- 1Department of Veterinary and Biomedical Sciences, University of Minnesota, St. Paul, MN, United States
- 2Department of Animal Sciences, The Ohio State University, Wooster, OH, United States
- 3Department of Food Science and Human Nutrition, Michigan State University, East Lansing, MI, United States
Temperature extremes can compromise livestock welfare and pose serious threats to both economic stability and global food security. In commercial poultry production, newly hatched birds are particularly vulnerable to thermal stress, with growth-selected species such as turkeys being at heightened risk. To cope with temperature challenges, poultry undergo metabolic, physiological, and behavioral adaptations—responses that may have lasting effects on muscle development and, ultimately, meat quality. This study examined transcriptional changes in the breast muscle of young commercial turkey poults exposed to acute thermal stress. Hatchlings were brooded for 3 days at one of three temperatures: control (35 °C), cold (31 °C), or heat (39 °C). Pectoralis major muscle samples were collected, RNA extracted, and transcriptomes were analyzed via deep sequencing. Both cold and heat exposure resulted in reduced body weight compared to control poults. Both thermal stress conditions produced significant differential gene expression. In commercial birds, affected genes were involved in muscle differentiation and development, stress adaptation and apoptosis/protein turnover, energy metabolism and nutrient processing, as well as mitochondrial function and oxidative stress response. Notably, cold stress altered genes related to lipid and glucose metabolism (PDK4, ANGPTL4 and DGAT2), while heat stress affected genes (C/EBPβ and MUSTN1) were associated with differentiation and development and intracellular lipid accumulation. These findings provide a foundation for further studies into the genetic mechanisms driving physiological responses to thermal challenge in poultry.
1 Introduction
Shifts in world climate have resulted in an increase in mean temperature and the frequency of extreme weather events (Karl and Trenberth, 2003). Extremes in hot and cold temperature affect livestock wellbeing and have significant economic and food security impacts (Schmidhuber and Tubiello, 2007). Although thermal stress affects all livestock, poultry are particularly susceptible, especially young birds with poorly developed thermal regulation (Modrey and Nichelmann, 1992; Shinder et al., 2007).
Muscle development during post-hatch is multifaceted and growth-selected poultry–particularly the turkey–have increased proliferation and differentiation of muscle stem cells (satellite cells, SCs), increased breast muscle mass, and increased muscle fiber diameter leading to reduced muscle capillary density (Velleman et al., 2000). As a result, muscle of growth-selected birds has a larger volume to surface area ratio, limiting heat dissipation and increasing the potential for adverse effects on muscle development (Yahav, 2005). In broilers, changes in muscle structure and composition from early post-hatch cold or heat stress include increased lipid deposition and damage to muscle fibers, leading to inferior meat quality with consequent economic losses to producers and processors (Piestun et al., 2017).
Commercial turkey husbandry potentially exposes hatchlings to extreme temperatures during transportation of hatchlings from hatcheries to grow-out facilities (Mitchell and Kettlewell, 2009). Under thermal challenge, poultry alter their physiology and behavior to aid thermoregulation. For example, when heat challenged, birds use vasodilation to reduce blood flow to heat-generating organs and increase blood flow to some areas of skin, particularly areas with less feather coverage (Wolfenson et al., 1981). Group-brooded birds respond to cold by huddling and feather piloerection and metabolically through increased catabolic metabolism and skeletal muscle thermogenesis (shivering and non-shivering) (Rowland et al., 2015). Ultimately, changes in cellular processes are driven by underlying changes in gene expression.
Prior studies by our group examined the effect of thermal stress on gene expression in newly hatched turkey poults of two select research lines; a comparatively slower growing random-bred control 2 (RBC2) line (Nestor, 1977), and the faster growing F-line derived from the RBC2 via single-trait selection for 16 weeks body weight (Nestor, 1984). In both lines, significant differential gene expression was observed in the breast muscle of 3-day-old poults exposed to thermal challenge with a more pronounced transcriptional response to heat than to cold (Barnes et al., 2019). Slower growing random-bred birds displayed modulation of lipid-related genes, suggesting reduction in lipid storage, transport, and synthesis, consistent with changes in energy metabolism required to maintain body temperature. In contrast, growth-selected birds displayed changes in genes predicted to have downstream transcriptional effects, putting them at greater risk for temperature-induced oxidative muscle damage, and reduced muscle growth.
These findings suggest that growth selection of turkeys alters thermal tolerance. Likewise, contemporary commercial turkey lines intensively selected for several traits including breast muscle mass, will show a similar sensitivity to thermal challenge as other growth-selected birds such as the F-Line. This study was designed to characterize the transcriptional profiles of breast muscle from thermally challenged and non-challenged post-hatch commercial turkey poults. We hypothesized that early post-hatch thermal challenge alters turkey growth by modifying breast muscle hypertrophy and increasing adipose tissue deposition through altered gene expression.
2 Materials and methods
2.1 Birds and sample collection
The turkeys used in this study were obtained as fertilized eggs from Select Genetics, Terre Haute, IN. Fertile eggs (200) were transported in an ice chest from the hatchery to the MSU Poultry Teaching and Research Center. Upon arrival, the eggs were stored at 16 °C for 15 h then placed in an incubator (Petersime, Model 5, Gettysburg, OH) and maintained at 38 °C and 60% relative humidity (RH). One cracked egg was discarded. Ten days after placement in the incubator, eggs were candled and 12 were removed as nonfertile/nonviable. Eggs were further incubated at 38 °C, 60% RH until hatch. At the completion of hatch (94.9% of the viable eggs), all birds were weighed and randomly assigned to one of three temperature treatments and brooded at 35 °C (control), 31 °C or 39 °C and transferred to a 4-tier Super Brooder (Alternative Design Mfg., Siloam Springs, AR). Each tier consisted of two cages. The top tier was maintained at 39 °C, the second and third tiers at 35 °C, and the bottom tier at 31 °C. After day 1, 3 hatchlings in the non-experimental group were removed because of leg problems.
After 3 days of thermal challenge, eight male birds from each treatment were randomly removed from one cage for each temperature, weighed and euthanized via cervical dislocation. The pectoralis major was removed, snap frozen in liquid nitrogen, and stored at −80 °C until RNA isolation. Statistical comparison of body weights was performed with one-way ANOVA with α < 0.05 considered as significant. The Institutional Animal Care and Use Committee of Michigan State University (#01/14-017-00) approved all animal care procedures.
2.2 RNA isolation, library preparation, and sequencing
Total RNA was isolated from each sample at the University of Minnesota by extraction with TRIzol (Ambion, Inc., Foster City, CA). Samples (4 per treatment group) were DNase treated (Turbo DNA-free TM Kit, Ambion, Inc., Foster City, CA) and stored at – 80 °C. Indexed libraries were constructed with the Illumina TruSeq Stranded Preparation Kit (Illumina, Inc., San Diego, CA) at the University of Minnesota Genomics Center. Libraries were pooled and converted to Aviti-compatible libraries using the Element Adept Rapid PCR + kit (Element Biosciences., San Diego, CA). Libraries were sequenced on an Aviti Cloudbreak Medium 2 × 150-bp run (Element Biosciences, San Diego, CA) to generate ≥500M pass-filter reads with all expected barcode combinations detected. Data are deposited in the NCBI’s Gene Expression Omnibus repository as SRA BioProject PRJNA1273166.
2.3 RNA sequence and gene expression analysis
Quality control, data alignment, and gene quantification were analyzed using the CHURP pipeline (Baller et al., 2019) at the University of Minnesota Supercomputing Institute (MSI). The FASTQ paired-end reads (2 × 150 bp) were trimmed using Trimmomatic (v0.33) enabled with the optional “headcrop −3” option, “-q” option; 3bp sliding-window trimming from 3′ end requiring minimum Q30. Quality control on raw sequence data for each sample was performed with FastQC. Read mapping was performed via HISAT2 (Kim et al., 2019) (v2.1.0) using the turkey genome (Meleagris_gallopavo.Turkey_5.1.104. gtf) as a reference and gene quantification was done via Feature Counts for raw read counts. Principal component analysis (PCA) was performed to assess data structure and sample variation. Differentially expressed genes (DEGs) in treatment comparisons were identified using the edgeR feature in CLC Genomics Workbench (CLCGWB, Qiagen, Inc.) using raw read counts as input. For all comparisons, p-values less than 0.05 were considered significant. Results in the generated list of DEGs were filtered based on a minimum |Log2fold change| > 2.0, and FDR p-value <0.05.
Gene IDs were obtained from Ensembl with supplementation from the NCBI database RefSeq RNA database. Gene ontology and functional enrichment analyses of the DEGs were conducted using DAVID (https://davidbioinformatics.nih.gov/home.jsp) and Panther19.0 (https://pantherdb.org/) using the Gallus gallus gene set.
3 Results
Mean weight of mixed-sex poults at the start of the experiment was 57.4 g. Following 3 days of treatment (Supplementary Table S1), mean body weights of males in the Hot and Cold treatment groups, were essentially identical (95.6 and 95.4g, respectively). However, these were on average 9.9 g lower than the mean body weight of the Control temperature birds (102.4 g). Overall, this difference was marginally significant (ANOVA F = 3.627, p-value = 0.044).
3.1 RNA sequencing and gene expression
Twelve libraries were paired-end sequenced and the resulting reads mapped to the turkey genome (UMD 5.1, ENSEMBL annotation 104). Reads from two libraries (35. T1 and 39. T4) displayed both biased read mapping and aberrant clustering, indicating failure during library prep or conversion. Data from these two libraries were subsequently removed from further analyses. The remaining 10 paired-end libraries averaged 44.5M reads/library with consistent Q scores and diversity scores ranging from 39.7% to 50.0% (Table 1). Principal component analysis (PCA) showed limited separation of samples by treatment (Figure 1).
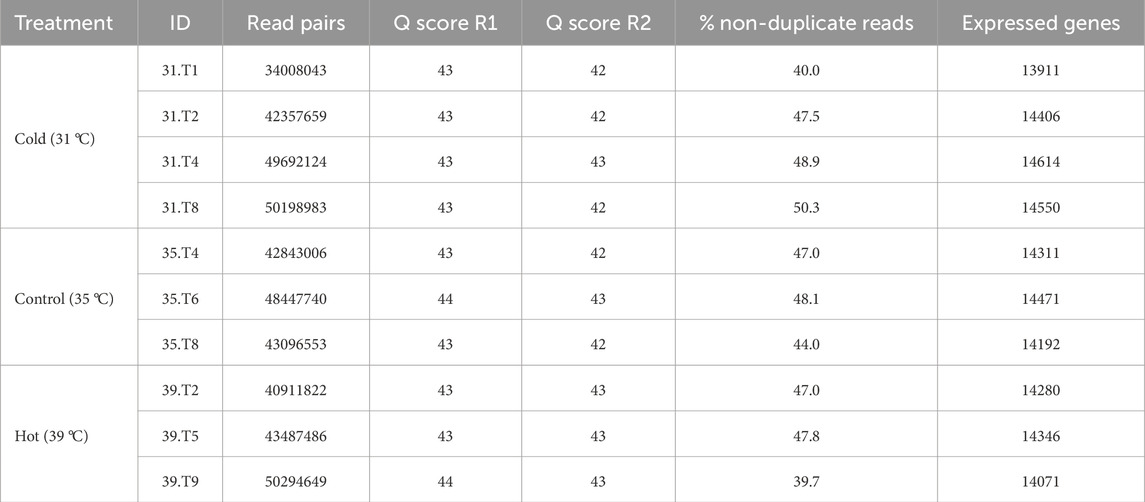
Table 1. Summary of RNA sequencing results. For each library the sample ID, number of read pairs per library, Average quality scores for forward and reverse reads, the percentage of non-duplicate reads and the number of expressed genes based on read alignments are given.
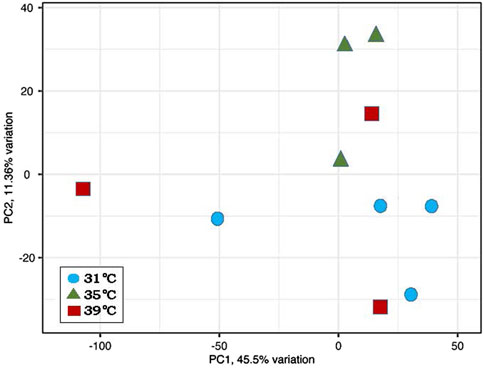
Figure 1. Principal component analysis (PCA) of normalized RNAseq read counts from turkey poults. Sample to sample distances are illustrated for each library dataset on the first two principal components with samples plotted by treatment.
Overall gene expression was similar between libraries with an average of 14,315.2 genes detected with a minimum of two mapped reads per sample (Table 1; Supplementary Table S2). The within-treatment group averages were also similar and averaged 14,309.08 expressed genes (Table 1). Genes with the highest experiment-wise read counts included, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), cytochrome c oxidase subunits I, II, and III (COX1, COX2, COX3), the and muscle-specific proteins actin (ACTA), myosin light chain (MYL1), troponin subunits (TNNT3, TNNI2, TNNC2) and a myosin motor domain-containing protein (ENSMGAG00000001884) (Supplementary Table S1).
3.2 Differential gene response to temperature
Temperature effects on gene expression in the breast muscle of the commercial poults were examined in two pairwise comparisons: 31 °C (cold) versus 35 °C (control), and 39 °C (hot) versus 35 °C (control). The cold-treatment comparison identified 67 significant DEGs (FDR adjusted P-value <0.05) with 55 having |Log2FC| > 2.0 (Table 2; Figure 2). Of those 55 DEGs, 35 were upregulated at 31 °C and 20 were downregulated. The gene showing the greatest upregulation was COL10A1 (collagen type X alpha one chain), a structural protein that is essential for the formation and maintenance of bone and may also be involved in the structure and remodeling of muscle extracellular matrix (Log2FC = 30.72), followed by ENSMGAG00000018664, a lncRNA (Log2FC = 15.85). The greatest downregulation was seen for U1 (U1 spliceosomal RNA), important in regulation of alternative polyA site selection (Log2FC = −7.18) and ENSMGAG00000021080, paramyosin-like (Log2FC = −5.025).
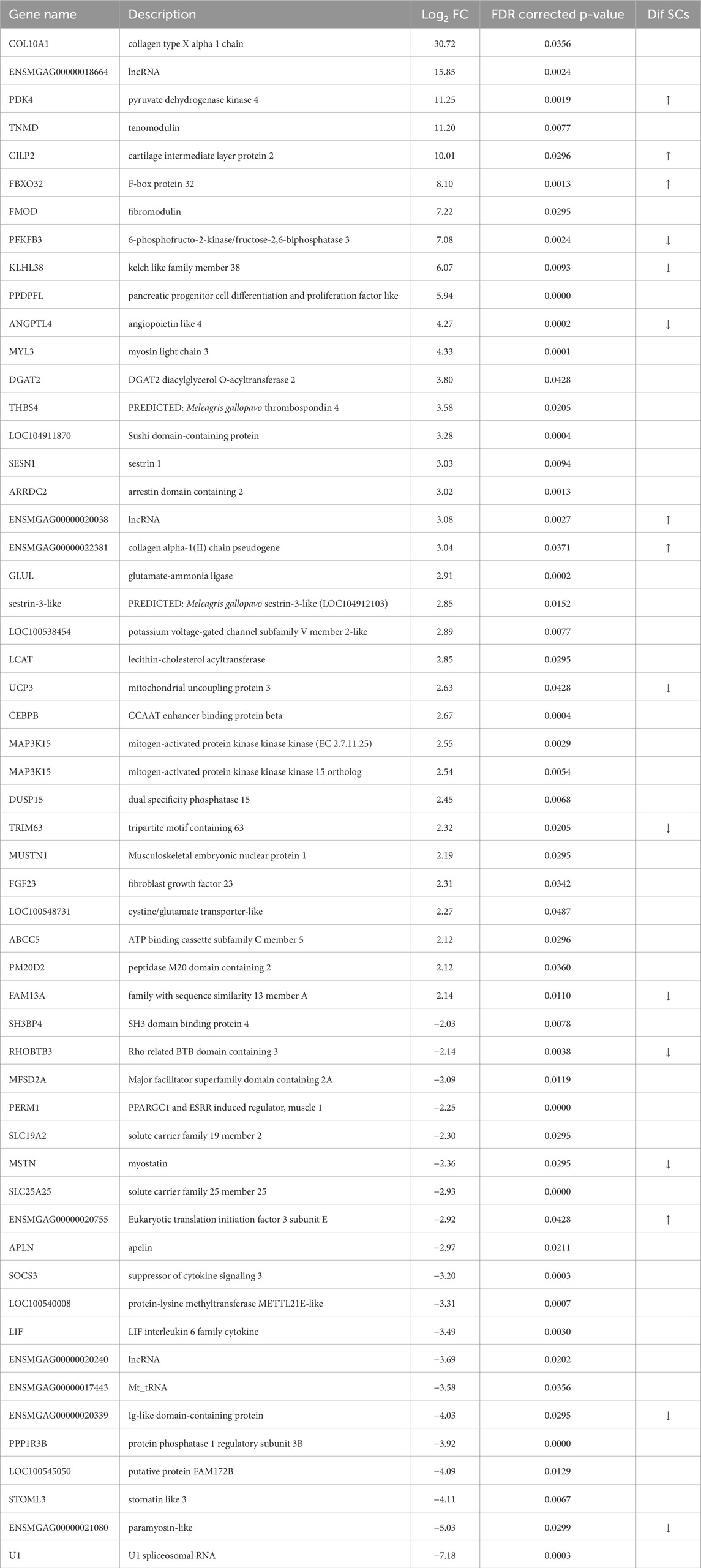
Table 2. Differentially expressed genes (DEGs) identified in comparison of RNAseq data from cold-treated (31 °C) 3 days turkey pouts compared to controls (35 °C). Arrows denote the directional change for genes that also had significant differential expression in differentiating muscle satellite cells (SCs) of commercial turkey following cold treatment (Reed et al., 2022b).
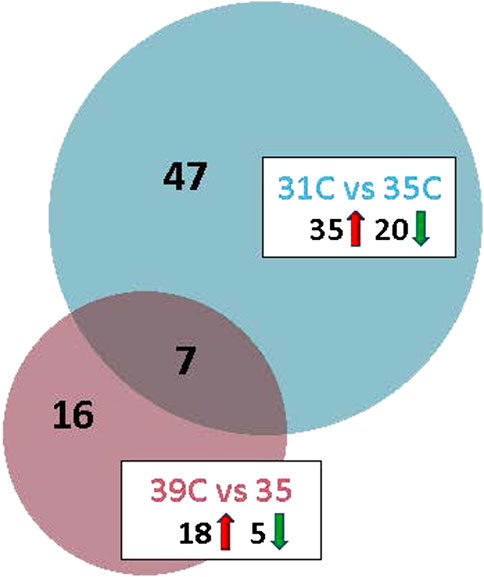
Figure 2. Venn diagram depicting the distribution of differentially expressed genes (DEGs) in the between thermal treatment comparisons. For each temperature comparison, the number of genes with FDR p-value <0.05 and |Log2FC| > 2.0 are given. The number of up and downregulated genes are indicated within the boxes.
Many of the genes that were differentially expressed in the cold-treated muscle group have been recognized for their key roles in muscle development/function. Upregulated genes include; ANGPTL4 (angiopoietin like 4), ARRDC2 (arrestin domain containing 2), C/EBPβ (CCAAT enhancer binding protein beta), DGAT2 (diacylglycerol O-acyltransferase 2), FBXO32 (F-box protein 32, Atogin-1), FMOD (fibromodulin), KLHL38 (kelch like family member 38), MUSTN1 (musculoskeletal embryonic nuclear protein 1), MYL3 (myosin light chain 3), PDK4 (pyruvate dehydrogenase kinase 4), SESN1 (sestrin 1), TRIM63 (tripartite motif containing 63, MuRF1), and UCP3 (mitochondrial uncoupling protein 3). Downregulated genes include LIF (interleukin 6 family cytokine), MSTN (myostatin), PERM1 (PPARGC1 and ESRR induced regulator, muscle 1), PPP1R3B (protein phosphatase one regulatory subunit 3B), SOCS3, (suppressor of cytokine signaling 3), and STOML3 (stomatin like 3). GO analysis for biological process of the 55 DEGs in DAVID found significant enrichment for metabolic regulation and development with several categories of negative regulation included in the top processes (Table 3).
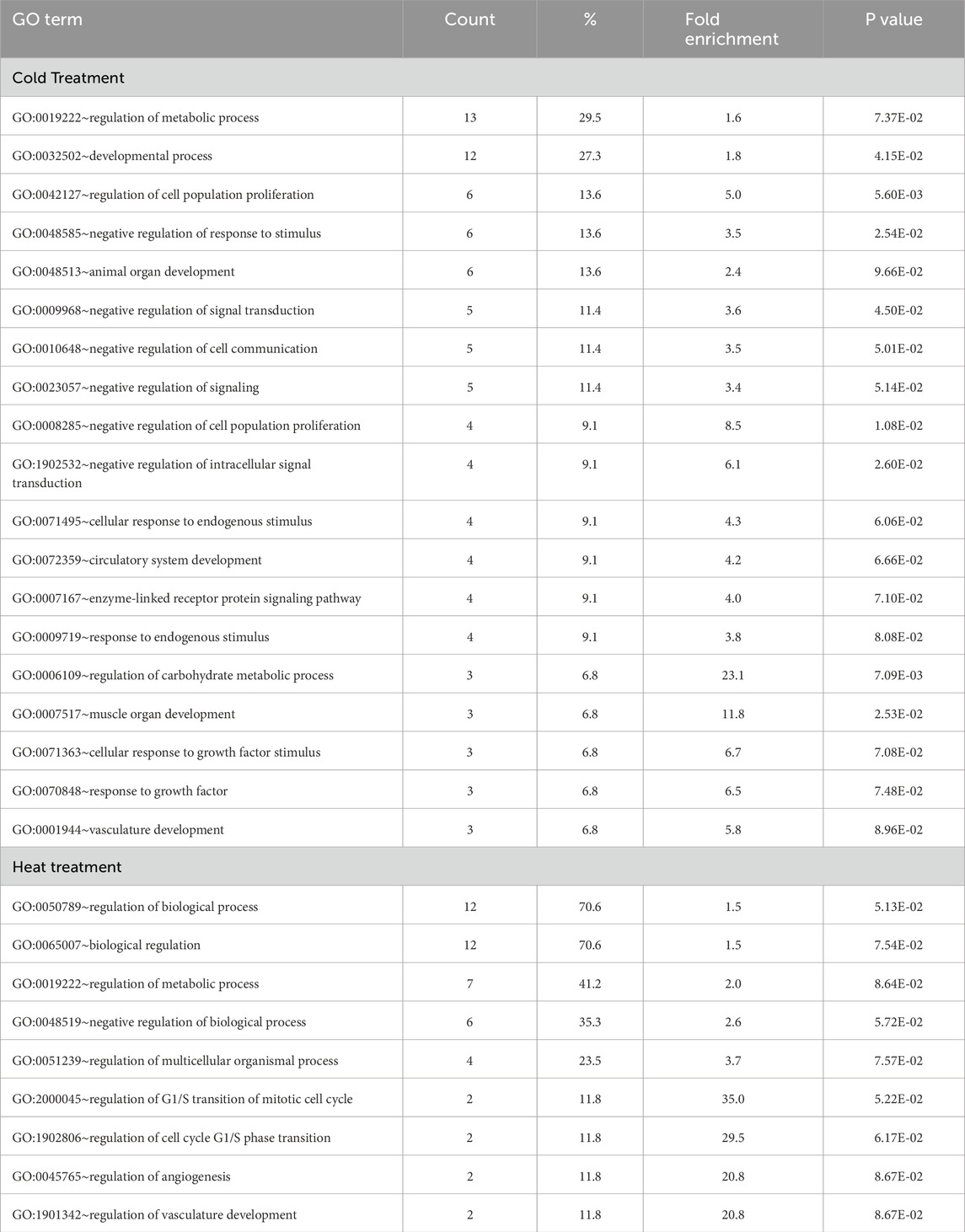
Table 3. Summary of GO analysis of genes differentially expressed in turkey poults. For each category, gene count % inclusion, fold enrichment and p value. Only processes with inclusion percentages >5.0 are shown.
The hot-treatment comparison identified 23 significant DEGs (FDR adjusted P-value <0.05) with all having |Log2FC| > 2.0 (Table 4; Figure 2). Of the 23 DEGs, 18 were upregulated at 39 °C, 5 were downregulated compared to control and 7 were shared with the cold-treatment comparison. Greatest upregulation was seen for the lncRNA, ENSMGAG00000018664 (Log2FC = 17.45) and LOC100548666, GTP-binding protein Rhes-like, (Log2FC = 13.83). Greatest downregulation was seen for ND4L, NADH dehydrogenase subunit 4L, involved in ATP synthesis coupled electron transport and proton transmembrane transport (Log2FC = −7.47), and CPNE4, copine 4, a calcium-dependent, phospholipid-binding protein (Log2FC = −5.00). Genes with functional importance in muscle include ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1), ARRDC2, CEBPβ, CSRP3 (cysteine and glycine rich protein 3), FHL1 (four and a half LIM domains 1), GLUL (glutamate-ammonia ligase), MUSTN1 and SESN1. One of the genes downregulated by heat is the micro-RNA, miR1-2, a muscle-specific microRNA critical for myogenesis. Interestingly, all of the 7 common DEGs (ARRDC2, CEBPB, GLUL, ENSMGAG00000018664, ENSMGAG00000020038, MUSTN1, SESN1) were upregulated in both comparisons relative to control. GO analysis for biological process of the 23 DEGs found significant enrichment for metabolic regulation (Table 3).
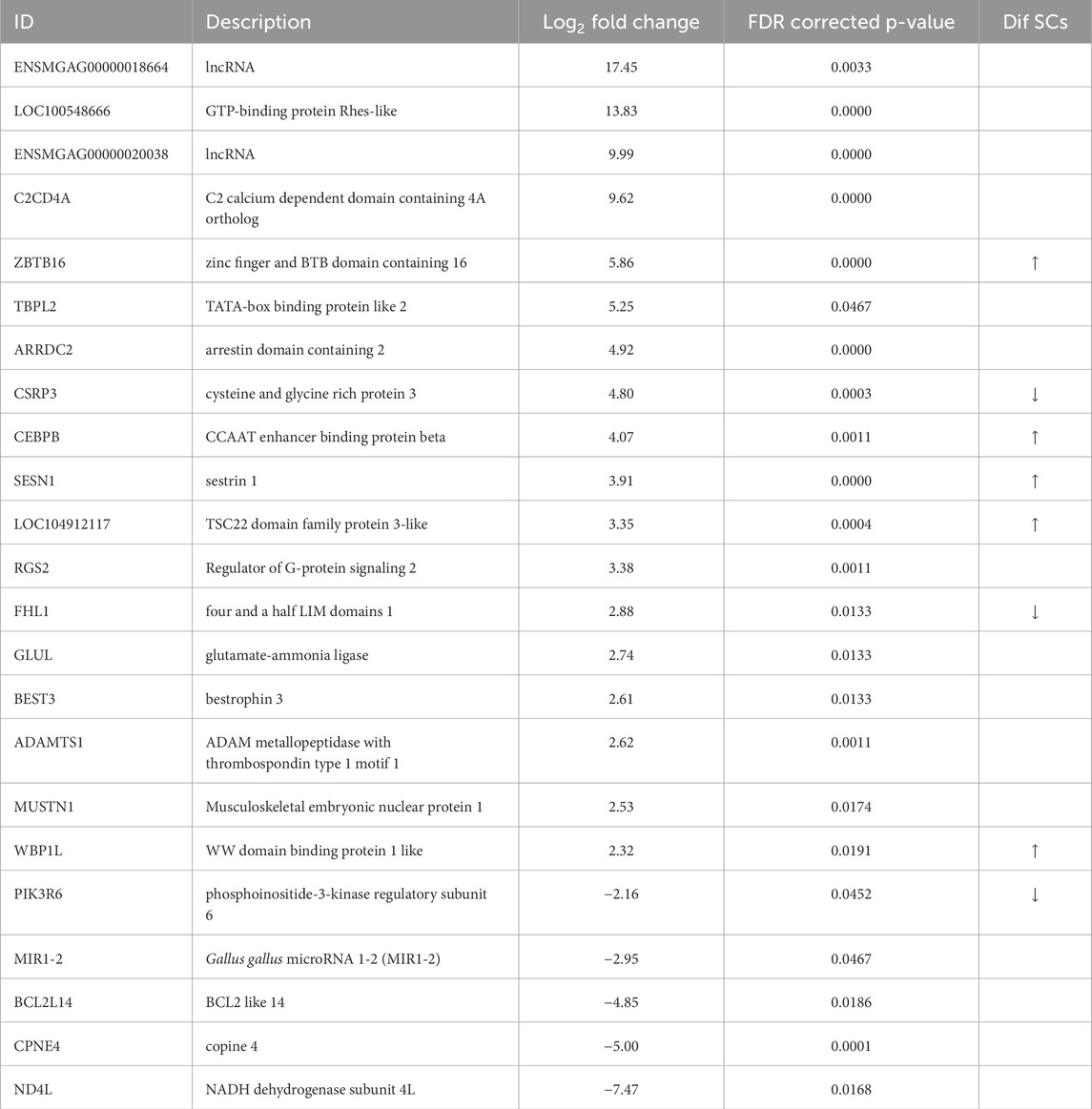
Table 4. Differentially expressed genes (GEDs) identified in comparison of RNAseq data from heat-treated (39 °C) 3 days turkey pouts compared to controls (35 °C). Arrows denote the directional change for genes that also had significant differential expression in differentiating muscle satellite cells (SCs) of commercial turkey following heat treatment (Reed et al., 2022b).
4 Discussion
Birds respond to ambient temperature changes through metabolic, physiologic, and behavioral processes and underlying these processes are cellular changes in gene expression. Although myogenesis begins during embryonic development, muscle stem cells (SCs, satellite cells) are most active in the days immediately following hatch (Halevy et al., 2004). Proliferating SCs (cells undergoing hyperplasia) fuse with existing muscle fibers to increase muscle fiber size (hypertrophy). As poults increase muscle use, synapses between motor neurons and muscle fibers are actively strengthened and muscle fibers increasingly become specialized. Associated with this muscle transition is the shift in metabolism from one that is primarily lipid-based from the yolk during embryonic development to that of carbohydrates in feed (Uni and Ferket, 2004). This shift from lipid to carbohydrate metabolism coupled with a concomitant increase in mitochondrial activity elevates muscle protein synthesis, enabling the rapid muscle growth seen in modern commercial poultry. It is during this dynamic point in development that thermal stress has significant effects that may have long-term impacts on muscle development and ultimately meat quality. As seen in this study, even short (3 days) thermal challenge altered the body weights of male birds compared to controls. This is consistent with earlier studies in chicken that found even short heat stress (Ernst et al., 1984) and cold stress (Ipek, 2006) can depress weight gains. The ability of poultry to regulate body temperature is primarily established during the initial 3-5 days post-hatch (Katz and Meiri, 2006). Studies in chickens have also shown that early thermal conditioning can improve tolerance in birds under subsequent heat stress (Ncho et al., 2021). This study did not evaluate the longer-term effect of early thermal challenge on turkey weights.
4.1 Effect of cold
Cold treatment significantly affected genes important in muscle differentiation and development. For example, musculoskeletal embryonic nuclear protein 1 (MUSTN1), an important regulatory protein for skeletal muscle (Kim and Hadjiargyrou, 2024), was significantly upregulated (Log2FC = 2.19) indicating ongoing proliferation and differentiation. MUSTN1 is primarily expressed in arterioles of the muscle microvasculature (Ducommun et al., 2024), is highly expressed during skeletal muscle development and is required for the differentiation of myoblasts into mature muscle fibers. It likely acts as a co-regulator of muscle-specific gene expression activating MyoD and MyoG as downstream targets (Liu et al., 2010). In the chicken (Hu et al., 2021), MUSTN1 influences the proliferation and differentiation of skeletal muscle SCs and its knockdown, downregulates proliferation and differentiation related genes. Studies comparing expression of MUSTN1 in broilers and layers showed increased expression in broilers, further supporting the importance of this gene in muscle hypertrophy (Zheng et al., 2009). In our previous studies of cultured differentiating commercial turkey SCs, MUSTN1 was slightly downregulated with cold treatment (Reed et al., 2022b).
Myostatin (MSTN) is a well-known negative regulator of muscle growth (Lee and McPherron, 2001), and loss of function mutation leads to increased muscle mass. Fibromodulin (FMOD), is a main regulator of myostatin activity during myoblast differentiation acting through the transforming growth factor-β signaling pathway (Yin et al., 2020) and was significantly upregulated (Log2FC = 7.22). FMOD also has a role in controlling the progression of SCs as it circumvents the inhibitory effects of MSTN triggering myoblast differentiation (Lee et al., 2016). Consistent with the upregulation of FMOD is the observed downregulation of MSTN (Log2FC = −2.36) in the cold-treated birds, which would promote muscle growth.
The transcription factor CCAAT/enhancer binding protein beta (C/EBPβ), involved as both an activator and a repressor in many regulatory and differentiation processes was upregulated in cold-treated birds. C/EBPβ expression is initially high in muscle SCs, but it is rapidly downregulated as myogenesis progresses (Marchildon et al., 2012). However, persistent expression of C/EBPβ constrains myogenic differentiation through inhibition of MyoD. The relatively higher expression of CEBPβ in cold-treated birds (Log2FC = 2.67) could reflect a delay in muscle development and differentiation. In contrast, in our study of differentiating commercial turkey SCs (Reed et al., 2022b), C/EBPB was significantly downregulated by cold treatment.
Protein recycling is an important aspect of muscle development and adaptation and several such genes were upregulated by cold treatment. FBXO32 (Atrogin-1) is a key regulator of skeletal muscle atrophy, involved in protein degradation and promotes the breakdown of proteins via the ubiquitin-proteasome system. A second protein also involved in ubiquitin-mediated protein degradation, KLHL38, shares regulatory elements with FBXO32 (Ehrlich et al., 2020). These genes target proteins like eIF3-f and MyoD for degradation (Tintignac et al., 2005; Lagirand-Cantaloube et al., 2008). Also upregulated was the E3 ubiquitin ligase (TRIM63, MuRF1), which tags muscle proteins for degradation critical for muscle protein turnover and atrophy (Perera et al., 2012). Short-term cold-stress in rats increased mRNA and protein levels of atrogin-1 and MuRF1 with increased atrophy (Manfredi et al., 2013). In differentiating turkey SCs, FBX032 was slightly upregulated whereas KLHL38 and TRIM63 were downregulated by cold treatment (Reed et al., 2022b). Significant upregulation of these genes in the muscle of turkey poults suggests increased muscle turnover.
The inhibitory factor (LIF) was downregulated in the cold-treated poults (Log2FC = −3.49). This gene is complex in that it can promote muscle regeneration by activating SCs to promote myoblast growth, but may also inhibit early differentiation (Jo et al., 2004). In rodents, recombinant LIF treatment promoted the proliferation of muscle SCs in vitro by maintaining their undifferentiated states (Spangenburg and Booth, 2002). Expression changes in genes like ARRDC2 and STOML3 also indicate altered muscle adaptation. ARRDC2 is a mechanosensitive gene known to regulate myotube diameter (Laskin et al., 2024) and STROML3 is essential in the formation of muscle proprioceptors, especially after injury, ensuring proper sensory perception and motor control (Haseleu et al., 2020). In the cold-treated birds, ARRDC2 was upregulated (Log2FC = 3.02) whereas STOML3 was downregulated (Log2FC = −4.11).
Cold exposure can have several direct effects on muscle metabolism. It can directly alter muscle contraction potentials and electrolyte balance (Klein et al., 1968), decrease protein metabolism and increase proteolysis, potentially leading to muscle breakdown, altered lipid and glucose metabolism and changes in fat deposition (Manfredi et al., 2013). Expression changes in several genes in the cold-treated poults are indicative of modified metabolism. In mammals, the mitochondrial uncoupling protein UCP3 is expressed in both skeletal muscle and brown adipose tissue and this gene was upregulated in the cold-treated poults. Although its function is not completely understood, UCP3 plays a role in skeletal muscle metabolism as a mediator of cold-induced thermogenesis (Riley et al., 2016) and may act to minimize oxidative damage (Reyb et al., 2010).
Three genes also upregulated by cold (PDK4, ANGPTL4 and DGAT2) are associated with lipid metabolism and intermuscular fat. In the turkey, pyruvate dehydrogenase kinase (PDK4) primarily functions to regulate glucose and lipid metabolism by modulating the activity of the pyruvate dehydrogenase complex. Upregulation of PDK4 indicates a shift in energy metabolism with cold treatment reducing glucose oxidation and increasing fatty acid oxidation and this gene was also upregulated in differentiating SCs (Reed et al., 2022b). Studies have also shown a significant decrease in PDK4 mRNA and protein levels associated with pale-soft-exudative (PSE) muscle condition the turkey (Malila et al., 2014). In skeletal muscle, ANGPTL4 regulates lipid uptake and metabolism (Son et al., 2023) and may influence muscle energy homeostasis by directing lipids to different tissues. It can also inhibit skeletal muscle differentiation by suppressing Wnt/β-catenin signaling. As demonstrated in pigeons, DGAT2 is important in control of intramuscular fat content and muscle fiber characteristics. Its involvement in the synthesis of triglycerides and the deposition of fat within muscle tissue directly influences meat quality (Mao et al., 2022).
Suppressor of cytokine signaling 3 (SOCS3) was significantly downregulated in cold-treated birds (Log2FC = −3.200). During development, SOC3 acts primarily to negatively regulate insulin and leptin signaling, affecting energy metabolism and both positive and negative aspects of muscle growth and myoblast differentiation (Caldow et al., 2011; Yang et al., 2012). Downregulation of SOC3 would potentially increase signaling, promoting glucose uptake, inhibiting glucose production, and influencing fat storage and breakdown. Also significantly downregulated was PPP1R3B (Log2FC = −3.92), also downregulated in differentiating turkey SCs (Reed et al., 2022b). This gene acts as a glycogen-targeting subunit for the enzyme protein phosphatase 1 (PP1) to facilitate regulation of glycogen metabolism and energy storage in muscle (Migocka-Patrzałek and Elias, 2021). Decreased PPP1R3B expression would lower glycogen synthesis and decrease glycogen storage.
Finally, cold treatment also affected genes with roles in oxidative stress. Sestrin 1 (SESN1) is involved in regulating mitochondrial function and calcium levels to protect skeletal muscle from damage by modulating oxidative stress (Jing et al., 2023). Upregulation under cold stress would be beneficial by reducing muscle atrophy and improving insulin sensitivity. A similar directional expression change occurred in differentiating SCs (Reed et al., 2022b). The regulatory gene PERM1 is also involved in mitochondrial biogenesis and oxidative metabolism. PERM1 functions in fatty acid metabolism and is essential for PGC-1α-induced mitochondrial biogenesis in skeletal muscle (Huang et al., 2022). In mammals, PERM1 is crucial for thermogenesis and is typically upregulated by cold treatment (Cho et al., 2013). Downregulation could diminish capacity for oxidative energy production, impair mitochondrial function, and reduce muscle metabolism.
4.2 Effect of heat
Depending on the duration and intensity of heat stress, the effect on muscle development can be either inhibiting or promoting. In activating signaling pathways, heat stress can stimulate muscle growth (Kim and Kim, 2023), while inhibition of protein synthesis and amino acid transportation has negative consequences (Ma et al., 2018). As discussed above, C/EBPβ is an important inhibitor of myogenic differentiation that must be downregulated for muscle cell development. In mammals, C/EBPβ expression increased in muscle during ER stress and proinflammatory conditions (van der Krieken et al., 2015). In previous studies, our group has shown that heat stress upregulates C/EBPβ expression in turkey muscle SCs altering intracellular lipid accumulation (Reed et al., 2022a; b; Xu et al., 2022). MUSTN1 was also upregulated in the heat-treated birds. This reflects its role in modulating skeletal muscle extracellular matrix in muscle plasticity, adaptive responses and repair (Kim and Hadjiargyrou, 2024). Thus, the combined increase of C/EBPβ and MUSTN1 expression in the heat-treated birds is an adaptation to high temperatures that may slow myogenic differentiation and increase intracellular lipids. MUSTN1 is also involved in regulation of adipogenesis and lipid deposition (Fu et al., 2025). The upregulation of this gene may account for increased lipid deposition reported in muscle as a response to heat or cold stress (Patael et al., 2019). Increased lipid accumulation in breast muscle is a hallmark of myopathies like Wooden Breast in broiler chickens (Papah and Abasht, 2019).
One of the genes downregulated by heat treatment corresponded to the muscle-specific microRNA, miR-1-2 (Log2FC = −2.95). MicroRNAs regulate gene expression by binding to messenger RNA to either inhibit translation or induce mRNA degradation (Saliminejad et al., 2019). In skeletal muscle, miR-1 promotes myogenesis by targeting myocyte enhancer factors that regulate proliferation and differentiation. Specifically, miR-1 inhibits histone deacetylase 4 (HDAC4) a repressor of muscle-specific transcription factors and inhibitor of differentiation (Güller and Russell, 2010). Downregulation of miR-1 would thus increase HDAC4 activity potentially decreasing differentiation. Analysis of proliferating and differentiating SCs in research-line birds also found significant downregulation of miR-1 with heat treatment (Reed et al., 2023).
FHL1 (Four and a half LIM domains protein 1) is a LIM protein that enhances myoblast fusion and myotube hypertrophy by promoting assembly of sarcomeres, the contractile units within muscle cells (Cowling et al., 2008). Muscle-specific FHL1 isoforms are highly expressed in skeletal muscle and interact directly with the NFATc1 transcription factor. There is significant correlation between FHL1 expression levels and muscle growth (McGrath et al., 2006). Upregulation of FHL1 in heat-treated turkeys would promote muscle maturation.
A second muscle LIM protein upregulated by heat treatment was cysteine and glycine-rich protein 3 (CSRP3, Log2FC = 4.80)). Important in muscle development, this gene is also involved in muscle structure and repair. As a positive regulator of myogenesis, CSRP3 interacts with muscle-specific transcription factors like MyoD, myogenin, and MRF4 to promote differentiation of myoblasts into myotubes and the organization of muscle cell structure (Germain et al., 2022). CSRP3 also regulates autophagy in component recycling and muscle cell remodeling (Rashid et al., 2015).
Increased expression of the metalloproteinase ADAMTS1 (Log2FC = 2.63) and the mechanosensitive gene ARRDC2 (Log2FC = 4.92) are additional evidence for extracellular matrix remodeling. While ADAMST1 can modify the extracellular matrix affecting cell behavior (Tan et al., 2013), it may also indirectly influence muscle development by promoting SC activation. After injury, macrophages release ADAMTS1 inducing SC activation by modulating the Notch1 signaling pathway (Du et al., 2017). As discussed above, AARDC2 plays a role in muscle adaptation (Laskin et al., 2024).
Evidence for heat-altered metabolism is present in the upregulation of SENS1 and GLUL (Log2FC = 3.91 and 2.74, respectively). As discussed above, SENS1 is important in modulating oxidative stress. Glutamate-ammonia ligase (GLUL) is an enzyme that catalyzes synthesis of glutamine a non-essential amino acid from glutamate and thus removes a toxic byproduct of protein metabolism (Newsholme and Parry-Billings, 1990). Glutamine is also important nitrogen carrier and fuel source for protein synthesis in growing muscle cells. The most significantly downregulated gene in the heat-treated birds was ND4L (Log2FC = −7.47). NDL4 subunit is part of Complex I, crucial in the process of oxidative phosphorylation, in generating ATP needed for muscle contraction and overall muscle function. Downregulation in poults is predicted to impair mitochondrial function and reduce energy production.
The importance of environmental changes (Halevy et al., 2006) and nutritional stresses (Velleman et al., 2010; Kornasio et al., 2011) on immediate post-hatch and long-term muscle development and growth are seen in broilers and turkeys. Our prior studies of muscle SCs from growth-selected birds (experimental and commercial) showed that these birds are more sensitive to temperature extremes than non-growth selected birds (Reed et al., 2017a; b, 2022a; b). Our study of turkey poults from select genetic research lines (Barnes et al., 2019) found the growth-selected F-line to have a more negative response to cold-treatment than the non-selected RBC2 line. Consistent with this result was the greater number of DEGs observed in the commercial poults following cold treatment compared to heat treatment.
4.3 Conclusion
Our long-term goal has been to elucidate the effects of temperature on body weight, muscle development, and fat deposition in growing turkeys. Results of this gene expression analysis are consistent with many of our previous transcriptome studies of skeletal muscle stem cells and developed/developing turkey muscle. Commercial turkey poults exposed to short-term thermal challenge displayed altered expression of genes responsible for key cellular processes in muscle. Both thermal challenges had direct effects on phenotype resulting in lower body weights after 3 days of treatment. Functional classification of altered genes suggests direct effects on muscle differentiation and development, adaptation and apoptosis/protein recycling, metabolism and nutrient utilization, and mitochondrial function and oxidative stress. Cold treatment had significant implications for altered lipid and glucose metabolism and heat treatment may increase intracellular lipids.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.ncbi.nlm.nih.gov/bioproject/1273166, PRJNA1273166.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of Michigan State University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KR: Formal Analysis, Writing – review and editing, Writing – original draft, Conceptualization, Funding acquisition. SV: Conceptualization, Funding acquisition, Resources, Writing – review and editing. GS: Resources, Funding acquisition, Conceptualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the United States Department of Agriculture, National Institute of Food Agriculture, AFRI competitive grant no. 2020-67015-30827 to GS, KR, and SV.
Acknowledgments
The authors thank Boon Hong Keng for assistance in animal husbandry, Kristelle Mendoza for technical laboratory assistance, and Juan Abrahante, University of Minnesota Informatics Institute, for assistance with data processing and analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI was used in initial queries of functional characterization of gene lists. The results of these queries were subsequently followed by direct review of primary literature to both confirm and expand on predictions produced by AI.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2025.1651079/full#supplementary-material
References
Baller J., Kono T., Herman A., Zhang Y. (2019) “CHURP: a lightweight CLI framework to enable novice users to analyze sequencing datasets in parallel,”96. New York, NY, USA: Association for Computing Machinery, 1–5. doi:10.1145/3332186.3333156
Barnes N. E., Mendoza K. M., Strasburg G. M., Velleman S. G., Reed K. M. (2019). Thermal challenge alters the transcriptional profile of the breast muscle in turkey poults. Poult. Sci. 98, 74–91. doi:10.3382/ps/pey401
Caldow M. K., Steinberg G. R., Cameron-Smith D. (2011). Impact of SOCS3 overexpression on human skeletal muscle development in vitro. Cytokine 55, 104–109. doi:10.1016/j.cyto.2011.03.012
Cho Y., Hazen B. C., Russell A. P., Kralli A. (2013). Peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-Induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J. Biol. Chem. 288, 25207–25218. doi:10.1074/jbc.M113.489674
Cowling B. S., McGrath M. J., Nguyen M. A., Cottle D. L., Kee A. J., Brown S., et al. (2013). Identification of FHL1 as a regulator of skeletal muscle mass: implications for human myopathy. J. Cell. Biol. 183, 1033–1048. doi:10.1083/jcb.200804077
Du H., Shih C. H., Wosczyna M. N., Mueller A. A., Cho J., Aggarwal A., et al. (2017). Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat. Commun. 8, 669. doi:10.1038/s41467-017-00522-7
Ducommun S., Jannig P. R., Cervenka I., Murgia M., Mittenbühler M. J., Chernogubova E., et al. (2024). Mustn1 is a smooth muscle cell-secreted microprotein that modulates skeletal muscle extracellular matrix composition. Mol. Metab. 82, 101912. doi:10.1016/j.molmet.2024.101912
Ehrlich K. C., Baribault C., Ehrlich M. (2020). Epigenetics of muscle- and brain-specific expression of KLHL family genes. Int. J. Mol. Sci. 21, 8394. doi:10.3390/ijms21218394
Ernst R. A., Weathers W. W., Smith J. (1984). Effects of heat stress on day-old broiler chicks. Poult. Sci. 63, 1719–1721. doi:10.3382/ps.0631719
Fu Y., Hao X., Nie J., Zhang H., Shang P., Zhang B., et al. (2025). MUSTN1 and FABP3 interact to regulate adipogenesis and lipid deposition. J. Lipid Res. 66, 100804. doi:10.1016/j.jlr.2025.100804
Germain P., Delalande A., Pichon C. (2022). Role of muscle LIM protein in mechanotransduction process. Int. J. Mol. Sci. 23, 9785. doi:10.3390/ijms23179785
Güller I., Russell A. P. (2010). MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J. Physiol. 588 (Pt 21), 4075–4087. doi:10.1113/jphysiol.2010.194175
Halevy O., Piestun Y., Allouh M. Z., Rosser B. W., Rinkevich Y., Reshef R., et al. (2004). Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev. Dyn. 231, 489–502. doi:10.1002/dvdy.20151
Halevy O., Yahav S., Rozenboim I. (2006). Enhancement of meat production by environmental manipulations in embryo and young broilers. World's Poult. Sci. J. 62, 485–497. doi:10.1079/wps2005110
Haseleu J., Walcher J., Lewin G. R. (2020). The mechanotransduction protein STOML3 is required for proprioceptor plasticity following peripheral nerve regeneration. bioRxiv 11 (10), 367748. doi:10.1113/EP092428
Hu Z., Xu H., Lu Y., He Q., Yan C., Zhao X., et al. (2021). MUSTN1 is an indispensable factor in the proliferation, differentiation and apoptosis of skeletal muscle satellite cells in chicken. Exp. Cell Res. 407, 112833. doi:10.1016/j.yexcr.2021.112833
Huang C.-Y., Oka S.-Y., Xu X., Chen C. F., Tung C. Y., Chang Y. Y., et al. (2022). PERM1 regulates genes involved in fatty acid metabolism in the heart by interacting with PPARα and PGC-1α. Sci. Rep. 12, 14576. doi:10.1038/s41598-022-18885-3
Ipek A. Ş. Ü., Sahan U. (2006). Effects of cold stress on broiler performance and ascites susceptibility. Asian-Australasian J An Sci. 19, 734–738. doi:10.5713/ajas.2006.734
Jing Y., Zuo Y., Sun L., Yu Z. R., Ma S., Hu H., et al. (2023). SESN1 is a FOXO3 effector that counteracts human skeletal muscle ageing. Cell Prolif. 56, e13455. doi:10.1111/cpr.13455
Jo C., Kim H., Jo I., Choi I., Jung S. C., Kim J., et al. (2004). Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim. Biophys. Acta 1743, 187–197. doi:10.1016/j.bbamcr.2004.11.002
Karl T. R., Trenberth K. E. (2003). Modern global climate change. Science 302, 1719–1723. doi:10.1126/science.1090228
Katz A., Meiri N. (2006). Brain-derived neurotrophic factor is critically involved in thermal-experience-dependent developmental plasticity. J. Neurosci. 26, 3899–3907. doi:10.1523/JNEUROSCI.0371-06.2006
Kim C. J., Hadjiargyrou M. (2024). Mustn1 in skeletal muscle: a novel regulator? Genes 15, 829. doi:10.3390/genes15070829
Kim W. S., Kim J. (2023). Exploring the impact of temporal heat stress on skeletal muscle hypertrophy in bovine myocytes. J. Therm. Biol. 117, 103684. doi:10.1016/j.jtherbio.2023.103684
Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Klein R., Haddow J. E., Kind C., Cockburn F. (1968). Effect of cold on muscle potentials and electrolytes. Metabolism 17, 1094–1103. doi:10.1016/0026-0495(68)90088-7
Kornasio R., Halevy O., Kedar O., Uni Z. (2011). Effect of in ovo feeding and its interaction with timing of first feed on glycogen reserves, muscle growth, and body weight. Poult. Sci. 90, 1467–1477. doi:10.3382/ps.2010-01080
Lagirand-Cantaloube J., Offner N., Csibi A., Leibovitch M. P., Batonnet-Pichon S., Tintignac L. A., et al. (2008). The initiation factor eIF3-f is a major target for Atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 27, 1266–1276. doi:10.1038/emboj.2008.52
Laskin G. R., Cabrera A. R., Greene N. P., Tomko R. J., Vied C., Gordon B. S. (2024). The mechanosensitive gene arrestin domain containing 2 regulates myotube diameter with direct implications for disuse atrophy with aging. Am. J. Physiol. Cell Physiol. 326, C768–C783. doi:10.1152/ajpcell.00444.2023
Lee S. J., McPherron A. C. (2001). Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. U. S. A. 98, 9306–9311. doi:10.1073/pnas.151270098
Lee E. J., Jan A. T., Baig M. H., Ashraf J. M., Nahm S. S., Kim Y. W., et al. (2016). Fibromodulin: a master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 30, 2708–2719. doi:10.1096/fj.201500133R
Liu C., Gersch R. P., Hawke T. J., Hadjiargyrou M. (2010). Silencing of Mustn1 inhibits myogenic fusion and differentiation. Am. J. Physiol. Cell Physiol. 298, C1100–C1108. doi:10.1152/ajpcell.00553.2009
Ma B., He X., Lu Z., Zhang L., Li J., Jiang Y., et al. (2018). Chronic heat stress affects muscle hypertrophy, muscle protein synthesis and uptake of amino acid in broilers via insulin like growth factor-mammalian target of rapamycin signal pathway. Poult. Sci. 97, 4150–4158. doi:10.3382/ps/pey291
Malila Y., Carr K. M., Ernst C. W., Velleman S. G., Reed K. M., Strasburg G. M. (2014). Deep transcriptome sequencing reveals differences in global gene expression between normal and pale, soft, and exudative Turkey meat. J. Anim. Sci. 92, 1250–1260. doi:10.2527/jas.2013-7293
Mao H., Yin Z., Wang M., Zhang W., Raza S. H. A., Althobaiti F., et al. (2022). Expression of DGAT2 gene and its associations with intramuscular fat content and breast muscle fiber characteristics in domestic pigeons (columba livia). Front. Vet. Sci. 9, 847363. doi:10.3389/fvets.2022.847363
Marchildon F., Lala N., Li G., St-Louis C., Lamothe D., Keller C., et al. (2012). CCAAT/Enhancer binding protein beta is expressed in satellite cells and controls myogenesis. Stem Cells 30, 2619–2630. doi:10.1002/stem.1248
Manfredi L. H., Zanon N. M., Garofalo M. A., Navegantes L. C., Kettelhut I. C. (2013). Effect of short-term cold exposure on skeletal muscle protein breakdown in rats. J. App.l Physiol. 115, 1496–505. doi:10.1152/japplphysiol.00474.2013
McGrath M. J., Cottle D. L., Nguyen M. A., Dyson J. M., Coghill I. D., Robinson P. A., et al. (2006). Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. J. Biol. Chem. 281, 7666–7683. doi:10.1074/jbc.M512552200
Migocka-Patrzałek M., Elias M. (2021). Muscle glycogen phosphorylase and its functional partners in health and disease. Cells 10, 883. doi:10.3390/cells10040883
Mitchell M. A., Kettlewell P. J. (2009) “Welfare of poultry during transport – a review,” in Proceedings of 8th poultry welfare symposium, cervia, Italy, 18-22 may 2009, 90–100.
Modrey P., Nichelmann M. (1992). Development of autonomic and behavioural thermoregulation in turkeys (meleagris gallopavo). J. Therm. Biol. 17, 287–292. doi:10.1016/0306-4565(92)90035-e
Ncho C. M., Gupta V., Goel A. (2021). Effect of thermal conditioning on growth performance and thermotolerance in broilers: a systematic review and meta-analysis. J. Therm. Biol. 98, 102916. doi:10.1016/j.jtherbio.2021.102916
Nestor K. E. (1977). The stability of two randombred control populations of turkeys. Poult. Sci. 56, 54–57. doi:10.3382/ps.0560054
Nestor K. E. (1984). Genetics of growth and reproduction in the Turkey. 9. Long-term selection for increased 16-week body weight. Poult. Sci. 63, 2114–2122. doi:10.3382/ps.0632114
Newsholme E. A., Parry-Billings M. (1990). Properties of glutamine release from muscle and its importance for the immune system. J. Parenter. Enter. Nutr. 14, 63S–67S. doi:10.1177/014860719001400406
Papah M. B., Abasht B. (2019). Dysregulation of lipid metabolism and appearance of slow myofiber-specific isoforms accompany the development of wooden breast myopathy in modern broiler chickens. Sci. Rep. 9, 17170. doi:10.1038/s41598-019-53728-8
Patael T., Piestun Y., Soffer A., Mordechay S., Yahav S., Velleman S. G., et al. (2019). Early posthatch thermal stress causes long-term adverse effects on pectoralis muscle development in broilers. Poult. Sci. 98, 3268–3277. doi:10.3382/ps/pez123
Perera S., Mankoo B., Gautel M. (2012). Developmental regulation of MURF E3 ubiquitin ligases in skeletal muscle. J. Muscle Res. Cell Motil. 33, 107–122. doi:10.1007/s10974-012-9288-7
Piestun Y., Patael T., Yahav S., Velleman S. G., Halevy O. (2017). Early posthatch thermal stress affects breast muscle development and satellite cell growth and characteristics in broilers. Poult. Sci. 96, 2877–2888. doi:10.3382/ps/pex065
Rashid M. M., Runci A., Polletta L., Carnevale I., Morgante E., Foglio E., et al. (2015). Muscle LIM protein/CSRP3: a mechanosensor with a role in autophagy. Cell Death Disc 1, 15014. doi:10.1038/cddiscovery.2015.14
Reed K. M., Mendoza K. M., Abrahante J. E., Barnes N. E., Velleman S. G., Strasburg G. M. (2017a). Response of Turkey muscle satellite cells to thermal challenge. I. Transcriptome effects in proliferating cells. BMC Genomics 18, 352. doi:10.1186/s12864-017-3740-4
Reed K. M., Mendoza K. M., Strasburg G. M., Velleman S. G. (2017b). Response of Turkey muscle satellite cells to thermal challenge. II. Transcriptome effects in differentiating cells. Front. Physiol. 8, 948. doi:10.3389/fphys.2017.00948
Reed K. M., Mendoza K. M., Strasburg G. M., Velleman S. G. (2022a). Transcriptome response of proliferating muscle satellite cells to thermal challenge in commercial Turkey. Front. Physiol. 13, 970243. doi:10.3389/fphys.2022.970243
Reed K. M., Mendoza K. M., Strasburg G. M., Velleman S. G. (2022b). Transcriptome response of differentiating muscle satellite cells to thermal challenge in commercial Turkey. Genes 13, 1857. doi:10.3390/genes13101857
Reed K. M., Mendoza K. M., Kono T., Powell A. A., Strasburg G. M., Velleman S. G. (2023). Expression of miRNAs in Turkey muscle satellite cells and differential response to thermal challenge. Front. Physiol. 14, 1293264. doi:10.3389/fphys.2023.1293264
Reyb R. D., Romestaing C., Belouze M., Rouanet J.-L., Desplanches D., Sibille B., et al. (2010). Upregulation of avian uncoupling protein in cold-acclimated and hyperthyroid ducklings prevents reactive oxygen species production by skeletal muscle mitochondria. BMC Physiol. 10, 5. doi:10.1186/1472-6793-10-5
Riley C. L., Dao C., Kenaston M. A., Muto L., Kohno S., Nowinski S. M., et al. (2016). The complementary and divergent roles of uncoupling proteins 1 and 3 in thermoregulation. J. Physiol. 594, 7455–7464. doi:10.1113/JP272971
Rowland L. A., Bal N. C., Periasamy M. (2015). The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol. Rev. 90, 1279–1297. doi:10.1111/brv.12157
Saliminejad K., Khorram Khorshid H. R., Soleymani Fard S., Ghaffari S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234, 5451–5465. doi:10.1002/jcp.27486
Schmidhuber J., Tubiello F. N. (2007). Global food security under climate change. Proc. Nat. Acad. Sci. U. S. A. 104, 19703–19708. doi:10.1073/pnas.0701976104
Shinder D., Rusal M., Tanny J., Druyan S., Yahav S. (2007). Thermoregulatory responses of chicks (Gallus domesticus) to low ambient temperatures at an early age. Poult. Sci. 86, 2200–2209. doi:10.1093/ps/86.10.2200
Son Y., Lorenz W. W., Paton C. M. (2023). Linoleic acid-induced ANGPTL4 inhibits C2C12 skeletal muscle differentiation by suppressing Wnt/β-catenin. J. Nutr. Biochem. 116, 109324. doi:10.1016/j.jnutbio.2023.109324
Spangenburg E. E., Booth F. W. (2002). Multiple signaling pathways mediate LIF-Induced skeletal muscle satellite cell proliferation. Am. J. Physiol. Cell Physiol. 283, C204–C211. doi:10.1152/ajpcell.00574.2001
Tan I. d. A., Ricciardelli C., Russell D. L. (2013). The metalloproteinase ADAMTS1: a comprehensive review of its role in tumorigenic and metastatic pathways. Int. J. Cancer 133, 2263–2276. doi:10.1002/ijc.28127
Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. (2005). Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 280, 2847–2856. doi:10.1074/jbc.M411346200
Uni Z., Ferket R. P. (2004). Methods for early nutrition and their potential. World’s Poult. Sci. J. 60, 101–111. doi:10.1079/WPS20038
van der Krieken S. E., Popeijus H. E., Mensink R. P., Plat J. (2015). CCAAT/Enhancer binding protein β in relation to ER stress, inflammation, and metabolic disturbances. Biomed. Res. Int. 2015, 324815. doi:10.1155/2015/324815
Velleman S. G., Liu X., Nestor K. E., McFarland D. C. (2000). Heterogeneity in growth and differentiation characteristics in Male and female satellite cells isolated from Turkey lines with different growth rates. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 125, 503–509. doi:10.1016/s1095-6433(00)00178-1
Velleman S. G., Nestor K. E., Coy C. S., Harford I., Anthony N. B. (2010). Effect of posthatch feed restriction on broiler breast muscle development and muscle transcriptional regulatory factor gene and heparan sulfate proteoglycan expression. Int. J. Poult. Sci. 9, 417–425. doi:10.3923/ijps.2010.417.425
Wolfenson D., Frei Y. F., Snapir N., Berman A. (1981). Heat stress effects on capillary blood flow and its redistribution in the laying hen. Pflugers Arch. 390, 86–93. doi:10.1007/BF00582717
Xu J., Strasburg G. M., Reed K. M., Velleman S. G. (2022). Thermal stress and selection for growth affect myogenic satellite cell lipid accumulation and adipogenic gene expression through mechanistic target of rapamycin pathway. J. Anim. Sci. 100, skac001. doi:10.1093/jas/skac001
Yahav S., Shinder D., Tanny J., Cohen S. (2005). Sensible heat loss: the broiler’s paradox. World’s Poult. Sci. J. 61, 419–434. doi:10.1079/wps200453
Yang Z., Hulver M., McMillan R. P., Cai L., Kershaw E. E., Yu L., et al. (2012). Regulation of insulin and leptin signaling by muscle suppressor of cytokine signaling 3 (SOCS3). PLoS One 7, e47493. doi:10.1371/journal.pone.0047493
Yin H., Cui C., Han S., Chen Y., Zhao J., He H., et al. (2020). Fibromodulin modulates chicken skeletal muscle development via the transforming growth factor-β signaling pathway. Anim. (Basel) 10, 1477. doi:10.3390/ani10091477
Keywords: RNAseq, Meleagris gallopavo, Pectoralis major, differential expression, thermal challenge
Citation: Reed KM, Velleman SG and Strasburg GM (2025) Thermal challenge significantly alters gene expression in breast muscle of commercial turkey poults. Front. Physiol. 16:1651079. doi: 10.3389/fphys.2025.1651079
Received: 20 June 2025; Accepted: 02 September 2025;
Published: 19 September 2025.
Edited by:
Gregoy Y. Bedecarrats, University of Guelph, CanadaReviewed by:
Qingwu Xin, Fujian Academy of Agriculture Sciences, ChinaJosephine Kwakye, University of Georgia, United States
Copyright © 2025 Reed, Velleman and Strasburg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kent M. Reed, a21yZWVkQHVtbi5lZHU=
 Kent M. Reed
Kent M. Reed Sandra G. Velleman
Sandra G. Velleman Gale M. Strasburg
Gale M. Strasburg