- 1College of Animal Science and Technology, Shandong Agricultural University, Tai'an, China
- 2Shandong Provincial Key Laboratory of Animal Biotechnology and Disease Control and Prevention, Shandong Agricultural University, Tai'an, China
- 3Shandong Provincial Engineering Technology Research Center of Animal Disease Control and Prevention, Shandong Agricultural University, Tai'an, China
Duck Tembusu virus (DTMUV), the causative agent of egg-drop syndrome, has caused substantial economic losses to duck industry. DTMUV infection leads to profound changes of host cells, including transcriptome and proteome. However, the lncRNA expression profile and the biological function of lncRNA have not been revealed. Therefore, DTMUV was used to inoculate duck embryo fibroblast cells (DEFs) for high-throughput RNA-sequencing (RNA-Seq). The results showed that 34 and 339 differently expressed lncRNAs were, respectively, identified at 12 and 24 h post-infection (hpi). To analyze their biological functions, target genes in cis were searched and the regulatory network was formed. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that the target genes were strongly associated with immune system, signaling molecular and interaction, endocrine system, and signal transduction. The differently expressed lncRNAs were selected and verified by quantitative real-time polymerase chain reaction (RT-qPCR). Our study, for the first time, analyzed a comprehensive lncRNA expression profile in DEFs following DTMUV infection. The analysis provided a view on the important roles of lncRNAs in gene regulation and DTMUV infection.
Introduction
In 2010, a novel disease characterized by a significant decline in egg-drop production broke out in many duck farms across Chain (1). The disease, diagnosed as duck hemorrhagic ovaritis, was finally proven to be caused by DTMUV (2–4). DTMUV, similar to other flaviviruses, is a single-stranded, positive-sense RNA virus with an approximately 11 kb genome. It can infect not only ducks but also geese (5), chickens (6), sparrows (5), pigeons (7), and mice (6). Interestingly, a wide spectrum of mammalian cells, including A549, BHK21, Hela, Vero, and SH-SY5Y, exhibit obvious cytopathic effects (CPEs) after DTMUV infection (8). Our previous study showed that CPEs were appeared on HEK293 when the cells were infected with DTMUV, which implies that the virus with the possibility to infect human can potentially threaten human health (9).
LncRNAs, more than 200 nucleotides in length, were recognized as pseudo-transcriptions due to the lack of protein-coding capacity (10). In recent years, they were implicated in complex biological processes through diverse mechanisms such as in gene regulation by titration of transcription factors, splicing alteration, sponging of microRNAs, and recruitment of chromatin modifying enzymes (11–14). Besides, they could function in cis to regulate expression of neighboring gene and in trans to impact gene expression across chromosomes (15). And emerging evidence uncovered that lncRNAs, induced by various viruses, were considered to regulate host innate immune response (16).
RNA-Seq, promising simultaneous transcript discovery and abundance estimation, is more powerful for revelation of transcriptome complexity and for identification of non-coding RNAs, new transcription units, and alternative splicing (17, 18). Recently, it has been widely used to reveal the expression levels of RNA transcripts in specific tissues or cells in different physiological states and cellular environments (19). Unquestionable, it provides important insights into the interaction mechanism between pathogen and host (20–22).
Up to now, extensive and in-depth researches about pathogen (5, 23, 24), pathogenicity (6, 25, 26), epidemiology (5, 27), rapid diagnosis (28–30), and vaccine (31) were carried out to prevent and control DTMUV infection. Recently, we revealed the expression profile and biological function of mRNAs in DTMUV-infected DEFs (8). However, the expression and function of lncRNAs in response to DTMUV infection remain superficial. Therefore, we analyzed a comprehensive lncRNA expression profile in DEFs following DTMUV infection. Besides, the cis target genes of differentially expressed lncRNAs were identified and then used for Gene Ontology (GO) and KEGG enrichment analyses to elucidate their biological processes and associated pathways. Our findings contribute to further understanding of the regulatory mechanism of lncRNAs. In addition, the analysis provides a new perspective for DTMUV-host interaction.
Materials and Methods
Cell Culture and Virus Infection
Freshly isolated DEFs were obtained from 10-day-old specific pathogen free (SPF) duck embryos (purchased from Harbin Veterinary Research Institute, Harbin, China). DEFs were cultured in Dulbecco's modified Eagle's medium (DMEM/F-121:1) (01-172-1ACS, BI, Kibbutz, Beit Haemek, Israel) supplemented with 10% fetal bovine serum (FBS) (04-001-1ACS, BI, Kibbutz, Beit Haemek, Israel), 100 μ/mL penicillin, and 100 μg/mL streptomycin (P1400, Solarbio Science & Technology Co., Ltd., Beijing, China) at 37°C with 5% CO2. When reaching 80–90% confluency, DEFs were mock-infected or infected with DTMUV (SDSM strain, GenBank Accession No. KC333867.1, which was obtained from our laboratory, the Poultry Disease Lab of Shandong Agricultural University) at a multiplicity of infection (MOI) of 3. After viral adsorption for 1.5 h at 37°C with 5% CO2, the inoculum was replaced with maintenance medium (DMEM/F-12 with 2% FBS) for further maintaining. The virus replication was detected at 12, 24, and 48 hpi. Each sample had three biological replicates.
This study was approved by the Committee on the Ethics of Animal of Shandong (permit number 20,156,681). All subjects gave informed consent for their participation in the study.
RNA Isolation and RNA-Seq
The total RNA was extracted using TRIzol reagent (Vazyme Biotech Company, China) according to the manufacturer's instructions. The concentration of total RNA was determined using a Nanodrop instrument (Thermo Fisher Scientific). RNA quality was assessed by the detection of the A260/A280 ratio, with a value of 1.8–2.0 indicating high quality. Ribo-zero-magnetic-kit (Epicenter, USA) was used to remove ribosomal RNA from the samples. RNA libraries were prepared using TruSeq RNA LT Sample Prep Kit v2 (Illumina, San Diego, CA, USA). Library sequencing was performed on an Illumina Hiseq3000 platform by the Shanghai Personal Biotechnology (Shanghai, China).
Bioinformatic Analyses
Clean data were obtained by removing adaptors, poly-N sequences, and poor-quality using Cutadapt software (http://cutadapt.readthedocs.io/en/stable/). Quality control analysis was performed on clean data using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). The filtered reads were then mapped to the Peking duck reference genome (duckbase.refseq.v4.fa, http://www.duckbase.org/Download) using TopHat2 software (http://tophat.cbcb.umd.edu/) (32, 33). The transcripts were assembled with the mapped reads using StringTie software (http://ccb.jhu.edu/software/stringtie/) (34).
Coding Potential Analyses and Differential Expression Analyses
The coding ability of lncRNAs was predicted using three tools, including Coding-Non-Coding-Index (CNCI) (https://github.com/www-bioinfo-org/CNCI) (35), Coding Potential Calculator (CPC) (http://cpc.cbi.pku.edu.cn/) (36), and Pfam-scan (http://www.ebi.ac.uk/Tools/pfa/pfamscan/help/) (37). The intersecting no-coding transcripts of the three tools were designated as credible lncRNAs. The DESeq software (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html) (38) was used to perform differential expression analyses. A p < 0.05 and |fold change| ≥ 2 were set as the threshold for significantly differential expression.
Target Gene Prediction and Function Analyses
In order to explore the role of differently expressed lncRNAs in regulating gene expression, cis analyses were implemented to predict the target genes. The known protein-encoding genes located within a 100-kb window upstream or downstream of lncRNAs were identified as cis target genes. To assess biological function of target genes, GO enrichment analysis basing on GO database (the date of the update of the database is January 1st, 2018) was performed using the topGO software (http://www.bioconductor.org/packages/release/bioc/html/RamiGO.html) (39). In this analysis, biological function was mainly classified into molecular function, biological process, and cellular component. Only categories with a p < 0.05 were considered significantly enriched. In addition, the associated pathways of cis target genes were predicted by KEGG database (the date of the update of the database is January 1st, 2018). The signal pathway terms with a p < 0.05 were considered significantly enriched.
RT-qPCR Analysis
The duck glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as the endogenous reference gene. All the primers, synthesized by TSINGKE Biological Technology (China), are listed in Table S1. RT-qPCR was carried out on a Light Cycler 480II instrument (Roche, Basel, Switzerland) using One Step TB Green PrimeScript™ RT-PCR Kit (TaKaRa, Dalian, China) according to the manufacturer's instructions, and melting curves were obtained. The relative expression levels of DTMUV and differentially expressed lncRNAs were calculated through 2−ΔΔCt method (40). Each sample had three biological replicates. Statistical analyses were performed using Student's t-tests. We performed correlation analysis between RNA-Seq and RT-qPCR with GraphPad Prism software, Version 7.0.
Results
Confirmation of DTMUV Infection in DEFs
DEFs were infected with DTMUV at a MOI of 3. The successful infection was verified by observation of CPE and determination of virus replication monitored by RT-qPCR at 12, 24, and 48 hpi. As is showed in Figure 1, no CPE was observed in mock-infected DEFs (Figures 1A–C), and CPE on DTMUV-infected DEFs at 12 hpi was not visible (Figure 1D). However, the pathological cellular state (the cells were shrinking and rounded) could be recognized at 24 hpi as early as possible (Figure 1E), and the pathological condition was more visible (the cells were shrinking, cracking, and suspending) at 48 hpi (Figure 1F). The result of virus replication was showed in Figure 2. The viral replication gradually increased at 12, 24, and 48 hpi, indicating the development of persistent infection. The DEFs at 12 and 24 hpi were harvested for RNA-Seq.
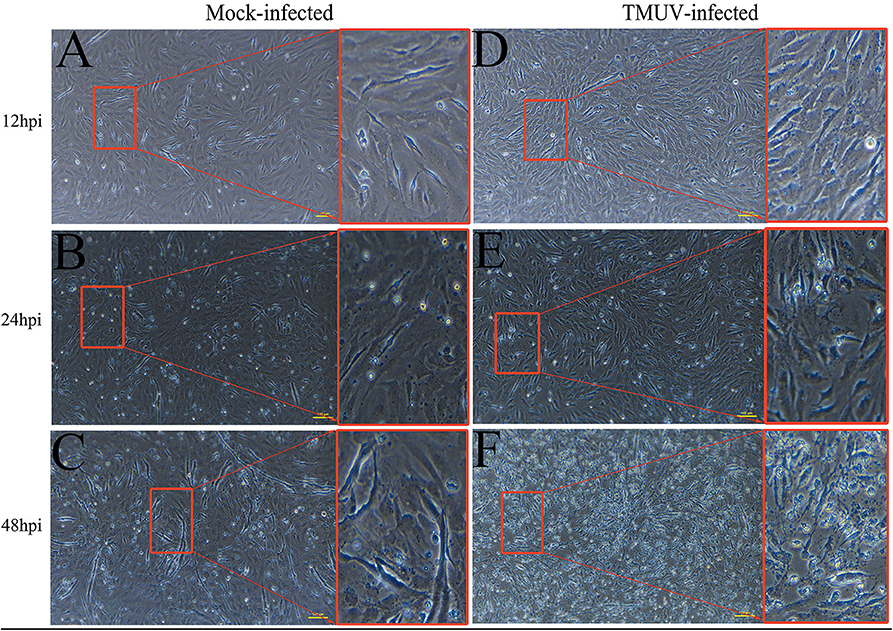
Figure 1. Cytopathic effects of duck embryo fibroblast cells following DTMUV infection at 12, 24, and 48 h post-infection. Each sample had three biological replicates. The yellow scale bar represents 100 μm. (A) The status of mock-infected DEFs at 12 hpi. (B) The status of mock-infected DEFs at 24 hpi. (C) The status of mock-infected DEFs at 48 hpi. (D) The status of TMUV-infected DEFs at 12 hpi. (E) The status of TMUV-infected DEFs at 24 hpi. (F) The status of TMUV-infected DEFs at 48 hpi.
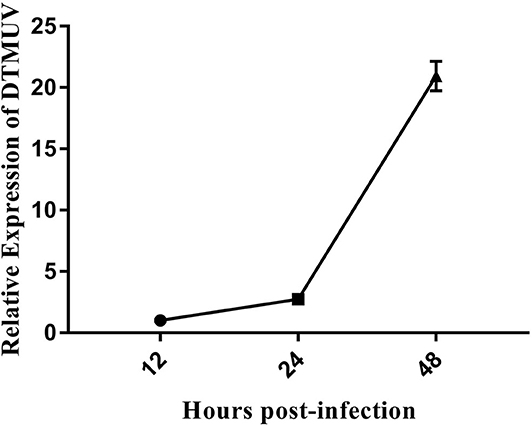
Figure 2. The DTMUV replication in duck embryo fibroblast cells following infection at 12, 24, and 48 h post-infection. Each sample had three biological replicates.
The Code Capacity, Length Distribution, and Density Distribution of lncRNAs
RNA-Seq was performed to determine the expression levels of lncRNAs in DEFs infected or uninfected with DTMUV. After removing adaptor and low-quality sequences, the ability of transcripts to encode protein was determined. The result showed that 1457 lncRNAs were filtrated by the three methods in common (Figure 3A). The details of the results are included in Table S2. The length and density distribution revealed that lncRNAs, 3,000–4,000 nucleotides in length and medium-expressed, occupy a dominant position in samples (Figures 3B,C).
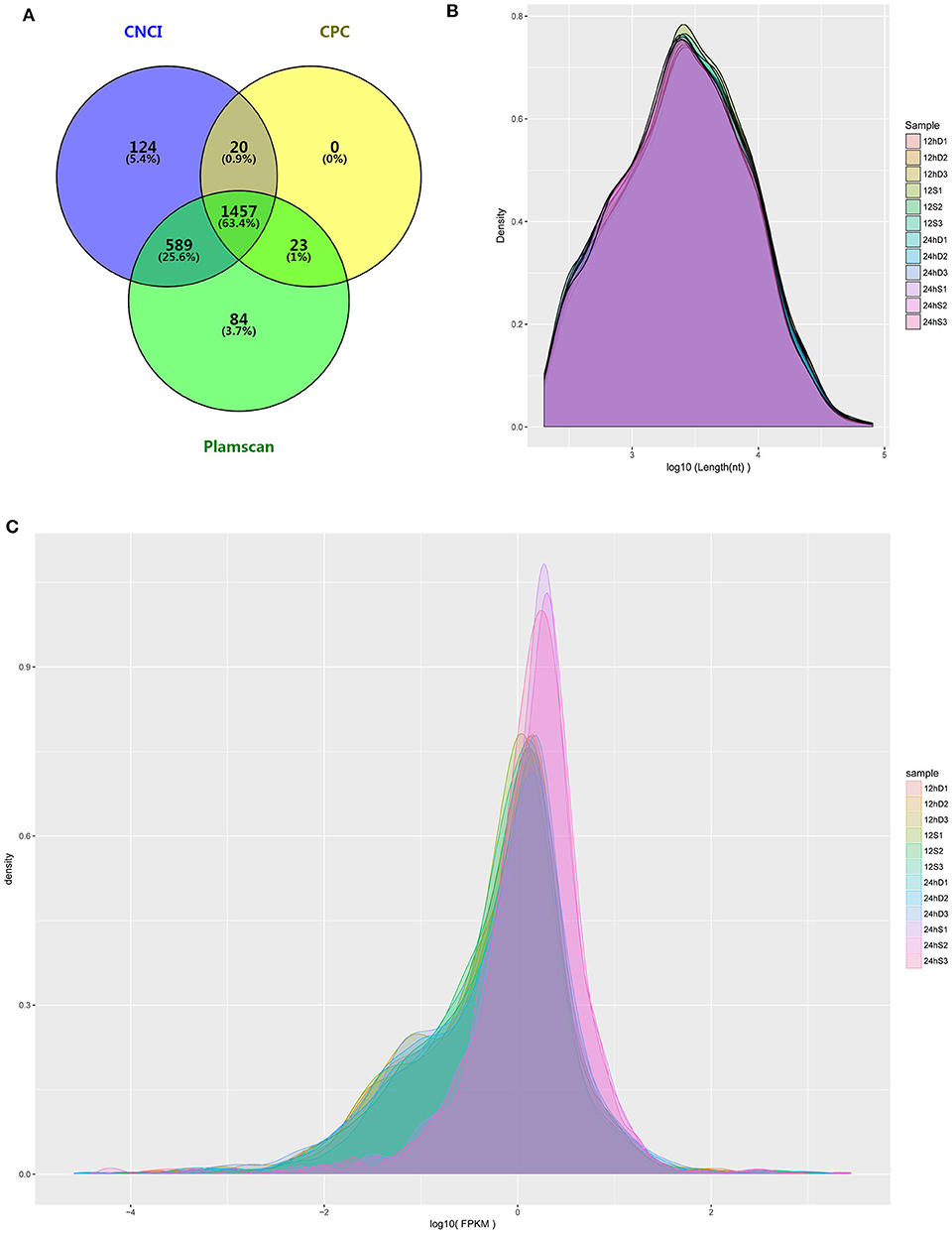
Figure 3. (A) The Venn diagrams showing the number of lncRNAs filtered by the three methods. (B) lncRNA length distribution. (C) lncRNA density distribution.
Differential Expression Analysis and Target Prediction
As is shown in Figure 4, 357 differently expressed lncRNAs were identified in DTMUV-infected DEFs compared with mock-infected DEFs, among which 34 lncRNAs were identified at 12 hpi with 21 up-regulated and 13 down-regulated (Figure 4A). Respectively, 339 lncRNAs were identified at 24 hpi with 317 up-regulated and 22 down-regulated (Figure 4B). The details of the information are included in Tables S3, S4. Notably, 16 lncRNAs expressed differently at both time points (Figure 5A). The differently expressed lncRNAs were then applied to a systematic cluster analysis. Obviously, lncRNA expression levels were significantly altered at 24 hpi, while the expression profile did not differ importantly at 12 hpi (Figure 5B). To better explore the regulatory function of differently expressed lncRNAs, cis target genes have been searched through location. The regulatory network is shown in Figure 6. The details of the information are included in Table S5.
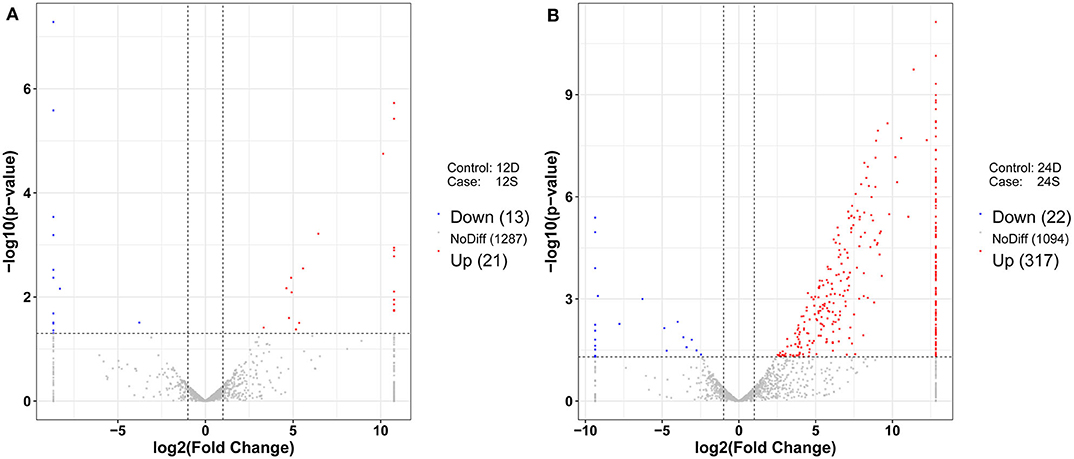
Figure 4. (A) Volcano chart of differently expressed lncRNAs at 12 hpi. (B) Volcano chart of differently expressed lncRNAs at 24 hpi. The x-axis shows the Log2 (fold change) and y-axis shows the –log10 (p-value). Red points represent the upregulated lncRNAs and green points represent the downregulated lncRNAs. The vertical line in the figure is a two-fold difference threshold, and the horizontal line is a p < 0.05 threshold.
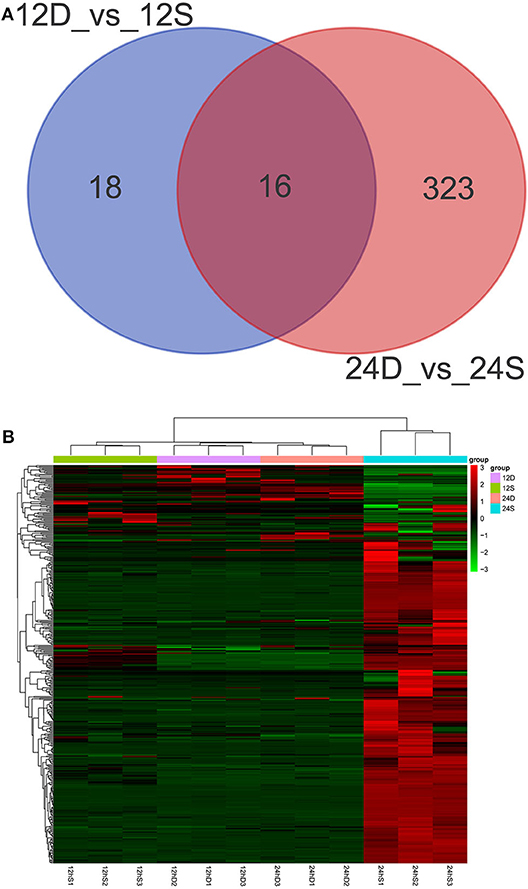
Figure 5. (A) Venn diagrams showing the numbers of differently expressed lncRNAs at 12 and 24 hpi. (B) The heat map showing the hierarchical clustering of altered lncRNAs. Red represents upregulation, and green represent downregulation.

Figure 6. The regulatory network of differently expressed lncRNAs and cis target genes. The blue dots represent differently expressed lncRNAs, and yellow dots represent target genes.
GO and KEGG Enrichment Analysis
To better understand the roles of differentially expressed lncRNAs in DTMUV-infected DEFs, GO and KEGG analyses were performed to explore the biological function. As is shown in Figure 7, the target genes were mainly related to biological regulation, cellular processes, single-organism processes, cell, cell part, and membrane at 12 hpi (Figure 7A). And organelle and binding were additionally enriched in at 24 hpi (Figure 7B). The details of the GO terms are included in Tables S6, S7. Besides, target genes were closely referred to the signal pathway categories of signal transduction, endocrine system, and cellular community at 12 hpi (Figure 8A). Additionally, immune system and signaling molecules and interaction were enriched in at 24 hpi (Figure 8B). The details of the KEGG terms are included in Tables S8, S9. The significantly enriched pathways are presented in Figure 9A at 12 hpi and Figure 9B at 24 hpi.
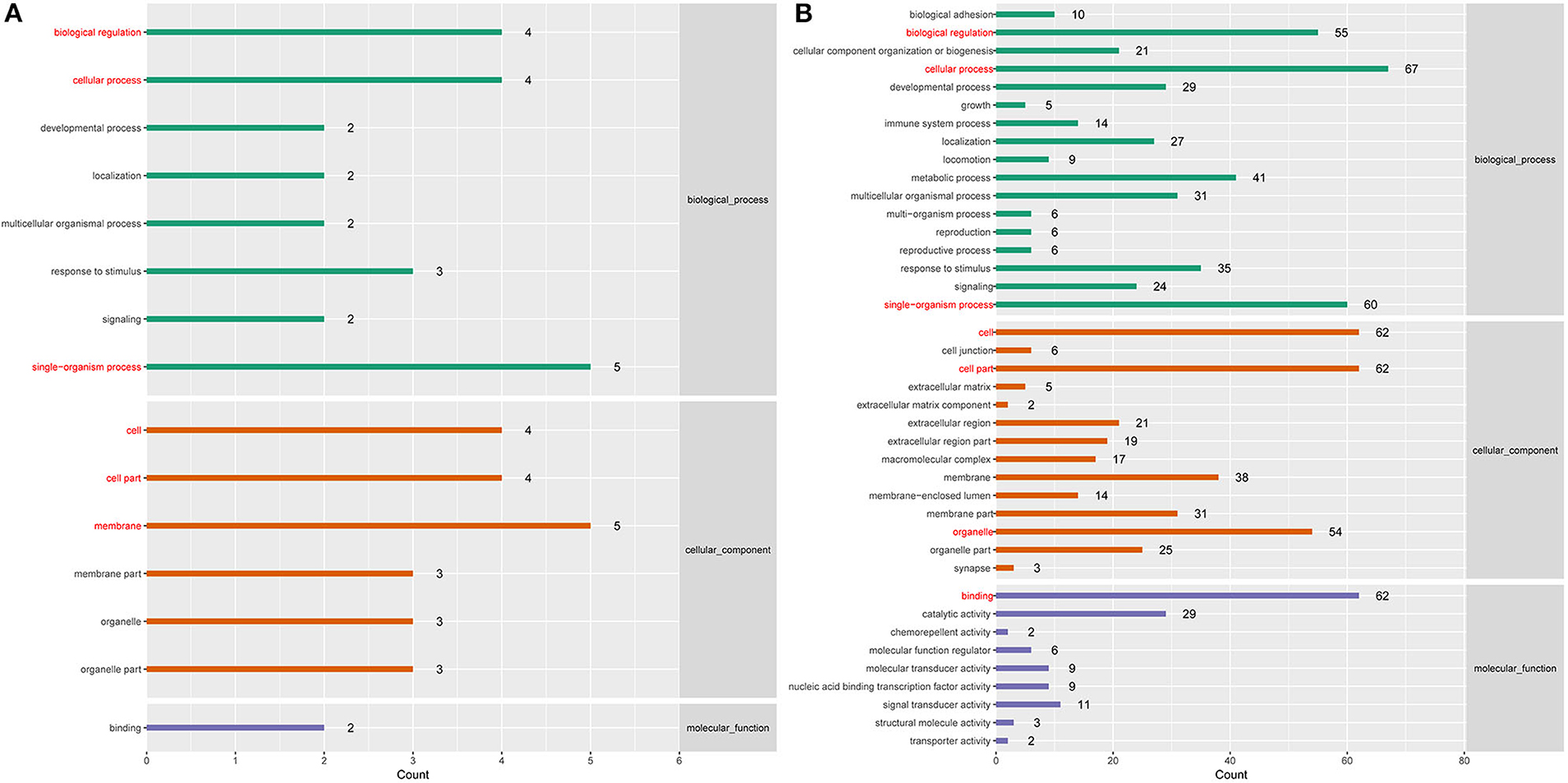
Figure 7. (A) GO enrichment analysis at 12 hpi. (B) GO enrichment analysis at 24 hpi. Each color represents a different biological process. The x-axis indicates the description and the y-axis indicates the –log10 (p-value).
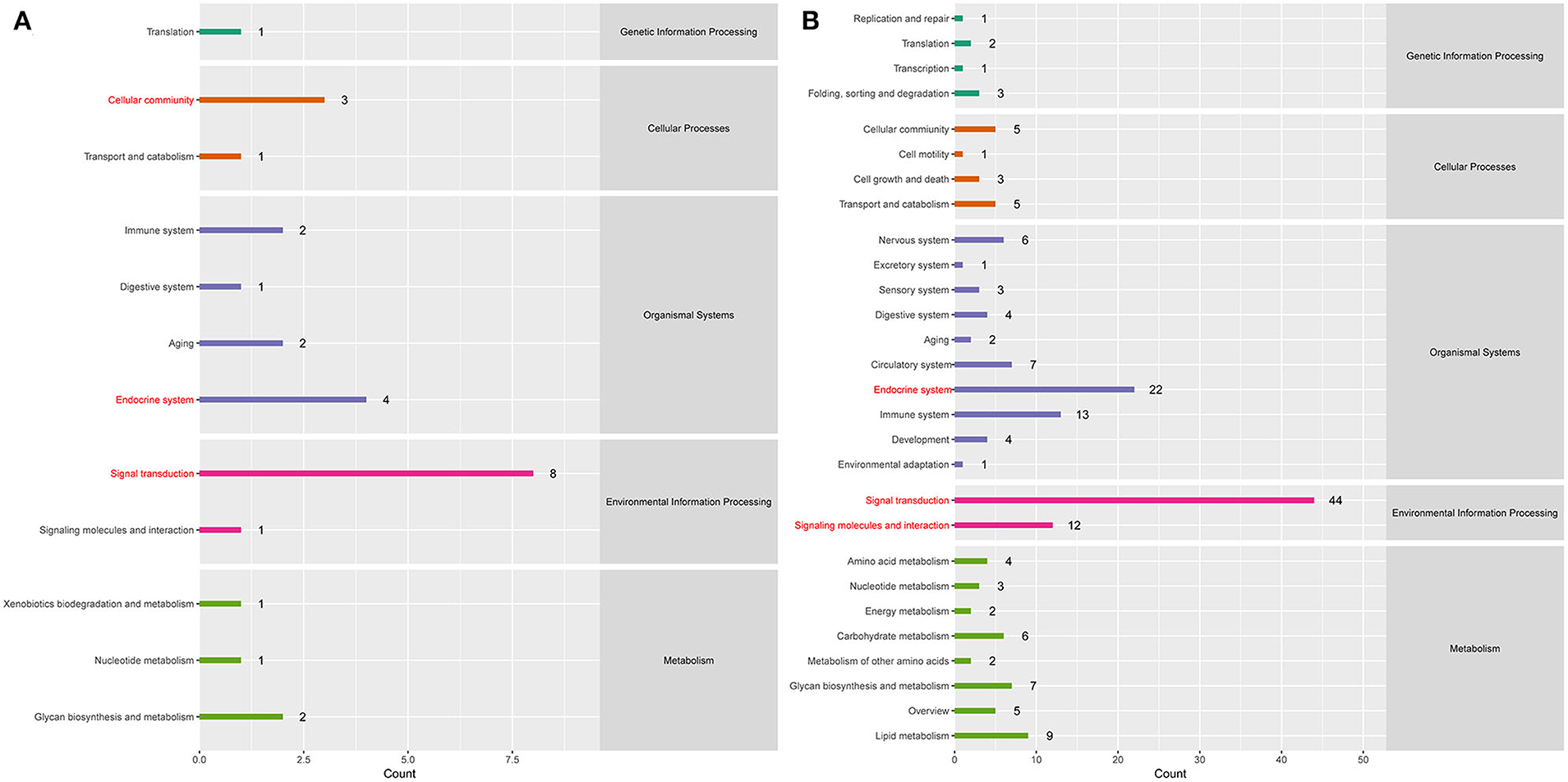
Figure 8. (A) KEGG pathway analysis at 12 hpi. (B) KEGG pathway analysis at 24 hpi. Each color represents a kind of signal pathway.
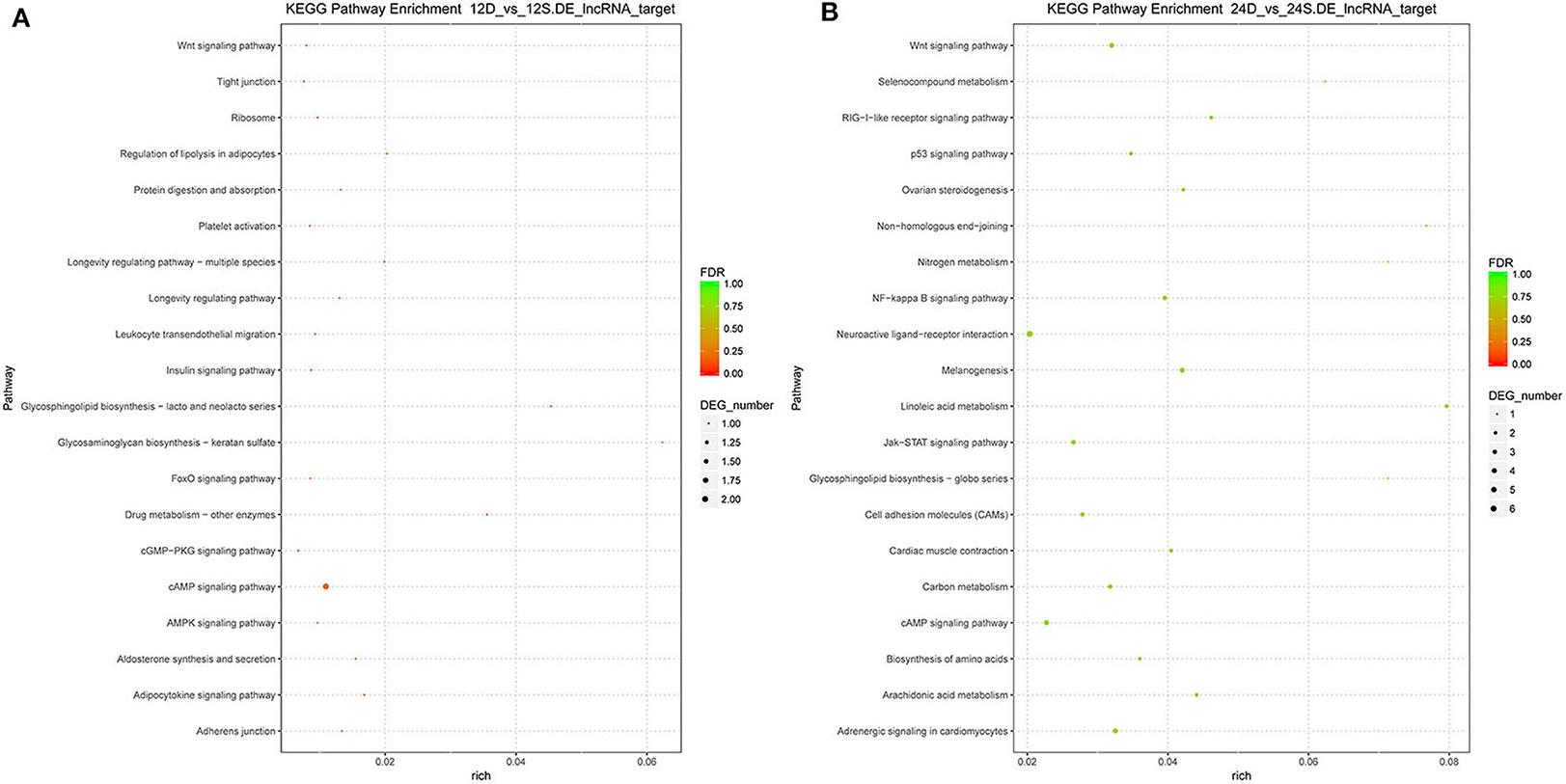
Figure 9. (A) The significantly enriched pathways at 12 hpi. (B) The significantly enriched pathways at 24 hpi. The x-axis and y-axis represent enrichment and pathway names, respectively. Point size represents the number of target genes.
Validation of Differentially Expressed lncRNAs by RT-qPCR
RT-qPCR was performed to further detect the expression changes of lncRNAs in RNA-Seq data. 4 and 11 lncRNAs at 12 and 24 hpi were validated by RT-qPCR. The results showed that expression changes confirmed by RT-qPCR were consistent with the RNA-Seq data (Figure 10). The details of the information are included in Table S10. Furthermore, the correlation analysis revealed that changes in lncRNA expression level were comparable between RNA-Seq and RT-qPCR, with the correlation coefficients of 0.8769 (P < 0.0001) (Figure 11). The results confirmed that the RNA-Seq data were relatively reliable and accurate.
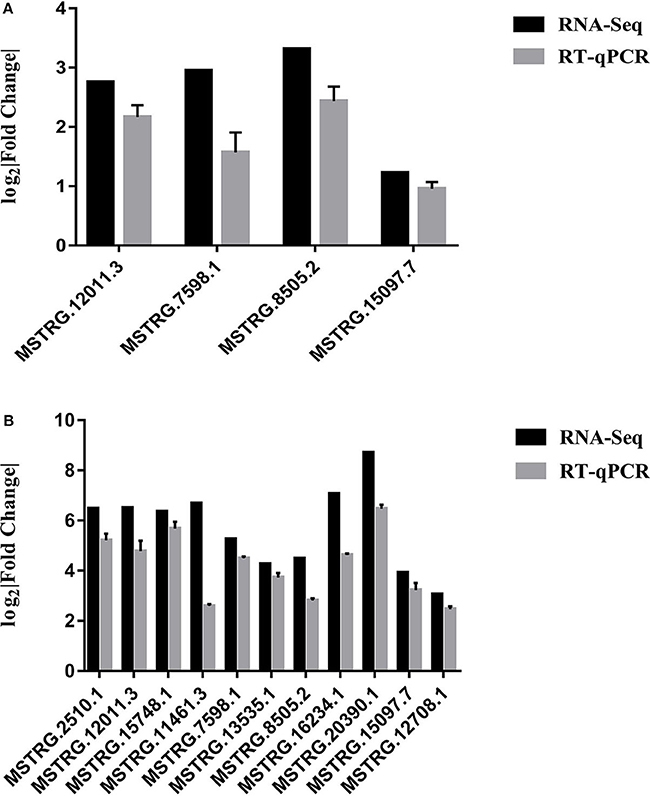
Figure 10. (A) Validation of differentially expressed lncRNAs by RT-qPCR at 12 hpi. (B) Validation of differentially expressed lncRNAs by RT-qPCR at 24 hpi. Each sample had three biological replicates.
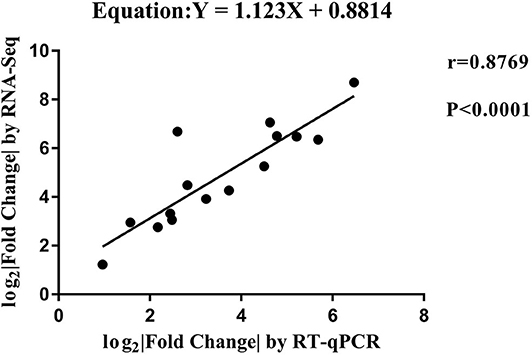
Figure 11. The correlation analyses of changes in lncRNA expression between RT-qPCR and RNA-Seq. The x-axis shows the Log2 (fold change) by RT-qPCR, and the y-axis is the Log2 (fold change) by RNA-Seq.
Discussion
DTMUV, an emerging member of flavivirus family, can cause acute anorexia, retarded growth, neurological dysfunction, and severe egg production drop, which results in large economic losses (41). To understand DTMUV infection deeply, the transcriptome and proteome in DTMUV-infected DEFs have been uncovered (8, 42, 43). However, little is known for lncRNA differential expression and the biological effect on gene regulation during DTMUV infection. In this study, expression changes of lncRNAs were investigated, potential regulatory network was formed, and biological function was predicted. The study provides significant insights into the deep exploration of pathogenic mechanism.
Accumulated studies demonstrated that viral infection can alter lncRNA expression profile of host (44), including infectious bronchitis virus infection of primary dendritic cells (45), H5N1 influenza viruses infection of ducks (46), and Porcine reproductive and respiratory syndrome virus infection of endometrial epithelial cells (47). In our study, the change of lncRNA expression profile was also observed in DEFs in response to DTMUV infection. To the best of our knowledge, this is the first report in which a complete lncRNA profile was provided in DTMUV-infected DEFs. The lncRNA profile showed that the expression levels of lncRNAs were specifically upregulated or downregulated in response to DTMUV infection.
Recent study showed that lncRNAs were able to cooperate with neighboring genes to perform cis regulatory function (48). For example, to control the lifespan of neutrophils, eosinophils and classical monocytes, lncRNA-Morrbid regulates the transcription of the neighboring pro-apoptotic gene, named Bcl2l11, by promoting the enrichment of the PRC2 complex at the Bcl2l11 promoter to maintain the gene in a poised state (49). Besides, LincRNA-p21, as a key modulator of gene expression in the p53 pathway, performs its cis control of p21 expression to influence the activation and chromatin state of hundreds of downstream genes (50). In our study, the cis target genes were searched and the cis regulatory network was formed.
Here, we performed GO and KEGG enrichment analysis to predict the biological function. Notably, many cis target genes were strongly associated with metabolism, such as lipid metabolism, glycan biosynthesis and metabolism, garbohydrate metabolism, and amino acid metabolism. To date, several studies have suggested that viruses regulate host metabolism to facilitate replication (51, 52). DTMUV also induced profound metabolic alterations in DEFs (8, 43). Emerging report showed that lncRNA regulates metabolic enzymes to regulate virus replication. The lncRNA ACOD1, induced in cells infected with various viruses, binds the metabolic enzyme glutamic-oxaloacetic transaminase and increases its catalytic activity to facilitate the production of metabolites that promote viral propagation (53). The findings suggested that differently expressed lncRNAs may regulate metabolism to affect the pathogenicity of DTMUV.
In addition to metabolism, the signal pathway categories of immune system were enriched in, such as RIG-I-like receptor signaling pathway. The genome-wide transcriptome analyses of DTMUV-infected macrophages revealed that the inductions of alpha interferon and beta interferon were blocked on transcription and translation levels in response to viral infection, despite the activation of major pattern recognition receptor signaling (54). Deep study showed that DTMUV non-structural protein 1 interacts with the adaptor protein mitochondrial antiviral signaling to inhibit the mitochondrial antiviral signaling pathway, resulting in the impaired induction of beta interferon (55). In addition, the non-structural protein 2B cleaves STING to inhibit interferon signaling (56). Recently, an increasing number of lncRNAs have been reported to play negative roles in innate immune response to virus infections. For example, lnc-Lsm3b, a mouse specific type I interferon-induced lncRNA, competes to bind RIG-I monomers with viral RNAs to prevent conformational changes of RIG-I and activation of its downstream signaling (57). NRAV, a down-regulated lncRNA in human epithelial cells upon influenza A virus infection, promotes virus replication by negatively regulating the transcription of multiple interferon-stimulated genes (58). Therefore, lncRNAs may participate in the invasion and infection of DTMUV through affecting the innate immunity.
Conclusions
In summary, we were the first to perform a comprehensive analysis of lncRNA expression profile in DEFs following DTMUV infection using RNA-Seq. We screened out numerous differently expressed lncRNAs, formed cis regulatory network, and conducted biological function analysis. Our results suggested that lncRNAs may participate in DTMUV-induced pathogenesis through affecting the metabolism and innate immunity of host cells, which provides a deeper insight into the pathogenic mechanism of DTMUV. Future investigations will be required to discover specific pathogenic mechanism and to identify novel and efficient strategies for DTMUV infection.
Data Availability Statement
The datasets generated for this study can be found in the Sequence Read Archive of the National Center for Biotechnology Information (accession number SRP150572).
Ethics Statement
The animal study was reviewed and approved by Committee on the Ethics of Animal of Shandong (permit number 20,156,681).
Author Contributions
YL and JY completed the experiments, analyzed the results, and drafted the work with the help of DH, XL, and JL. YT and YD provided the materials, designed the experiments, and reviewed the manuscript. All authors have read and approved the manuscript for publication.
Funding
This research was funded by the National Key Research and Development Program of China (2016YFD0500107-2, 2018YFD0501506); the National Natural Science Foundation of China (31872500); the Natural Science Foundation of Shandong Province (ZR2019YQ17); Youth Innovation and Technology Support Project of Shandong Provincial Colleges and Universities (2019KJF022); the China Agriculture Research System (CARS-42-19).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01729/full#supplementary-material
References
1. Su J, Li S, Hu XX, Yu X, Wang Y, Liu P, et al. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS ONE. (2011) 6:e18106. doi: 10.1371/journal.pone.0018106
2. Wang Y, Yuan X, Li Y, Yu K, Yang J, Xu H, et al. Rapid detection of newly isolated Tembusu-related Flavivirus by reverse-transcription loop-mediated isothermal amplification assay. Virol J. (2011) 8:553–9. doi: 10.1186/1743-422X-8-553
3. Yan P, Zhao Y, Zhang X, Xu D, Dai X, Teng Q, et al. An infectious disease of ducks caused by a newly emerged Tembusu virus strain in mainland China. Virology. (2011) 417:1–8. doi: 10.1016/j.virol.2011.06.003
4. Cao Z, Zhang C, Liu Y, Ye W, Han J, Ma G, et al. Tembusu virus in ducks, China. Emerg Infect Dis. (2011) 17:1873–5. doi: 10.3201/eid1710.101890
5. Yu G, Lin Y, Tang Y, Diao Y. Evolution of tembusu virus in ducks, chickens, geese, sparrows, and mosquitoes in Northern China. Viruses. (2018) 10:485–93. doi: 10.3390/v10090485
6. Thivalai C, Lertwatcharasarakul P, Jala S, Phattanakunanan S, Chakritbudsabong W, Saengnual P, et al. Studies on the pathogenicity of duck tembusu virus strain KPS54A61 using mice and chickens. Jpn J Vet Res. (2019) 171:238–41. doi: 10.14943/jjvr.67.4.295
7. Liu P, Lu H, Li S, Moureau G, Deng YQ, Wang Y, et al. Genomic and antigenic characterization of the newly emerging Chinese duck egg-drop syndrome flavivirus: Genomic comparison with Tembusu and Sitiawan viruses. J Gen Virol. (2012) 93(Pt 10):2158–70. doi: 10.1099/vir.0.043554-0
8. Yu G, Lin Y, Tang Y, Diao Y. Comparative transcriptomic analysis of immune-related gene expression in duck embryo fibroblasts following duck tembusu virus infection. Int J Mol Sci. (2018) 19:2328–56. doi: 10.3390/ijms19082328
9. Tang Y, Gao X, Diao Y, Feng Q, Chen H, Liu X, et al. Tembusu Virus in Human, China. Transbound Emerg Dis. (2013) 60:193–6. doi: 10.1111/tbed.12085
10. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. (2007) 14:103–5. doi: 10.1038/nsmb0207-103
11. Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell. (2019) 177:639–53. doi: 10.1016/j.cell.2019.03.008
12. Xu X, Wang K, Zha X. An antisense lncRNA functions in alternative splicing of Bmdsx in the silkworm, Bombyx mori. Biochem Biophys Res Commun. (2019) 516:639–44. doi: 10.1016/j.bbrc.2019.06.107
13. D'Angelo D, Mussnich P, Sepe R, Raia M, del Vecchio L, Cappabianca P, et al. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J Mol Med. (2019) 97:1019–32. doi: 10.1007/s00109-019-01789-7
14. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. (2017) 3:eaao2110. doi: 10.1126/sciadv.aao2110
15. Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
16. Liu S, Liu X, Li J, Zhou H, Carr MJ, Zhang Z, et al. Long noncoding RNAs: Novel regulators of virus-host interactions. Rev Med Virol. (2019) 29:e2046. doi: 10.1002/rmv.2046
17. Xin XL, He D, Wren D, Wen ZY, Zhong SJ. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. (2010) 11:472–92. doi: 10.1186/1471-2164-11-472
18. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. (2008) 320:1344–9. doi: 10.1126/science.1158441
19. Yin F, Gao Q, Tang B, Sun P, Han K, Huang W. Transcriptome and analysis on the complement and coagulation cascades pathway of large yellow croaker (Larimichthys crocea) to ciliate ectoparasite Cryptocaryon irritans infection. Fish Shellfish Immunol. (2016) 50:127–41. doi: 10.1016/j.fsi.2016.01.022
20. Chen G, He L, Luo L, Huang R, Liao L, Li Y, et al. Transcriptomics sequencing provides insights into understanding the mechanism of grass carp reovirus infection. Int J Mol Sci. (2018) 19:488–503. doi: 10.3390/ijms19020488
21. Zhao P, Liu S, Zhong Z, Jiang T, Weng R, Xie M, et al. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci Rep. (2018) 8:11858. doi: 10.1038/s41598-018-30359-z
22. Zhang Y, Yu T, Ding Y, Li Y, Lei J, Hu B, et al. Analysis of expression profiles of long noncoding RNAs and mRNAs in A549 cells infected with H3N2 swine influenza virus by RNA Sequencing. Virol Sin. (2019) 35:171–80. doi: 10.1007/s12250-019-00170-9
23. Li X, Shi Y, Liu Q, Wang Y, Li G, Teng Q, et al. Airborne transmission of a novel tembusu virus in ducks. J Clin Microbiol. (2015) 53:2734–6. doi: 10.1128/jcm.00770-15
24. Ti J, Li Z, Li X, Lu Y, Diao Y, Li F. Identification of one B-cell epitope from NS1 protein of duck Tembusu virus with monoclonal antibodies. PLoS ONE. (2017) 12:e0181177. doi: 10.1371/journal.pone.0181177
25. Lv C, Li R, Liu X, Li N, Liu S. Pathogenicity comparison of duck Tembusu virus in different aged Cherry Valley breeding ducks. BMC Vet Res. (2019) 15:282–98. doi: 10.1186/s12917-019-2020-8
26. Liang T, Liu X, Qu S, Lv J, Yang L, Zhang D. Pathogenicity of egg-type duck-origin isolate of Tembusu virus in Pekin ducklings. BMC Vet Res. (2019) 15:362–70. doi: 10.1186/s12917-019-2136-x
27. Zhou X, Zhang T, Song D, Huang T, Peng Q, Chen Y, et al. Whole-genome sequence of duck tembusu virus strain DTMUV/CH/2014, isolated in China. Genome Announc. (2016) 4:e01657–15. doi: 10.1128/genomea.01657-15
28. Tang Y, Yeh YT, Chen H, Yu C, Gao X, Diao Y. Comparison of four molecular assays for the detection of Tembusu virus. Avian Pathol. (2015) 44:379–85. doi: 10.1080/03079457.2015.1061650
29. Zhou Q, Bi Z, Yin D, Gu X, Xu Z, Huang R, et al. Development and application of an indirect ELISA for the serological detection of duck Tembusu virus infection based on the NS1 protein antigen. Arch Virol. (2019) 165:709–14. doi: 10.1007/s00705-019-04495-4
30. Deng J, Liu Y, Jia R, Wang M, Chen S, Zhu D, et al. Development of an immunochromatographic strip for detection of antibodies against duck Tembusu virus. J Virol Methods. (2017) 249:137–42. doi: 10.1016/j.jviromet.2017.08.022
31. He D, Zhang X, Chen L, Tang Y, Diao Y. Development of an attenuated live vaccine candidate of duck Tembusu virus strain. Vet Microbiol. (2019) 231:218–25. doi: 10.1016/j.vetmic.2019.03.022
32. Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. (2009) 25:1105–11. doi: 10.1093/bioinformatics/btp120
33. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. (2013) 14:R36. doi: 10.1186/gb-2013-14-4-r36
34. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. (2015) 33:290–5. doi: 10.1038/nbt.3122
35. Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. (2013) 41:e166. doi: 10.1093/nar/gkt646
36. Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. (2007) 35(Web Server issue):W345–9 doi: 10.1093/nar/gkm391
37. Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. (2016) 44:D279–85. doi: 10.1093/nar/gkv1344
38. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. (2010) 11:R106. doi: 10.1186/gb-2010-11-10-r106
39. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
40. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[−ΔΔ C(T)] method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
41. Li N, Lv C, Yue R, Shi Y, Wei L, Chai T, et al. Effect of age on the pathogenesis of duck tembusu virus in Cherry Valley ducks. Front Microbiol. (2015) 6:581–9. doi: 10.3389/fmicb.2015.00581
42. Zhang J, Huang Y, Li L, Dong J, Liao M, Sun M. Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain. Int J Mol Sci. (2020) 21:2402–18. doi: 10.3390/ijms21072402
43. Hu F, Li Y, Yu K, Huang B, Ma X, Liu C, et al. ITRAQ-based quantitative proteomics reveals the proteome profiles of primary duck embryo fibroblast cells infected with duck tembusu virus. Biomed Res Int. (2019) 2019:1–14. doi: 10.1155/2019/1582709
44. Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, et al. Unique signatures Of long noncoding RNA expression in response to virus infection And altered innate immune signaling. MBio. (2010) 1:e00206–10. doi: 10.1128/mBio.00206-10
45. Lin J, Wang Z, Wang J, Yang Q. Microarray analysis of infectious bronchitis virus infection of chicken primary dendritic cells. BMC Genomics. (2019) 20:557–70. doi: 10.1186/s12864-019-5940-6
46. Lu C, Xing Y, Cai H, Shi Y, Liu J, Huang Y. Identification and analysis of long non-coding RNAs in response to H5N1 influenza viruses in duck (Anas platyrhynchos). BMC Genomics. (2019) 20:36–45. doi: 10.1186/s12864-018-5422-2
47. Zhang K, Ge L, Dong S, Liu Y, Wang D, Zhou C, et al. Global miRNA, lncRNA, and mRNA transcriptome profiling of endometrial epithelial cells reveals genes related to porcine reproductive failure caused by porcine reproductive and respiratory syndrome virus. Front Immunol. (2019) 10:1221. doi: 10.3389/fimmu.2019.01221
48. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. (2020) 21:102–17. doi: 10.1038/s41576-019-0184-5
49. Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. (2016) 537:239–43. doi: 10.1038/nature19346
50. Dimitrova N, Zamudio J, Jong R, Soukup D, Resnick R, Sarma K, et al. LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. (2014) 54:777–90. doi: 10.1016/j.molcel.2014.04.025
51. Vijayan M, Hahm B. Influenza viral manipulation of sphingolipid metabolism and signaling to modulate host defense system. Scientifica (Cairo). (2014) 2014:793815. doi: 10.1155/2014/793815
52. Gualdoni GA, Mayer KA, Kapsch AM, Kreuzberg K, Puck A, Kienzl P, et al. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc Natl Acad Sci USA. (2018) 115:e7158–65. doi: 10.1073/pnas.1800525115
53. Wang P, Xu J, Wang Y, Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. (2017) 358:1051–5. doi: 10.1126/science.aao0409
54. Ma Y, Liang Y, Wang N, Cui L, Chen Z, Wu H, et al. Avian flavivirus infection of monocytes/macrophages by extensive subversion of host antiviral innate immune responses. J Virol. (2019) 93:e00978–19. doi: 10.1128/jvi.00978-19
55. Wang J, Lei C-Q, Ji Y, Zhou H, Ren Y, Peng Q, et al. Duck tembusu virus nonstructural protein 1 antagonizes ifn-β signaling pathways by targeting VISA. J Immunol. (2016) 197:4704–13. doi: 10.4049/jimmunol.1502317
56. Wu Z, Zhang W, Wu Y, Wang T, Wu S, Wang M, et al. Binding of the duck tembusu virus protease to STING is mediated by NS2B and is crucial for STING cleavage and for impaired induction of IFN-β. J Immunol. (2019) 203:3374–85. doi: 10.4049/jimmunol.1900956
57. Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. (2018) 173:906–19. doi: 10.1016/j.cell.2018.03.064
Keywords: DTMUV infection, DEFs, lncRNA expression, gene regulation, RNA-Seq
Citation: Lin Y, Yang J, He D, Li X, Li J, Tang Y and Diao Y (2020) Differently Expression Analysis and Function Prediction of Long Non-coding RNAs in Duck Embryo Fibroblast Cells Infected by Duck Tembusu Virus. Front. Immunol. 11:1729. doi: 10.3389/fimmu.2020.01729
Received: 11 September 2019; Accepted: 29 June 2020;
Published: 04 August 2020.
Edited by:
Xiaofeng Yang, Lewis Katz School of Medicine at Temple University, United StatesReviewed by:
Xiao-Feng Li, Beijing Institute of Microbiology and Epidemiology, ChinaYafeng Li, Shanxi Provincial People's Hospital, China
Copyright © 2020 Lin, Yang, He, Li, Li, Tang and Diao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Tang, tyck288@163.com; Youxiang Diao, yxdiao@126.com
†These authors have contributed equally to this work
 Yun Lin
Yun Lin Jing Yang
Jing Yang Dalin He
Dalin He Xudong Li
Xudong Li Jing Li
Jing Li Yi Tang
Yi Tang Youxiang Diao
Youxiang Diao