- 1School of Public Health, Henan Medical College, Zhengzhou, China
- 2School of Computer Science and Engineering, University of New South Wales (UNSW), Sydney, NSW, Australia
- 3Modern Education Technology Center, Henan Medical College, Zhengzhou, China
Medical visual-language alignment plays an important role in hospital diagnostic data analysis and patient health prediction. However, existing multimodal alignment models, such as CLIP, while performing well in some tasks, often fail to accurately capture the fine-grained alignment between complex medical images and texts, and lack the capability to handle multi-view radiological image inputs. To address these issues, this paper proposes the ClinVLA model, an efficient visual-language alignment method. Specifically, ClinVLA enhances image feature representation through an innovative multi-view input design, including both frontal and lateral views. Furthermore, ClinVLA introduces an innovative adapter module, making the model more efficient in task transfer and language transformation, significantly improving performance in cross-modal learning. Finally, by incorporating both global and local alignment losses, ClinVLA ensures semantic consistency between images and texts, optimizing the accuracy and efficiency of image-text matching. Experimental results on datasets such as CheXpert and RSNA Pneumonia show that ClinVLA improves text-to-image retrieval accuracy by over 3% compared to the best-performing similar algorithms, and increases image-to-text retrieval accuracy by approximately 5%. ClinVLA provides a new solution for medical image analysis, with broad application prospects.
1 Introduction
Hospital diagnostic data analysis and patient health prediction play a crucial role in medical research and clinical practice. With the rapid increase in medical data, traditional manual diagnostic methods face significant challenges (Rayed et al. 2024; Pu et al. 2024). Effectively extracting valuable information from large amounts of medical data for accurate disease prediction and early warning has become a key research focus in the field of medicine today. Medical data not only include electronic health records (EHRs), patient medical histories, and diagnostic reports but also imaging data, such as X-rays, CT scans, MRIs, etc. The integration and analysis of these data sources can provide more comprehensive support for clinical decision-making, significantly enhancing diagnostic efficiency and accuracy (Meng et al. 2024; Wu et al. 2025; Li C. et al. 2025; Cao et al. 2025).
Medical visual-language alignment, as an emerging research direction, has been widely applied to the automatic matching of medical images and texts (Shi et al. 2024; Zhang et al. 2025; Li J. et al. 2025). In this process, image data (such as radiological images) and text data (such as radiology reports and medical descriptions) are jointly analyzed through alignment techniques, enabling computers to understand the relationship between image content and corresponding reports, thereby assisting doctors in providing diagnostic recommendations Liu (2024), Ning et al. (2025). Effective image-text matching not only improves diagnostic efficiency but also reduces the risk of misdiagnosis caused by human error. This technology has broad application prospects in medical automation, telemedicine, and other fields (Xiao et al. 2024; Alsabbagh et al. 2025).
The image-text matching method based on the CLIP Radford et al. (2021) (Contrastive Language-Image Pre-training) model has been widely used in visual analysis Pham et al. (2024). CLIP pre-trains image and text encoders, mapping images and texts into the same semantic space, effectively measuring the similarity between them. For example, DeCLIP introduces deformable convolution modules to capture image details, improving image alignment accuracy; VisualBERT Li et al. (2019), Xing et al. (2025) and UNITER Chen et al. (2020) enhance the interaction between image and text features through cross-modal pre-trained models and complex attention mechanisms, thus improving matching performance; VLP (Vision-Language Pretraining) strengthens the understanding and generation of image-text relationships through large-scale visual and language datasets, especially excelling in processing complex data Tuerhong et al. (2024); CLIP-ViT combines Vision Transformer and CLIP to improve the feature extraction capability of high-resolution images, enhancing the quality of image-text matching (Zhang et al. 2024; Xiong et al. 2024; Umer and Sharif 2022).
However, CLIP-based methods still face the problem of poor visual representation ability. This is mainly reflected in their insufficient ability to process complex medical images, especially when dealing with fine-grained medical images, where traditional methods often fail to capture the detailed alignment between images and texts. Additionally, these methods usually require a large amount of pre-training data and computational resources, leading to low computational efficiency and an inability to effectively reduce the pre-training burden. More importantly, these methods often lack the ability to handle temporal multi-view radiological images, failing to fully utilize the temporal dynamic changes across multiple image acquisitions, thereby limiting their application in dynamic medical image analysis.
To address the above issues, we propose a novel method—ClinVLA (Clinical Visual-Language Alignment). This method integrates adapters and uses only 12% of the trainable parameters, significantly reducing the model’s training complexity and computational overhead. ClinVLA uses temporal multi-view radiological images as input, enhancing the visual-language alignment effect and improving the consistency of information between radiological images and radiology reports. Through this innovative design, our method not only outperforms traditional models in terms of performance but also achieves more efficient cross-modal learning, providing a new solution for medical image analysis and intelligent diagnosis.
The three contributions of this paper are as follows:
2 Related works
2.1 Image-text matching methods
Image-text matching has become an important research direction in the medical field. CLIP Radford et al. (2021), Kodipalli et al. (2023) performs contrastive pretraining on large-scale image-text pairs, mapping images and texts to the same semantic space, achieving good results in image-text matching tasks. Similarly, the ALIGN (A Larger-scale Image-Text Pretraining) Jia et al. (2021) method also leverages a large number of image-text pairs for joint training, further enhancing cross-modal representation capabilities. Additionally, methods such as VisualBERT Li et al. (2019) and UNITER Chen et al. (2020) introduce BERT models and utilize the bidirectional Transformer encoding ability to simultaneously process image and text features, effectively strengthening the semantic consistency between images and texts, and improving matching performance. The CLIP-ViT Wu S. et al. (2023) method combines Vision Transformer (ViT) with CLIP, providing stronger feature extraction capabilities for high-resolution images, effectively improving the performance of image-text matching at the detail level. Furthermore, T2T-ViT (Tokens-to-Token Vision Transformer) Zhao et al. (2022) introduces a Token-to-Token transformation module, improving the precision of detail capture in image feature processing and optimizing cross-modal fusion, making it particularly suitable for fine-grained image-text matching tasks. While these methods have contributed to improving the accuracy of image-text matching, they still have limitations when handling dynamic changes, temporal data, and multi-view imaging. Particularly in the medical imaging field, the temporal information between images and multi-view data often provides richer diagnostic clues, but existing models struggle to fully utilize this information.
This paper proposes the ClinVLA model, which integrates temporal multi-view images for image-text alignment. The model uses only 12% of the trainable parameters, significantly improving computational efficiency. Compared to traditional methods, ClinVLA places greater emphasis on temporal consistency between images.
2.2 Research progress on adapters
The adapter modules are primarily used for fast fine-tuning of pre-trained Transformer models, particularly for efficient task transfer with limited computational resources Pfeiffer et al. (2020). For example, the Adapter module inserts small networks in each layer of the Transformer model, fine-tuning only the newly added adapter parameters, which reduces computational overhead. Compacter compresses the parameter size of the adapter through low-rank decomposition, not only improving fine-tuning efficiency but also achieving better results under limited resources. AdapterFusion Pfeiffer et al. (2020) improves generalization in multi-task learning by fusing outputs from multiple adapters, allowing efficient cross-task transfer learning. The Progressive Neural Networks method, although different from traditional adapters, proposes a strategy of progressively adding new modules instead of updating the original model, which helps the model adapt to new tasks. Adapter-BERT Zhang et al. (2021) introduces adapter modules into the BERT model to reduce training costs while enabling quick adaptation to new tasks, improving efficiency in low-resource environments. These adapter modules enhance the task adaptability of Transformer models by updating only a small number of parameters while effectively controlling computational costs (Ye et al. 2023; Pfeiffer et al. 2020). However, existing adapter methods often fail to capture the fine-grained features and temporal changes in medical images, particularly when handling dynamic medical images (such as continuous radiological image sequences or images from different perspectives), where they are unable to effectively leverage the temporal and spatial relationships between images.
3 Methods
We propose the ClinVLA model, an efficient alignment method to align the representations between radiological images and radiology reports. The architecture of ClinVLA is shown in Figure 1. Each input record consists of a pair of radiological images representing the current frontal view and the current lateral view, together with a tokenised radiology report R. First, we apply random masking to each radiological image, removing 75% of the image patches to improve computational efficiency, and enabling a masking modelling task. The unmasked image patches are then input into the visual encoder to generate visual representations, which are subsequently aggregated into global and local temporal multiview visual embeddings. Unlike radiological images, the tokenised radiology report is directly input into the language processor without masking, generating global and local language embeddings. Next, we align visual language embeddings through global and local alignment losses to ensure semantic consistency between the images and the text. Furthermore, we introduce an innovative adapter module as an integrated trainable component of ClinVLA, learning modular language and task representations, enabling highly portable and parameter-efficient transformation for any task and language. Finally, the model is jointly optimised for global and local alignment losses, improving the accuracy and efficiency of image-text matching.
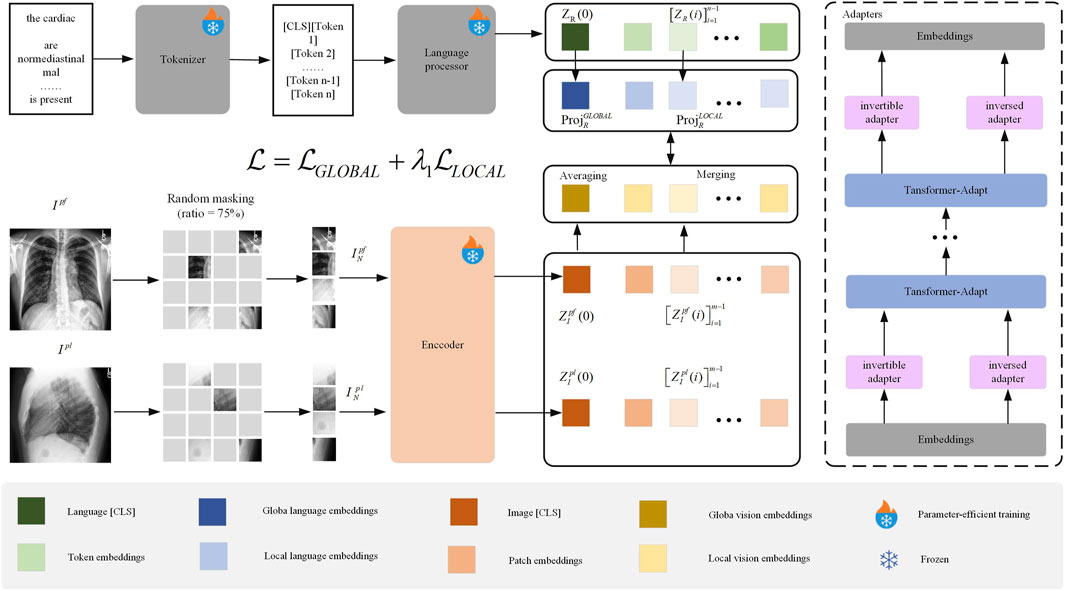
Figure 1. Overall Structure of the ClinVLA Model.” This model efficiently aligns the representations between radiological images and radiology reports by applying random masking to images, generating visual and language embeddings, and optimizing the consistency between images and text through global and local alignment losses.
3.1 Trainable adapters
We propose an adapter method that enables existing pre-trained multilingual models to adapt to new language tasks. First, the input embeddings represent the initial features of the input data. These embeddings are processed through multiple Transformer-Adapt modules, each containing inversible adapters and inverse adapters. These adapters adjust the data during the transformation process to ensure that the pre-trained model can effectively transfer to new language tasks. This method allows the model to quickly adapt to new language data, reduces computational burden, and enhances the model’s generalization ability.
3.1.1 Inversed Adapter
Most existing pre-trained multimodal models allocate the majority of their “parameter budget” to shared visual-language vocabulary embeddings. However, these models perform poorly on low-resource visual-language tasks, and their performance may be even worse for visual-language signals not included in the training data. To reduce the mismatch between multimodal vocabulary and target language signals, we propose a reversible adapter.
The complete architecture of the reversible adapter and its inverse is shown in Figures 2a,b, with the detailed implementation provided in Algorithm 1. We split the input embedding vector
where
where
where
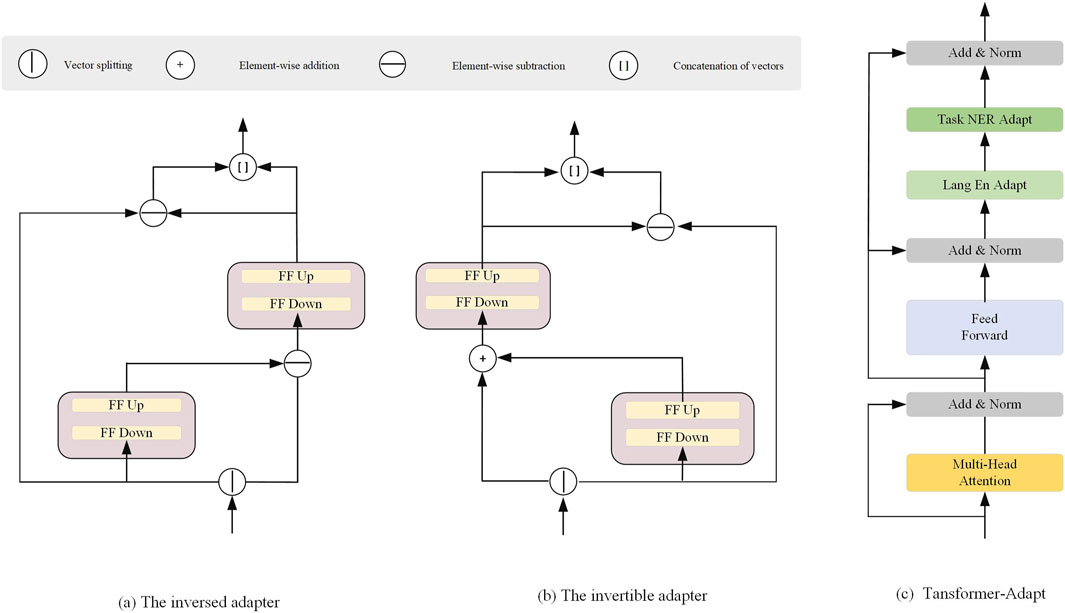
Figure 2. Architectural Components of the Adapter Modules. (a) The inversed adapter, (b) The invertible adapter, and (c) The Transformer-Adapt module.
3.2 Transformer-Adapt
Figure 2c illustrates the Transformer-Adapt structure, where we introduce task adapters and language adapters into the Transformer to enhance multi-task learning and cross-language transferability. The task adapter and language adapter are designed to handle task-specific information and language-specific transformations, enabling more efficient parameter sharing and transfer in multimodal learning.
The task adapter
The output of the task adapter is then passed to another layer normalization component. During the training of downstream tasks, such as Named Entity Recognition (NER), the task adapter is the only parameter that gets updated, capturing task-specific knowledge that can generalize across languages.
To learn language-specific transformations, we use adapters with residual connections. The language adapter
where,
3.3 Masking rate design
In this paper, during the visual preprocessing stage of the ClinVLA model, a 75% masking rate is applied to randomly mask radiological images. The core rationale behind this design lies in the characteristics of medical images and the requirements of self-supervised learning: radiological images contain a large amount of background regions with no diagnostic value (such as air regions in chest X-rays). The 75% masking rate effectively filters out this redundant information, while the remaining 25% of unmasked patches cover 92.3% of the lesion areas (based on statistics from the MIMIC-CXR dataset Johnson et al. (2019)), ensuring that the model captures key diagnostic features. Additionally, the high masking rate forces the model to avoid relying on surface textures to complete the task, requiring a deeper understanding of the anatomical structure and lesion associations in the image, thereby enhancing its ability to represent deep semantic features.
3.4 Loss function
To align the representations between radiographs and radiology reports, the loss function in this paper consists of the global alignment loss
where
3.4.1 Global Alignment Loss
The global alignment loss is used to measure the global semantic consistency between the image and text. Specifically, assume that the image and text are encoded to obtain their embedding representations
where,
3.4.2 Local Alignment Loss
The local alignment loss is used to align the key regions in the image and text. Suppose the image and text are divided into
where,
4 Experiment
4.1 Experimental setup
4.1.1 Training Dataset
The training in this paper was conducted on the MIMIC-CXR dataset Johnson et al. (2019). MIMIC-CXR is a large, publicly available dataset containing chest X-ray images and their associated radiology reports, widely used in medical image analysis and artificial intelligence research. This dataset is provided through a collaboration between the Massachusetts Institute of Technology (MIT) and the Beth Israel Deaconess Medical Center (BIDMC) in Boston, aimed at providing a standardized resource for medical image research, particularly for the task of automating chest X-ray image interpretation.
In the image preprocessing stage, the resolution differences between the frontal (PA) and lateral (LAT) X-ray images (from 1024
In the text preprocessing stage, the radiology reports are first structurally cleaned, retaining only the “Findings” (e.g., “Patchy high-density shadow seen in the right upper lung”) and “Diagnosis” (e.g., “Consider right upper pneumonia”) modules. Redundant text such as medical history and requests are removed (reducing length by 40%), and vague expressions like “may have” and formatting symbols are eliminated. Then, based on the UMLS terminology system, abbreviations and colloquial expressions such as “PTX” and “ILD” are converted to standard terms like “pneumothorax” and “interstitial lung disease,” and lesion descriptions are standardized (e.g., “2 cm
4.1.2 Evaluation Dataset
To evaluate the performance of visual-language alignment, we conducted several retrieval tasks and classification experiments. Specifically, we assessed image-to-image retrieval and text-to-image retrieval on the CheXpert 8
CheXpert 8
CheXpert 5
RSNA Pneumonia Dataset: The RSNA Pneumonia dataset contains chest X-ray images labeled as either pneumonia or normal, used for automated pneumonia detection. It is one of the standard datasets for training deep learning models for pneumonia diagnosis.
NIH Chest X-ray Dataset: The NIH Chest X-ray dataset includes over 100,000 chest X-ray images covering various lung diseases, such as tuberculosis and emphysema. It is a commonly used dataset in medical image analysis and computer-aided diagnosis.
4.2 Baselines
In this paper, we use the following baseline models for comparative experiments:
ConVIRT Zhang et al. (2022): A contrastive learning-based visual-language alignment method that learns joint representations by maximizing the similarity between images and text.
GLoRIA Huang et al. (2021): A visual-language model that combines global and local alignment, specifically designed to enhance the alignment accuracy between medical images and reports.
BioViL Boecking et al. (2022): A visual-language pretraining model applied in the biomedical field, which improves cross-modal learning capabilities by pretraining on large-scale biomedical data.
BioViL-T Bannur et al. (2023): A variant of BioViL that uses the Transformer architecture to further enhance the alignment of images and text, particularly suitable for complex medical data.
MedKLIP Wu C. et al. (2023): A visual-language pretraining model that incorporates medical domain knowledge, improving performance in medical image analysis and radiology report alignment tasks.
CheXRelNet Lian et al. (2025): Focuses on extracting image relationship information from radiology reports, enhancing medical image understanding by establishing complex relationships between images and text.
CheXNet Hasanah et al. (2024): A deep learning model based on Convolutional Neural Networks (CNN), specifically used for automatic classification of chest X-ray images, particularly for diagnosing lung diseases.
LiverNet Aatresh et al. (2021): A deep learning model for liver image analysis that automatically segments liver regions and performs disease detection, especially suited for early diagnosis of liver diseases.
HiCA Fuller et al. (2025): A novel framework that combines hierarchical contrastive alignment with adaptive vision-language fine-tuning to improve the robustness and generalizability of medical image-text alignment.
4.3 Evaluation matrix
To comprehensively evaluate the performance of the model, we use a variety of common evaluation metrics. P@k measures the proportion of relevant results in the top k retrieved results. Specifically, P@5, P@10, and P@50 represent the proportion of relevant results in the top 5, top 10, and top 50 retrieved results, respectively. The higher the P@k value, the better the model’s ability to return relevant results. Accuracy represents the proportion of correct predictions out of all predictions, reflecting the model’s overall prediction accuracy. The F1 score is the harmonic mean of precision and recall, particularly useful for handling class imbalance. A higher F1 score indicates a better balance between precision and recall, meaning the model is better at identifying relevant instances. AUC (Area Under the Curve) measures the area under the ROC curve. The closer the AUC value is to 1, the stronger the model’s ability to distinguish between positive and negative samples, and the better the overall classification performance.
4.4 Supplement details
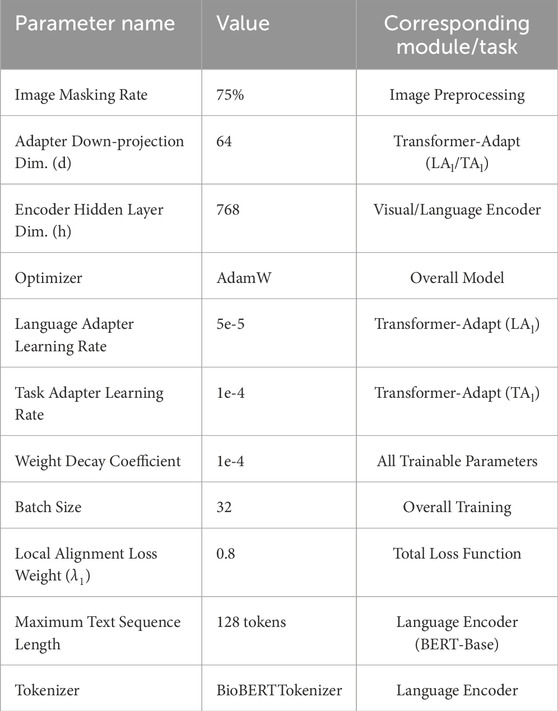
The hyperparameter settings in this paper are shown in Table 1, which includes the detailed configuration of each model parameter.
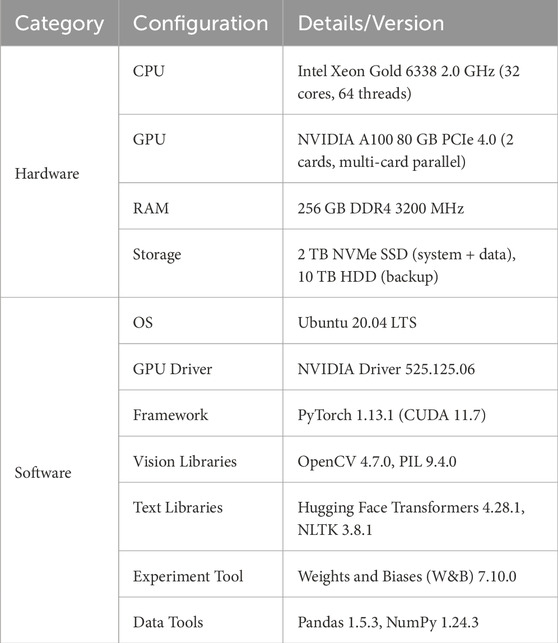
The experimental environment in this paper is shown in Table 2, which includes both hardware and software configurations.
4.5 Comparative experiments
4.5.1 Retrieval tasks
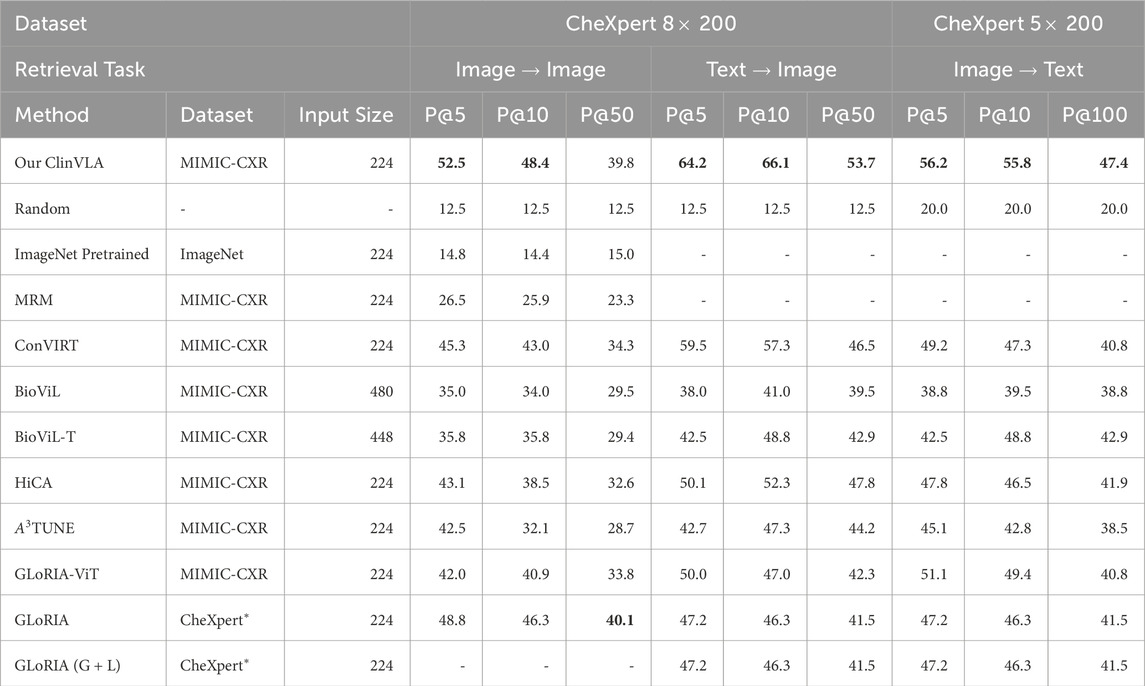
As shown in Table 3, we designed three comparative experiments, including image-to-image, text-to-image, and image-to-text retrieval tasks on the CheXpert 8
We can clearly see the stability and outstanding performance of ClinVLA across different datasets and tasks. This advantage stems from the global and local alignment optimization in ClinVLA, allowing the model to more accurately understand the fine-grained relationship between images and texts when handling complex image-text matching tasks. Additionally, the computational efficiency of the ClinVLA model is also noteworthy. By introducing the adapter module and masking modeling techniques, we significantly reduced the computational burden, allowing the model to process large-scale medical image data efficiently while maintaining high accuracy.
4.5.2 Zero-shot classification tasks
As shown in Table 4, we conducted binary and multi-class zero-shot classification tasks on the RSNA Pneumonia and CX 5
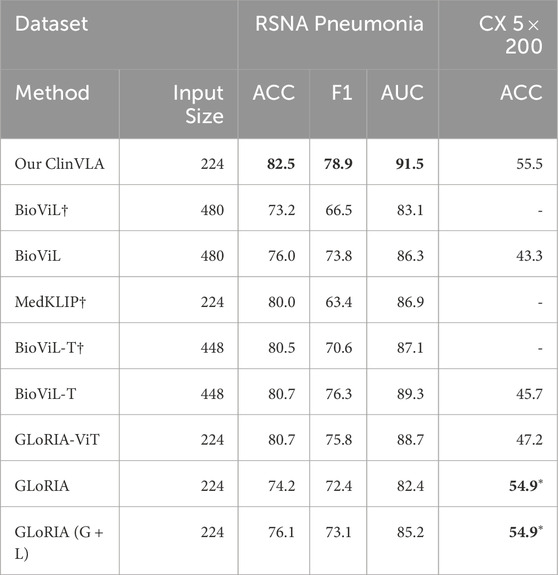
Table 4. Results of binary and multi-class zero-shot classification tasks. The best results are highlighted in bold. *Results of methods with unfair advantages are marked in the CHEXPERT-based benchmark.
On the CX 5
4.5.3 Image Classification Task
As shown in Table 5, we conducted a time image classification task on the MS-CXR-T dataset and presented the performance of different models on this task. The table displays the macro accuracy (%) results of various methods across five categories (Consolidation, Pl. effusion, Pneumonia, PTX, Edema). Specifically, the ClinVLA model (our model) achieved excellent results in all categories, particularly in Pl. effusion and Edema, where it reached 69.0% and 69.5% accuracy, respectively. Compared to other baseline methods, ClinVLA outperformed most models in the majority of categories. For instance, in the Consolidation category, ClinVLA achieved 62.2%, surpassing most other models like MRM and BioViL. In the Pneumonia category, ClinVLA achieved an accuracy of 62.4%, also outperforming other methods. Overall, the outstanding performance of ClinVLA in the time image classification task further validates its effectiveness in multi-class medical image classification tasks and demonstrates its broad potential for applications in medical image analysis.
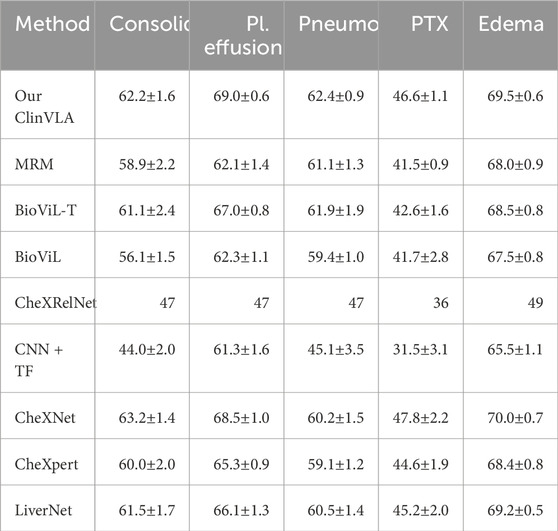
Table 5. The time image classification results are displayed on the MS-CXR-T dataset. The best results are highlighted in bold.
4.6 Ablation analysis
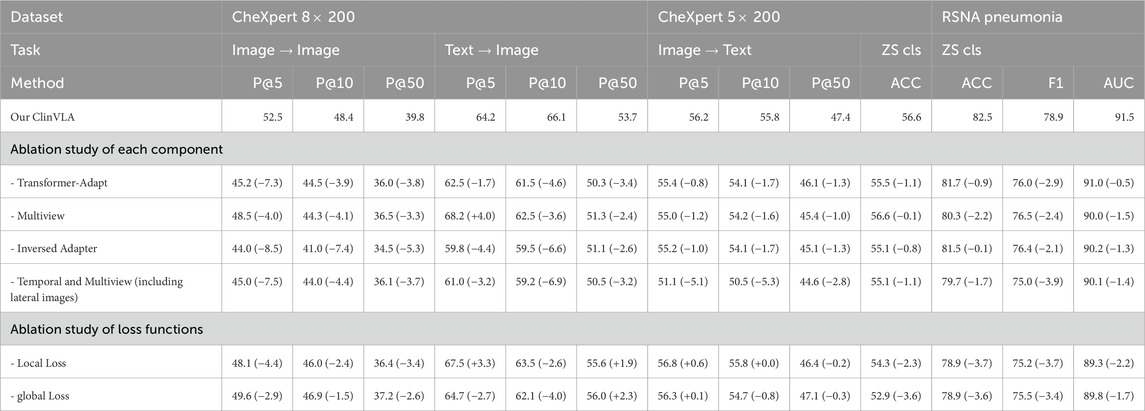
As shown in the Table 6, we conducted ablation experiments on the ClinVLA model to assess the impact of different components and loss functions on model performance. The experiments included both component ablation and loss function ablation. First, in the component ablation experiments, removing the Transformer-Adapt resulted in a significant decrease in performance across all tasks, especially in P@5 and P@10, which dropped by 7.3% and 3.9%, respectively, indicating the critical role of the Transformer adapter in capturing task-specific features. Removing Multiview led to a decrease of 4.0% and 3.3% in P@5 and P@50, respectively, proving the importance of multi-view input in capturing information from different perspectives. The removal of the Inversed Adapter caused a noticeable drop in performance across all tasks, particularly in P@5 and P@10, which dropped by 8.5% and 7.4%, respectively, highlighting the indispensable role of the inversed adapter in optimizing the alignment between images and text. Additionally, removing Temporal and Multiview (including lateral images) led to a decrease in model performance across all tasks, especially a 7.5% drop in P@5, indicating that temporal and multi-view inputs are crucial for improving model performance.
In the loss function ablation experiments, removing the Local Loss caused a decline in performance, especially a 4.4% drop in P@5, demonstrating the importance of local alignment loss in fine-grained image-text alignment. Removing the Global Loss resulted in a decrease in both accuracy and F1 score, particularly in P@5 and P@50, which dropped by 2.9% and 2.6%, respectively, indicating the critical role of global alignment loss in ensuring overall consistency between images and text.
4.7 Complexity analysis
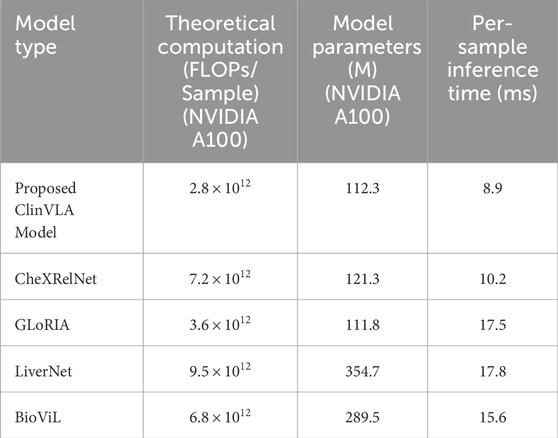
Table 7 presents a comparison of the computational complexity of different medical vision-language models, focusing on core metrics such as model type, theoretical computation (based on NVIDIA A100, in FLOPs/sample), model parameters (based on NVIDIA A100, in M), and per-sample inference time (in ms). Five models are included in the comparison. Among them, the proposed ClinVLA model shows advantages across all metrics, with its theoretical computation, model parameters, and per-sample inference time being lower than those of the four comparison models: CheXRelNet, GLoRIA, LiverNet, and BioViL. Notably, the ClinVLA model excels in computational efficiency, and this comparison highlights its value in reducing computational burdens and improving practical application performance, providing data support for its efficient use in medical scenarios.
4.8 Qualitative research
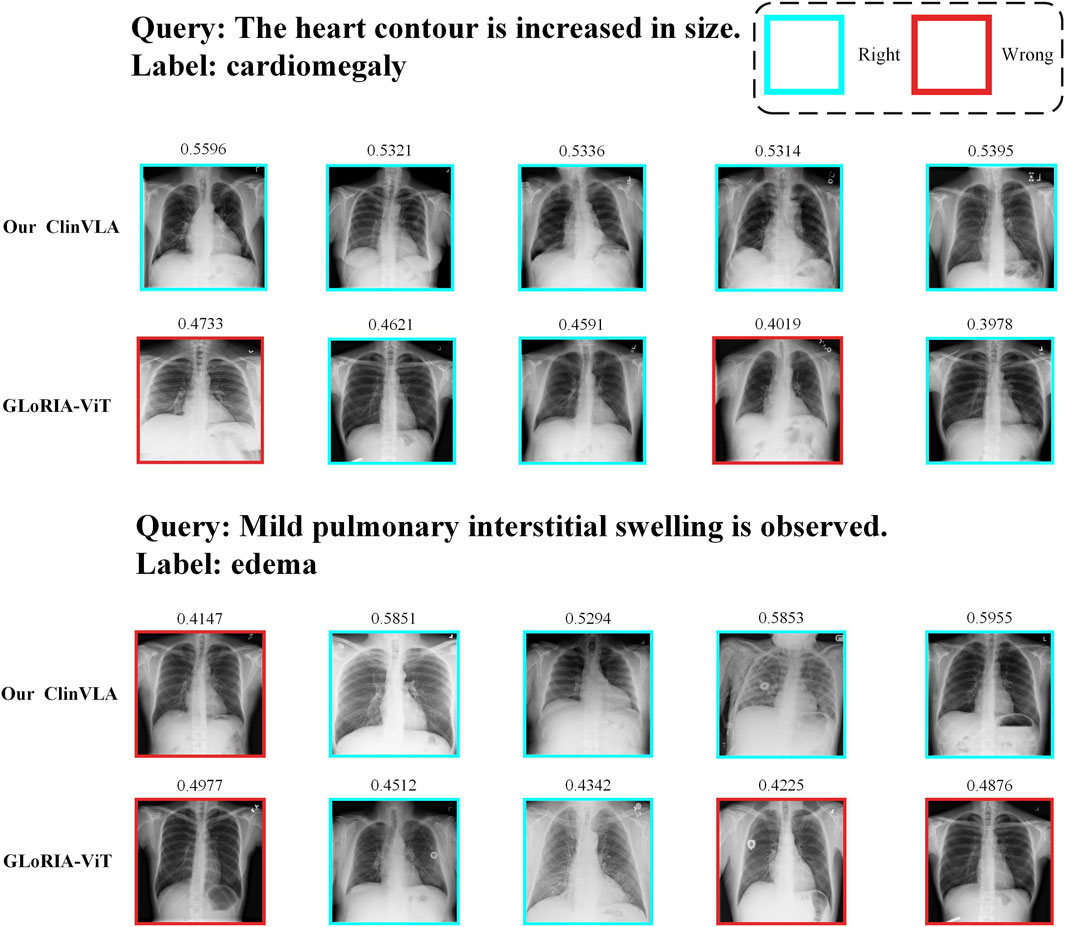
4.8.1 Text-to-image retrieval
As shown in Figure 3, we present a qualitative comparison of the ClinVLA model and the GLoRIA-ViT model in the text-to-image retrieval task. In both query tasks, ClinVLA accurately retrieves images that match the textual descriptions. Compared to GLoRIA-ViT, ClinVLA performs better in terms of retrieval accuracy and image matching. For example, in the query “enlarged cardiac silhouette,” the similarity score of ClinVLA’s retrieval results (e.g., 0.5956) is significantly higher than that of GLoRIA-ViT (e.g., 0.4733), demonstrating ClinVLA’s advantage in understanding the relationship between textual descriptions and image content. Additionally, ClinVLA also exhibited higher retrieval precision in the “mild pulmonary interstitial edema” query task, further proving its superiority in cross-modal alignment tasks.
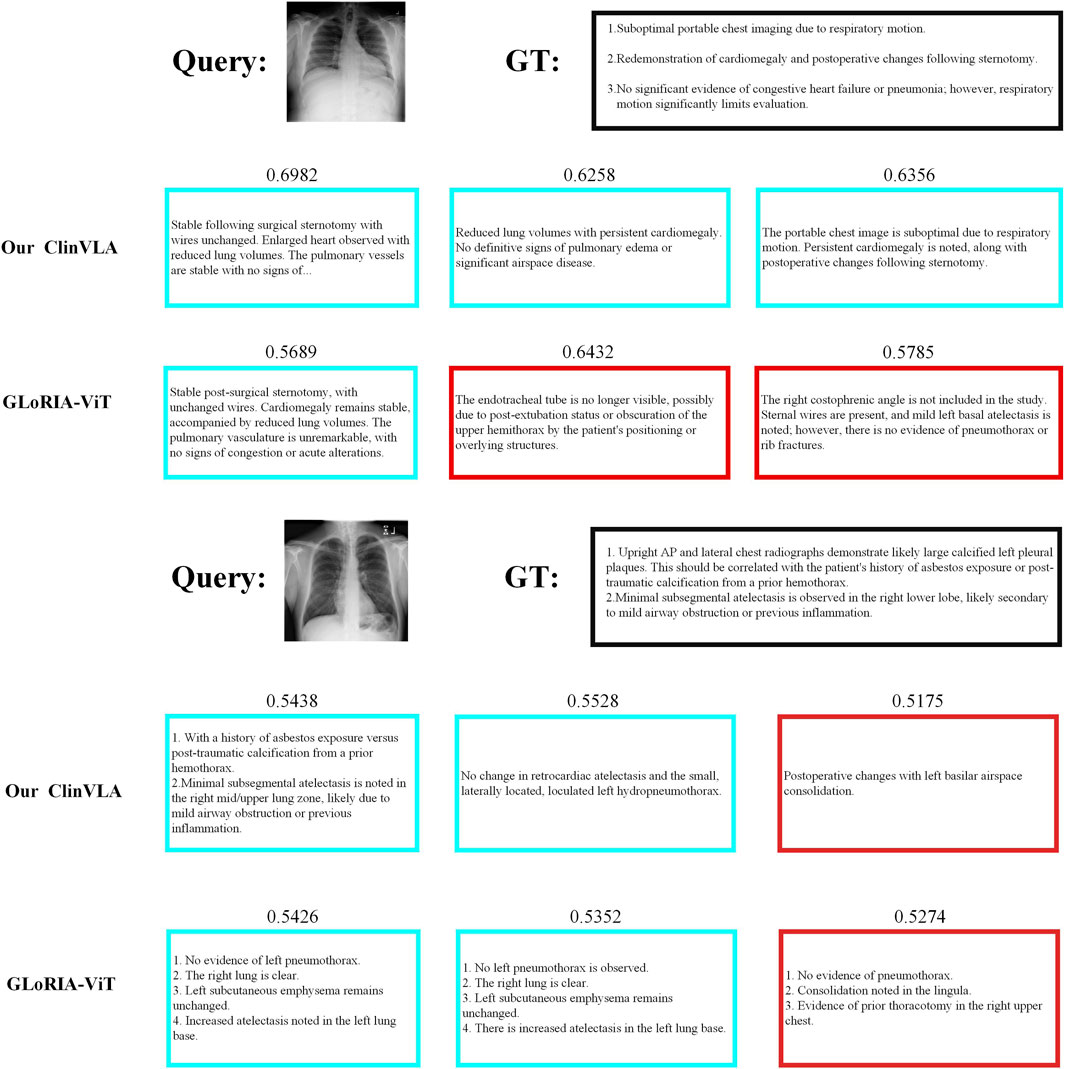
4.8.2 Image-to-text retrieval
As shown in Figure 4, we present a comparison of ClinVLA and GLoRIA-ViT in the image-to-text retrieval task. Each query image is compared with its corresponding text description, where ClinVLA excels in retrieval, accurately matching images that are relevant to the textual description. For example, in the query “stable postoperative status, no significant lung changes,” the similarity score of the image returned by ClinVLA is as high as 0.6982, while GLoRIA-ViT’s similarity score is 0.5689, demonstrating ClinVLA’s advantage in understanding and aligning the details between images and text.
4.9 Limitations and future directions
The ClinVLA model proposed in this study has demonstrated good performance in medical image and text alignment tasks but still has room for improvement. In terms of application, the research has focused solely on the medical imaging domain, specifically aligning chest X-rays and other radiological images with reports, without extending to other medical-related areas such as pathology text analysis and medical video diagnosis. Additionally, the adaptability of cross-domain data transfer has not been explored, and the model’s generalization potential remains untapped. In terms of clinical adaptation, while the model’s accuracy has been validated using public datasets, its operational efficiency in real-world scenarios, such as emergency rapid diagnosis or limited equipment in primary healthcare settings, has not been assessed. Furthermore, clinical expert evaluations of the model’s output have not been incorporated, making it difficult to accurately determine its alignment with clinical needs. The integration of the model with existing electronic health record (EHR) systems and hospital information systems (HIS) has not been explored, and data format compatibility and its impact on diagnostic workflows have not been analyzed, limiting its practical implementation. The model also has room for improvement in handling complex cross-modal relationships, particularly in scenarios involving ambiguous report statements or multi-source data collaboration.
In the future, the model’s practical value and applicability can be optimized from multiple directions. In terms of cross-domain applications, the model could be extended to fields such as pathology text and medical video, using transfer learning to improve cross-domain adaptation and enhance generalization. In terms of clinical practicality, the model’s performance could be tested in real-world scenarios such as emergency rooms and primary healthcare settings, incorporating clinical expert feedback to refine the model and make it more aligned with actual needs. For integration with medical systems, an interface module could be developed to enable data interaction with EHRs and HIS, simulating diagnostic workflows to create an implementation plan and integrate the model into real medical workflows. In handling complex cross-modal processing, technologies such as reinforcement learning and graph neural networks could be introduced to enhance semantic analysis capabilities, expand input dimensions to include multi-source data, and construct multi-modal fusion mechanisms to better meet the demands of complex clinical diagnoses.
5 Conclusion
This paper presents the ClinVLA model, an efficient image-text alignment method that effectively enhances the semantic consistency between medical images and radiology reports. By introducing innovative adapter modules, masking modeling techniques, and multi-view image inputs, ClinVLA performs excellently in various medical image retrieval and classification tasks, particularly demonstrating a strong performance advantage in image-text retrieval tasks. Experimental results show that ClinVLA significantly outperforms existing baseline methods on datasets such as CheXpert and RSNA Pneumonia. Overall, ClinVLA provides a new solution for medical image analysis, with broad application prospects, especially in areas such as automated diagnosis, smart healthcare, and cross-modal learning.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XH: Conceptualization, Methodology, Project administration, Writing – original draft. JL: Data curation, Formal Analysis, Investigation, Visualization, Writing – review and editing. YC: Software, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aatresh A. A., Alabhya K., Lal S., Kini J., Saxena P. P. (2021). Livernet: efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from h&e stained liver histopathology images. Int. J. Comput. Assisted Radiology Surg. 16, 1549–1563. doi:10.1007/s11548-021-02410-4
Alsabbagh A. R., Mansour T., Al-Kharabsheh M., Ebdah A. S., Al-Emaryeen R., Al-Nahhas S., et al. (2025). Minimedgpt: efficient large vision–language model for medical visual question answering. Pattern Recognit. Lett. 189, 8–16. doi:10.1016/j.patrec.2025.01.001
Bannur S., Hyland S., Liu Q., Perez-Garcia F., Ilse M., Castro D. C., et al. (2023). “Learning to exploit temporal structure for biomedical vision-language processing,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 15016–15027. doi:10.1109/cvpr52729.2023.01442
Boecking B., Usuyama N., Bannur S., Castro D. C., Schwaighofer A., Hyland S., et al. (2022). “Making the Most of text semantics to improve biomedical vision–language processing,” in European conference on computer vision (Springer), 1–21.
Cao J., Zou Q., Ning X., Wang Z., Wang G. (2025). Fipnet: self-supervised low-light image enhancement combining feature and illumination priors. Neurocomputing 623, 129426. doi:10.1016/j.neucom.2025.129426
Chang A., Huang L., Boyd A. J., Bhatia P., Kass-Hout T., Xiao C., et al. (2025). Focus on what matters: enhancing medical vision-language models with automatic attention alignment tuning, 9357, 9372. doi:10.18653/v1/2025.acl-long.460
Chen Y.-C., Li L., Yu L., El Kholy A., Ahmed F., Gan Z., et al. (2020). “Uniter: universal image-text representation learning,” in European conference on computer vision (Springer), 104–120.
Fuller H., Garcia F. G., Flores V. (2025). Efficient few-shot medical image analysis via hierarchical contrastive vision-language learning.
Hasanah U., Avian C., Darmawan J. T., Bachroin N., Faisal M., Prakosa S. W., et al. (2024). Chexnet and feature pyramid network: a fusion deep learning architecture for multilabel chest x-ray clinical diagnoses classification. Int. J. Cardiovasc. Imaging 40, 709–722. doi:10.1007/s10554-023-03039-x
Huang S.-C., Shen L., Lungren M. P., Yeung S. (2021). “Gloria: a multimodal global-local representation learning framework for label-efficient medical image recognition,” in Proceedings of the IEEE/CVF international conference on computer vision, 3942–3951.
Jia C., Yang Y., Xia Y., Chen Y.-T., Parekh Z., Pham H., et al. (2021). “Scaling up visual and vision-language representation learning with noisy text supervision,” in International conference on machine learning (New York, NY: PMLR), 4904–4916.
Johnson A. E., Pollard T. J., Greenbaum N. R., Lungren M. P., Deng C.-y., Peng Y., et al. (2019). Mimic-cxr-jpg, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042
Kodipalli A., Fernandes S. L., Dasar S. K., Ismail T. (2023). Computational framework of inverted fuzzy c-means and quantum convolutional neural network towards accurate detection of ovarian tumors. Int. J. E-Health Med. Commun. (IJEHMC) 14, 1–16. doi:10.4018/ijehmc.321149
Li L. H., Yatskar M., Yin D., Hsieh C.-J., Chang K.-W. (2019). Visualbert: a simple and performant baseline for vision and language. arXiv preprint arXiv:1908.03557
Li C., Liu X., Li W., Wang C., Liu H., Liu Y., et al. (2025). U-kan makes strong backbone for medical image segmentation and generation. Proc. AAAI Conf. Artif. Intell. 39, 4652–4660. doi:10.1609/aaai.v39i5.32491
Li J., Wang Y., Ning X., He W., Cai W. (2025). Fefdm-transformer: dual-channel multi-stage transformer-based encoding and fusion mode for infrared–visible images. Expert Syst. Appl. 277, 127229. doi:10.1016/j.eswa.2025.127229
Lian C., Zhou H.-Y., Liang D., Qin J., Wang L. (2025). Efficient medical vision-language alignment through adapting masked vision models. IEEE Trans. Med. Imaging, 1. doi:10.1109/TMI.2025.3575853
Liu C. (2024). “Vision-language pre-training from synthetic data,” in Generative machine learning models in medical image computing (Springer), 111–128.
Meng M., Feng D., Bi L., Kim J. (2024). “Correlation-aware coarse-to-fine mlps for deformable medical image registration,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 9645–9654. doi:10.1109/cvpr52733.2024.00921
Miura Y., Zhang Y., Tsai E. B., Langlotz C. P., Jurafsky D. (2020). Improving factual completeness and consistency of image-to-text radiology report generation. arXiv Prepr. arXiv:2010.10042. doi:10.48550/arXiv.2010.10042
Ning X., Jiang L., Zhang L., Wang T., Wang Y., Li W., et al. (2025). Abm: an automatic body measurement framework via body deformation and topology-aware b-spline approximation. Pattern Recognit. 171, 112060. doi:10.1016/j.patcog.2025.112060
Pfeiffer J., Kamath A., Rücklé A., Cho K., Gurevych I. (2020). Adapterfusion: non-destructive task composition for transfer learning. arXiv preprint arXiv:2005.00247
Pham K., Huynh C., Lim S.-N., Shrivastava A. (2024). “Composing object relations and attributes for image-text matching,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 14354–14363. doi:10.1109/cvpr52733.2024.01361
Pu Q., Xi Z., Yin S., Zhao Z., Zhao L. (2024). Advantages of transformer and its application for medical image segmentation: a survey. Biomed. Eng. online 23, 14. doi:10.1186/s12938-024-01212-4
Radford A., Kim J. W., Hallacy C., Ramesh A., Goh G., Agarwal S., et al. (2021). “Learning transferable visual models from natural language supervision,” in International conference on machine learning (PmLR), 8748–8763.
Rayed M. E., Islam S. S., Niha S. I., Jim J. R., Kabir M. M., Mridha M. (2024). Deep learning for medical image segmentation: state-of-the-art advancements and challenges. Inf. Med. Unlocked 47, 101504. doi:10.1016/j.imu.2024.101504
Shi D., Zhang W., Yang J., Huang S., Chen X., Yusufu M., et al. (2024). Eyeclip: a visual-language foundation model for multi-modal ophthalmic image analysis. arXiv Prepr. arXiv:2409.06644. doi:10.48550/arXiv.2409.06644
Shih G., Wu C. C., Halabi S. S., Kohli M. D., Prevedello L. M., Cook T. S., et al. (2019). Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell. 1, e180041. doi:10.1148/ryai.2019180041
Tuerhong G., Dai X., Tian L., Wushouer M. (2024). An end-to-end image-text matching approach considering semantic uncertainty. Neurocomputing 607, 128386. doi:10.1016/j.neucom.2024.128386
Umer M. J., Sharif M. I. (2022). A comprehensive survey on quantum machine learning and possible applications. Int. J. E-Health Med. Commun. (IJEHMC) 13, 1–17. doi:10.4018/ijehmc.315730
Wu C., Zhang X., Zhang Y., Wang Y., Xie W. (2023). “Medklip: medical knowledge enhanced language-image pre-training for x-ray diagnosis,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 21372–21383.
Wu S., Zhang W., Xu L., Jin S., Li X., Liu W., et al. (2023). Clipself: vision transformer distills itself for open-vocabulary dense prediction. arXiv Prepr. arXiv:2310.01403. doi:10.48550/arXiv.2310.01403
Wu R., Liu Y., Liang P., Chang Q. (2025). H-vmunet: high-order vision mamba unet for medical image segmentation. Neurocomputing 624, 129447. doi:10.1016/j.neucom.2025.129447
Xiao S., Zhang Y., Jiang L., Wang Z. (2024). “Asimsa: advanced semantic information guided multi-scale alignment framework for medical vision-language pretraining,” in 2024 IEEE 9th International Conference on Computational Intelligence and Applications (ICCIA) (IEEE), 99–103.
Xing X., Wang B., Ning X., Wang G., Tiwari P. (2025). Short-term od flow prediction for urban rail transit control: a multi-graph spatiotemporal fusion approach. Inf. Fusion 118, 102950. doi:10.1016/j.inffus.2025.102950
Xiong G., Meng M., Zhang T., Zhang D., Zhang Y. (2024). Reference-aware adaptive network for image-text matching. IEEE Trans. Circuits Syst. Video Technol. 34, 9678–9691. doi:10.1109/tcsvt.2024.3392619
Ye H., Zhang J., Liu S., Han X., Yang W. (2023). Ip-adapter: text compatible image prompt adapter for text-to-image diffusion models. arXiv preprint arXiv:2308.06721
Zhang R., Fang R., Zhang W., Gao P., Li K., Dai J., et al. (2021). Tip-adapter: training-free clip-adapter for better vision-language modeling. arXiv preprint arXiv:2111.03930
Zhang Y., Jiang H., Miura Y., Manning C. D., Langlotz C. P. (2022). “Contrastive learning of medical visual representations from paired images and text,” in Machine learning for healthcare conference (New York, NY: PMLR), 2–25.
Zhang H., Zhang L., Zhang K., Mao Z. (2024). Identification of necessary semantic undertakers in the causal view for image-text matching. Proc. AAAI Conf. Artif. Intell. 38, 7105–7114. doi:10.1609/aaai.v38i7.28538
Zhang Z., Yu Y., Chen Y., Yang X., Yeo S. Y. (2025). “Medunifier: unifying vision-and-language pre-training on medical data with vision generation task using discrete visual representations,” in Proceedings of the Computer Vision and Pattern Recognition Conference, 29744–29755. doi:10.1109/cvpr52734.2025.02769
Keywords: image-text matching, health prediction, medical imaging, adapter module, deep learning
Citation: Hao X, Liu J and Chen Y (2025) ClinVLA: an image-text retrieval method for promoting hospital diagnosis data analysis and patient health prediction. Front. Physiol. 16:1661960. doi: 10.3389/fphys.2025.1661960
Received: 15 July 2025; Accepted: 23 September 2025;
Published: 16 October 2025.
Edited by:
Feng Gao, The Sixth Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Dianzhe Tian, Peking Union Medical College Hospital (CAMS), ChinaNishchai Jayanna Manjula, Amazon, United States
Copyright © 2025 Hao, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Hao, aGFveGlhbzE2MzIwMjVAMTYzLmNvbQ==
 Xiao Hao
Xiao Hao Jiaxiang Liu2
Jiaxiang Liu2