- 1Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Hainan Grand Suntop Ocean Breeding Co., Ltd., Wenchang, China
- 5Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
Improvements of growth traits are always the focus in selective breeding programs for the Pacific white shrimp Litopenaeus vannamei (L. vannamei). Identification of growth-related genes or markers can contribute to the application of modern breeding technologies, and thus accelerate the genetic improvement of growth traits. The aim of this study was to identify the genes and molecular markers associated with the growth traits of L. vannamei. A population of 200 individuals was genotyped using 2b-RAD techniques for genome-wide linkage disequilibrium (LD) analysis and genome-wide association study (GWAS). The results showed that the LD decayed fast in the studied population, which suggest that it is feasible to fine map the growth-related genes with GWAS in L. vannamei. One gene designated as LvSRC, encoding the class C scavenger receptor (SRC), was identified as a growth-related candidate gene by GWAS. Further targeted sequencing of the candidate gene in another population of 322 shrimps revealed that several non-synonymous mutations within LvSRC were significantly associated with the body weight (P < 0.01), and the most significant marker (SRC_24) located in the candidate gene could explain 13% of phenotypic variance. The current results provide not only molecular markers for genetic improvement in L. vannamei, but also new insights for understanding the growth regulation mechanism in penaeid shrimp.
Introduction
Litopenaeus vannamei (L. vannamei), as one of the most economically important marine aquaculture species, is playing an important role in fulfilling the increased requirement for high quality animal proteins consumption. It is estimated that L. vannamei provided approximately 70% of the total shrimp production in the world (Li et al., 2018). The continuous development of shrimp industry drives the genetic improvement of important economic traits. During the past decade, large efforts have been put to improve the key economic traits, including growth traits and disease resistance (Argue et al., 2002; Huang et al., 2011; Andriantahina et al., 2013). Among these traits, growth is always the focus of breeders because it directly contributes to the shrimp production. In general, the growth trait, such as body weight, presents a moderate to high heritability (Sui et al., 2016; Nolasco-Alzaga et al., 2017), and the genetic gain per generation reached 10.7% (Andriantahina et al., 2012), which is higher than that of farmed terrestrial species. At present, broodstocks with high and stable growth traits are urgently needed to meet the requirement of shrimp culture industry. Modern molecular breeding technologies, including marker assisted selection (MAS), gene assisted selection (GAS), and gene editing technology, etc., are promising methods for accelerating the genetic improvement of growth traits (Gjedrem and Baranski, 2010). Till present, several growth-related genes involved in molting and muscle development such as molt-inhibiting hormone (MIH), crustacean hyperglycaemic hormone (CHH), ecdysteroid receptor (EcR), actin and myostatin differential factor 11 (MSTN), etc., were identified (Li et al., 2011; Jung et al., 2013). However, the growth traits are likely to be highly polygenic, and the underlying physiological bases may involve complex regulatory networks of many interacting genes with different effects. Although QTL mapping analysis of growth traits has been conducted in L. vannamei (Yu et al., 2015), limited number of markers makes the fine mapping of QTLs difficult. Hence, new methods are urgently required to localized the major genes or markers related to growth traits in shrimp.
Genome-wide association study (GWAS) have been successfully performed to identify genes participating in the regulation of complex traits in human (Mccarthy et al., 2008), livestock (Zhang et al., 2013), and crop (Huang and Han, 2014). Recently, with the development of high-throughput sequencing technologies and the successive decoding of aquatic animal genomes, GWAS is becoming a powerful tool to analyze the genetic basis of complex traits, and some candidate genes associated with growth traits or disease resistance were reported in a number of aquatic animals, including Atlantic salmon (Sodeland et al., 2013; Gutierrez et al., 2015; Correa et al., 2017), rainbow trout (Vallejo et al., 2014; Gonzalez-Pena et al., 2016), and catfish (Geng et al., 2015; Jin et al., 2016). However, there is no relevant study in L. vannamei. In the present study, we aimed to identify growth-related loci or genes in diverse population by using GWAS integrated with candidate gene association study, and provide a convinced result for revealing the molecular mechanism of growth traits in L. vannamei.
Materials and Methods
Animals and Genotyping
Two populations, designated as A16 and B2016_13, had been used in this study. These two populations were created and cultured at Guangtai Marine Breeding Company in Hainan province, China. The population A16 was established in 2015 as previously described (Wang et al., 2017). Briefly, it was composed of 200 individuals from 13 full-sib families (offsprings of 13 dams and 13 sires). Each full-sib family was cultured separately in the 5 m2 tank before their body length reached 3 cm, and then 50 individuals from each family were transferred to a 10 m2 pond for culture. At the harvest, two hundred individuals were randomly collected for the phenotyping and genotyping. For population B2016_13, it was constructed in 2016 and the individuals from multiple full-sib families were mixed after spawn, a total of 322 individuals were collected and phenotyped. The sex of all individuals from these two populations was determined by sex-associated marker (Yu et al., 2017). The average body weight for A16 population was 5.56 ± 2.16 g and that for B2016_13 population was 9.51 ± 3.30 g.
Total DNA of each sample was extracted from the muscle of shrimp using Plant Genomic DNA Kit (TIANGEN, Beijing, China) according to the manual instruction. The purity and integrity of the extracted DNA was determined by using a NanoDrop 1000 Spectrophotometer (NanoDrop, Wilmington, DE, United States) and electrophoresis on 1% agarose gel. Qualified genomic DNA was stored at -20°C.
All individuals from A16 population were genome-widely genotyped using 2b-RAD method (Wang et al., 2012), which was carried out by OE Biotech Company (OE Biotech, Shanghai, China). The reference genome were de novo assembled using the reads from the 10 individuals with high sequencing depth, and the genotyping of each individuals were conducted using RADtyping program (Fu et al., 2013). The shrimp from B2016_13 population were genotyped for the targeted locus of the candidate genes by using PCR-based sequencing.
Genome-Wide Linkage Disequilibrium Analysis
The physical position of SNPs was identified by blasting the 2b-RAD marker to the assembled reference genome of L. vannamei (Zhang et al., 2019). LD was estimated by using SNPs genotyping and physical position information. The squared correlation of allele frequencies (r2) was used as a measure of LD (Hill, 1974). The r2 between each pair of SNPs on the same chromosome was calculated using “genetics” package in R (R Core Team, 2018). The decay of the r2 with distance was fitted using the expected value of r2 under drift-recombination equilibrium that had previously been implemented (Remington et al., 2001; Marroni et al., 2011).
Genome-Wide Association Study
Genome-wide association study for body weight were performed using the egscore function in the R package GenABEL (Aulchenko et al., 2007). The potential bias in association caused by hidden population stratification was corrected by principal components (PCs) of genomic kinship matrix (Price et al., 2006). Via inspecting the eigenvalues of the kinship matrix, the first four PCs were selected to adjusting the genotypes and phenotypes. Sex was selected as fixed factor. Besides, adjusting with PCs did not remove all population stratification, hence a further genomic control correction of the obtained P values was performed using the inflation factor. Considering the small sample population size and the sparse marker density, the significance level for genome-wide significance was set as P = 0.01 ( = 2).
Candidate Genes Study
The sequences of SNPs associated with body weight were compared by BLAST against the genome sequence of the L. vannamei (Zhang et al., 2019). Given the rapid LD decay rate (Figure 1), the genes within the 18 kb upstream and downstream of the significant SNPs were considered as candidate genes. Then, the SNPs in the coding region of candidate genes were detected by PCR-based sequencing. The non-synonymous SNPs were genotyped in all the individuals from B2016_13 population and tested for association with body weight. The association test of candidate genes was performed by using linear model in R software, and sex was selected as covariate. According to the principle of variance decomposition in linear model (Ho and Lin, 2003), the ratio of phenotypic variance (Var) explained by the SNP, significantly associated with the body weight of L. vannamei, was calculated as following:
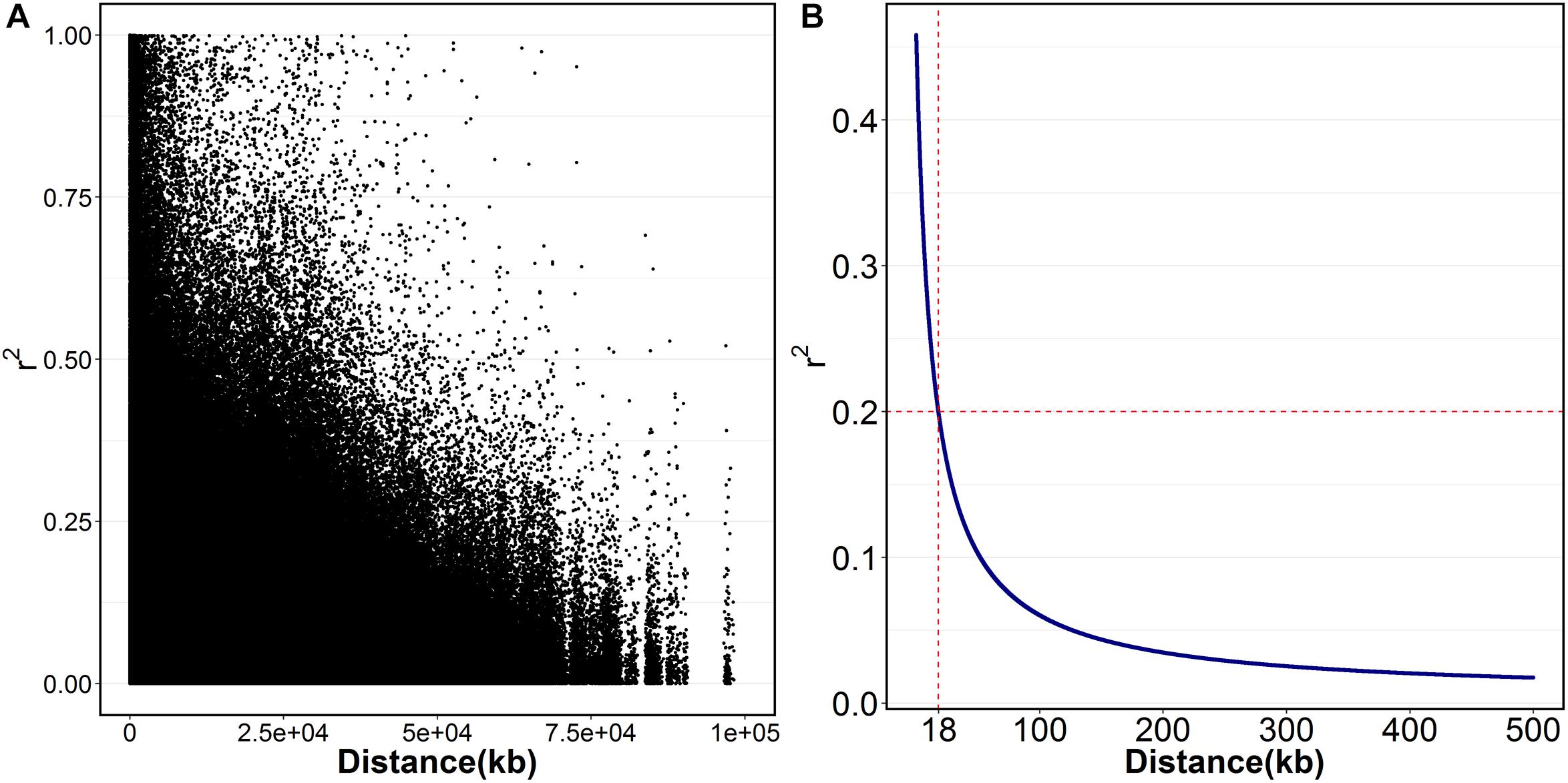
Figure 1. The extent of LD (r2) in Litopenaeus vannamei. (A) The scatter plot of r2 against the physical distance; (B) the fitted decay curve of r2 within 500 kb.
where SSR is the sum of squares produced by the SNP; SSS is the sum of squares produced by the sex; SSE is the residual sum of squares.
Results
Genome-Wide Linkage Disequilibrium
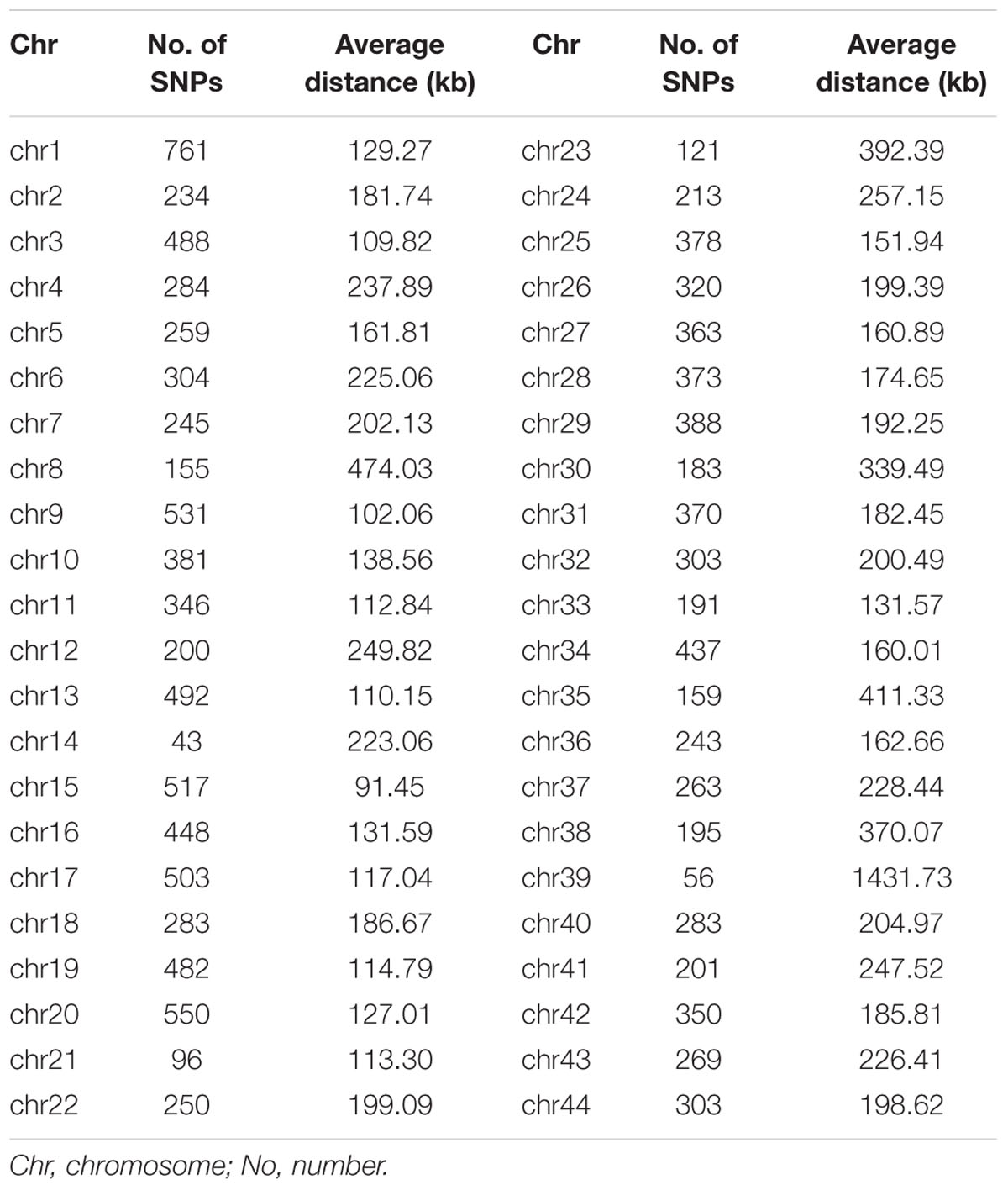
A total of 23,049 single nucleotide polymorphism (SNP) markers were obtained after quality control that SNPs with missing rate at more than 5% across samples and minor allele frequency less than 0.05 were removed. By blasting these markers to the assembled reference genome of L. vannamei, 13,814 SNPs were successfully mapped onto chromosomes. These SNPs were located on 44 chromosomes (Chrs) with a median distance between adjacent markers of 226.12 kb and an average of 314 SNP markers per chromosome (Table 1). The number of SNPs varied among Chrs, from 43 on chr14 to 761 on chr1. The average distance between the adjacent SNPs pairs within Chr was also different, ranging from 91.45 kb on chr15 to 1431.73 kb on chr39. A total of 2,579,595 paired SNPs had been used to calculate the r2. The r2 with distance was plotted in Figure 1A. The overall LD across the genome between all paired SNPs was 0.06 and only few values (0.4%) of r2 > 0.6 were found. A rapid decay of LD was presented in Figure 1B, where r2 decreased to 0.2 at SNP marker interval of 18 kb.
Genome-Wide Association Study
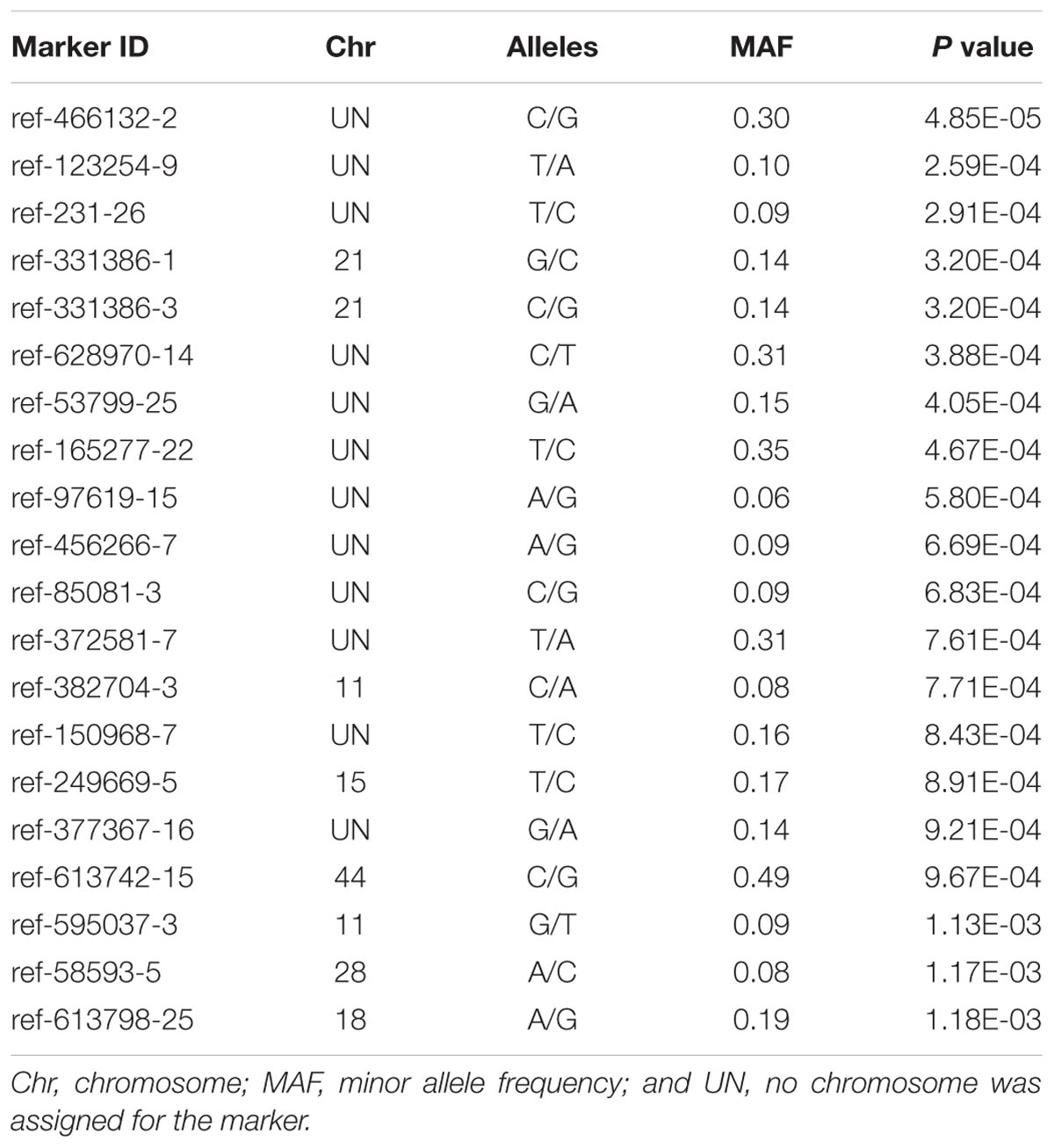
The Manhattan plot of all SNPs is shown in Figure 2. A total of 226 SNPs significantly associated with body weight were identified at a threshold of P < 0.01 ( > 2). Among the 226 significant SNPs, 84 SNPs are currently unassigned to chromosomes, and the remaining 142 SNPs were successfully mapped to 39 chromosomes. Given the large number of significant markers, the first twenty significant markers were used for subsequent analysis. Of these, 12 SNPs are currently unassigned to chromosomes, and the remaining 8 SNPs were successfully mapped to 6 chromosomes (Table 2). Gene annotation showed that only the marker ref-613798-25 was located in the coding region of one gene which can encode the class C scavenger receptor (SRC). Therefore, the gene, referred to hereafter as LvSRC, was considered as the most likely candidate gene for body weight in L. vannamei.
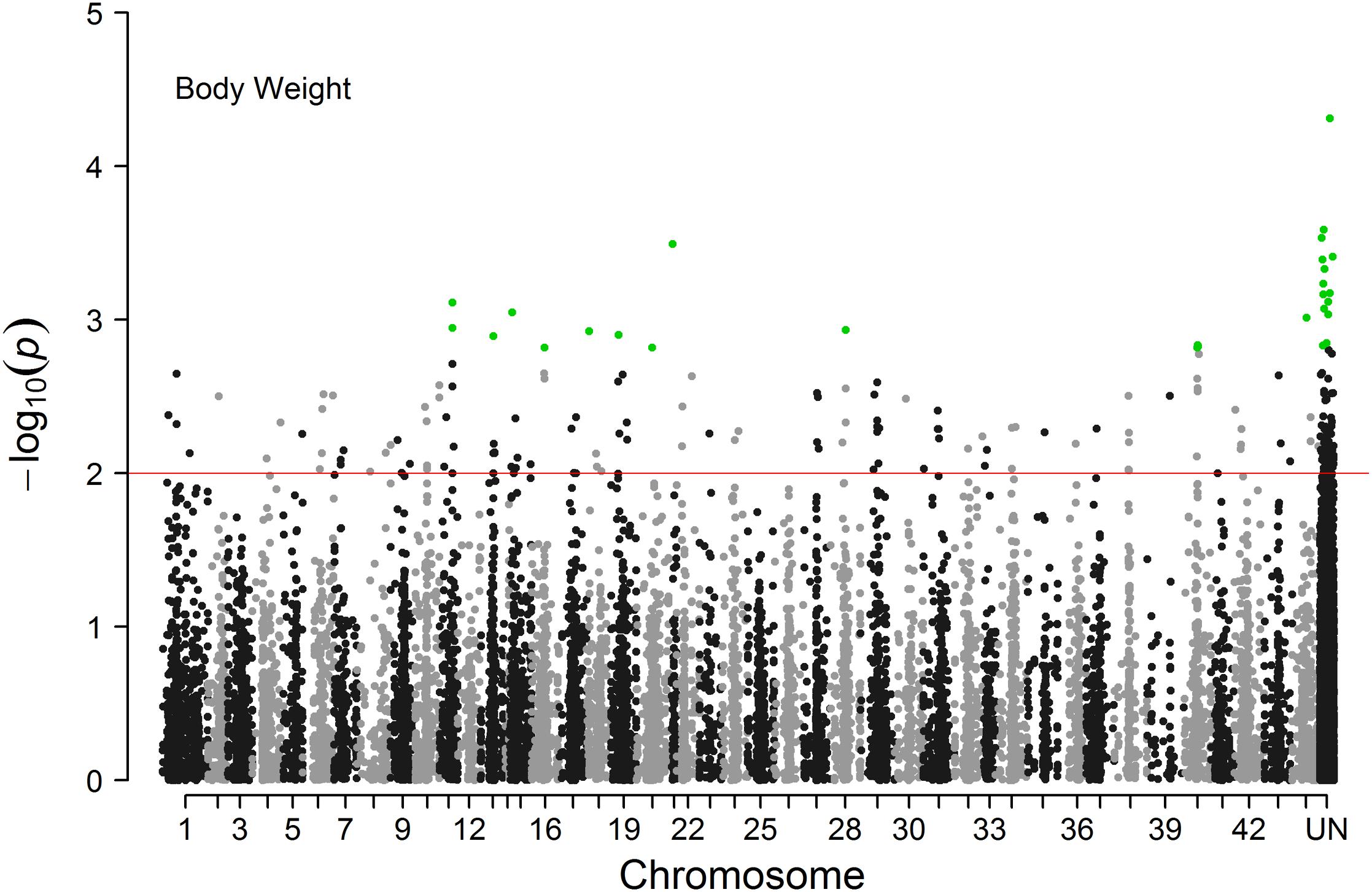
Figure 2. Manhattan plot of genome-wide association analysis for body weight of Litopenaeus vannamei.
Candidate Gene Association Study
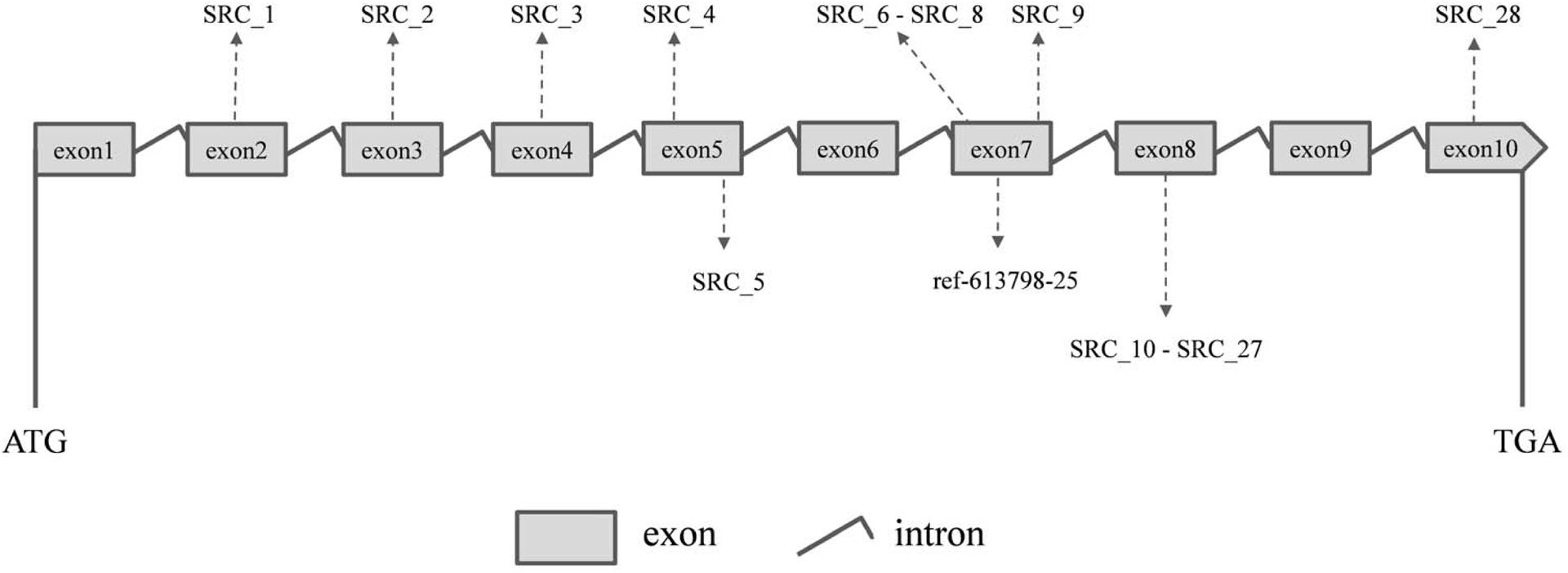
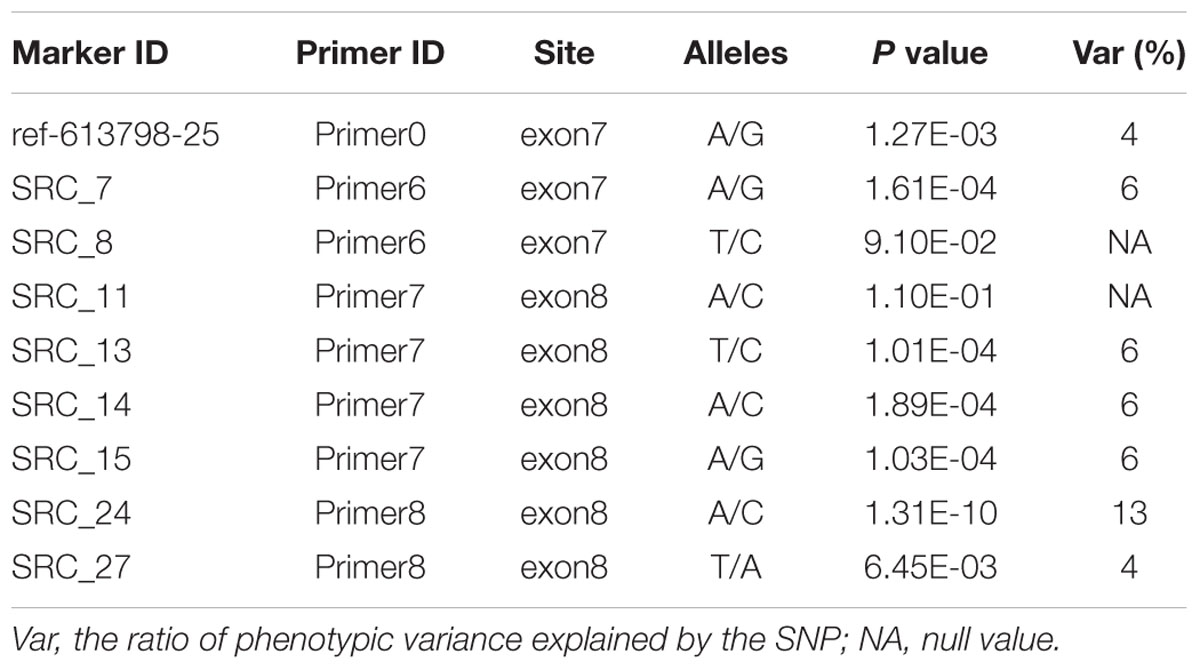
Ten PCR primers (Table 3) were developed from the targeted genome sequences of LvSRC and then used to amplify for a specific locus. A total of 29 SNPs (including ref-613798-25) were identified in the coding region of LvSRC (Figure 3). Among these, 20 SNPs are synonymous mutation (Supplementary Table S1), and 9 SNPs are non-synonymous mutations (Table 4). All these non-synonymous mutations were examined for association with body weight in B2016_13 population. The statistical results showed that 7 SNPs presented significant association (P < 0.01) with body weight, and the SNP (SRC_24) contributed most significantly to the trait and it could explain 13% of phenotypic variance, followed by SRC_13 (6%), SRC_15 (6%), SRC_7 (6%), SRC_14 (6%), ref-613798-25 (4%), and SRC_27 (4%).
Discussion
To our knowledge, this is the first report about LD pattern and GWAS in L. vannamei. Overall, the decay of LD in this population is rapid, which suggested it is feasible to perform the fine mapping of growth-related genes with GWAS. However, it is worth noting that high-density markers will be required to increase the power of GWAS. An average r2 greater than 0.2 has been proposed to be the desirable requirement for GWAS in previous studies (Meuwissen et al., 2001; Mckay et al., 2007). Considering a genome length of 2.64 Gb in L. vannamei(Yu et al., 2015), ∼150 K fully informative markers would be needed to saturate the requirement of GWAS at an average resolution of 18 kb.
Although only ∼23 K markers were used for GWAS in this study, a large number of markers significantly associated with body weight were identified (P < 0.01). This result may confirm the previous speculation that the shrimp growth is highly polygenic, and regulated by complex regulatory networks of many interacting genes (Moss and Moss, 2009). The current identified LvSRC gene may be one of those interacting genes and play an important role in the regulation of shrimp growth.
Scavenger receptors (SRs) comprise a large family of structurally diverse transmembrane cell surface glycoproteins and nine heterogeneous subclasses (A-I) were classified in accordance with their multidomain structures (Canton et al., 2013). As one member of SRs, SRC has only been identified in a few invertebrates, including Drosophila melanogaster, Aedes aegypti, and Marsupenaeus japonicus. Especially, previous studies only reveal the function of SRC in immunological process (Rämet et al., 2001; Yang et al., 2016, 2017), and whether SRC participates in growth regulation remains largely unknown. Indeed, although SRs family encompasses a wide range of molecules with little structural homology (Canton et al., 2013), almost all of them have been characterized in vertebrates by the common feature to bind modified low density lipoproteins (LDLs), such as oxidized LDL (OxLDL) and acetylated LDL (AcLDL). Therefore, SRs can play a central role in lipid metabolism. The similar function of SRs was also revealed in invertebrates. For example, in Macrobrachium nipponense, the expression of gene encoding the class B SR can be regulated by dietary lipid sources including soybean and linseed oils (Ding et al., 2016). Therefore, it’s interesting to note that SRC may be related to the body weight of shrimp by participating in lipid metabolism.
The significant SNPs in the coding region of LvSRC, especially the marker SRC_24, could be promising candidates for marker assisted breeding of growth traits in L. vannamei. Nevertheless, it is still uncertain that which mutations within the LvSRC gene are the causative loci associated with growth of shrimp. Therefore, gene editing technology will be a powerful tool to determine the causative locus in the future. Besides, it is important to note that the phenotypic variation of complex traits can be affected by the mutations in the non-coding region of genes, including untranslated region (Si et al., 2016) or promoter region (Wang et al., 2016). Therefore, it should be further investigated that whether the causative loci located in the non-coding region of LvSRC.
In addition, it is important to note that a number of significant markers from GWAS failed to be annotated. There may be two reasons for this result. Firstly, parts of the reference genome was not fully assembled which result in the difficulty of gene annotation. Secondly, the region of candidate genes was determined based on the average LD decay rate in this study; however, the LD decay of different genomic regions might be quite different (Lu et al., 2012; Kawakami et al., 2017). Therefore, in the future, more growth-related genes would be revealed with the increase of genome information and the detail survey of LD decay of different genome regions.
Conclusion
In this study, the LD decay of the studied population is rapid with an average r2 (0.2) values at 18 kb, which suggested that it is feasible to fine map the growth-related genes by using this population. By using GWAS integrated with candidate gene association study, the LvSRC was proved to be associated with growth traits. This result not only provides molecular markers that may contribute to accelerate the genetic improvement for penaeid shrimp, but also provides new insights to help understand regulatory mechanism of shrimp growth. Further studies are needed to fine mapping the causative mutation in the LvSRC and investigate its regulatory mechanism on shrimp growth.
Author Contributions
QW and YY conducted the experiment and data processing. JX and FL conceived and supervised the project. QZ, XZ, and JY contributed to prepare the genomic DNA for SNP genotyping. HH prepared and cultured the experimental animals. QW, YY, and FL wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by National Key R&D Program of China (2018YFD0901301), National Natural Science Foundation of China (31830100), and China Agriculture Research System-48 (CARS-48).
Conflict of Interest Statement
HH was employed by company Hainan Grand Suntop Ocean Breeding Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00520/full#supplementary-material
References
Andriantahina, F., Liu, X., Huang, H., and Xiang, J. (2012). Response to selection, heritability and genetic correlations between body weight and body size in Pacific white shrimp. Litopenaeus vannamei. Chin. J. Oceanol. Limnol. 30, 200–205. doi: 10.1007/s00343-012-1066-2
Andriantahina, F., Liu, X., Huang, H., and Xiang, J. (2013). Selection for growth performance of tank-reared Pacific white shrimp. Litopenaeus vannamei. Chin. J. Oceanol. Limnol. 31, 534–541. doi: 10.1007/s00343-013-2220-1
Argue, B. J., Arce, S. M., Lotz, J. M., and Moss, S. M. (2002). Selective breeding of Pacific white shrimp (Litopenaeus vannamei) for growth and resistance to taura syndrome virus. Aquaculture 204, 447–460. doi: 10.1016/s0044-8486(01)00830-4
Aulchenko, Y. S., Ripke, S., Isaacs, A., and van Duijn, C. M. (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296. doi: 10.1093/bioinformatics/btm108
Canton, J., Neculai, D., and Grinstein, S. (2013). Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634. doi: 10.1038/nri3515
Correa, K., Lhorente, J. P., Bassini, L., López, M. E., Di Genova, A., Maass, A., et al. (2017). Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50K SNP genotyping array. Aquaculture 472, 61–65. doi: 10.1016/j.aquaculture.2016.04.008
Ding, Z., Luo, N., Kong, Y., Li, J., Zhang, Y., Cao, F., et al. (2016). Scavenger receptor class B, Type I, a CD36 related protein in macrobrachium nipponense: characterization, RNA interference, and expression analysis with different dietary lipid sources. Int. J. Genomics 2016:6325927. doi: 10.1155/2016/6325927
Fu, X., Dou, J., Mao, J., Su, H., Jiao, W., Zhang, L., et al. (2013). RADtyping: an integrated package for accurate de novo codominant and dominant RAD genotyping in mapping populations. PLoS One 8:e79960. doi: 10.1371/journal.pone.0079960
Geng, X., Sha, J., Liu, S., Bao, L., Zhang, J., Wang, R., et al. (2015). A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance. BMC Genomics 16:196. doi: 10.1186/s12864-015-1409-4
Gjedrem, T., and Baranski, M. (2010). Selective Breeding In Aquaculture: an Introduction. Berlin: Springer Science & Business Media.
Gonzalez-Pena, D., Gao, G., Baranski, M., Moen, T., Cleveland, B. M., Kenney, P. B., et al. (2016). Genome-wide association study for identifying loci that affect fillet yield. carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front. Genet. 7:203. doi: 10.3389/fgene.2016.00203
Gutierrez, A. P., Yáñez, J. M., Fukui, S., Swift, B., and Davidson, W. S. (2015). Genome-wide association study (gwas) for growth rate and age at sexual maturation in atlantic salmon (Salmo salar). PLoS One 10:e0119730. doi: 10.1371/journal.pone.0119730
Hill, W. G. (1974). Estimation of linkage disequilibrium in randomly mating populations. Heredity 33:229. doi: 10.1038/hdy.1974.89
Ho, C., and Lin, Z. (2003). Analysis and application of grey relation and ANOVA in chemical–mechanical polishing process parameters. Int. J. Adv. Manuf. Technol. 21, 10–14. doi: 10.1007/s001700300001
Huang, X., and Han, B. (2014). Natural variations and genome-wide association studies in crop plants. Ann. Rev. Plant Biol. 65, 531–551. doi: 10.1146/annurev-arplant-050213-035715
Huang, Y.-C., Yin, Z.-X., Ai, H.-S., Huang, X.-D., Li, S.-D., Weng, S.-P., et al. (2011). Characterization of WSSV resistance in selected families of Litopenaeus vannamei. Aquaculture 311, 54–60. doi: 10.1016/j.aquaculture.2010.11.032
Jin, Y., Zhou, T., Geng, X., Liu, S., Chen, A., Yao, J., et al. (2016). A genome-wide association study of heat stress-associated SNPs in catfish. Animal Genet. 48:233. doi: 10.1111/age.12482
Jung, H., Lyons, R. E., Hurwood, D. A., and Mather, P. B. (2013). Genes and growth performance in crustacean species: a review of relevant genomic studies in crustaceans and other taxa. Rev. Aquac. 5, 77–110. doi: 10.1111/raq.12005
Kawakami, T., Mugal, C. F., Suh, A., Nater, A., Burri, R., Smeds, L., et al. (2017). Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine-scale recombination rate variation in birds. Mol. Ecol. 26, 4158–4172. doi: 10.1111/mec.14197
Li, E., Xu, C., Wang, X., Wang, S., Zhao, Q., Zhang, M., et al. (2018). Gut microbiota and its modulation for healthy farming of pacific white shrimp Litopenaeus vannamei. Rev. Fisher. Sci. Aquac. 26, 1–19.
Li, X., Xin, J., Feng, T., and Liu, X. (2011). Single nucleotide polymorphism discovery of molt Inhibiting hormone gene 3 exons and its association with growth traits in white Shrimp (Litopenaeus vannamei). J. Anim. Vet. Adv. 10, 2856–2858.
Lu, D., Sargolzaei, M., Kelly, M., Li, C., Vander, G. V., Wang, Z., et al. (2012). Linkage disequilibrium in angus. charolais, and crossbred beef cattle. Front. Genet. 3:152. doi: 10.3389/fgene.2012.00152
Marroni, F., Pinosio, S., Zaina, G., Fogolari, F., Felice, N., Cattonaro, F., et al. (2011). Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet. Genomes 7, 1011–1023. doi: 10.1007/s11295-011-0391-5
Mccarthy, M. I., Abecasis, G. R., Cardon, L. R., Goldstein, D. B., Little, J., Ioannidis, J. P., et al. (2008). Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369. doi: 10.1038/nrg2344
Mckay, S. D., Schnabel, R. D., Murdoch, B. M., Matukumalli, L. K., Aerts, J., Coppieters, W., et al. (2007). Whole genome linkage disequilibrium maps in cattle. BMC Genet. 8:74. doi: 10.1186/1471-2156-8-74
Meuwissen, T. H., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genet. 157, 1819–1829.
Moss, S. M., and Moss, D. R. (2009). “Selective breeding of penaeid shrimp,” in Shellfish Safety and Quality, eds S. Sandra and R. Gary (Cambridge: Woodhead Publishing), 425–452. doi: 10.1533/9781845695576.3.425
Nolasco-Alzaga, H. R., Perez-Enriquez, R., Enez, F., Bestin, A., Palacios-Mechetnov, E., Haffray, P., et al. (2017). Quantitative genetic parameters of growth and fatty acid content in the hemolymph of the Whiteleg shrimp Litopenaeus vannamei. Aquaculture 482, 17–23. doi: 10.1016/j.aquaculture.2017.09.015
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., and Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. https://www.r-project.org/
Rämet, M., Pearson, A., Manfruelli, P., Li, X., Koziel, H., Göbel, V., et al. (2001). Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity 15:1027. doi: 10.1016/s1074-7613(01)00249-7
Remington, D. L., Thornsberry, J. M., Matsuoka, Y., Wilson, L. M., Whitt, S. R., Doebley, J., et al. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. U.S.A. 98, 11479–11484. doi: 10.1073/pnas.201394398
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sodeland, M., Gaarder, M., Moen, T., Thomassen, M., Kjøglum, S., Kent, M., et al. (2013). Genome-wide association testing reveals quantitative trait loci for fillet texture and fat content in Atlantic salmon. Aquaculture 408, 169–174. doi: 10.1016/j.aquaculture.2013.05.029
Sui, J., Luan, S., Luo, K., Meng, X., Lu, X., Cao, B., et al. (2016). Genetic parameters and response to selection for harvest body weight of pacific white shrimp. Litopenaeus vannamei. Aquac. Res. 47, 2795–2803. doi: 10.1111/are.12729
Vallejo, R. L., Palti, Y., Liu, S., Marancik, D. P., and Wiens, G. D. (2014). Validation of linked QTL for bacterial cold water disease resistance and spleen size on rainbow trout chromosome Omy19. Aquaculture 432, 139–143. doi: 10.1016/j.aquaculture.2014.05.003
Wang, Q., Yu, Y., Yuan, J., Zhang, X., Huang, H., Li, F., et al. (2017). Effects of marker density and population structure on the genomic prediction accuracy for growth trait in Pacific white shrimp Litopenaeus vannamei. BMC Genet. 18:45. doi: 10.1186/s12863-017-0507-5
Wang, S., Meyer, E., Mckay, J. K., and Matz, M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9:808. doi: 10.1038/nmeth.2023
Wang, X., Wang, H., Liu, S., Ferjani, A., Li, J., Yan, J., et al. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48:1233. doi: 10.1038/ng.3636
Yang, M., Shi, X., Yang, H., Sun, J., Xu, L., Wang, X., et al. (2016). Scavenger receptor C mediates phagocytosis of white spot syndrome virus and restricts virus proliferation in shrimp. PLoS Pathog. 12:e1006127. doi: 10.1371/journal.ppat.1006127
Yang, M., Yang, H., Li, J., Sun, J., Bi, W., Niu, G., et al. (2017). Scavenger receptor C promotes bacterial clearance in kuruma shrimp Marsupenaeus japonicus by enhancing hemocyte phagocytosis and AMP expression. Fish Shellfish Immunol. 67:254. doi: 10.1016/j.fsi.2017.06.003
Yu, Y., Zhang, X., Yuan, J., Li, F., Chen, X., Zhao, Y., et al. (2015). Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific White Shrimp Litopenaeus vannamei. Sci. Rep. 5:15612. doi: 10.1038/srep15612
Yu, Y., Zhang, X., Yuan, J., Wang, Q., Li, S., Huang, H., et al. (2017). identification of sex-determining loci in pacific white shrimp Litopeneaus vannamei using linkage and association analysis. Mar. Biotechnol. 19, 277–286. doi: 10.1007/s10126-017-9749-5
Zhang, H., Wang, Z., Wang, S., and Li, H. (2013). Progress of genome wide association study in domestic animals. J. Anim. Sci. Biotechnol. 4, 1–1.
Keywords: penaeid shrimp, growth traits, GWAS, candidate gene, class C scavenger receptor
Citation: Wang Q, Yu Y, Zhang Q, Zhang X, Yuan J, Huang H, Xiang J and Li F (2019) A Novel Candidate Gene Associated With Body Weight in the Pacific White Shrimp Litopenaeus vannamei. Front. Genet. 10:520. doi: 10.3389/fgene.2019.00520
Received: 04 November 2018; Accepted: 13 May 2019;
Published: 31 May 2019.
Edited by:
Gen Hua Yue, Temasek Life Sciences Laboratory, SingaporeReviewed by:
Zhe Zhang, South China Agricultural University, ChinaQi Cun Zhou, Ningbo University, China
Copyright © 2019 Wang, Yu, Zhang, Zhang, Yuan, Huang, Xiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuhua Li, ZmhsaUBxZGlvLmFjLmNu
†These authors have contributed equally to this work
 Quanchao Wang
Quanchao Wang Yang Yu
Yang Yu Qian Zhang1,3
Qian Zhang1,3 Xiaojun Zhang
Xiaojun Zhang Jianbo Yuan
Jianbo Yuan Jianhai Xiang
Jianhai Xiang Fuhua Li
Fuhua Li