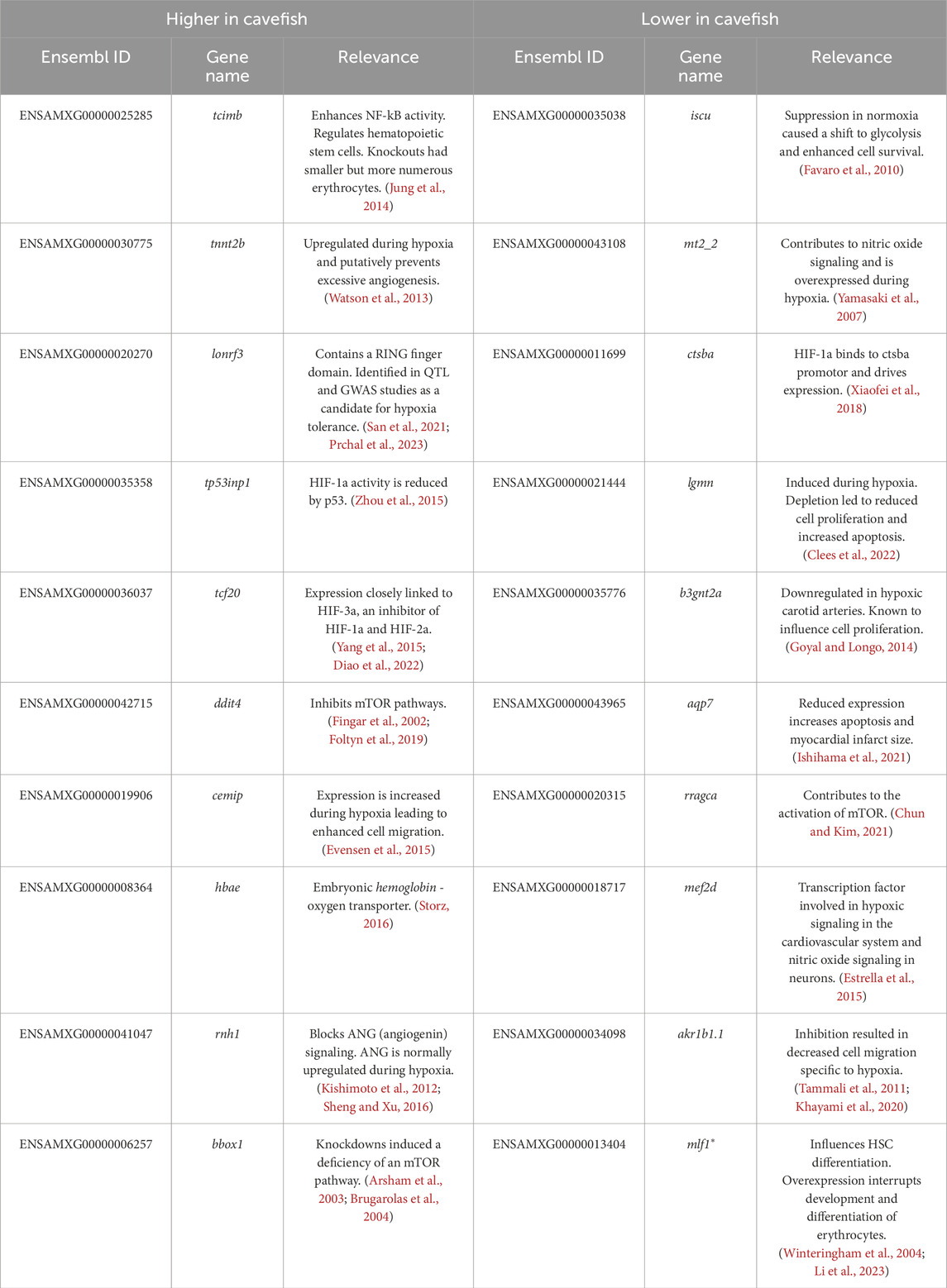
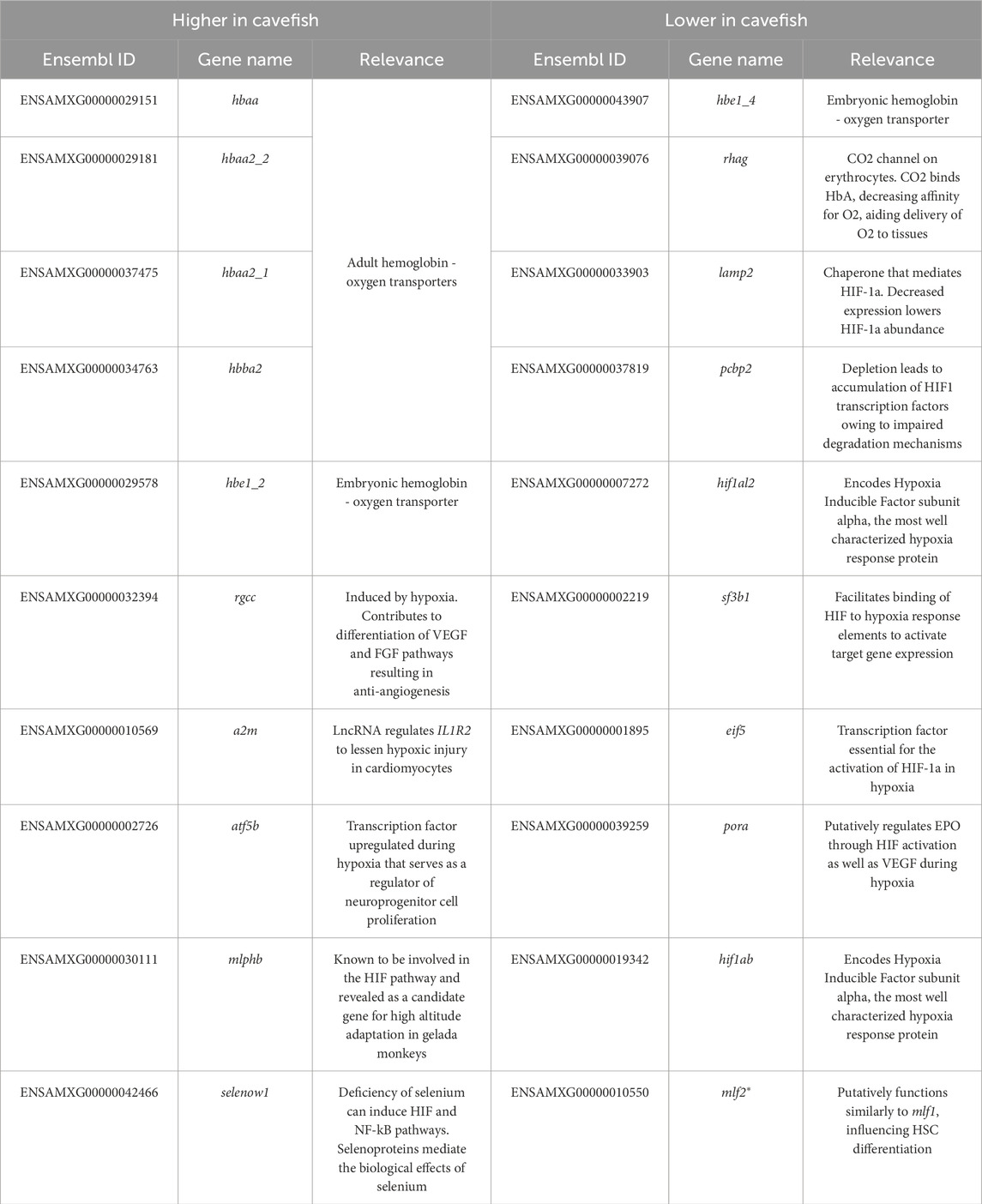
We further aimed to investigate genes of potential interest that may not have been detected in these GO analyses. Accordingly, we created four lists representing genes that are biologically-relevant to hypoxia including: genes expressed higher or lower in all examined cave populations compared to surface fish (Table 1) and genes expressed higher and lower in Tinaja and Pachón compared to surface fish (Table 2), while excluding Chica, given their similarity in hemoglobin expression. A literature search for each of these genes was conducted to provide any potential relevance to adaptation to hypoxic caves.
Consistent with prior findings (Boggs and Gross, 2025), many hemoglobin genes were expressed higher in cavefish compared to surface fish (Tables 1 and 2) with the vast majority expressed higher only in Tinaja and Pachón (relative to Chica and Surface). We also identified numerous genes associated with canonical hypoxia response pathways. Interestingly, many genes normally activating these pathways were expressed lower in cavefish compared to surface fish, and genes typically suppressing these pathways were expressed higher in cavefish (Tables 1, 2). Notably, two hypoxia inducible factor (hif) genes (hif1al2 and hif1ab) and multiple genes contributing to HIF signaling, including cathepsin Ba (ctsba), lysosomal associated membrane protein 2 (lamp2), splicing factor 3b subunit 1 (sf3b1), eukaryotic translation initiation factor 5 (eif5), and p450 (cytochrome) oxidoreductase a (pora) were expressed lower in cavefish compared to surface fish. Additionally, two genes known to suppress HIF signaling (tp53inp1 and tcf20) were expressed higher in cavefish compared to surface fish.
In normoxic conditions, hif is continuously transcribed, but is controlled post-translationally by prolyl hydroxylase (PHD) and von Hippel-Lindau (VHL) proteins. During hypoxia, PHD activity is inhibited and Hif is not degraded. Thus, hif transcript abundance is not necessarily representative of Hif activity in mammals (Semenza and Wang, 1992; Maxwell et al., 1999; Ivan et al., 2001; Jaakkola et al., 2001; Bruick and McKnight, 2001; Epstein et al., 2001). In the Chinese sucker (Myxocyprinus asiaticus), a study revealed increased hif transcription is required to prevent degradation of Hif during hypoxia (Chen et al., 2012). Having said this, a recent study uncovered diverse reports of hifa mRNA abundance in fish exposed to hypoxia, as a likely function of varying methodologies (Murphy and Rees, 2024). Nevertheless, elasmobranch fish conditioned to hypoxia express hif higher than individuals that have not experienced hypoxia (Rytkönen et al., 2012). Additionally, certain hif family members are expressed higher in Pachón cavefish embryos (after normoxic rearing or exposure to hypoxia) than in surface fish (van der Weele and Jeffery, 2022). Thus, we were initially surprised to find that adult cavefish express two hif family members much lower than surface fish and express other known hypoxia response genes in similar, counterintuitive, patterns. In light of varying reports of hifa transcription in fish (Murphy and Reese, 2024), it will be essential to better characterize protein levels of hif1a in forthcoming studies through the use of Western blot analyses.
One explanation for these observed patterns may be the negative consequences associated with prolonged expression of hif. Hif is linked to many human pathologies including tumorigenesis, cardiovascular, metabolic, and reproductive diseases [reviewed in Chen et al. (2020)]. In mice, pharmacological knock-down of Hif protein relieved symptoms of rheumatoid arthritis (Hu et al., 2020). Hif pathways can also impair major histocompatibility complex function in culture, leading to an inability to recognize and eliminate cancerous and other harmful cells (Sethumadhavan et al., 2017). Additionally, Hif proteins influence ion fluctuations and homeostasis in fish, a well-characterized mechanism to conserve energy during hypoxia [reviewed in Pelster and Egg (2018)]. Thus, future work in Astyanax may determine if downregulation of hif and other known hypoxia response pathways are advantageous in cavefish to save energy, maintain proper immune function, and prevent disease and inflammation.
Prior work in Astyanax revealed that oxygen levels are a good deal lower in cave waters compared to surface waters (Boggs and Gross, 2025). An important consideration for this study is the fact that all experimental animals were reared in normoxic conditions. Indeed, our putative Chica cavefish were acquired from a commercial vendor, and therefore it is not possible for us to determine the extent to which transcriptomic changes are a function of assimilation to captivity. Interestingly, a number of cave populations maintain significantly elevated levels of hemoglobin despite rearing in normoxia for generations (Boggs and Gross, 2025). Given that the transcriptome can change markedly when comparing captive-bred versus wild-caught individuals (Krishnan et al., 2020), an essential future direction for this research includes examination of the blood transcriptome from individuals drawn from the natural population.
Materials and methods
Animal husbandry and tissue collection
Astyanax cave and surface fish were reared in a satellite aquatic facility at the University of Cincinnati within a custom-designed reverse osmosis husbandry unit comprised of 5- and 10- gallon continuous flow tanks (Aquaneering, San Diego, CA). Animals were exposed to a 12:12 h light: dark cycle and fed a slurry of dry flake food (TetraMin Pro) and system water daily. Water in this system is processed through a series of filters including UV, 25-micron polypropylene felt, activated carbon, and dense particulate. Additionally, water conditions were adjusted using real-time dose monitoring of sodium bicarbonate and Instant Ocean sea salt to conductivity of 750 μS/cm (±50 μS/cm) and pH of 7.4 (±0.2). Water temperature was kept at 24°C (±2°C). Importantly, dissolved oxygen was not manipulated for this study meaning all fish were exposed to ample oxygen.
The surface fish, Pachón cavefish and Tinaja cavefish used in this study were derived from breeding adults originally provided to our lab by Dr. Richard Borowsky (New York University). Specifically, the pedigrees used included Asty-152 and Asty-155 (surface fish), Asty-163 and Asty-138 (Pachón cavefish), and Asty-19 (Tinaja cavefish). Surface fish are descended from wild-caught individuals from the Río Sabinas and Río Valles drainages near Ciudad Valles in San Luis Potosí, Mexico. All Chica cavefish were acquired from the commercial pet trade. We extracted whole blood from (n = 4) surface, Pachón, Tinaja, and Chica populations (total n = 16) via the caudal vein using 31G syringes (BD Ultra-Fine™, BD Biosciences, San Jose, CA). In order to limit any potential effects outside the scope of this study, two male and two female fish were used from each population, fish were post-breeding age, and whole blood extractions were completed between 12:00 p.m - 1:00 p.m. All procedures were conducted in accordance with University of Cincinnati IACUC (Protocol# 22-01-06-01).
RNA isolation, sequencing, and read processing
Immediately following whole blood extraction, whole RNA was isolated using an RNeasy Universal Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s directions. All RNA samples were subjected to quantification using a Nanodrop Lite spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). The purity of samples was estimated based on the A260/A280 ratio, and only RNA samples of ∼2.0 were submitted for sequencing. Owing to the technical requirements of RNA-sequencing, samples had to be pooled by population. To mitigate potential effects of sequencing error, each pool was sub-aliquoted into three technical replicates with each replicate (n = 12) containing the same volume of RNA. Pools were submitted to the DNA Core at Cincinnati Childrens’ Hospital and Medical Center. There, additional RNA QC was conducted, polyA stranded libraries were generated, QC was conducted on the libraries, and they were subject to sequencing using an Illumina HiSeq 2,500 sequencer. This resulted in twenty-million 125bp paired end reads per sample. Raw reads were assessed for quality and length using FastQC (Wingett and Andrews, 2018) (version 0.11.8) and adapters were trimmed using Trimmomatic (Bolger et al., 2014) (version 0.39).
RNA sequencing
Analysis of gene expression was conducted by running a reference based analysis in CLC Genomics (Qiagen, Germantown, MD, version 12.0.1) using manufacturer recommended parameters. The latest Astyanax genome (AstMex3_surface, GCA_023375975.1) was used as the reference sequence. We used the latest annotation file for this reference from NCBI RefSeq (GCF_023375975.1, NCBI annotation release 103).
In order to increase efficiency of downstream transcriptome-wide analysis, we conducted a second RNA sequencing experiment in CLC Genomics using the “Astyanax-mexicanus-2.0” genome retrieved from Ensembl [GCA_000372685.2 (Warren et al., 2021)] as the reference and annotations from Ensembl release 106 were used to identify genes and determine expression. RNA-sequencing was validated using qPCR for five genes [see Boggs and Gross (2025)].
Gene ontology enrichment and candidate gene nomination
To investigate transcriptome-wide patterns of gene expression, we conducted a Gene Ontology (GO) Statistical Overrepresentation Test. Each cave population was assessed independently against surface fish. Thus, we created seven gene lists, one list representing genes expressed higher in a cave population versus surface fish, one list representing genes expressed lower in a cave population versus surface, and a list containing all genes detectable in this assay [noise threshold surpassed with TPM value of at least 2 (Wagner et al., 2013)]. Each list representing a comparison between a cave and surface population contained genes detectable for at least one of the two populations and with a fold change of at least 2x (any gene with a TPM value of 0 was substituted with the lowest TPM value in the entire dataset - 0.00192,433 in Tinaja fat1a–so that a fold change value could be calculated). We used PANTHERdb (Mi et al., 2019; Thomas et al., 2022) (version 17.0) to conduct a statistical overrepresentation test. Because Astyanax GO terms are not available in PANTHER, IDs in each list were converted to orthologous Danio rerio IDs by using BioMart (Smedley et al., 2009). We successfully converted 6,882 of 8,550 (∼80%) IDs from our and used these as our reference (Aleksander et al., 2023) for the statistical overrepresentation test. We used a Fisher’s Exact text to calculate p-values which were corrected using false discovery rate to determine statistical significance. Each of three categories of GO terms were assessed: biological process, molecular function, and cellular component. Results from each cave-to-surface analysis were then compared to determine convergence/divergence between cave populations.
In addition, we investigated genes of potential interest that may have been missed in the GO analysis. Thus, we compiled four additional lists of genes: two lists representing genes of putative biological relevance that are either expressed higher or lower in Chica, Tinaja, and Pachón cavefish compared to surface fish as well as two lists expressed higher or lower in Tinaja and Pachón compared to surface. Expression data derived from Chica cavefish was omitted from these lists owing to the difference in expression of hemoglobin compared to Tinaja and Pachón. Genes were ranked according to putative biological relevance. Rank was determined by subtracting the fold change (cavefish expression value divided by surface fish expression value) of a gene from each cavefish expression value and summing the absolute values from each cave population. Genes that have not been characterized were removed and the remaining genes were filtered for relevance to hypoxia using literature searches.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject PRJNA1079358.
Ethics statement
The animal study was approved by University of Cincinnati IACUC. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
TB: Investigation, Writing – review and editing, Methodology, Software, Supervision, Conceptualization, Writing – original draft, Validation, Data curation, Visualization, Formal Analysis, Project administration. LB: Formal Analysis, Data curation, Writing – original draft, Investigation. JG: Writing – review and editing, Funding acquisition, Supervision, Writing – original draft, Resources, Conceptualization, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. JBG is supported by the following grants from the National Science Foundation: IOS-2205928 and DEB-2343857.
Acknowledgments
The authors wish to thank members of the Gross lab for assistance with animal husbandry, especially Danny Berning and Kaitlyn Reimer. Additionally, we are grateful for helpful discussions of these results with Alyssa Hamm.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aleksander S. A., Balhoff J., Carbon S., Cherry J. M., Drabkin H. J., Ebert D., et al. (2023). The gene ontology knowledgebase in 2023. Genetics 224, iyad031. doi:10.1093/genetics/iyad031
Arsham A. M., Howell J. J., Simon M. C. (2003). A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278, 29655–29660. doi:10.1074/jbc.M212770200
Basu B., Wang W., Ruppin R., Hannenhalli H. (2021). Predicting tissue-specific gene expression from whole blood transcriptome. Sci. Adv. 7, eabd6991. doi:10.1126/sciadv.abd6991
Boggs T. E., Friedman J. S., Gross J. B. (2022). Alterations to cavefish red blood cells provide evidence of adaptation to reduced subterranean oxygen. Sci. Rep. 12, 3735. doi:10.1038/s41598-022-07619-0
Boggs T. E., Gross J. B. (2025). Elevated blood hemoglobin in different cavefish populations evolves through diverse hemoglobin gene expression patterns. J. Exp. Zoology Part B Mol. Dev. Evol. 344, 175–181. doi:10.1002/jez.b.23289
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., et al. (2004). Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes and Dev. 18 (23), 2893–2904. doi:10.1101/gad.1256804
Bruick R. K., McKnight S. L. (2001). A conserved family of Prolyl-4-Hydroxylases that modify HIF. Science 294, 1337–1340. doi:10.1126/science.1066373
Chen N., Chen L. P., Zhang J., Chen C., Wei X. L., Gul Y., et al. (2012). Molecular characterization and expression analysis of three hypoxia-inducible factor alpha subunits, HIF-1α/2α/3α of the hypoxia-sensitive freshwater species, Chinese sucker. Gene. 498, 81–90. doi:10.1016/j.gene.2011.12.058
Chen P., Chiu W., Hsu P., Lin S., Peng I., Wang C., et al. (2020). Pathophysiological implications of hypoxia in human diseases. J. Biomed. Sci. 27, 63. doi:10.1186/s12929-020-00658-7
Chun Y., Kim J. (2021). AMPK–mTOR signaling and cellular adaptations in hypoxia. Int. J. Mol. Sci. 22, 9765. doi:10.3390/ijms22189765
Clees A., Stolp V., Häupl B., Fuhrmann D. C., Wempe F., Seibert M., et al. (2022). Identification of the cysteine protease legumain as a potential chronic hypoxia-specific multiple myeloma target gene. Cells 11, 292. doi:10.3390/cells11020292
Cohen I., Huang Y., Chen J., Benesty J., Benesty J., Chen J., et al. (2009). “Pearson correlation coefficient,” in Noise reduction in speech processing, 1–4.
Diao X., Ye F., Zhang M., Ren X., Tian X., Lu J., et al. (2022). Identification of oleoylethanolamide as an endogenous ligand for HIF-3α. Nat. Commun. 13 (1), 2529. doi:10.1038/s41467-022-30338-z
Elliott W. R. (2018). The astyanax caves of Mexico: Cavefishes of Tamaulipas, San Luis Potosí, and Guerrero. Assoc. Mexican Cave Stud. 26, 1–325.
Epstein A. C. R., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 107, 43–54. doi:10.1016/s0092-8674(01)00507-4
Estrella N. L., Desjardins C. A., Nocco S. E., Clark A. L., Maksimenko Y., Naya F. J. (2015). MEF2 transcription factors regulate distinct gene programs in Mammalian skeletal muscle differentiation. J. Biol. Chem. 290, 1256–1268. doi:10.1074/jbc.M114.589838
Evensen N. A., Li Y., Kuscu C., Liu J., Cathcart J., Banach A., et al. (2015). Hypoxia promotes Colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP). Oncotarget 6, 20723–20739. doi:10.18632/oncotarget.3978
Fago A., Jensen F. B. (2015). Hypoxia tolerance, nitric oxide, and nitrite: lessons from extreme animals. Physiology 30, 116–126. doi:10.1152/physiol.00051.2014
Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M., et al. (2010). MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. Plos One 5, e10345. doi:10.1371/journal.pone.0010345
Fingar D. C., Salama S., Tsou C., Harlow E. D., Blenis J. (2002). Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487. doi:10.1101/gad.995802
Foltyn M., Luger A. L., Lorenz N. I., Sauer B., Mittelbronn M., Harter P. N., et al. (2019). The physiological mTOR complex 1 inhibitor DDIT4 mediates therapy resistance in glioblastoma. Br. J. Cancer 120 (5), 481–487. doi:10.1038/s41416-018-0368-3
Fumey J., Hinaux H., Noirot C., Thermes C., Rétaux S., Casane D. (2018). Evidence for late Pleistocene origin of Astyanax mexicanus cavefish. BMC Evol. Biol. 18, 43. doi:10.1186/s12862-018-1156-7
Garduño-Sánchez M., Hernández-Lozano J., Moran R. L., Miranda-Gamboa R., Gross J. B., Rohner N., et al. (2023). Phylogeographic relationships and morphological evolution between cave and surface Astyanax mexicanus populations (de filippi 1853) (actinopterygii, characidae). Mol. Ecol. doi:10.1111/mec.17128
Goyal R., Longo L. D. (2014). Acclimatization to long-term hypoxia: gene expression in ovine carotid arteries. Physiol. Genomics 46, 725–734. doi:10.1152/physiolgenomics.00073.2014
Herman A., Brandvain Y., Weagley J., Jeffery W. R., Keene A. C., Kono T. J. Y., et al. (2018). The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra, Astyanax mexicanus. Mol. Ecol. 27, 4397–4416. doi:10.1111/mec.14877
Hu Y., Zhang T., Chen J., Cheng W., Chen J., Zheng Z., et al. (2020). Downregulation of hypoxia-inducible Factor-1α by RNA interference alleviates the development of collagen-induced arthritis in rats. Mol. Ther. - Nucleic Acids 19, 1330–1342. doi:10.1016/j.omtn.2020.01.014
Ishihama S., Yoshida S., Yoshida T., Mori Y., Ouchi N., Eguchi S., et al. (2021). LPL/AQP7/GPD2 promotes glycerol metabolism under hypoxia and prevents cardiac dysfunction during ischemia. FASEB J. 35, e22048. doi:10.1096/fj.202100882R
Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., et al. (2001). HIFalpha targeted for VHL-Mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. doi:10.1126/science.1059817
Jaakkola P., Mole D. R., Tian Y., Wilson M. I., Gielbert J., Gaskell S. J., et al. (2001). Targeting of HIF-α to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science. 292, 468–472. doi:10.1126/science.1059796
Jung Y., Kim M., Soh H., Lee S., Kim J., Park S., et al. (2014). TC1(C8orf4) regulates hematopoietic stem/progenitor cells and hematopoiesis. Plos One 9, e100311. doi:10.1371/journal.pone.0100311
Khayami R., Hashemi S. R., Kerachian M. A. (2020). Role of aldo-keto reductase family 1 member B1 (AKR1B1) in the cancer process and its therapeutic potential. J. Cell Mol. Med. 24, 8890–8902. doi:10.1111/jcmm.15581
Kishimoto K., Yoshida S., Ibaragi S., Yoshioka N., Okui T., Hu G., et al. (2012). Hypoxia-induced up-regulation of angiogenin, besides VEGF, is related to progression of oral cancer. Oral Oncol. 48, 1120–1127. doi:10.1016/j.oraloncology.2012.05.009
Krishnan J., Persons J. L., Peuß R., Hassan H., Kenzior A., Xiong S., et al. (2020). Comparative transcriptome analysis of wild and lab populations of Astyanax mexicanus uncovers differential effects of environment and morphotype on gene expression. J. Exp. Zool. Mol. Dev. Evol. 334, 530–539. doi:10.1002/jez.b.22933
Krishnan J., Wang Y., Kenzior O., Hassan H., Olsen L., Tsuchiya D., et al. (2022). Liver-derived cell lines from cavefish Astyanax mexicanus as an in vitro model for studying metabolic adaptation. Sci. Rep. 12, 10115. doi:10.1038/s41598-022-14507-0
Li Z., Yang Y., Wu K., Li Y., Shi M. (2023). Myeloid leukemia factor 1: a “double-edged sword” in health and disease. Front. Oncol. 13, 1124978. doi:10.3389/fonc.2023.1124978
Mandic M., Joyce W., Perry S. F. (2021). The evolutionary and physiological significance of the hif pathway in teleost fishes. J. Exp. Biol. 224, jeb231936. doi:10.1242/jeb.231936
Maxwell P. H., Wiesener M. S., Chang G., Clifford S. C., Vaux E. C., Cockman M. E., et al. (1999). The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275. doi:10.1038/20459
Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., et al. (2019). Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v. 14.0). Nat. Protoc. 14, 703–721. doi:10.1038/s41596-019-0128-8
Miranda-Gamboa R., Espinasa L., de los Angeles Verde-Ramírez M., Hernández-Lozano J., Lacaille J. L., Espinasa M., et al. (2023). A new cave population of Astyanax mexicanus from Northern Sierra de El Abra, Tamaulipas, Mexico. Subterr. Biol. 45, 95–117. doi:10.3897/subtbiol.45.98434
Moran R. L., Richards E. J., Ornelas-García C. P., Gross J. B., Donny A., Wiese J., et al. (2023). Selection-driven trait loss in independently evolved cavefish populations. Nat. Commun. 14, 2557. doi:10.1038/s41467-023-37909-8
Murphy T. E., Rees B. B. (2024). Diverse responses of hypoxia-inducible factor alpha mRNA abundance in fish exposed to low oxygen: the importance of reporting methods. Front. Physiology 15, 1496226. doi:10.3389/fphys.2024.1496226
Nikinmaa M., Rees B. B. (2005). Oxygen-dependent gene expression in fishes. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 288, R1079–R1090. doi:10.1152/ajpregu.00626.2004
Pelster B., Egg M. (2018). Hypoxia-inducible transcription factors in fish: expression, function and interconnection with the circadian clock. J. Exp. Biol. 221, jeb163709. doi:10.1242/jeb.163709
Prchal M., D'Ambrosio J., Lagarde H., Lallias D., Patrice P., François Y., et al. (2023). Genome-wide association study and genomic prediction of tolerance to acute hypoxia in rainbow trout. Aquaculture 565, 739068. doi:10.1016/j.aquaculture.2022.739068
Riddle M. R., Aspiras A. C., Damen F., Hutchinson J. N., Chinnapen D. J.-, Tabin J., et al. (2020). Genetic architecture underlying changes in carotenoid accumulation during the evolution of the blind Mexican cavefish, Astyanax mexicanus. J. Exp. Zool. Mol. Dev. Evol. 334, 405–422. doi:10.1002/jez.b.22954
Rytkönen K. T., Renshaw G. M., Vainio P. P., Ashton K. J., Williams-Pritchard G., Leder E. H., et al. (2012). Transcriptional responses to hypoxia are enhanced by recurrent hypoxia (Hypoxic preconditioning) in the epaulette shark. Physiol. genomics 44 (22), 1090–1097. doi:10.1152/physiolgenomics.00081.2012
San L., Liu B., Liu B., Zhu K., Guo L., Guo H., et al. (2021). Genome-wide association study reveals multiple novel SNPs and putative candidate genes associated with low oxygen tolerance in golden pompano Trachinotus ovatus (linnaeus 1758). Aquaculture 544, 737098. doi:10.1016/j.aquaculture.2021.737098
Semenza G. L., Wang G. L. (1992). A Nuclear Factor Induced by Hypoxia via de novo Protein Synthesis Binds to the Human Erythropoietin Gene Enhancer at a Site Required for Transcriptional Activation. Mol. Cell Biol. 12, 5447–5454. doi:10.1128/mcb.12.12.5447
Sethumadhavan S., Silva M., Philbrook P., Nguyen T., Hatfield S. M., Ohta A., et al. (2017). Hypoxia and hypoxia-inducible factor (HIF) downregulate antigen-presenting MHC class I molecules limiting tumor cell recognition by T cells. Plos One 12, e0187314. doi:10.1371/journal.pone.0187314
Sheng J., Xu Z. (2016). Three decades of research on angiogenin: a review and perspective. Acta Biochim. Biophys. Sin. 48, 399–410. doi:10.1093/abbs/gmv131
Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., et al. (2009). BioMart–biological queries made easy. BMC Genomics 10, 22–12. doi:10.1186/1471-2164-10-22
Storz J. F. (2016). Gene duplication and evolutionary innovations in hemoglobin-oxygen transport. Physiology 31 (3), 223–232. doi:10.1152/physiol.00060.2015
Storz J. F. (2018). Hemoglobin: insights into protein structure, function, and evolution. Oxford University Press.
Tammali R., Saxena A., Srivastava S. K., Ramana K. V. (2011). Aldose reductase inhibition prevents hypoxia-induced increase in hypoxia-inducible factor-1alpha (HIF-1alpha) and vascular endothelial growth factor (VEGF) by regulating 26 S proteasome-mediated protein degradation in human Colon cancer cells. J. Biol. Chem. 286, 24089–24100. doi:10.1074/jbc.M111.219733
Thomas P. D., Ebert D., Muruganujan A., Mushayahama T., Albou L., Mi H. (2022). PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22. doi:10.1002/pro.4218
van der Weele C. M., Jeffery W. R. (2022). Cavefish cope with environmental hypoxia by developing more erythrocytes and overexpression of hypoxia-inducible genes. eLife 11, e69109. doi:10.7554/eLife.69109
Wagner G. P., Kin K., Lynch V. J. (2013). A model based criterion for gene expression calls using RNA-Seq data. Theory Biosci. 132, 159–164. doi:10.1007/s12064-013-0178-3
Warren W. C., Boggs T. E., Borowsky R., Carlson B. M., Ferrufino E., Gross J. B., et al. (2021). A chromosome-level genome of Astyanax mexicanus surface fish for comparing population-specific genetic differences contributing to trait evolution. Nat. Commun. 12, 1447. doi:10.1038/s41467-021-21733-z
Watson O., Novodvorsky P., Gray C., Rothman A. M. K., Lawrie A., Crossman D. C., et al. (2013). Blood flow suppresses vascular notch signalling via dll4 and is required for angiogenesis in response to hypoxic signalling. Cardiovasc Res. 100, 252–261. doi:10.1093/cvr/cvt170
Wijesooriya K., Jadaan S. A., Perera K. L., Kaur T., Ziemann M. (2022). Urgent need for consistent standards in functional enrichment analysis. PLOS Comput. Biol. 18, e1009935. doi:10.1371/journal.pcbi.1009935
Wingett S. W., Andrews S.(2018). FastQ Screen: A tool for multi-genome mapping and quality control. F1000 Research 7, 1338.
Winteringham L. N., Kobelke S., Williams J. H., Ingley E., Klinken S. P. (2004). Myeloid leukemia factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27Kip1 accumulation. Oncogene 23, 5105–5109. doi:10.1038/sj.onc.1207661
Xiao W. (2015). The hypoxia signaling pathway and hypoxic adaptation in fishes. Sci. China Life Sci. 58, 148–155. doi:10.1007/s11427-015-4801-z
Xiaofei C., Yanqing L., Dongkai Z., Dong C., Feng Z., Weilin W. (2018). Identification of cathepsin B as a novel target of hypoxia-inducible factor-1-alpha in HepG2 cells. Biochem. Biophysical Res. Commun. 503 (2), 1057–1062. doi:10.1016/j.bbrc.2018.06.116
Yamasaki M., Nomura T., Sato F., Mimata H. (2007). Metallothionein is up-regulated under hypoxia and promotes the survival of human prostate cancer cells. Oncol. Rep. 18, 1145–1153. doi:10.3892/or.18.5.1145
Yang S. L., Wu C., Xiong Z. F., Fang X. (2015). Progress on hypoxia-inducible factor-3: its structure, gene regulation and biological function (review). Mol. Med. Rep. 12 (2), 2411–2416. doi:10.3892/mmr.2015.3689
Keywords: hypoxia, subterranean, GO terms, enrichment analysis, normoxia
Citation: Boggs TE, Bucher LR and Gross JB (2025) RNA-seq analysis of blood from cave- and surface-dwelling Astyanax morphs reveal diverse transcriptomic responses to normoxic rearing. Front. Physiol. 16:1617136. doi: 10.3389/fphys.2025.1617136
Received: 23 April 2025; Accepted: 07 July 2025;
Published: 17 July 2025.
Edited by:
David Cristóbal Andrade, University of Antofagasta, ChileReviewed by:
Bernard B. Rees, University of New Orleans, United StatesWeiLiang Shen, Ningbo Academy of Oceanology and Fishery, China
Copyright © 2025 Boggs, Bucher and Gross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joshua B. Gross, Z3Jvc3NqYUB1Y21haWwudWMuZWR1
 Tyler E. Boggs
Tyler E. Boggs Joshua B. Gross
Joshua B. Gross