- International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India
Peanut allergy is one of the serious health concern and affects more than 1% of the world’s population mainly in Americas, Australia, and Europe. Peanut allergy is sometimes life-threatening and adversely affect the life quality of allergic individuals and their families. Consumption of hypoallergen peanuts is the best solution, however, not much effort has been made in this direction for identifying or developing hypoallergen peanut varieties. A highly diverse peanut germplasm panel was phenotyped using a recently developed monoclonal antibody-based ELISA protocol to quantify five major allergens. Results revealed a wide phenotypic variation for all the five allergens studied i.e., Ara h 1 (4–36,833 µg/g), Ara h 2 (41–77,041 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (829–103,892 µg/g), and Ara h 8 (0.01–70.12 µg/g). The hypoallergen peanut genotypes with low levels of allergen proteins for Ara h 1 (4 µg/g), Ara h 2 (41 µg/g), Ara h 3 (22 µg/g), Ara h 6 (829 µg/g), and Ara h 8 (0.01 µg/g) have paved the way for their use in breeding and genomics studies. In addition, these hypoallergen peanut genotypes are available for use in cultivation and industry, thus opened up new vistas for fighting against peanut allergy problem across the world.
Introduction
Food allergy causes severe health issues throughout the globe and the incidences are increasingly recorded across the globe. Even though, approximately 5% of young kids and 4% of adult in western countries are affected by food allergens (Sicherer et al., 2010), the problem has now become more common in developing countries (Liew et al., 2013; Leung et al., 2018). About 40% of the food allergies occur due to the consumption of plants and plant-derived products. Peanut is identified as one of the major sources of food allergy in addition to milk, egg, dry fruits (almonds, cashews, hazelnuts, pistachios, pecans, and walnuts), fish, shellfish, soy, and wheat, with ∼90% cumulative contribution among food allergies in human (Hefle et al., 1996). A large number of population across the world are affected by peanut allergy and several reports are coming more frequently. For instance, the 1% population of Canadian children are allergic to peanuts (Ben-Shoshan et al., 2009) while the prevalence of peanut-based allergy in France and Denmark ranged between 0.3-0.75% and 0.2-0.4%, respectively (Morisset et al., 2005; Osterballe et al., 2005). About 3% of Australians are allergic to peanuts and peanut-based products (Sicherer and Sampson, 2007; Sicherer and Sampson, 2014). It is a big problem in the United Kingdom (UK) as well, and the prevalence of sensitization increased from 1.3 to 3.2% in 3 years old kids (Grundy et al., 2002). Importantly, the prevalence of peanut-based allergy in United States of America (USA) has been increased from 0.4 to 1.4% between 1997 to 2008 (Sicherer et al., 2010). Similarly, peanut allergy has also been reported in the Asian countries such as Singapore and Philippines where 0.47 and 0.43% school children, respectively, were found allergic to peanuts (Shek et al., 2010). Although information from China is not available, however, the situation in China may be similar to Singapore as 76.8% of Singapore residents are Chinese in origin (http://www.singstat.gov.sg/). Although there may be several cases of food allergy in India (Mahesh et al., 2016), however, not much information is available from India on peanut allergy. Keeping in mind the interdependence and trade among countries for producing raw material, processing, and consumption of peanut, such health-concerning features of the crop reduces its importance in international trade and commerce (Pandey et al., 2012; Pandey and Varshney 2018; Varshney et al., 2018;Varshney et al., 2019). Therefore, the countries producing the peanuts and peanut based product with the most safe, nutritious, and healthy features will get a competitive advantage over other producing countries.
All the major food allergies, including peanut, may induce anaphylaxis leading to life-threatening reactions (Dodo et al., 2005; Sicherer and Sampson, 2010) and it is almost impossible to avoid accidental ingestion of peanut-based products (Berger and Smith, 1998; Kagan et al., 2003). Remarkably, food-based allergies cause around 150–200 deaths per year (http://www.startribune.com/peanut-allergy-kills-22-year-old-twin-cities-man/366152021/), largely due to the consumption of peanuts (50–62%) and tree nuts (15–30%) in USA (Lanser et al., 2015). Proteins are the major cause of food allergy, and these proteins are usually highly resistant to heat and proteolysis (Cabanillas et al., 2012). Peanut is the largest source of the immunoglobulin E (IgE)-mediated food allergies and there is no effective treatment due to which the allergic person is forced to avoid consuming peanut or peanut-based products (Wen et al., 2007). However, the peanut being a common food ingredient in many food preparations, it is very challenging for the allergic person to know the composition of these preparations to avoid consumption (Maleki et al., 2000). The threshold of allergen levels differ among the allergic population and even a minute dose of 100 µg of Ara h 1 can trigger an allergic reaction (Warner, 1999). The diagnosis of peanut allergy can be done using different methods such as double-blind, placebo-controlled food challenge (DBPCFC), the basophil activation test, the specific skin prick test (SPT), and the measurement of specific IgE (Hamilton et al., 2010; Lieberman and Sicherer, 2011; Nicolaou et al., 2011).
Of the 32 different types of proteins present in peanut seeds (Pele, 2010), 18 of these proteins show the allergic property (Iqbal et al., 2016). Further, out of 18 peanut allergen proteins mainly Ara h 1, Ara h 2, Ara h 3, and Ara h 6 are considered as major allergens due to their life-threatening reactions recognized by the IgE leading to anaphylaxis (Krause et al., 2010). The remaining allergen proteins are considered as minor allergens as they don’t cause life-threatening allergic reactions (anaphylaxis). Nevertheless, if a person is already sensitive to Bet v 1 allergen caused due to birch pollen, then one of these minor peanut allergens, Ara h 8, shows cross-reactivity with IgE antibodies causing oral allergy syndrome (OAS) (Mittag et al., 2004; Riecken et al., 2008; Kondo and Urisu, 2009). Allergic protein belongs to different protein families namely cupin (vicilin-type, 7S globulin, legumin-type, 11S globulin, glycinin), conglutin (2S albumin), profilin, nonspecific lipid-transfer protein 1, pathogenesis-related protein (PR-10) 14 kDa, oleosin (16 kDa), and seed storage proteins particularly Ara h 1, Ara h 2, Ara h 3, and Ara h 6 (Pele, 2010). Many studies have shown that the most abundant peanut-based allergens (Ara h 1 and Ara h 3 but, Ara h 2 and Ara h 6) bind strongly with peanut allergic IgE and release basophils mediators, which were confirmed in vitro (de Jong et al., 1998; Koppelman et al., 2005; Palmer et al., 2005; Porterfield et al., 2009) and in vivo (Koppelman et al., 2003; Koppelman et al., 2005; Peeters et al., 2007) with regards to food allergy (Porterfield et al., 2009). Although all the five peanut allergens (Ara h 1 and Ara h 3 but, Ara h 2 and Ara h 6) show IgE reactivity to these peanut allergens, however, the Ara h 2 and Ara h 6 allergens are more commonly recognized in children (Flinterman et al., 2007).
Possible solutions to peanut allergy include the development of vaccine or development of allergen-free peanut varieties. Much research has been focussed on diagnosis and cure to minimize the impact of allergens in the human population, however, reducing allergen proteins in peanut varieties and their products can be the best solution. Unfortunately, insufficient scientific information on a total number of allergen genes in the peanut genome and level of phenotypic variability in existing peanut germplasm hinders further research in this area. Therefore, the main objective of this research was to identify hypoallergen peanut lines by screening a large number of diverse germplasm in the peanut reference set (Upadhyaya, 2009). The hypoallergen peanut varieties that have been identified will promote their commercialization and use in the peanut-based industry. These lines and information generated out of this work can be of great importance in efforts toward fighting peanut allergy and ensuring food safety across the world.
Materials and Methods
Plant Materials
The peanut “reference set” consisting of 300 diverse accessions representing 51 countries (Upadhyaya, 2009) (Supplementary Table 1 and Figure 1A) were selected from the composite collection. The reference set included the 184 accessions of the peanut mini core collection (Upadhyaya et al., 2002) that represented diversity of the core collection (1,704 accessions) which in turn represents the diversity of entire International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) peanut genebank collection (14,310 accessions) (Upadhyaya et al., 2003). This set comprised of 264 cultivated species (Arachis hypogaea) and 36 wild species and has fair representation for its two subspecies namely fastigiata (154) and hypogaea (95). The subspecies fastigiata was further classified into four botanical varieties namely Fastigiata (70 accessions), Peruviana (5 accessions), Vulgaris (78 accessions), and Aequitoriana (1 accession). Similarly, the other subspecies hypogaea was classified into two botanical varieties namely Hirsuta (2 accessions) and Hypogaea (93 accessions) (Holbrook and Stalker 2003). These cultivated genotypes can also be classified into four agronomic types based on their growth habit namely Spanish bunch (73), Valencia bunch (70), Virginia bunch (51), and Virginia runner (33). The seeds for cultivated genotypes (264) were collected from two seasons (Rainy 2016 and Post-rainy 2016–17) for estimating allergen content. Since many of the wild accessions have annual growth period, seeds from two different lots were taken for allergen estimation.
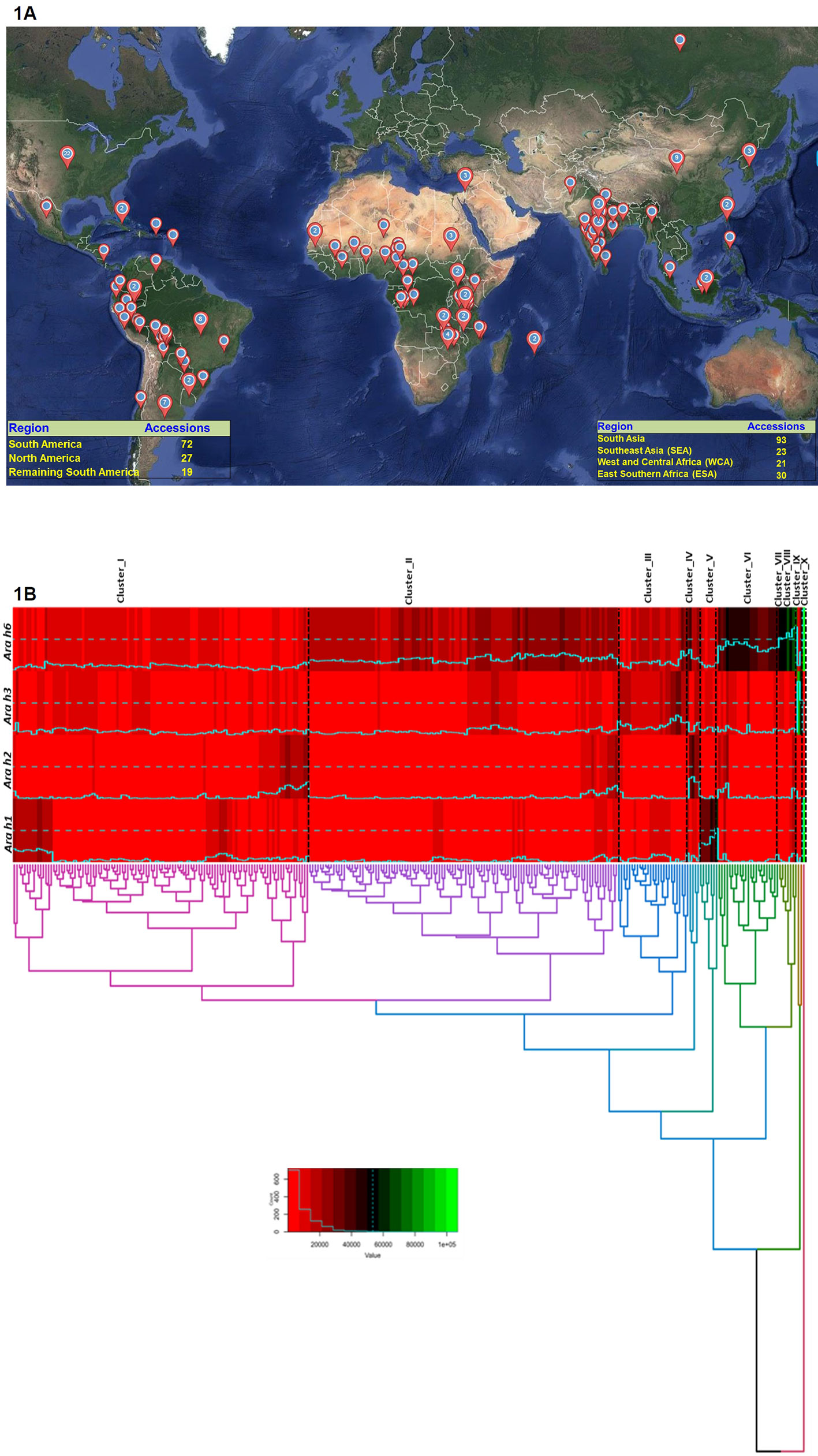
Figure 1 Graphical representation showing global coverage of peanut “reference set” across 51 countries. (1A) The world map was constructed by using Maptive software (http://fortress.maptive.com). Out of 300 accessions of the ICRISAT peanut reference set, 93 belongs to South Asia, 23 from Southeast Asia, 21 from West and Central Africa 30 from East and Southern Africa, 72 from South America, 27 from North America, and 19 from remaining South America. (1B) Hierarchical clustering on the basis of phenotyping data grouped 300 accessions in 10 cluster. Cluster 1 having 112 accessions, cluster 2 having 117 accessions while cluster 3, and 6 having 27 and 23 accessions respectively. Cluster 7, 8, 9, and 10 showing a high amount of Ara h 6 in their germplasm lines. In cluster 1 all the accessions having a lower amount of allergen content.
Sample Preparation and Protein Isolation
Sample preparation and protein isolation were performed following the protocol mentioned in (Pandey et al., 2019). In brief, 2 g of seeds were grinded to make a fine powder and then dissolved in 40 ml of PBS-T (0.05% Tween in phosphate buffered saline, pH 7.4) containing 1 M NaCl in 50 ml falcon tubes (Sarstedt No: 55.476). After 2 h of gentle stirring at room temperature on the rocking platform, the aqueous phase was collected by centrifugation at 2,500 rpm at 4°C for 20 min. The aqueous phase was subsequently centrifuged to remove residual traces and insoluble particles at 3,500 rpm for 10 min at room temperature. Until use, extracts of proteins were stored at −20°C.
Allergens Estimation Using Enzyme-Linked Immunosorbent Assay
Sandwich format ELISA was used in the study. The peanut allergen proteins were first sandwiched between two antibodies, and then streptavidin-peroxidase was captured. Each peanut sample contains a different quantity of allergen proteins which makes very difficult to estimate the accurate amount of allergen proteins present in seed samples. Dilution is a vital step for ELISA experiment which in turn determines the values of detection range for antibody and target antigen concentrations.
The estimation of peanut allergen through sandwich ELISA were performed according to the recently published protocol (Pandey et al., 2019). Each allergen protein was estimated at different dilution factors (DF). We used a number of dilutions in the peanut samples to detect the specific allergic protein in seeds. The Ara h 1 was detected on three serial doubling dilutions, 1:1,000, 1:2,000, and 1: 4,000 while Ara h 2 and Ara h 3 detected on same dilution 1: 5,000, 1:10,000, and 1: 20,000. In peanut seeds, the Ara h 6 was detected in the high range (1: 40,000, 1: 80,000, and 1:160,000) DF while Ara h 8 detected in a low range of dilution, i.e., 1:10, 1:20, and 1:40.
Cluster and Data Analysis on the Basis of Allergen Content
Statistical analysis was performed to identify the wide variation of peanut allergens among samples using SigmaPlot (http://www.sigmaplot.co.uk/products/sigmaplot/sigmaplot-details.php). Hierarchical clustering was done using average allergens content of five major allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8) on the basis of similarity matrix generated using HCA (hierarchical cluster analysis). Dendextend statistical package (Galili, 2015) was used for clustering the genotypes on the basis of similarity of average allergen content. This software provides a set of functions for cluster analysis and construction of dendo-gram. The heat map was generated using R package gplots (Warnes et al., 2016) for allergen content of 300 samples.
Results
Phenotypic Variation for Allergens Between Cultivated and Wild Gene Pool
Phenotyping of 300 diverse accessions (264 cultivated and 36 wild accessions) showed wide phenotypic variations for all the five allergens. Among cultivated accessions, the phenotypic variation was very high for all five major allergens namely Ara h 1 (4–36,833 µg/g), Ara h 2 (52–77,042 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (829–103,892 µg/g), and Ara h 8 (0.01–70 µg/g) as compared to the wild relatives i.e., Ara h 1 (28–1,293 µg/g), Ara h 2 (516–33,055 µg/g), Ara h 3 (1,185–20,474 µg/g), Ara h 6 (4,381–56,858 µg/g), and Ara h 8 (0.3–11 µg/g) (Table 1). Except Ara h 3 (6,857 µg/g), the average content of Ara h 1 (404 µg/g), Ara h 2 (606 µg/g), Ara h 6 (13,420 µg/g), and Ara h 8 (2 µg/g) was found lower in wild accessions compared to cultivated genotypes (Table 1 and Figure 2A).
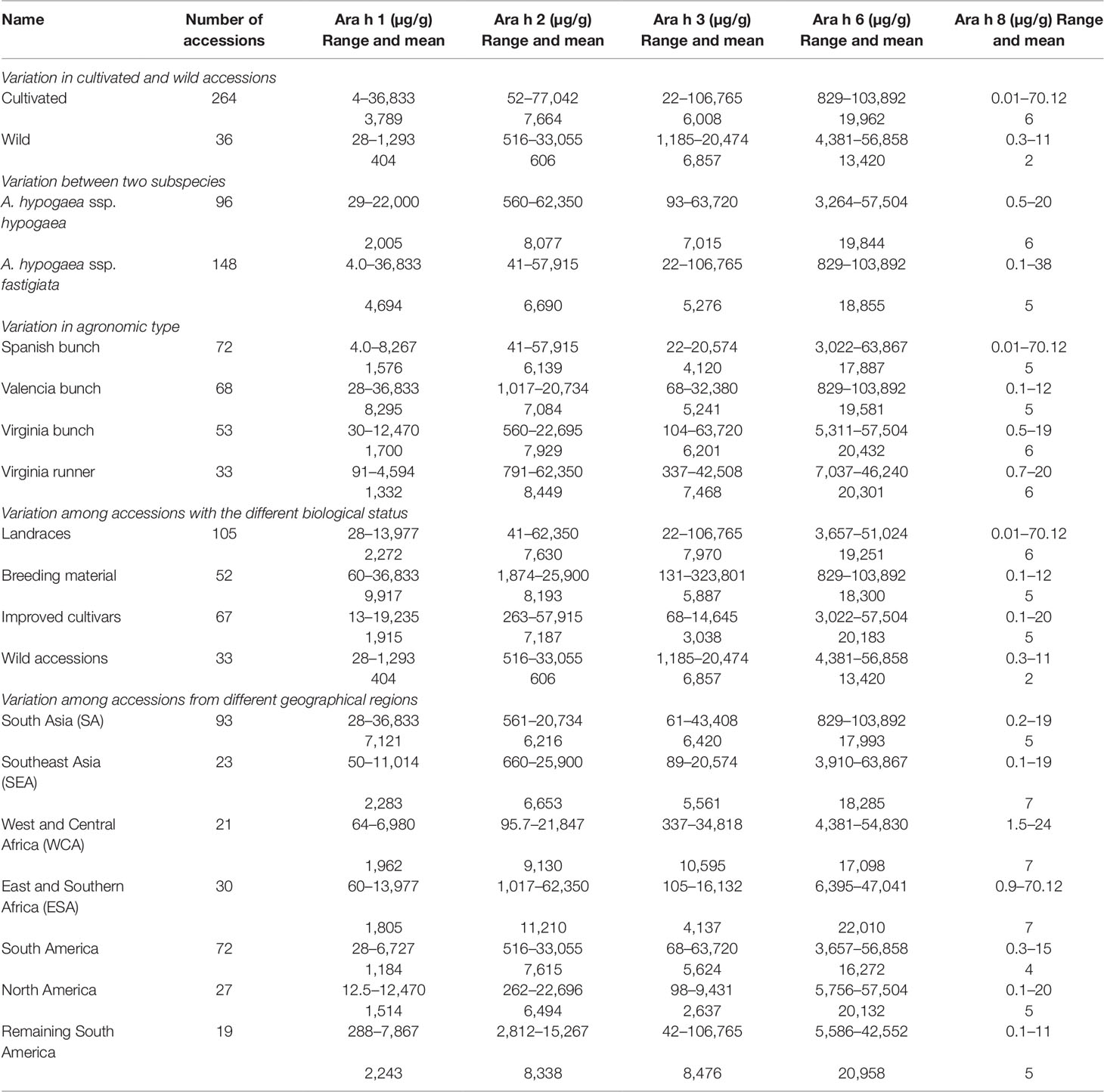
Table 1 Range and mean value for five major peanut allergens in peanut germplasm lines of the reference set in the rainy 2016 and 2016–17 postrainy seasons, International Crops Research Institute for the Semi-Arid Tropics, Patancheru, India.
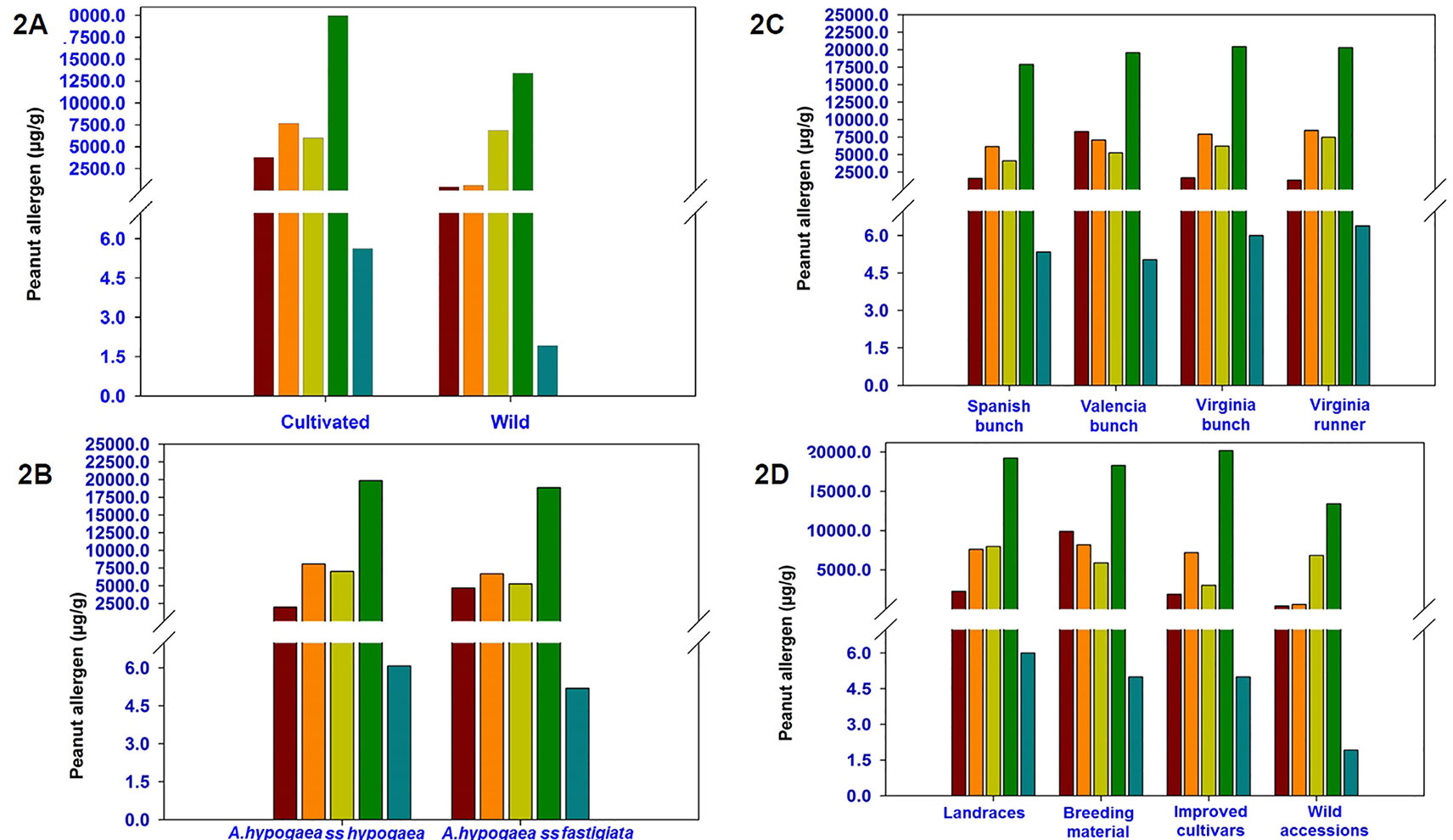
Figure 2 Phenotypic variation for five major peanut allergens (2A) between cultivated and wild germplasm accessions (2B) between subspecies hypogaea and fastigiata (2C) among different agronomic types (2D) and among the biological types of peanut reference set collection.
Phenotypic Variation for Allergens Between Subspecies hypogaea and fastigiata
Phenotyping result showed a wide variation for all the five allergens between two subspecies of cultivated peanut i.e., A. hypogaea ssp. hypogaea (96 accessions) and A. hypogaea ssp. fastigiata (148 accessions). In ssp. hypogaea, the allergen content ranged from 29 to 22,000 µg/g for Ara h 1, 560 to 62,350 µg/g for Ara h 2, 93 to 63,720 µg/g for Ara h 3, 3,264 to 57,504 µg/g for Ara h 6, and 0.5 to 20 µg/g for Ara h 8 whereas in ssp. fastigiata, the allergen content varied from Ara h 1 (4–36,833 µg/g), Ara h 2 (41–57,915 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (829–103,892 µg/g), and Ara h 8 (0.3–11 µg/g) (Table 1). Excluding Ara h 1 (2,005 µg/g), the average amount of Ara h 2 (8,007 µg/g), Ara h 3 (7,015 µg/g), Ara h 6 (19,844 µg/g), and Ara h 8 (6 µg/g) was higher in ssp. hypogaea as compared to ssp. fastigiata i.e., Ara h 1 (4,694 µg/g), Ara h 2 (6,690 µg/g), Ara h 3 (5,276 µg/g), Ara h 6 (18,855 µg/g), and Ara h 8 (5 µg/g) (Table 1 and Figure 2B). Even the average amount of allergens was detected high in ssp. hypogaea, the hypoallergen lines i.e., ICG 1534 (4 µg/g) for Ara h 1, ICG 13491 (41 µg/g) for Ara h 2, ICG 6375 (22 µg/g) for Ara h 3, and ICGV 1328 (829 µg/g) for Ara h 6 were identified from ssp. fastigiata (Table 1). The above results were also supported by HCA. In HCA analysis, cluster 1 formed on the basis of the low quantity of allergens in their accessions. In cluster 1 the allergen content ranged from 12.5 to 27,375 µg/g for Ara h 1, 40.5 to 20,600 µg/g for Ara h 2, 55.43 to 23,306 µg/g for Aar h 3, 829 to 16,483 µg/g for Ara h 6, and 0.067 to 70 µg/g for Ara h 8 (Supplementary Table 2 and Figure 1B).
Phenotypic Variation for Allergens Among Different Peanut Agronomic Types
Phenotyping of accessions from different agronomic types namely Spanish bunch (72 accessions), Valencia bunch (68 accessions), Virginia bunch (53 accessions), and Virginia runner (33 accessions) indicated significant variation (Table 1 and Figure 2C). Although the average amount of Ara h 1 was lowest in Virginia runner (1,332 µg/g) followed by Spanish bunch (1,576 µg/g), Virginia bunch (1,700 µg/g), and Valencia bunch (8,295 µg/g), however, the most hypoallergen line, ICG 1534 (4 µg/g) belongs to Spanish bunch (Table 1 and Figure 2C). Likewise, Spanish bunch (17,887 µg/g) agronomic type had the lowest average for Ara h 6 as compared to other agronomic types but the most hypoallergen line was identified from Valencia bunch, ICGV 1328 (829 µg/g) (Table 1). Interestingly for Ara h 2 (6,139 µg/g), Ara h 3 (4,120 µg/g), and Ara h 8 (5 µg/g), the accessions with minimum average as well most hypoallergen line were detected in Spanish bunch agronomic type (Table 1).
Phenotypic Variation for Allergens Among Peanut Accessions of Cultivated Gene Pool With Different Biological Status
The reference set included 105 traditional cultivars/landraces, 52 breeding/research material, 67 advanced/improved cultivars, and 36 wild accessions. Except for Ara h 3 (6,857 µg/g), the average amount of Ara h 1 (404 µg/g), Ara h 2 (606 µg/g), Ara h 6 (13,420 µg/g), and Ara h 8 (2 µg/g) was lower in wild accessions as compared to all the biological status groups of cultivated gene pool (Table 1 and Figure 2D). Interestingly, most hypoallergen lines were detected in different biological status groups of cultivated gene pool. For example, Ara h 1 in advanced/improved cultivars (13–19,235 µg/g), Ara h 2 in traditional cultivars/landraces (41–62,350 µg/g), Ara h 6 in breeding/research material (829–103,892 µg/g), and Ara h 8 in traditional cultivars/landraces (0.01–70 µg/g). Similarly, the average amount of Ara h 3 allergens was found lowest in advanced/improved cultivars (3,038 µg/g) but the hypoallergen line was identified in traditional cultivars/landraces (41–62,350 µg/g) (Table 1).
Phenotypic Variation for Allergens Among Peanut Accessions Representing Different Geographical Regions
The phenotyped set of 300 accessions represent different geographical regions namely South Asia (SA, 93 accessions), Southeast Asia (SEA, 23 accessions), West and Central Africa (WCA, 21 accessions), East and Southern Africa (ESA, 30 accessions), South America (72 accessions), North America (27 accessions), and 19 accessions from remaining South America. The average amount for Ara h 1 was lowest in accession representing South America (1,184 µg/g) followed by North America (1,514 µg/g), ESA (1,805 µg/g), WCA (1,962 µg/g), remaining South America (2,243 µg/g), SEA (2,283 µg/g), and SA (7,121). Likewise, the average amount of Ara h 2, Ara h 3, Ara h 6, and Ara h 8 were identified in SA (6,216 µg/g), North America (2,637 µg/), WCA (17,098), and South America (4 µg/g), respectively (Table 1 and Figure 3A). The genotypes available in North America region especially in the USA had low amount of Ara h 1 (12.5–12,470 µg/g) as compared to genotypes grown in other parts of the world while the average content for Ara h 2 was low in WCA (41–21,847 µg/g) (Table 1 and Figure 3A). The genotypes from South America had low amount of Ara h 3 (68–63,720 µg/g). In general, the allergen content for Ara h 6 was high and low for Ara h 8 across geographical regions. The most hypoallergen line for Ara h 6 (829 µg/g) was identified from the SA region (Table 1 and Figure 3A). Cluster analysis also revealed that the North America region having low allergen lines for Ara h 1 (4–21,999 µg/g) and Ara h 2 (40.5–20,600 µg/g) while South America region having hypoallergen lines for Ara h 3 (53.43–23,306 µg/g) (Supplementary Table 2 and Figure 1B).
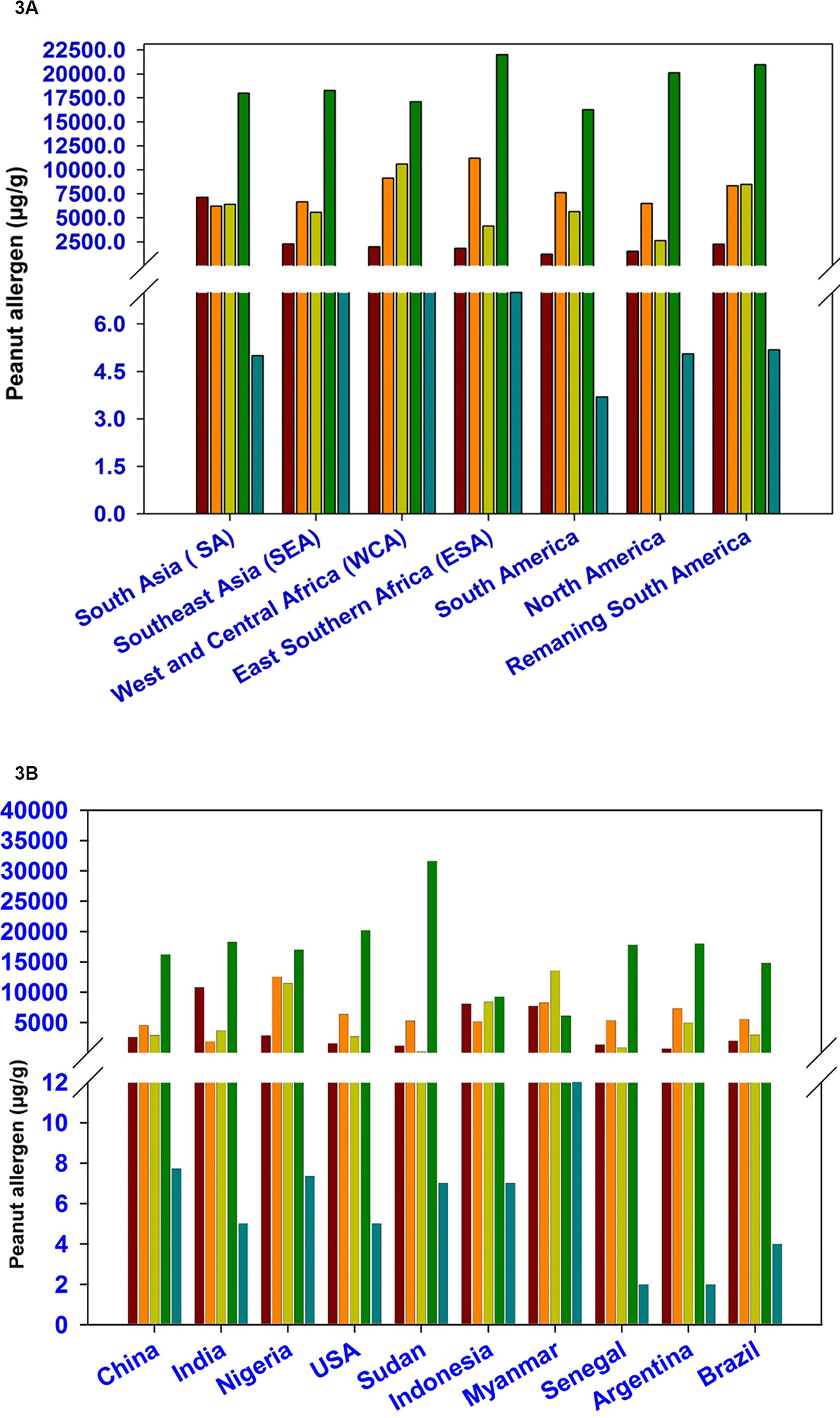
Figure 3 Phenotypic variation for five major peanut allergens among peanut germplasm lines representing various geographical regions (3A) and top 10 peanut producing countries (3B).
Phenotypic Variation for Allergens Among Peanut Accessions Representing Top 10 Peanut Producing Countries
Top 10 peanut producing countries include China, India, Nigeria, USA, Sudan, Indonesia, Myanmar, Senegal, Argentina, and Brazil which together contributed more than 81% of total global peanut production. Lowest average amount of Ara h 1 was found in Argentina (673 µg/g) followed by Sudan (1,174 µg/g), Senegal (1,383 µg/g), USA (1,554 µg/g), Brazil (1,968 µg/g), Nigeria (2,874 µg/g), Indonesia (4,037 µg/g), China (4,138 µg/g), Myanmar (7,714 µg/g), and India (Table 2; Supplementary Table 3). But the germplasm used in the USA had comparatively lower for Ara h 1 (13–5,023 µg/g) (Table 2 and Figure 3B), followed by India (28–36,833 µg/g) and China (51–63,910 µg/g). Likewise for Ara h 2, USA (263–22,969 µg/g) (Table 2 and Figure 3B) showed hypoallergen lines even the average content of Ara h 2 was lower in Indian germplasm line (1,876 µg/g). The hypoallergen lines for Ara h 3 (61–43,408 µg/g) and Ara h 6 (829–103,892 µg/g) were identified in Indian germplasm lines even though the average content of Ara h 3 (183 µg/g) and Ara h 6 (6,087 µg/g) was lower in Sudan and Myanmar germplasm lines. (Table 2, Supplementary Table 3 and Figure 3B).
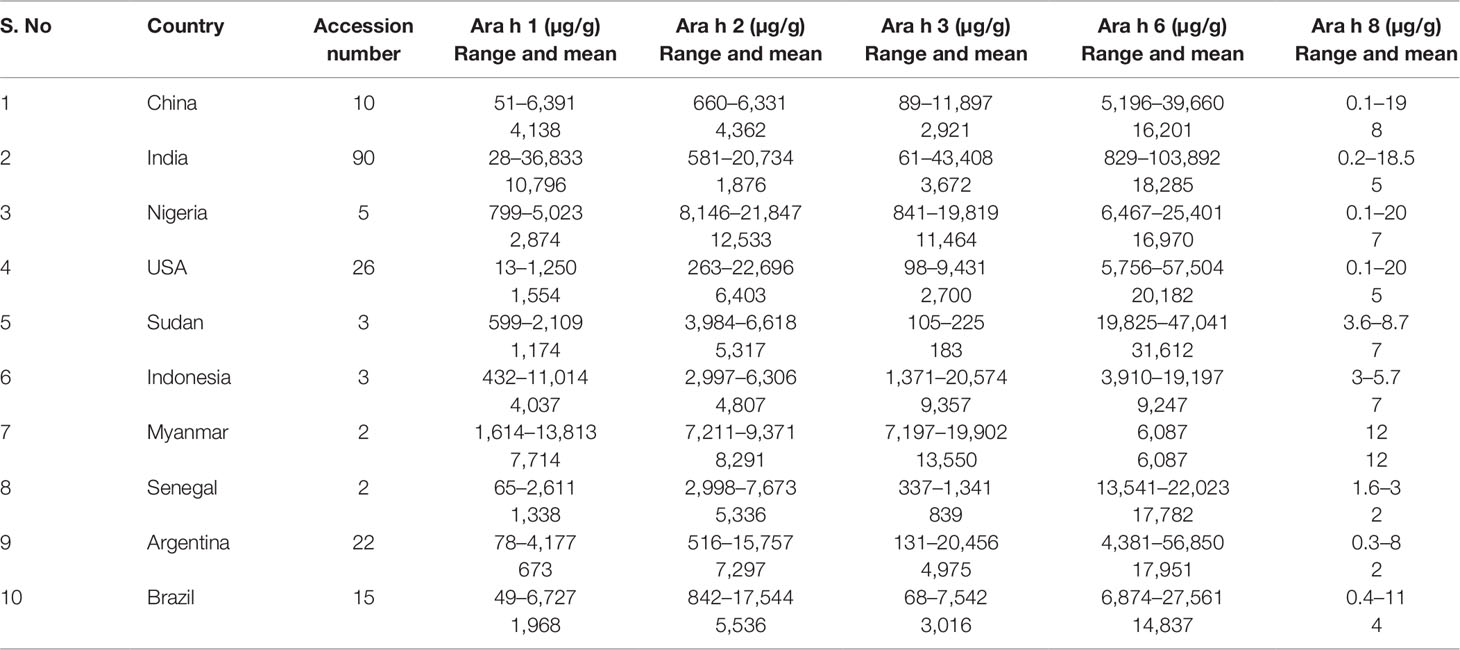
Table 2 Range and mean value for 5 major peanut allergens in peanut germplasm lines of the reference set representing top 10 peanut producing countries.
Hypoallergen Lines for Five Major Peanut Allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8)
A wide variation among the 300 germplasm lines was observed for the presence of five major allergens i.e., Ara h 1 (4–36,833 µg/g), Ara h 2 (41–77,041 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (829–103,892 µg/g), and Ara h 8 (0.01–70.12 µg/g). Some germplasm lines showed low allergen content for combinations of two to three allergens.
For Ara h 1, the phenotypic variation ranged from 4 µg/g (ICG 1534) to 36,833 µg/g (ICGV 02038) among cultivated accessions while it ranged from 28 µg/g (ICG 8124) to 1,293 µg/g (ICG 11555) among wild accessions (Table 1 and Figure 4A). The most hypoallergen lines for Ara h 1 included ICG 1534 (4 µg/g) followed by ICG 311 (12.5 µg/g), ICG 81 (17.6 µg/g), ICG 442 (22.8 µg/g), and ICG 115 (27.9 µg/g) (Table 3 and Figure 4A). All these hypoallergen lines belong to Spanish bunch types of A. hypogaea ssp. fastigiata except ICG 115 which belongs to Valencia bunch of the same subspecies (Table 3 and Figure 4A). The best low allergen lines can be used for conducting further research on genomics and breeding.
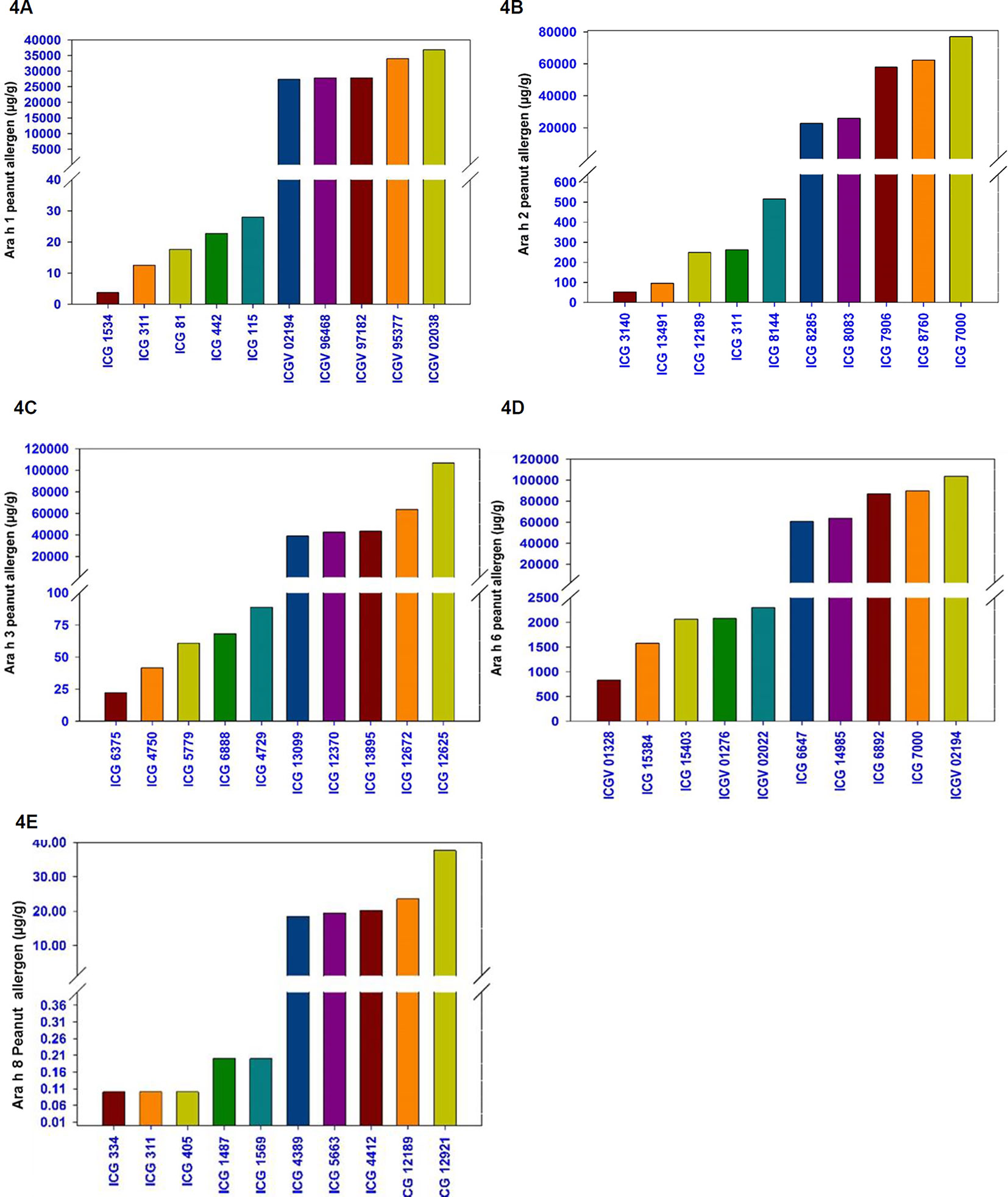
Figure 4 Selected germplasm lines with minimum and the maximum amount of allergen protein. (4A) Five peanut lines with highest and five lines with lowest allergen content for Ara h1 (4B) Ara h 2, (4C) Ara h 3, (4D) Ara h 6, and (4E) Ara h 8.
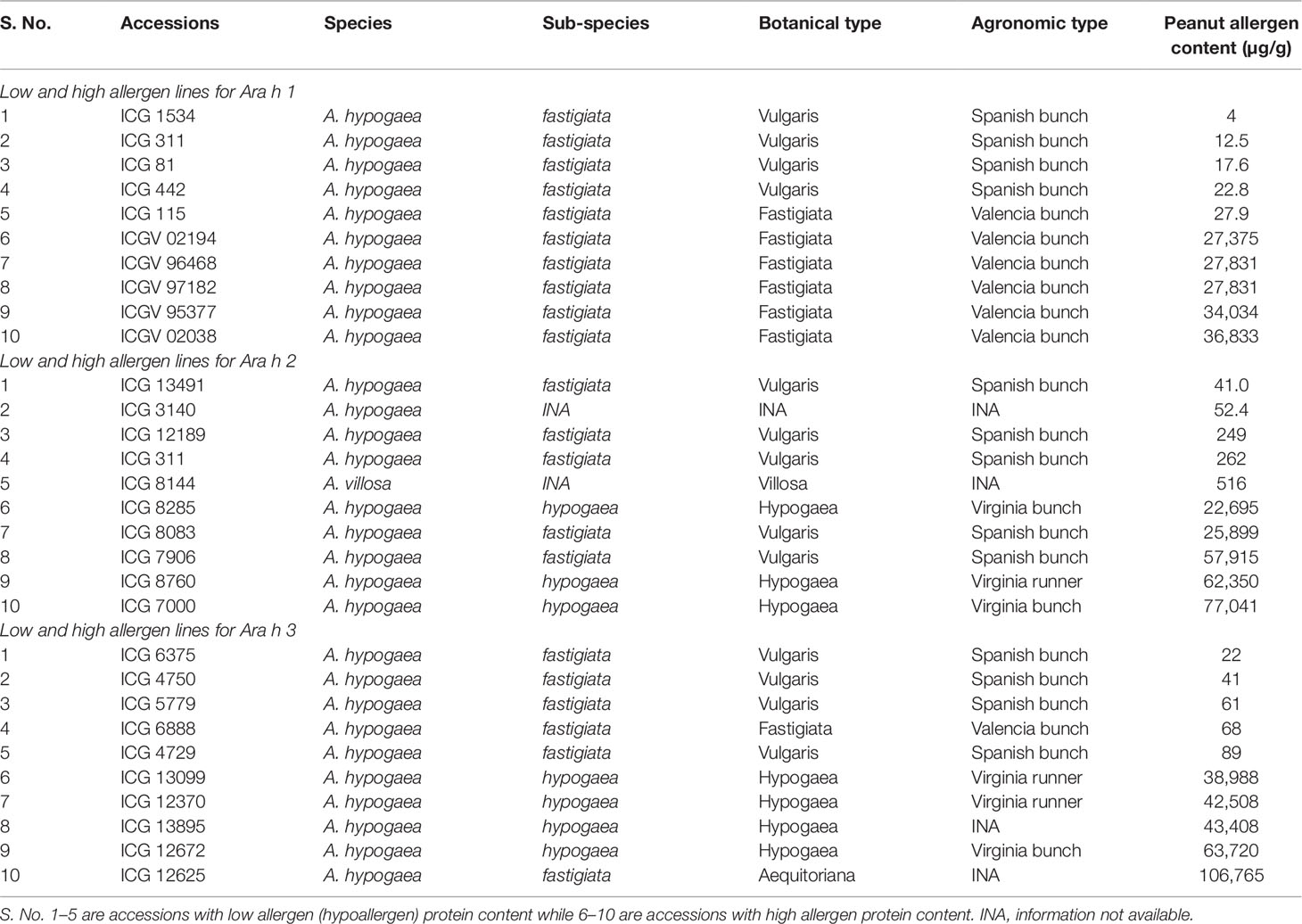
Table 3 List of accessions having minimum and maximum allergen protein content for Ara h 1, Ara h 2, and Ara h 3 from peanut germplasm lines.
A wide variation also observed in the presence of Ara h 2 and it ranged from 52 µg/g (ICG 3140) to 77,042 µg/g (ICGV 7000) among cultivated accessions (264) while it ranged from 516 µg/g (ICG 8144) to 15,757 µg/g (ICG 13218) among wild germplasm accessions (Table 1 and Figure 2A). The most hypoallergen lines for Ara h 2 were ICG 13491 (41 µg/g), followed by ICG 3140 (52 µg/g), ICG 12189 (249 µg/g), ICG 311 (262 µg/g), and ICG 8144 (516 µg/g) (Table 3 and Figure 4B). All these hypoallergen lines belonging to Spanish bunch types of A. hypogaea ssp. fastigiata. Interestingly ICG 311 belonging to Spanish bunch types of A. hypogaea ssp. fastigiata also had low Ara h 1 (12.5 µg/g) (Table 3).
The phenotypic variation of Ara h 3 in cultivated genotypes ranged from 22 µg/g (ICG 6375) to 106,765 µg/g (ICG 12625) while in wild accessions ranged from 1,185 µg/g (ICG 8135) to 20,474 µg/g (ICG 1156) (Table 1). The most hypoallergen lines for Ara h 3 included ICG 6375 (22 µg/g) followed by ICG 4750 (41 µg/g), ICG 15779 (61 µg/g), ICG 6888 (68 µg/g), and ICG 4729 (89 µg/g) (Table 3 and Figure 4C). All these hypoallergen lines belong to Spanish bunch types of A. hypogaea ssp. fastigiata except ICG 6888 (68 µg/g) which belongs to Valencia bunch of the same subspecies (Table 3 and Figure 4C).
For Ara h 6, the phenotypic wide variation identified in cultivated genotypes, and it ranged from 829 µg/g (ICGV 01328) to 103,892 µg/g (ICGV 02194) compared to wild accessions ranged from 4,381 µg/g (ICG 8123) to 56,857 µg/g (ICG 8195) (Table 1). The most hypoallergen lines for Ara h 6 identified in cultivated genotypes namely ICG 01328 (829 µg/g) followed by ICG 15384 (1,577 µg/g), ICG 15403 (2,066 µg/g), ICGV 01276 (2,078 µg/g), and ICGV 02022 (2,298 µg/g) (Table 4 and Figure 4D). All these hypoallergen lines belong to Valencia bunch type of A. hypogaea ssp. fastigiata.
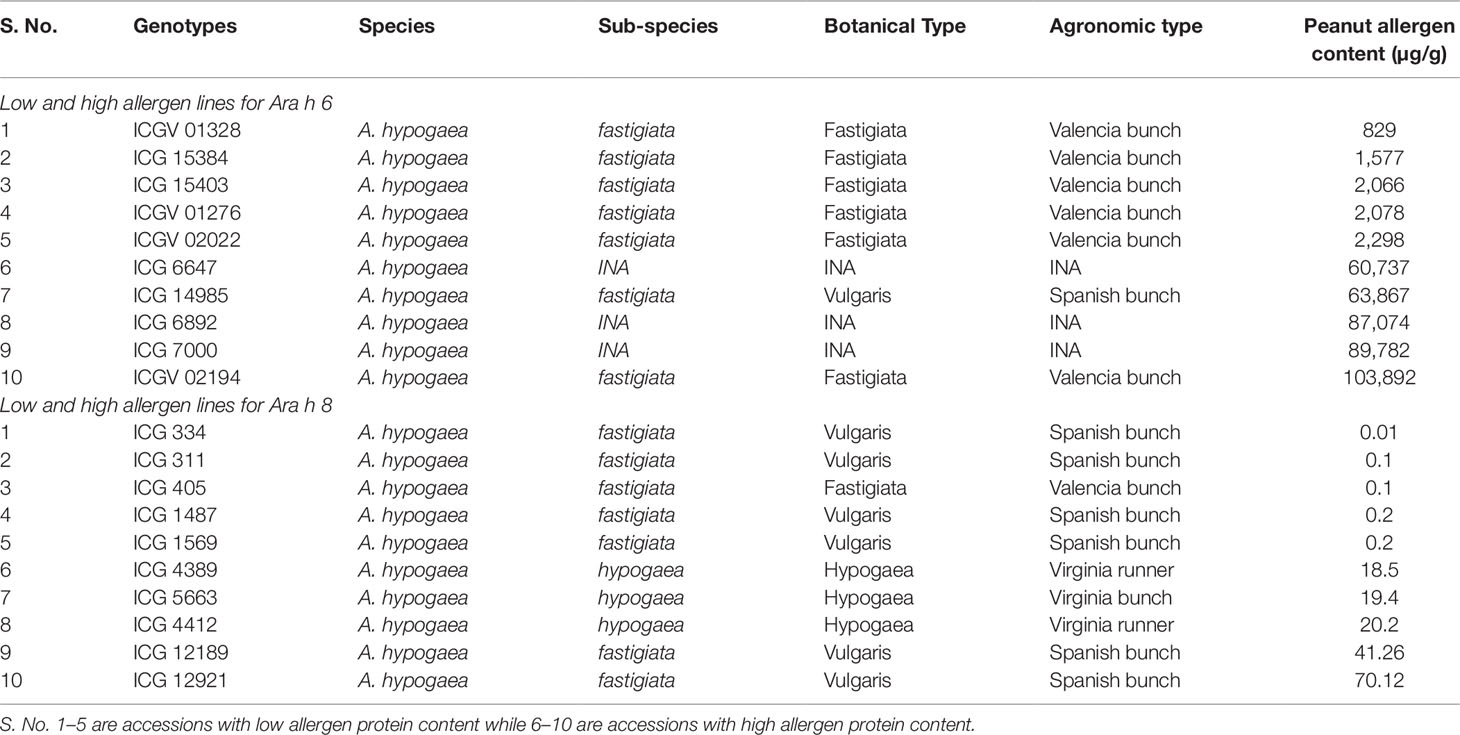
Table 4 List of accessions having minimum and maximum allergen protein content for Ara h 6 and Ara h 8 from peanut germplasm lines.
The phenotypic variation for Ara h 8 was very narrow in cultivated accessions and it ranged from 0.01 µg/g (ICG 334) to 70 µg/g (ICG 12921) compared to wild accessions ranging from 0.3 µg/g (ICG 13206) to 11.27 µg/g (ICG 8973) (Table 1). The best low allergen lines for Ara h 8 included ICG 334 (.01 µg/g) followed by ICG 311(0.1 µg/g), ICG 405(0.1 µg/g), ICG 1487 (0.2 µg/g), and ICG 1569 (0.2 µg/g) (Table 4 and Figure 4D). All these hypoallergen lines belong to Spanish bunch type of A. hypogaea ssp. fastigiata.
Discussion
Peanut allergy is now a global health problem and so far no permanent solution is available to deal with this menace. More importantly, the peanut is consumed in the form of several peanut-based products, therefore, making the life of an allergic person more complicated and difficult. Hypoallergen lines provide an alternative approach to avoid these adverse reaction caused by IgE (Tscheppe and Breiteneder, 2017; Satitsuksanoa et al., 2018). The skin, the respiratory tract, and the gastrointestinal tract are allergic to the peanut and peanut-based product (Sicherer et al., 1998) and cute urticaria, acute vomiting, laryngeal oedema, hypotension, and dysrhythmia are the common symptoms (Bock et al., 2001; Sampson et al., 2017). Peanut-based allergy is very risky, and even the ingestion of trace amounts of peanut can cause life threats in minutes (Bock et al., 2001).
The Stand-Alone Effort for Phenotyping Large-Scale, Diverse Germplasm Set for Major Peanut Allergens Using Most Sensitive Enzyme-Linked Immunosorbent Assay Protocol
Not much efforts have been done toward phenotyping a large peanut germplasm collections in the world. This is majorly due to lack of robust and high-throughput analytical assays to quantify major allergen proteins in peanut seeds. Recently our lab developed an ELISA based protocol to estimate major peanut allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8) using peanut seeds (Pandey et al., 2019). By using this protocol, we phenotyped 300 germplasm lines to quantify major peanut allergens. This study successfully identified hypoallergen lines for all the five allergens and this genetic variation for allergens can be exploited in crop improvement for developing improved hypoallergen lines (Figure 5). Using a pool of human serum from patients, a sample ELISA protocol was used to identify antigens in the peanut seed (Dodo et al., 2002) which reported no significant difference in the allergen content. Another such study on 53 Chinese peanut cultivars revealed that the allergenicity was caused by the allergen composition rather than a single allergen (Wu et al., 2016). This study also reported that the allergen content was high in all the peanut cultivars, however, the peanut allergen content could not be quantified in peanut seeds due to unavailability of antibodies. Hence our study is the first of its kind and identified a low/hypoallergen lines from peanut reference set to ensure food safety and security.

Figure 5 Plant and seed features of hypoallergen and high allergen peanut lines identified for five major peanut allergens (Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8).
Diverse Germplasm Set Representing 51 Countries Showed Wide Phenotypic Variation for Allergens
Results confirmed a wide variation of five major peanut allergens in the peanut reference set. This study used monoclonal antibodies for each allergen for phenotyping of ICRISAT reference set representing global diversity. These monoclonal antibodies also used to observe differences in specific peanut allergen profile in peanut flour and peanut-based products such as peanut butter, flour, and other confectionary preparations for clinical use (Filep et al., 2018). Screening of ICRISAT peanut reference set showed wide range of variation for all the five allergens i.e., Ara h 1 (4–36,833 µg/g), Ara h 2 (41–77,041 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (829–103,892 µg/g), and Ara h 8 (0.01–70 µg/g). Similar wide variation was also identified for Ara h 1, Ara h 2, and Ara h 3 in peanut butter, peanut powder, and peanut flour (Filep et al., 2018). An earlier study reported screening of 34 peanut accessions through patient sera, but no significant difference was observed for allergen content (Dodo et al., 2002). The other study also reported not much variation among 53 Chinese peanut cultivars (Wu et al., 2016) which may be due to the use of human sera to estimate the allergen content in their cultivars. These circumstances encouraged us to develop ELISA based protocol which can be used for quantifying allergen content in peanut kernels. Furthermore, we used the most diverse panel “reference set” consists of 300 genotypes which geographically represents 51 countries (Upadhyaya et al., 2003; Upadhyaya et al., 2010) and showed wide variation for all the five major allergens. The sensitivity of peanut allergens varied among populations in different geographical regions (Vereda et al., 2011). In USA and Sweden, the Ara h 1, Ara h 2, and Ara h 3 cause majority of the peanut allergenic reactions leading to serious illnesses. Similar trend has also been observed in 11 European countries (Ballmer-Weber and Beye, 2018). In contrast, the Spanish patients have less sensitivity to Ara h 1, Ara h 2, and Ara h 3 allergens and have shown more sensitivity to Ara h 9, lipid transfer protein. Similarly, the Spanish patients had the highest level of sensitivity rate to birch pollen allergen, Ara h 8, a cross-reactive homolog Bet v 1. It is important to note that despite few reports, not much have been reported from different Asian and African countries. The above difference in allergen sensitivity among countries and continents may have resulted due to several factors, including genetic makeup, environmental factors, and food habits.
Hypoallergen Lines Identified for Major Peanut Allergens
Previous limited efforts in phenotyping closely related germplasm lines have not yielded in the identification of hypo-allergen peanut lines. Keeping in mind this fact, we explored a large number of diverse germplasm lines for phenotyping using newly developed very precise protocol (Pandey et al., 2019). As a result, this study reports low or hypoallergen lines for five major peanut allergens for the first time. For Ara h 1, five hypoallergen lines ranged between 4 and 28 µg/g and the ICG 1534 (4 µg/g) and ICGV 02038 (36,833 µg/g) had minimum Ara h 1 allergen protein. ICG 1534 belongs to Spanish bunch while ICGV 02038 to Valencia bunch. Similarly, the best hypoallergen lines for Ara h 2 were ICG 3140 with just 52.4 µg/g allergen protein. The accessions ICG 6375 (Spanish bunch), ICGV 01328 (Valencia Bunch), and ICG 334 (Spanish Bunch) were identified as best hypoallergen lines for Ara h 3, Ara h 6, and Ara h 8, respectively. The screening of 53 Chinese peanut cultivars through human sera, the Spanish bunch type having low peanut allergen content than the other agronomic type (Wu et al., 2016). They also reported that the Virginia type (Xinxiandahuasheng), Valencia type (Bangjihonghuasheng), Spanish type (Mangdou), and Peruvian type (Yaoshangxiaomake) are low allergen cultivars. Another study screened 35 US peanut cultivars using human antisera of the allergic patient but could not detect any significant variation (Dodo et al., 2002) which may be due to the narrow genetic base of these US cultivars derived from just two founder parents (Isleib and Wynne, 1992).
Landraces Conserve Higher Diversity for Major Peanut Allergens
The landraces have shown less allergen protein accumulation for Ara h 1 (28–13,977 µg/g), Ara h 2 (41–62,350 µg/g), Ara h 3 (22–106,765 µg/g), Ara h 6 (3,657–51,024 µg/g), and Ara h 8 (0.01–70.12 µg/g) as compared to other biological groups i.e., breeding/research material, advanced/improved cultivar, and wild accessions. The quantification of five major allergens through immunological assay showed that the landraces conserved hypoallergen feature. These accessions are ICG 442 (22.7 µg/g) for Ara h 1, ICG 13491 (41 µg/g) for Ara h 2, ICG 6375 (22 µg/g) for Ara h 3, ICG 15405 (3,657 µg/g), and ICG 334 (0.01 µg/g). These accessions mostly belong to fastigiata subspecies and Spanish bunch types and can be used for developing hypoallergen lines through marker-assisted selection (MAS) or clusters of regularly interspaced short palindromic repeats (CRISPR)/Cas9 approach. One previous study reported that the landraces conserved genetic variation for edible oil properties and also suitable for biodiesel production in Algerian peanut landraces (Giuffre et al., 2016). This finding provides hope to use either directly cultivating or further improvement through breeding for developing hypoallergen lines. Some of the hypoallergen lines identified in this study have also been reported having resistance to multiple stresses, e.g., ICG 442, a Spanish hypoallergen line for Ara h 1 was reported resistant to multiple abiotic stresses such as drought, salinity, and phosphorus deficiency (Upadhyaya et al., 2014).
A Sound Basis for Further Research and Cultivation of Hypoallergen Lines to Ensure Human Health From a Peanut Allergy
The development and release of several improved cultivars with high yield potential, biotic and abiotic stresses resistance, and enhanced/improved nutritional quality features in peanut has successfully been developed by combining the plant breeding techniques and efficient phenotyping methods. One of the previous studies reported that there are no significant differences in the allergen content among different peanut agronomic types consumed in western countries (Koppelman et al., 2016). However, that particular study involved very few numbers of genotypes representing various agronomic types. In our study, we used a large diverse peanut germplasm set and reporting that there are wide variation for allergen content among different agronomic types such as Spanish bunch, Valencia bunch, Virginia bunch, and Virginia runner. This study will provide hope to food industries to use hypoallergen lines in their food product preparations. Genetic improvement can be done using various modern tools and techniques through genomic research (Guo et al., 2012). Functional genomics and biotechnological techniques help discover and characterize agriculturaly important genes through deep analysis of the transcriptome, and their direct transfer to chosen cultivars (Brasileiro et al., 2014). Genes which encode storage protein, metabolic enzyme genes, genes involved in oil metabolism, and differentially expressed genes in response to pathogen stress, were identified and cloned in peanut by expressed sequence tag sequencing and are used to improve peanut production.
Wide varieties of peanut are grown to meet need of oil, food, and industries. The identified hypoallergen peanut lines can directly be used for cultivation and use in industry. Further, the identification of functional variation through genomics will facilitate the development of diagnostic markers for different allergens. The diagnostic markers can be used for improving varieties through MAS while the genes can be now edited through CRISPR/Cas9. CRISPR/Cas9 system has proven to be successful in various crop species over past years including wheat, tobacco, rice, potato, tomato sorghum, orange, and maize (Bortesi and Fischer, 2015). Although in peanut, there were no reports to implement genome editing, however, several reports of MAS and marker-assisted backcrossing (MABC) are available (Chu et al., 2011; Varshney et al., 2014; Janila et al., 2016; and Bera et al., 2018). CRISPR/Cas9 is able to introduce homozygous mutations into rice and tomato potentially accelerating crop improvement in the first generation of the transformants (Shen et al., 2014; Zhang et al., 2014). The elimination of allergen through genome editing technology would be useful for a specfiic group of customers. Silencing of Mal d 1 has decreased the allergenicity of apple, which may enhance the consumption without allergic reactions (Dubois et al., 2015). The immune dominant Ara h 2 peanut allergen successfully reduced the allergenicity in peanut through RNA interference technology (Dodo et al., 2008). All allergens coding genes should be silenced or removed in order to develop hypoallergen peanut that are safe for consumption by many patients, and the genome editing provide offers to do so effectively. The availability of hypoallergen lines will impact the peanut industry as well as contribute toward fighting the peanut allergy menace globally.
Summary
The study identified several hypoallergen peanut lines for further study. These hypoallergen lines can be directly used for commercial cultivation in addition to further breeding research for developing improved peanut varieties by combining several other agronomic traits. The output of this study also encourages researchers to identify functional variation so that molecular breeding through MAS, MABC, and genome editing can be deployed for developing new hypoallergen lines in peanut. The results have shown great hope toward fighting peanut allergy and ensuring enhanced food safety and security for humans as well as promises good opportunity for economic gains by producers, processors, and industry.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MP conceived the idea. MP, AP, HS, HU, and RV designed the experiments. AP performed the experiment. AP and HS analyzed the Data. AP, RV, HS, and MP wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to partial financial assistance from Indian Council of Agricultural Research (ICAR) and Department of Biotechnology (DBT), India. We acknowledge Sunil S. Gangurde for performing the cluster analysis. The work reported in this article was undertaken as part of the CGIAR Research Program on Grain Legumes and Dryland Cereals (CRP-GLDC). ICRISAT is a member of CGIAR Consortium.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01177/full#supplementary-material
References
Ballmer-Weber, B. K., Beye, K. (2018). Food challenges. J. Allergy Clin. Immunol. 141, 69–71. doi: 10.1016/j.jaci.2017.06.038
Ben-Shoshan, M., Kagan, R. S., Alizadehfar, R., Joseph, L., Turnbull, E., St Pierre, Y., et al. (2009). Is the prevalence of peanut allergy increasing? A 5-year follow-up study in children in Montreal. J. Allergy Clin. Immunol. 123, 783–788. doi: 10.1016/j.jaci.2009.02.004
Bera, S., Kamdar, J., Kasundra, S., Dash, P., Maurya, A., Patel, S., et al. (2018). Improving oil quality by altering levels of fatty acids through marker-assisted selection of ahfad2 alleles in peanut (Arachis hypogaea L.) varieties. Euphytica 214–162. doi: 10.1007/s10681-018-2241-0"10.1007/s10681-018-2241-0
Berger, Smith. (1998). Science commentary: why do some children grow out of peanut allergy? Br. Med. J. 316, 1275.
Bock, S. A., Muñoz-Furlong, A., Sampson, H. A. (2001). Fatalities due to anaphylactic reactions to foods. J. Allergy Clin. Immunol. 107, 91–93. doi: 10.1067/mai.2001.112031
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Brasileiro, A. C. M., Araujo, A. C. G., Leal-Bertioli, S. C., Guimaraes, P. M. (2014). Genomics and genetic transformation in Arachis. Int. J. Plant Biol. Res. 2, 1017.
Cabanillas, B., Maleki, S. J., Rodríguez, J., Burbano, C., Muzquiz, M., Jiménez, M. A., et al. (2012). Heat and pressure treatments effects on peanut allergenicity. Food Chem. 132, 360–366. doi: 10.1016/j.foodchem.2011.10.093
Chu, Y., Wu, C. L., Holbrook, C. C., Tillman, B. L., Person, G., Ozias-Akins, P. (2011). Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 4, 110–117. doi: 10.3835/plantgenome2011.01.0001
de Jong, E. C., van Zijverden, M., Spanhaak, S., Koppelman, S. J., Pellegrom, H., Penninks, A. H. (1998). Identification and partial characterization of multiple major allergens in peanut proteins. Clin. Exp. Allergy 28, 743–751. doi: 10.1046/j.1365-2222.1998.00301.x
Dodo, H., Marsic, D., Callender, M., Cebert, E., Viquez, O. (2002). Screening 34 Peanut introductions for allergen content using ELISA. Food Agric. Immunol. 14, 147–154. doi: 10.1080/09540100220145179
Dodo, H., Konan, K., Viquez, O. M. (2005). A genetic engineering strategy to eliminate peanut allergy. Curr. Allergy Asthm. R. 5, 67–73. doi: 10.1007/s11882-005-0058-0
Dodo, H. W., Konan, K. N., Chen, F. C., Egnin, M., Viquez, O. M. (2008). Alleviating peanut allergy using genetic engineering: the silencing of the immuno dominant allergen Ara h 2 leads to its significant reduction and a decrease in peanut allergenicity. Plant Biotechnol. J. 6, 135–145. doi: 10.1111/j.1467-7652.2007.00292.x
Dubois, A. E. J., Pagliarani, G., Brouwer, R. M., Kollen, B. J., Dragsted, L. O., Eriksen, F. D., et al. (2015). First successful reduction of clinical allergenicity of food by genetic modification: Mal d 1-silenced apples cause fewer allergy symptoms than the wild-type cultivar. Allergy 70, 1406–1412. doi: 10.1111/all.12684
Filep, S., Block, D. S., Smith, B. R. E., King, E. M., Commins, S., Kulis, M., et al. (2018). Specific allergen profiles of peanut foods and diagnostic or therapeutic allergenic products. J. Allergy Clin. Immunol. 141, 626–663. doi: 10.1016/j.jaci.2017.05.049
Flinterman, A. E., van Hoffen, E., Hartog Jager, C. F., Koppelman, S., Pasmans, S. G., Hoekstra, M. O., et al. (2007). Children with peanut allergy recognize predominantly Ara h2 and Ara h6, which remains stable over time. Clin. Exp. Allergy 37, 1221–1228. doi: 10.1111/j.1365-2222.2007.02764.x
Galili, T. (2015). Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinf. 31, 3718–3720. doi: 10.1093/bioinformatics/btv428
Giuffrè, A. M., Tellah, S., Capocasale, M., Zappia, C., Latati, M., Badiani, M., et al. (2016). Seed oil from ten Algerian peanut landraces for edible use and biodiesel production. J. Oleo Sci. 65, 9–20. doi: 10.5650/jos.ess15199
Grundy, J., Matthews, S., Bateman, B., Dean, T., Arshad, S. H. (2002). Rising prevalence of allergy to peanut in children: Data from 2 sequential cohorts. J. Allergy Clin. Immunol. 110, 784–789. doi: 10.1067/mai.2002.128802
Guo, B. Z., Chen, C. Y., Chu, Y., Holbrook, C. C., Ozias-Akins, P., Stalker, H. T. (2012). Advances in genetics and genomics for sustainable peanut production, in Sustainable Agriculture and New Biotechnologies (Boca Raton, Fla, USA: CRC Press), 341–367. doi: 10.1201/b10977-16
Hamilton, R. G., MacGlashan, D. W., Saini, S. S. (2010). IgE antibody-specific activity in human allergic disease. Immunol. Res. 47, 273–284. doi: 10.1007/s12026-009-8160-3
Hefle, S. L., Nordlee, J. A., Taylor, S. L. (1996). Allergenic foods. Crit. Rev. Food Sci. Nutr. 36, S6989. doi: 10.1080/10408399609527760
Holbrook, C. C., Stalker, H. T. (2003). Peanut breeding and genetic resources. Plant Breed. Rev. 22, 297–356. doi: 10.1002/9780470650202.ch6
Iqbal, A., Shah, F., Hamayun, M., Ahmad, A., Hussain, A., Waqas, M., et al. (2016). Allergens of Arachis hypogaea and the effect of processing on their detection by ELISA. Food Nutr. Res. 60, 28945. doi: 10.3402/fnr.v60.28945
Isleib, T. G., Wynne, J. C. (1992). Use of plant introduction in peanut improvement,” in Use of Plant Introduction to Cultivar Development, Part 2 (MADISON, WI, USA: CSSA Spec. publication 20 of Crop Science Society of America).
Janila, P., Pandey, M. K., Shasidhar, Y., Variath, M. T., Sriswathi, M., Khera, P., et al. (2016). Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 242, 203–213. doi: 10.1016/j.plantsci.2015.08.013
Kagan, R. S., Joseph, L., Dufresne, C., ray-Donald, K., Turnbull, E., St Pierre, Y., et al. (2003). Prevalence of peanut allergy in primary-school children in Montreal, Canada. J. Allergy Clin. Immuno. 112, 1223–1228. doi: 10.1016/j.jaci.2003.09.026
Kondo, Y., Urisu, A. (2009). Oral allergy syndrome. Allergol Int. 58, 485–491. doi: 10.2332/allergolint.09-RAI-0136
Koppelman, S. J., Knol, E. F., Vlooswijk, R. A. A., Wensing, M., Knulst, A. C., Hefle, S. L., et al. (2003). Peanut allergen Ara h 3: Isolation from peanuts and biochemical characterization. Allergy 58, 1144–1151. doi: 10.1034/j.1398-9995.2003.00259.x
Koppelman, S. J., de Jong, G. A., Laaper-Ertmann, M., Peeters, K. A. B. M., Knulst, A. C., Hefle, S. L., et al. (2005). Purification and immunoglobulin E-binding properties of peanut allergen Ara h 6: Evidence for cross-reactivity with Ara h 2. Clin. Exp. Allergy 35, 490–497. doi: 10.1111/j.1365-2222.2005.02204.x
Koppelman, S. J., Jayasena, S., Luykx, D., Schepens, E., Apostolovic, D., De Jong, G. A., et al. (2016). Allergenicity attributes of different peanut market types. Food Chem. Toxicol. 1, 82–90. doi: 10.1016/j.fct.2016.02.016
Krause, S., Latendorf, T., Schmidt, H., Darcan-Nicolaisen, Y., Reese, G., Petersen, A., et al. (2010). Peanut varieties with reduced Ara h 1 content indicating no reduced allergenicity. Mol. Nutr. Food Res. 54, 3817. doi: 10.1002/mnfr.200900072
Lanser, B. J., Wright, B. L., Orgel, K. A., Vickery, B. P., Fleischer, D. M. (2015). Current options for the treatment of food allergy. Pediatr. Clin. 62, 1531–1549. doi: 10.1016/j.pcl.2015.07.015
Leung, A. S. Y., Leung, N. Y. H., Wai, C. Y. Y., Leung, T. F., Wong, G. W. K. (2018). Allergen immunotherapy for food allergy from the Asian perspective: key challenges and opportunities. Expert Rev. Clin. Immunol. 15, 153–164. doi: 10.1080/1744666X
Lieberman, J. A., Sicherer, S. H. (2011). Diagnosis of food allergy: epicu-taneous skin tests, in vitro tests, and oral food challenge. Curr. Allergy Asthma Rep. 11, 58–64. doi: 10.1007/s11882-010-0149-4
Liew, W. K., Chiang, W. C., Goh, A. E., Lim, H. H., Chay, O. M., Chang, S., et al. (2013). Paediatric anaphylaxis in a Singaporean children cohort: changing food allergy triggers over time. Asia Pac. Allergy 3, 29–34. doi: 10.5415/apallergy.2013.3.1.29
Mahesh, P. A., Wong, G. W. K., Ogorodova, L., Potts, J., Leung, T. F., Fedorova, O., et al. (2016). Prevalence of food sensitization and probable food allergy among adults in India: the EuroPrevall INCO study. Allergy 71, 1010–1019. doi: 10.1111/all.12868
Maleki, S. J., Chung, S. Y., Champagne, E. T., Raufmann, J. P. (2000). The effects of roasting on the allergenic properties of peanut proteins. J. Allergy Clin. Immunol. 106, 763–768. doi: 10.1067/mai.2000.109620
Mittag, D., Akkerdaas, J., Ballmer-Weber, B. K., Vogel, L., Wensing, M., Becker, W. M., et al. (2004). Ara h 8, a Bet v 1-homologous allergen from peanut, is a major allergen in patients with combined birch pollen and peanut allergy. J. Allergy Clin. Immunol. 114, 1410–1417. doi: 10.1016/j.jaci.2004.09.014
Morisset, M., Moneret-Vautrin, D. A., Kanny, G. (2005). Prevalence of peanut sensitization in a population of 4,737 subjects: an Allergo-Vigilance Network enquiry carried out in 2002. Eur. Ann. Allergy Clin. Immunol. 37, 54–57.
Nicolaou, N., Murray, C., Belgrave, D., Poorafshar, M., Simpson, A., Custovic, A. (2011). Quantification of specific IgE to whole peanut extract and peanut components in prediction of peanut allergy. J. Allergy Clin. Immunol. 127, 684–685. doi: 10.1016/j.jaci.2010.12.012
Osterballe, M., Hansen, T. K., Mortz, C. G., Host, A., Bindslev-Jensen, C. (2005). The prevalence of food hypersensitivity in an unselected population of children and adults. Pediatr. Allergy Immunol. 16, 567–573. doi: 10.1111/j.1399-3038.2005.00251.x
Palmer, G. W., Dibbern, D. A., Jr., Burks, A. W., Bannon, G. A., Bock, S. A., Porterfield, H. S., et al. (2005). Comparative potency of Ara h 1 and Ara h 2 in immunochemical and functional assays of allergenicity. Clin. Immunol. 115, 302–312. doi: 10.1016/j.clim.2005.02.011
Pandey, M. K., Varshney, R. K. (2018). Groundnut entered post-genome sequencing era: opportunities and challenges in translating genomic information from genome to field, in Biotechnologies in Crop Improvement, vol. 3 . Eds. Gosal, S. H., Wani, S. S. (New York City: Springer International Publishing). doi: 10.1007/978-3-319-94746-4_9
Pandey, M. K., Monyo, E., Ozias-Akins, P., Liang, X., Guimarães, P., Nigam, S. N., et al. (2012). Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 30, 639–651. doi: 10.1016/j.biotechadv.2011.11.001
Pandey, A. K., Varshney, R. K., Sudini, H. K., Pandey, M. K. (2019). An improved enzyme-linked immunosorbent assay (ELISA) based protocol using seeds for detection of five major peanut allergens Ara h 1, Ara h 2, Ara h 3, Ara h 6 and Ara h 8. Front. Nutr. 6, 68. doi: 10.3389/fnut.2019.00068
Peeters, K. A., Koppelman, S. J., Van Hoffen, E., Van Der Tas, C. W., Hartog Jager, C. F., Penninks, A. H., et al. (2007). Does skin prick test reactivity to purified allergens correlate with clinical severity of peanut allergy? Clin. Exp. Allergy 37, 108–115. doi: 10.1111/j.1365-2222.2006.02628.x
Porterfield, H. S., Murray, K. S., Schlichting, D. G., Chen, X., Hansen, K. C., Duncan, M. W., et al. (2009). Effector activity of peanut allergens: a critical role for Ara h 2, Ara h 6, and their variants. Clin. Exp. Allergy 39, 1099–1108. doi: 10.1111/j.1365-2222.2009.03273.x
Riecken, S., Lindner, B., Petersen, A., Jappe, U., Becker, W. M. (2008). Purification and characterization of natural Ara h 8, the Bet v 1 homologous allergen from peanut, provides a novel isoform. Biol. Chem. 389, 415–423. doi: 10.1515/BC.2008.038
Sampson, H. A., Shreffler, W. G., Yang, W. H., Sussman, G. L., Brown-Whitehorn, T. F., Nadeau, K. C., et al. (2017). Effect of varying doses of epicutaneous immunotherapy vsplacebo on reaction to peanut protein exposure among patients with peanut sensitivity: a randomized clinical trial. JAMA 318, 1798–1809. doi: 10.1001/jama.2017.16591
Satitsuksanoa, P., Globinska, A., Jansen, K., de Veen, W., Akdis, M. (2018). Modified allergens for immunotherapy. Curr. Allergy Asthma Rep. 18, 9. doi: 10.1007/s11882-018-0766-x
Shek, L. P., Cabrera-Morales, E. A., Soh, S. E., Gerez, I., Ng, P. Z., Yi, F. C., et al. (2010). A population-based questionnaire survey on the prevalence of peanut, tree nut, and shellfish allergy in 2 Asian populations. J. Allergy Clin. Immunol. 126, 24–31. doi: 10.1016/j.jaci.2010.06.003
Shen, B., Zhang, W. S., Zhang, J., Zhou, J., Wang, J., Chen, L., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Meth. 11, 399–402. doi: 10.1038/nmeth.2857
Sicherer, H. S., Sampson, A. H. (2007). Peanut allergy: emerging concepts and approaches for an apparent epidemic. J. Allergy Clin. Immunol. 120, 491–503. doi: 10.1016/j.jaci.2007.07.015
Sicherer, S.H., Sampson, H.A. (2010). Food allergy. J. Allergy Clin. Immunol. 125, 116–125. doi: 10.1016/j.jaci.2009.08.028
Sicherer, H. S., Sampson, A. H. (2014). Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J. Allergy Clin. Immunol. 133, 291–307. doi: 10.1016/j.jaci.2013.11.020
Sicherer, S. H., Burks, A. W., Sampson, H. A. (1998). Clinical features of acute allergic reactions to peanut and tree nuts in children. Pediatr. 102, e6. doi: 10.1542/peds.102.1.e6
Sicherer, S. H., Munoz-Furlong, A., Godbold, J. H., Sampson, H. (2010). US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J. Allergy Clin. Immunol. 125, 1322–1326. doi: 10.1016/j.jaci.2010.03.029
Tscheppe, A., Breiteneder, H. (2017). Recombinant allergens in structural biology, diagnosis, and immunotherapy. Int. Arch. Allergy Immunol. 172, 187–202. doi: 10.1159/000464104
Upadhyaya, H. D., Bramel, P. J., Ortiz, R., Singh, S. (2002). Developing a mini core of peanut for utilization of genetic resources. Crop Sci. 42, 2 150–22156. doi: 10.2135/cropsci2002.2150
Upadhyaya, H. D., Dwivedi, S. L., Vadez, V., Hamidou, F., Singh, S., Varshney, R. K., et al. (2014). Multiple resistant and nutritionally dense germplasm identified from mini core collection in peanut. Crop Sci. 54, 679–693. doi: 10.2135/cropsci2013.07.0493
Upadhyaya, H. D., Ortiz, R., Bramel, P. J., Singh, S. (2003). Development of a groundnut core collection using taxonomical, geographical and morphological descriptors. Genet. Resour. Crop Evol. 50, 139–148. doi: 10.1023/A:1022945715628
Upadhyaya, H. D., Yadav, D., Dronavalli, N., Gowda, C. L. L., Singh, S. (2010). Mini core germplasm collections for infusing genetic diversity in plant breeding programs. Electron J. Plant Breed. 1, 1294–1309.
Varshney, R. K., Pandey, M. K., Janila, P., Nigam, S. N., Sudini, H., Gowda, M. V. C., et al. (2014). Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor. Appl. Genet. 127, 1771–1781. doi: 10.1007/s00122-014-2338-3
Varshney, R. K., Pandey, M. K., Bohra, A., Singh, V. K., Thudi, M., Saxena, R. K. (2019). Toward sequence-based breeding in legumes in post-genome sequencing era. Theor. Appl. Genet. doi: 10.1007/s00122-018-3252-x
Varshney, R. K., Thudi, M., Pandey, M. K., Tardieu, F., Ojiewo, C., Vadez, V., et al. (2018). Accelerating genetic gains in legumes for prosperous smallholder agriculture: integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 69, 3293–3312. doi: 10.1093/jxb/ery088
Vereda, A., van Hage, M., Ahlstedt, S., Ibañez, M. D., Cuesta-Herranz, J., van Odijk, J., et al. (2011). Peanut allergy: Clinical and immunologic differences among patients from 3 different geographic regions. J. Allergy Clin. Immunol. 127, 603–607. doi: 10.1016/j.jaci.2010.09.010
Warner, J. O. (1999). Peanut allergy: A major public health issue. Pediatr. Asthma Allergy Immunol. 10, 14–20. doi: 10.1034/j.1399-3038.1999.101011.x
Warnes, G. R., Bolker, B., Lodewijk, B., Gentleman, R., Liaw, W. H. A., Lumley, T., et al. (2016). Package 'gplots "Various R Programming Tools for Plotting Data" Repository CRAN, Vol. 21. balloonplot pp. 04–18.
Wen, H. W., Borejsza-Wysocki, W., DeCory, T. R., Durst, R. A. (2007). Peanut allergy, peanut allergens, and methods for the detection of peanut contamination in food products. Compr. Rev. Food Sci. Food Saf. 6, 47–58. doi: 10.1111/j.1541-4337.2007.00017.x
Wu, Z., Zhou, N., Xiong, F., Li, X., Yang, A., Tong, P., et al. (2016). Allergen composition analysis and allergenicity assessment of Chinese peanut cultivars. Food Chem. 196, 459–465. doi: 10.1016/j.foodchem.2015.09.070
Keywords: reference set, peanut allergens, Ara h 1, Ara h 2, Ara h 3, Ara h 6, Ara h 8, enzyme-linked immunosorbent assay
Citation: Pandey AK, Sudini HK, Upadhyaya HD, Varshney RK and Pandey MK (2019) Hypoallergen Peanut Lines Identified Through Large-Scale Phenotyping of Global Diversity Panel: Providing Hope Toward Addressing One of the Major Global Food Safety Concerns. Front. Genet. 10:1177. doi: 10.3389/fgene.2019.01177
Received: 29 May 2019; Accepted: 24 October 2019;
Published: 27 November 2019.
Edited by:
Suresh T. Mathews, Samford University, United StatesReviewed by:
Hongbing Chen, Nanchang University, ChinaGuang Ren, University of Alabama at Birmingham, United States
Copyright © 2019 Pandey, Sudini, Upadhyaya, Varshney and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manish K. Pandey, bS5wYW5kZXlAY2dpYXIub3Jn
 Arun K. Pandey
Arun K. Pandey Hari K. Sudini
Hari K. Sudini Hari D. Upadhyaya
Hari D. Upadhyaya Rajeev K. Varshney
Rajeev K. Varshney Manish K. Pandey
Manish K. Pandey