- 1Department of Animal Sciences, The Ohio State University, Columbus, OH, USA
- 2Department of Animal Sciences, Ohio Agriculture Research and Development Center, Wooster, OH, USA
Poultry litter is a mixture of bedding materials and enteric bacteria excreted by chickens, and it is typically reused for multiple growth cycles in commercial broiler production. Thus, bacteria can be transmitted from one growth cycle to the next via litter. However, it remains poorly understood how litter reuse affects development and composition of chicken gut microbiota. In this study, the effect of litter reuse on the microbiota in litter and in chicken gut was investigated using 2 litter management regimens: fresh vs. reused litter. Samples of ileal mucosa and cecal digesta were collected from young chicks (10 days of age) and mature birds (35 days of age). Based on analysis using DGGE and pyrosequencing of bacterial 16S rRNA gene amplicons, the microbiota of both the ileal mucosa and the cecal contents was affected by both litter management regimen and age of birds. Faecalibacterium, Oscillospira, Butyricicoccus, and one unclassified candidate genus closely related to Ruminococcus were most predominant in the cecal samples, while Lactobacillus was predominant in the ileal samples at both ages and in the cecal samples collected at day 10. At days 10 and 35, 8 and 3 genera, respectively, in the cecal luminal microbiota differed significantly in relative abundance between the 2 litter management regimens. Compared to the fresh litter, reused litter increased predominance of halotolerant/alkaliphilic bacteria and Faecalibacterium prausnitzii, a butyrate-producing gut bacterium. This study suggests that litter management regimens affect the chicken GI microbiota, which may impact the host nutritional status and intestinal health.
Introduction
The gastrointestinal (GI) tract of chickens harbors a complex microbiota that plays an essential role in nutrient digestion and absorption, immune system development, and pathogen exclusion (Yeoman et al., 2012; Pan and Yu, 2014). Previous studies have demonstrated that diet (Jia et al., 2009; Hammons et al., 2010) and feed additives (Amerah et al., 2011; Danzeisen et al., 2011; Rodriguez et al., 2012) can impact the chicken GI microbiota with respect to diversity, composition, and structure. Understandably, most of the previous studies focused on how feed and feed additives affect the prevalence of enteric pathogens, such as Salmonella (Santos et al., 2008; Peinado et al., 2012), Clostridium perfringens (Si et al., 2009; Wei et al., 2013a), and Campylobacter jejuni (Chinivasagam et al., 2010; Ridley et al., 2011). However, prevalence of these pathogens and the risk of associated diseases can be lowered by a healthy GI microbiota through colonization resistance and competitive exclusion (Wagner, 2006; Callaway et al., 2008; Kerr et al., 2013). Some studies have also suggested that certain commensal bacteria can positively affect the efficiency of feed utilization by broiler chickens (Stanley et al., 2012, 2013). These studies have advanced our understanding on how diet, feed additives, and antimicrobial growth promoters (AGP) modulate the GI microbiota of chickens (Wise and Siragusa, 2007; Gong et al., 2008; Santos et al., 2008; Danzeisen et al., 2011). Similar to what is observed in mammals, the GI microbiota of chickens develops in the early stage of life (particularly within the first 2 weeks). When young chicks are delivered from the hatchery to a chicken house (typically at the age of 1–2 days), their initial GI microbiota is very simple containing a very small number of bacteria belonging to a few species (Fonseca et al., 2011; Cox et al., 2012; Hiett et al., 2013). After being placed in commercial chicken houses where litter serves as the bedding material, chicks are exposed to several sources of bacteria that can gain entry into the immature gut. These exogenous sources of bacteria include litter materials, feed, water, and ambient air. Because there is little colonization resistance in the young GI tract, many bacteria can readily colonize therein. As young chicks grow, their GI microbiota undergoes a series of temporal successions (van der Wielen et al., 2002; Lu et al., 2003a) and becomes increasingly diverse and complex (Wei et al., 2013b). Beginning from approximately day one, chicks begin pecking at and consuming litter materials, inoculating their young GI tract with bacteria present in the litter. Therefore, litter can have a significant effect on the development process of GI microbiota and its eventual composition and structure in chickens (Garrido et al., 2004; Torok et al., 2009).
Commercial broiler production involves fairly short growth cycles (about 6–7 weeks per growth cycle). Each growth cycle begins with placement of young chicks at 1 day of age at a high density (< 0.1 m2) in chicken houses with litter as bedding materials on the floor. Poultry litter is a mixture of bedding materials (e.g., pine shavings) and chicken excreta that contain chicken GI bacteria, undigested feed, uric acid, and other substances of host origin. Several studies have documented that poultry litter contains a complex and dynamic microbiota, composed primarily of GI and environmental bacteria depending on the litter management regimens (Lu et al., 2003b; Lovanh et al., 2007). The composition and structure of litter microbiota can be affected by the bedding materials used (Torok et al., 2009). In the US, broiler chicken litter (primarily pine shavings) is commonly reused for 6 or more consecutive growth cycles before a thorough cleanout to reduce the cost of fresh litter materials and disposal of reused litter (Coufal et al., 2006). Repeated use of poultry litter results in considerable changes in the chemical and microbiological conditions of the litter, and poor litter management can lead to increased litter moisture with concomitant increases in ammonia, pH, and increased density and diversity of microbes (primarily bacteria) (Omeira et al., 2006; Cressman et al., 2010). Changes in the litter microbiota brought about by repeated litter reuse can serve as a driving force that shapes the chicken GI microbiota (Cressman et al., 2010) because exposure of young chicks to different bacterial inocula can profoundly affect GI microbiota development (Yin et al., 2010). In addition, reused litter was shown to harbor less Salmonella (Roll et al., 2011) and Clostridium perfringens (Wei et al., 2013a) but to enable Campylobacter jejuni and C. coli to survive longer compared to fresh litter (Kassem et al., 2010). Moreover, 2 recent studies have shown that reused litter can affect the immune system of chickens (Lee et al., 2011; Shanmugasundaram et al., 2012), which suggests that litter conditions can also affect the GI microbiota of chickens indirectly through their immune system.
Numerous studies have examined the microbiota in the GI tract of chickens or in poultry litter. However, only one study examined the microbiota both in the GI tract and in the poultry litter with an focus on the interaction between the 2 microbiotas (Cressman et al., 2010). That study revealed that the litter microbiota and the GI microbiota affected each other in a reciprocal manner and fresh litter resulted in increased diversity and predominance of environmental bacteria in the GI microbiota of young chicks, while reused litter increased the bacteria of gut origin (Cressman et al., 2010). While that study pioneered a new area of research, only a limited number of bacteria were identified because it used sequencing of clone libraries of 16S rRNA gene amplicons. Therefore, the extent to which the microbiotas of litter and of GI tract of chickens affect each other remains to be determined. The objective of the present study was to further investigate the reciprocal effect of the litter and GI microbiotas of broiler chickens using pyrosequencing. The results could be useful in understanding the relationship between the litter and gut microbiotas of chickens as it relates to improving the health and well-being of chickens through litter management.
Materials and Methods
Litter Management
The experiment was conducted over 6 consecutive growing cycles with each cycle lasting 6 weeks. There was a 2-week down period between 2 consecutive cycles. Broiler chicks at 1 day of age were purchased from a commercial hatchery, and 50 chicks were randomly placed in each of 16 floor pens (4.73 m2 floor area per pen) with pine shavings as litter. This placement density, 0.095 m2 per chicks, is similar to that at commercial chicken houses. The pens were allocated into 1 of 2 management groups. For one group (8 pens), the litter was cleaned out after each growth cycle, and fresh pine shavings were placed prior to the arrival of new chicks for the next growth cycle (referred to as fresh litter, FL). For the reused litter (RL) group (8 pens), the litter was piled up in the center of each pen at the end of each growth cycle and stored for 10 days (without mixing or turning) before being redistributed within the same pen, as it is commonly practiced in broiler houses in the U.S. About 5 cm of fresh pine shavings were “top-dressed” or added on top of the used litter 2–3 days prior to the arrival of new chicks for the next growth cycle. Six growth cycles were performed for both the FL and the RL groups in parallel so that RL was generated and differential seasonal impact on FL and RL was avoided. All pens were physically separated by 24-inch-high plastic barriers to prevent litter contamination between pens. Within each pen, there was a 0.28 m2 concrete floor section without litter near the entry to the pen so that daily watering and feeding management could be performed without stepping onto the litter. Water was provided by bell-shaped poultry Plasson drinkers, and feed was provided by trough-type feeders. Disposable shoe covers were used before entering each pen to minimize potential cross contamination between pens. All the birds were fed the same commercial type of corn-soybean meal-based diet that met the nutrient levels recommended by NRC (NRC, 1994). The chicks/chickens were cared and handled following the animal use protocols approved specifically for this study by The Ohio State University Institutional Animal Care and Use Committee.
Sample Collection
Samples of Ileal mucosa and cecal digesta were collected from 4 randomly selected chickens from each pen at days 10 and 35 of the 6th growth cycle as described previously (Cressman et al., 2010). The 2 samples days were chosen to represent young and mature broiler chickens. Briefly, the ileum between the Meckles diverticulum and the ileocecal junction was removed. After the digesta was flushed out with sterile buffered saline, the ileal mucosa was scraped off using sterilized microscope slides. The cecal luminal content was squeezed out of the cecum. Both the ileal mucosa and the cecal luminal content were collected from each of the sampled birds. At the end of 6th growth cycle, 6 litter samples were also collected from each pen from the area around the drinkers and feeders and along the side of each pen. The litter samples from each pen were thoroughly mixed using a blender to reduce heterogeneity. All the samples were stored at −80°C until further analysis. The ileal mucosa was chosen because it is the interface between the host and the small intestinal bacteria.
DNA Extraction, PCR, and DGGE Analysis
The samples of ileal mucosa and of cecal content from the 4 sampled chickens of each pen at each age (day 10 and day 35) were pooled based on the same wet weight. Metagenomic DNA was extracted from each pen-based composite GI sample and from each pen-based composite litter sample using the repeated bead beating and column purification method (Yu and Morrison, 2004b). The V3 region of 16S rRNA gene was PCR amplified using bacteria-specific primers (357F: CCT ACG GGA GGC AGC AG and 518R: ATT ACC GCG GCT GCT GG) with the forward primer having a 40 bp GC clamp attached to its 5′ end (Yu and Morrison, 2004a). The confirmed amplicons were subsequently analyzed using DGGE with a 40–60% denaturing gradient as described previously (Yu and Morrison, 2004a). The DGGE profiles were analyzed using BioNumerics (V.5.1; Applied Maths, Inc., Austin, TX). The DGGE banding patterns were transformed into a binary (presence and absence of bands) correlation cross-product matrix and then subjected to principle component analysis (PCA) using the PC-ORD software (V.5.0, MJM Software, Gleneden Beach, OR) as described previously (Cressman et al., 2010).
Pyrosequencing and Data Analysis
Preparation of amplicon libraries and pyrosequencing were done at the Research and Testing Laboratories (Lubbock, TX) as described previously (Kim and Yu, 2014). Briefly, amplicon libraries of the V1–V3 region was prepared using the primers Gray28F (5′-GAGTTTGATCNTGGCTCAG-3′) and Gray-519R (5′-GTNTTACNGCGGCKGCTG-3′). The amplicons were sequenced when pyrosequencing was the only next-generation sequencing technology that could produce about 500 bp reads. The 8 pen-based composite samples within each litter management regimen were pooled (the same DNA quantity from each DNA sample) into one litter type-based composite sample for the same sample type (ileal mucosa, cecal content, and litter) to reduce cost, increase depth coverage, and obtain an “average” appraisal of the microbiota of each sampling location at each age. Each composite sample (it either represented ileal or cecal samples from 32 chickens or represented 48 litter samples), one unique barcode was added between the primers and the adaptors A and B that are required by the Roche 454 FLX Titanium system. The pyrosequencing data were processed using the Qiime pipeline (Caporaso et al., 2010) for denoising, removal of chimeric sequences, and quality checking as described previously (Kim and Yu, 2014), except more stringent criteria at the split_library.py step (--min_seq_length 200, --max_seq_length 600, --min_qual_score 25, maximum number of ambiguous bases 6, maximum length of homopolymer run 6, maximum number of primer mismatches 1, maximum number of errors in barcode 0, sliding window test of quality scores 50). The quality-checked sequences were aligned against the Greengenes core set Gg_13_5_99, and the sequences that failed to align with the reference sequence set were excluded from further analysis. The aligned sequences were grouped into operational taxonomic units (OTU) using de novo OTU picking at a distance of 0.04, which allows similar clustering of species-equivalent OTU based on the V1–V3 region as a distance 0.03 based on full-length of 16S rRNA genes (Kim et al., 2011). The same number of sequences was used for each sample type (8470 sequences for the ileal mucosa samples, 3078 sequences for the cecal content samples, and 4752 sequences for the litter samples) to avoid impact from different numbers of sequences from different samples. The OTUs were classified to species or a higher taxon using the RDP Naïve Bayesian Classifier (Wang et al., 2007) implemented in Qiime against the default taxonomy file (gg_13_5_otus/taxonomy/97_otu_taxonomy.txt) and the reference sequence file (gg_13_5_otus/rep_set/97_otus.fasta) at the default confidence level (80%). Pair-wise comparison between the 2 litter management regimens was performed using the Library Compare function at RDP with a significant level of p < 0.001. Library Compare was used because it estimates the likelihood that individual taxa differ in frequency between 2 libraries using a statistical test that can compare transcript levels in “digital Northern” analysis (Audic and Claverie, 1997). Good's estimate of coverage was calculated for each sample using Qiime.
The distributions of the OTUs obtained were visualized using the heatmap and clustering method implemented in the software GAP (http://gap.stat.sinica.edu.tw/Software/GAP/) as described previously (Li et al., 2014, 2015). Briefly, the abundance (number of sequences) of these OTUs was first log transformed for normalization. Pearson's correlation coefficients were calculated to examine the community similarity among the samples. Hierarchical clustering trees were generated using the rank-two ellipse seriation method (Chen, 2002; Wu et al., 2010) to grouping of the microbiotas.
Data Availability
The pyrosequencing data are available in the MG-RAST database (http://metagenomics.anl.gov/) under the project ID #9131.
Results
DGGE Profiles of the Microbiota
All samples produced many DGGE bands (data not shown). PCA analysis of the DGGE profiles grouped the samples based on sampling types (litter vs. GI) or locations (ileal mucosa vs. cecal digesta), litter conditions (fresh vs. reused), and age (day 10 vs. day 35) (Figure 1). As shown with superposed sample symbols, some samples with each sample type (8 samples per sample type) had identical DGGE profiles. Small variance was explained by PC1 (28.6% of total variation) and PC2 (15.8%), reflecting relatively small differences in microbiota among the 8 pens of each sample type. For both the ileal mucosal and the cecal digesta samples, greater variability among replicate pens within each litter management regimen was noted at day 35 than at day 10.
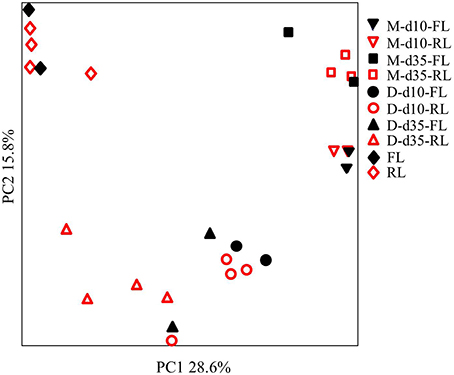
Figure 1. Principal component analysis (PCA) plot of the DGGE profiling of the microbiome. M, ileal mucosa; D, cecal digesta; d10 and d35, samples collected at 10 and 35 days, respectively, of bird age; FL, fresh litter; RL, reused litter. Some samples were superposed due to identical DGGE profiles.
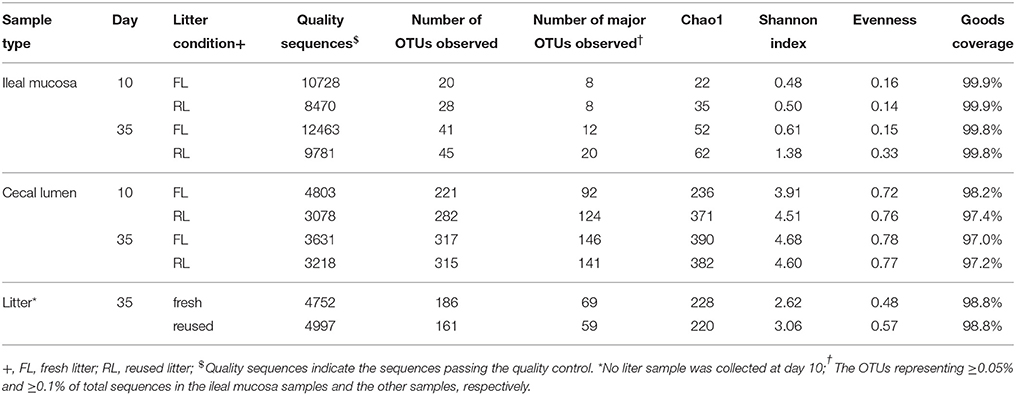
Alpha Diversity of Individual Samples
In total, 147,670 quality-checked sequences were obtained and subjected to subsequent analysis. Estimate of Good's coverage reached >97.8% for all the samples, with higher coverage being achieved for the ileal mucosa samples (Table 1). The ileal mucosal bacterial community had much fewer species-equivalent OTUs, and a few of them dominated, resulting in low Shannon diversity index and evenness index. Compared to the cecal digesta samples, the litter samples had lower OTU richness. The phylum Firmicutes was dominant in all the samples at both ages. However, in the ileal mucosal samples, Lactobacillales was the most predominant order, while in the cecal digesta samples Clostridiales was most predominant (Supplementary Figures S1, S2). In the litter samples, the orders Actinomycetales, Bacillales, and Lactobacillales were more predominant than any other order (Supplementary Figure S3). More taxonomic orders of bacteria were also noted in the cecal and in the litter samples than in the ileal mucosal samples. More bacterial orders were identified in the fresh litter than in the reused litter.
Beta Diversity Among Different Samples
The microbiotas were compared based on unweighted UniFrac analysis of the pyrosequencing data, and the comparison was visualized using a tree based on the unweighted pair group method with arithmetic mean (UPGMA). Similar to the comparison based on PCR-DGGE profiles, the pyrosequencing data also showed that the microbiotas were clustered in accordance with sample types, sampling locations, and the ages of birds (Figure 2). The fresh litter and the reused litter resulted in bifurcation of microbiota irrespective of sample type, sampling location, or age.
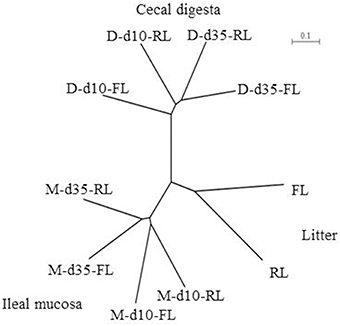
Figure 2. Unweighted pair group method with arithmetic mean (UPGMA) tree based on a β-diversity distance matrix calculated using unweighted UniFrac metrics of the pyrosequencing data. M, illeal mucosa; D, cecal digesta; d10 and d35, samples collected at 10 and 35 days, respectively, of bird age; FL, fresh litter; RL, reused litter.
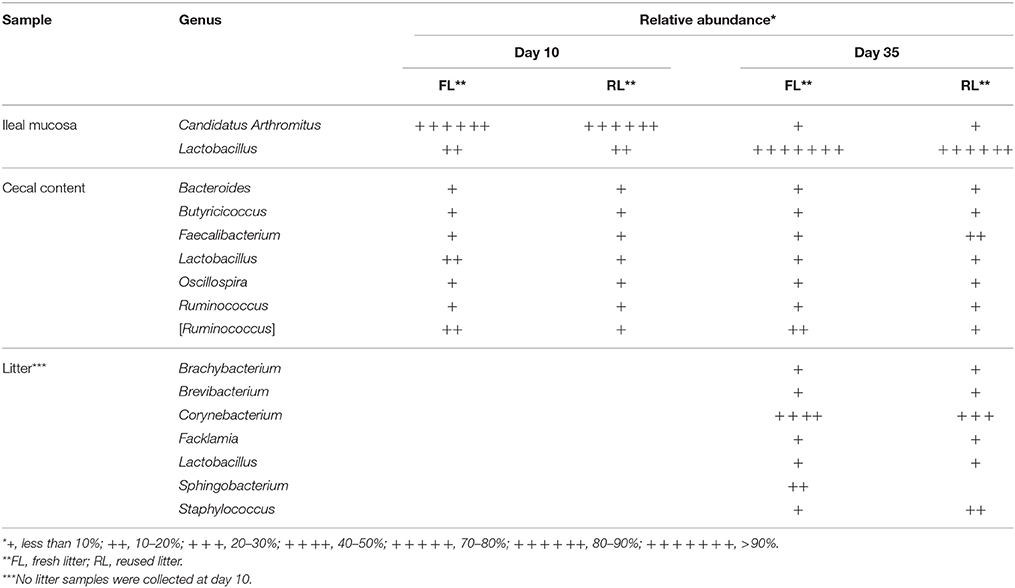
Major Taxa Found in the Samples
Numerous genera were found in the cecal digesta and the litter samples (Supplementary Table S1). Interestingly, Candidatus Arthromitus, a new proposed genus of segmented filamentous bacteria (Thompson et al., 2012), was the most predominant in the ileal mucosa at day 10 but gave way to Lactobacillus at day 35 (Table 2). The genera Faecalibacterium, Butyricicoccus, and an undefined candidate genus closely related to Ruminococcus (referred to [Ruminococcus] as used in the RDP database) were most prevalent in the cecal samples (Supplementary Table S1). The most predominant genus in both the fresh and the reused litters was Corynebacterium, while Staphylococcus was also predominant in the reused litter. Most members of these genera are either aerobic or facultatively anaerobic.
The relative abundance of the major genera was visualized using heatmap (Figure 3) and their occurrence was listed in Supplementary Table S2 (the Genera spreadsheet). Consistent with the UniFrac analysis (Figure 2), the microbiotas were grouped, based on Pearson's coefficients (Figure 3B), primarily by sample types and to a lesser extent by litter management regimens. Based on distribution patterns, these genera were clustered into 5 groups (Figure 3D). Group L1 contained 8 genera with Sphingobacterium being most predominant, and they were found only in the fresh litter, while Group L2a (11 genera) was shared by both the fresh and the reused litter samples (Supplementary Table S2). Group L2b only had Dialister and Oceanobacillus, and it was only found in the reused litter. Overall, many genera found in the litter samples were not common or predominant GI bacteria. Group M contained Enterococcus, Lactobacillus, and Candidatus Arthromitus, and Candidatus Arthromitus was most predominant in the ileal mucosa at day 10. Group D1 contained 3 genera found only in the cecal digesta collected at day 35 from the chickens reared on the fresh litter (referred to as fresh-litter chickens). Group D2 contained 3 subgroups that were found almost exclusively in the cecal digesta samples. Group D2a (only 1 genus) was found in the cecal digesta collected at day 35 from the chickens reared on the reused litter (referred to as reused-litter chickens). Group D2b (12 genera) was shared by all the cecal digesta samples, while Group D2c (1 genus) was found in the cecal digesta samples of the fresh-litter chickens collected at day 10. All the genera found in the cecal digesta were common and/or predominant GI bacteria, such as Ruminococcus, Clostridium, Blautia, Bacteroides, and Faecalibacterium.
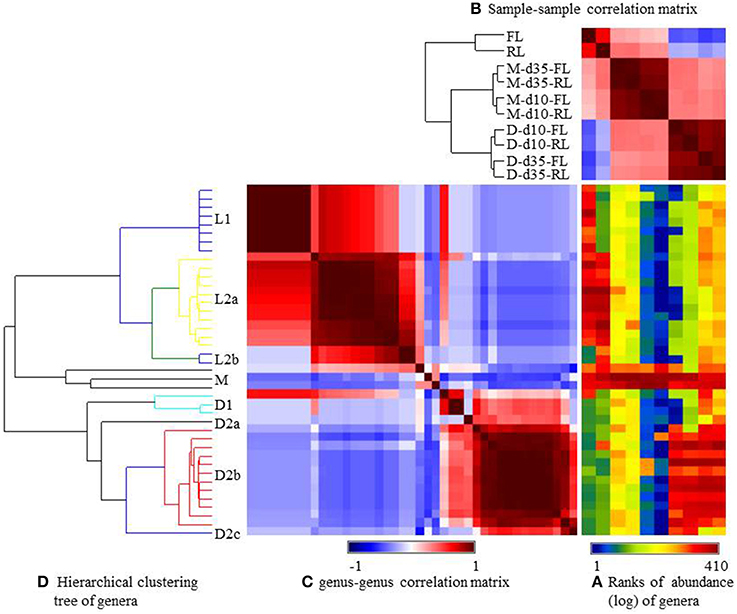
Figure 3. Generalized association plots for the major genera identified in the 10 datasets. (A) Genus abundance heatmap, (B) The sample-sample correlation map, (C) The genus-genus correlation map, and (D) The hierarchical clustering tree for sorting the genus-genus correlation map in (C). The 10 datasets and the genera were sorted based on corresponding sample-sample correlation and genus-genus correlation, respectively. M, illeal mucosa; D, cecal digesta; d10 and d35, samples collected at 10 and 35 days, respectively, of bird age; FL, fresh litter; RL, reused litter.
To identify the cecal bacterial genera that significantly differed between the 2 litter management regimens, the sequences representing known genera were compared between the 2 litter management regimens using the RDP Library Compare function, which can detect differentially represented taxa between 2 samples (Cole et al., 2009). Numerous genera differed in relative abundance between the 2 litter management regimens, including Lactobacillus, the Escherichia/Shigella group, Bacteroides, Subdoligranulum, and Clostridium XIVb for the litter samples, and Blautia, Faecalibacterium, and Anaerotruncus for the cecal digesta samples (Table 3). Overall, more cecal bacterial genera differed in relative abundance at day 10 than at day 35. Faecalibacterium and Oscillibacter were more predominant in cecum of the reused-litter chickens at day 35, while Subdoligranulum was more predominant in the fresh-litter chickens at day 10. The reused litter increased the relative abundance of Enterococcus but decreased that of Lactobacillus in the ileal mucosa samples. The 2 litter conditions also affected the relative abundance of some genera of bacteria in the litter samples, particularly Corynebacterium, Facklamia, Escherichia/Shigella, which were more predominant in the fresh litter, and Yaniella, Staphylococcus, Brevibacterium, Salinicoccus, which were more predominant in the reused litter.
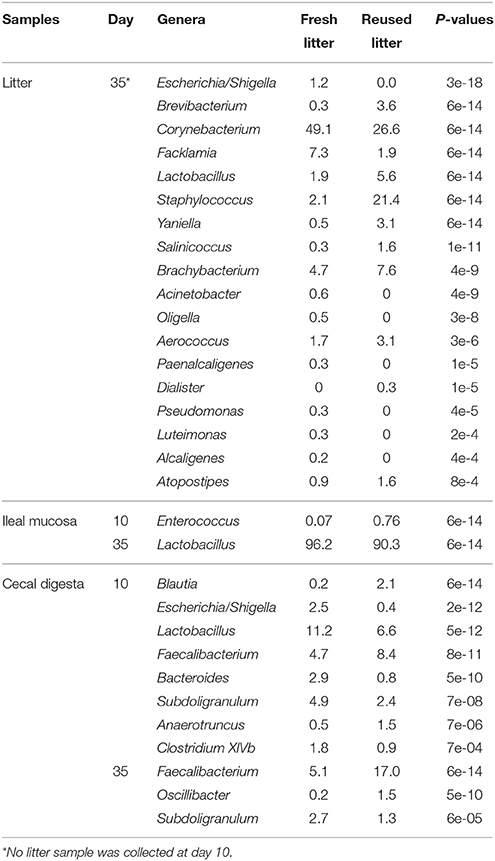
Table 3. Genera that differed in relative abundance (% of total sequences) between the fresh and the reused litter conditions.
The relative abundance of the major OTUs was visualized using heatmap (Figure 4) and their occurrence was listed in Supplementary Table S2 (the OTUs spreadsheet). Similar as at genus level, the microbiotas was mostly influenced by sample types and to a lesser extent by litter management regimens (Figure 3B). Three large groups of OTUs were found, each corresponding to a sample type: cecal digesta (designated as D), litter (designated as L), and ileal mucosa (designated as M) (Figure 4D). Group D1a contained 25 OTUs, including 17 being found only in the reused-litter chickens at day 10 and 8 also being found in the fresh-litter chickens at day 35 (Supplementary Table S2). Containing 34 OTUs, Group D1b was found only in the fresh-litter chickens at day 35, except 7 and 10 OTUs that were also found in the reused-litter chickens at day 35 and in the fresh-litter chickens at day 10, respectively. Group D1c contained the most number of OTUs (85 in total), and most of them were shared by all the cecal digesta samples. Consisting of 44 OTUs, Group D1d was found in the reused-litter chickens at day 35, with some OTUs being also found in the fresh-litter chickens at day 35. Group D2 contained 20 OTUs, which were found primarily in the fresh-litter chickens at day 10. The genus Ruminococcus, order Clostridiales, and family Lachnospiraceae were represented by the most numbers of OTUs of the cecal digesta.
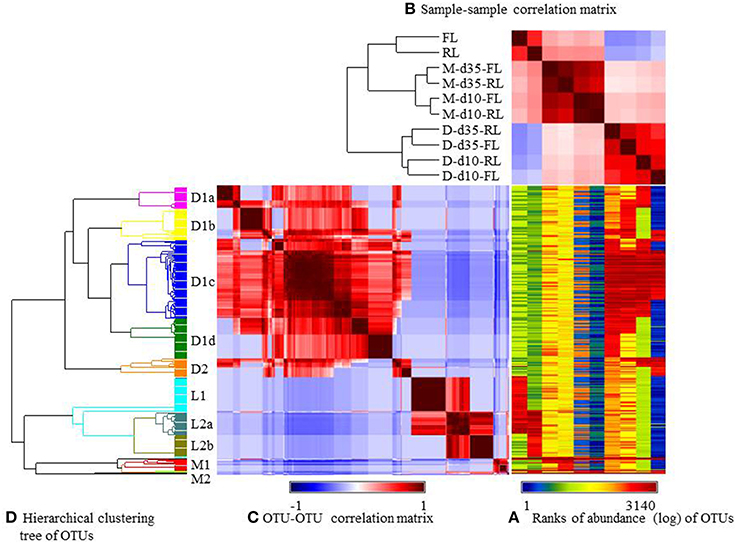
Figure 4. Generalized association plots for the major OTUs identified in the 10 datasets. (A) OUT abundance heatmap, (B) The sample-sample correlation map, (C) The OTU-OTU correlation map, and (D) The hierarchical clustering tree for sorting the OTU-OTU correlation map in (C). The 10 datasets and the OTUs were sorted based on corresponding sample-sample correlation and OUT-OUT correlation, respectively. M, illeal mucosa; D, cecal digesta; d10 and d35, samples collected at 10 and 35 days, respectively, of bird age; FL, fresh litter; RL, reused litter.
The OTUs found in the litter samples had distinct distribution patterns, with Group L1 (37 OTUs) being exclusively found in the fresh litter, Group L2b (25 OTUs) exclusively in the reused litter, and Group L2a (26 OTUs) shared by both types of litter. The family Sphingobacteriaceae and the genera Trichococcus, Acinetobacter, Corynebacterium, Leucobacter, and Facklamia were the largest taxa represented by the Group L1 OTUs. The Group L2a OTUs were primarily assigned to the genera Staphylococcus and Brachybacterium and the family Bacillaceae, while those of Group L2b were mainly assigned to Bacillaceae, Actinomycetales, and Corynebacterium. Group M1 contained many OTUs assigned to Lactobacillus, while Group M2 contained only 2 OTUs assigned to Enterococcus and Candidatus Arthromitus. Three OTUs assigned to Lactobacillus and one OTU of Candidatus Arthromitus in these 2 OTU groups were found in all the samples, while the remaining OTUs were mostly found in the ileal mucosal samples.
The representative sequences of the 314 major OTUs (representing >0.05% and >0.1% of total sequences in at least one ileal mucosal sample and one cecal or litter sample, respectively) were classified using the Greengenes taxonomy database included in Qiime. In total, 133 OTUs were assigned to species within 41 genera, including 35 assigned to 19 known species within 15 genera. Amongst these species, 1, 4, 2, and 1 were assigned to Bacteroides, Lactobacillus, Staphylococcus, and Faecalibacterium, respectively. Most of these species, including Bacteroides fragilis, Butyricicoccus pullicaecorum, F. prausnitzii, L. salivarius, L. vaginalis, Staphylococcus equorum, and S. sciun, differed significantly in relative abundance between the 2 litter management regimens (Figure 5). Of the identified species, L. salivarius is the most dominant in the ileal mucosa samples, which increased from about 0.1% of total sequence reads at day 10 to >10% at day 35, suggesting temporal shift of mucosal bacteria as chickens grew. Although, the order Clostridiales was dominant in the cecal digesta samples (Supplementary Figure S2), no known species was predominant except F. prausnitzii and B. pullicaecorum, indicating greater diversity of unknown species in the cecal digesta than in the ileal mucosa.
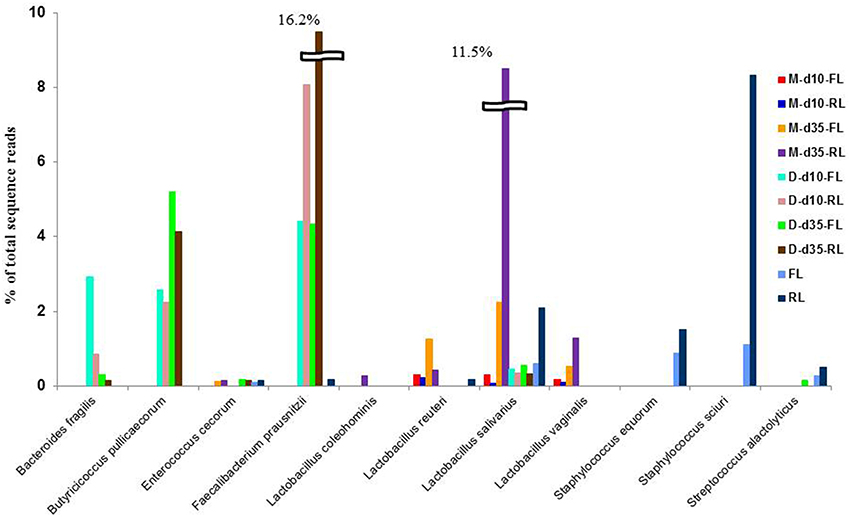
Figure 5. Prevalence of some predominant bacterial species in the samples. M, illeal mucosa; D, cecal digesta; d10 and d35, samples collected at 10 and 35 days, respectively, of bird age; FL, fresh littler; RL, reused litter.
Discussion
Understanding the reciprocal impact between litter microbiota and chicken GI microbiota is important to guide proper management of poultry litter and bird health, especially as antibiotic growth promoters will be phased out in the US. This study for the first time used 16S rRNA gene-based metagenomic analysis in examining such reciprocal impact in a comprehensively manner. Consistent with a previous study (Cressman et al., 2010), fresh litter and reused litter differed in microbiota, but the present study revealed much greater bacterial diversity (21 vs. 2 genera) and more detailed differences in microbiota composition between the two litter management regimens. Five of the 8 major genera that were found only in the fresh litter (Group L1 genus) belong to Proteobacteria. Although, the predominance of Proteobacteria and occurrence of Pseudomonas (not a major genus) was also reported in the fresh litter in the previous study (Cressman et al., 2010), the present study documented a greater diversity of environmental bacteria in the fresh litter. Interestingly, Oceanobacillus and Dialister, which are alkaliphilic (Heyrman and Vos, 2015) and bile-tolerant (Wade, 2015), respectively, were found only in the reused litter, and Salinicoccus, another halophilic genus (Ventosa, 2015), was found more predominant (near by 1 log) in the reused than in the fresh litter. These results suggest that the reused litter might have increased salt content due to repeated reuse and composting. Although the litter samples were not chemically analyzed and salt content in poultry litter has not been reported in the literature, salt can accumulate in reused litter, especially when reused litter is composted as done in the present study. It should be noted, the litter in the previous study was sampled after 2 years, while in the present study the litter was sampled after about 1 year reuse. The length of litter reuse might probably have affected the effect of litter management regimens.
Achieving coverage >97%, this study identified nearly all the bacteria present in the samples. A close examination of the lineage of the major OTUs showed that although some genera were shared between the fresh and the reused litter, some OTUs were found only in one of the two litter types. These results suggest impact of the litter management regimens on litter microbiota at species level. The fresh litter contained bacteria that are not commonly found in GI tract but in other environments, such as the OTUs (Group L1 OTUs) assigned to Acinetobacter, Devosia, Luteimonas, Trichococcus, and Yaniella. These bacteria were probably derived from the fresh litter material or acquired during the growth cycles, and they were not competitive in the reused litter. On the other hand, some OTUs (Group L2b OTUs) were only found in the reused liter and they are members of halo- or bile-tolerant genera (e.g., Salinicoccus, Oceanobacillus, and Dialister). These OTUs might represent bacteria adapted to the reused litter. Among the OTUs (Group L2a) that were shared between the two types of litter, some represent common genera found in GI tract, such as Enterococcus, Streptococcus, Facklamia, and Brachybacterium, while others represent genera that were also found in fresh litter (Group L1 OTUs). These latter bacteria might be from the initial litter material and/or the environment but had become adapted to the reused litter conditions. Nevertheless, Salmonella, Clostridium perfringens, or Campylobacter jejuni was not detected in the fresh or the reused litter. This concurs with the similar mortality rates between the 2 litter management regimens (data not shown). Future studies using qPCR are needed to determine if litter management regimens can significantly affect the prevalence and abundance of these enteric pathogens.
This study is congruent with several previous studies with respect to revelation of the major groups of GI bacteria, such as Lactobacillus in ileal mucosa and Clostridia in cecal lumen (Gong et al., 2002, 2007; Lu et al., 2003b; Choi et al., 2014). The predominance of Facklamia, Salinicoccus, and Corynebacterium also corroborates the finding of a previous study (Lu et al., 2003a). However, this study revealed impact of litter management (fresh vs. reused) on the microbiota in both ileal mucosa and cecal digesta. First, no genus was only found in the ileal mucosa corresponding to either litter type. However, the litter management regimens affected the predominance of Enterococcus and Lactobacillus, with the reused litter favoring Enterococcus in the ileal mucosa at day 10 but Lactobacillus at day 35 (Table 3), suggesting that reused litter may serve as a source of Enterococcus for young chicks. Second, Lactobacillus and Candidatus Arthromitus showed opposite temporal trends in predominance between the 2 ages irrespective of the litter management regimens, with the former being more predominant at day 35, while the latter more predominant at day 10. Candidatus Arthromitus was first proposed by Snel et al. (1995) to include segmented filamentous bacteria that have been reported in trout, mice, and rats. It has been reported 3 times in chickens, once in cecal content (Snel et al., 1995) and twice in mucosa (Gong et al., 2007). The Candidatus Arthromitus sequences we found and those identified in the gut of chicken, turkey, and rat (Snel et al., 1995; Gong et al., 2007; Danzeisen et al., 2013) are very similar. Having close contact with gut epithelial wall (Thompson et al., 2012), Candidatus Arthromitus was thought to modulate host immune response (Bolotin et al., 2014), but there is also a conflicting report (Thompson et al., 2013). Future studies are warranted to verify this dynamic trend and the potential biological importance to host.
No litter-specific genus was found, suggesting minor impact of the litter management on the bacteria present in ileal mucosa at genus level. However, OTUs specific to age or litter types were found, including 4 OTUs only found at day 35 and 7 OTUs found only at 35 in reused-liter chickens, and most of these OTUs belong to Lactobacillus (Supplementary Table S2). It is also of interest to note that the reused litter corresponded to more OTUs in the ileal mucosa than the fresh litter at day 35. These results suggest that litter management regimens can affect some of the ileal mucosal bacteria residing in an age-dependent manner. The association between Lactobacillus and chicken performance has been mixed. In one study, one bacterial OTU distantly related to L. crispatus was negatively associated with feed conversion efficiency (Stanley et al., 2012), but in another study, 2 OTUs related to L. coleohominis were positively associated with feed conversion efficiency (Stanley et al., 2013). In a third study (Torok et al., 2011), bacterial phylotypes related to L. salivarius, L. aviarius, and L. crispatus were found to be associated with decreased bird performance. Because litter management can affects occurrence of Lactobacillus, the indirect effect of litter management through effect on Lactobacillus warrants further research.
In the cecal digesta samples, the reused litter resulted in greater species richness than the fresh litter at day 10 but not at day 35. In addition, more genera in the cecal digesta differed in relative abundance at day 10 than at day 35 (Table 3, Figure 3). These results suggest that litter management regimens can have more profound impact to the GI microbiota of young chicks than to that of mature birds. The decrease in the litter effect with age might best be explained by the increasing species richness, and thus colonization resistance (Spees et al., 2013), in mature birds. It may also be a reflection of the accumulation of fecal bacteria in the fresh litter at day 35. Blautia, Faecalibacterium, and Anaerotruncus, all of which are common fecal bacteria, were more predominant in the cecal digesta of the reused-litter young chickens, suggesting that reused litter may expedite bacterial colonization of GI tract, and thus colonization resistance, in young chicks. On the other hand, the fresh litter resulted in more Escherichia/Shigella, Lactobacillus, Bacteroides, and Subdoligranulum in the cecal digesta (Table 3). These genera were probably less competitive in reused litter than in fresh litter. The previous study also noted decreased litter effect on the microbiota of cecal digesta than that of the ileal mucosa (Cressman et al., 2010). Given that the ileum is upstream of the cecum and that the ileum has lower microbial diversity than the cecum, this reduction in the litter effect along the GI tract is expected.
Greater litter effect on the cecal bacterial community was noted at species-equivalent OTU level than at genus level. Most of these OTUs were assigned to Ruminococcaceae, Lachnospiraceae, and Clostridiales. Although some of these OTUs were shared between the 2 litter management regimens (e.g., most of the Group D1c OTUs), many of them were found only at one of the 2 ages or corresponded to only one of the 2 litter types (Figure 3, Supplementary Table S2). Because most of the OTUs were not assigned to known species genera, their differential occurrence with respect to either age or litter type cannot be explained.
Butyrate is anti-inflammatory (Van Immerseel et al., 2010; Celasco et al., 2014), and some of the identified cecal bacteria were assigned to known butyrate-producing genera, such as Faecalibacterium, Subdoligranulum, and Butyricicoccus (Louis and Flint, 2009). Faecalibacterium was more predominant in the cecal digesta of reused-litter chickens, while Subdoligranulum showed the opposite trend at both ages (Table 3). A recent study showed that Faecalibacterium and Subdoligranulum constituted a single major group of cecal bacteria in conventionally reared chickens (Lund et al., 2010). F. prausnitzii has been reported to be anti-inflammatory in humans (Sokol et al., 2008), but it is not known if members of Subdoligranulum are anti-inflammatory. Because reused litter can induce inflammatory response in the intestine of chickens (Shanmugasundaram et al., 2012), future research is needed to determine if these butyrate producers contribute to the immune response induced by reared litter.
Collectively, halotolerant/alkaliphilic bacteria tended to increase in reused litter. Ileal mucosal bacterial community was affected more profoundly than cecal luminal bacterial community. Litter management regimens also had greater impact on gut bacterial community in young chicks than in mature birds. Some butyrate-producing bacteria and Lactobacillus were affected by litter management regimens, potentially affecting host health and feed conversion efficiency. Concurrent chemical analysis of litter material and bird performance is needed in future studies to determine how litter management affects bird health and performance.
Author Contributions
ML, ZY conceived the study and designed the experiments. LW did the data analysis and wrote the paper. ZY, ML revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This material is based upon work that was partially supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2008-35204-18845.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00593
References
Amerah, A. M., Péron, A., Zaefarian, F., and Ravindran, V. (2011). Influence of whole wheat inclusion and a blend of essential oils on the performance, nutrient utilisation, digestive tract development and ileal microbiota profile of broiler chickens. Br. Poult. Sci. 52, 124–132. doi: 10.1080/00071668.2010.548791
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Bolotin, A., de Wouters, T., Schnupf, P., Bouchier, C., Loux, V., Rhimi, M., et al. (2014). Genome sequence of “Candidatus Arthromitus” sp. strain SFB-mouse-NL, a commensal bacterium with a key role in postnatal maturation of gut immune functions. Genome Announ. 2:e00705-14. doi: 10.1128/genomeA.00705-14
Callaway, T. R., Edrington, T. S., Anderson, R. C., Harvey, R. B., Genovese, K. J., Kennedy, C. N., et al. (2008). Probiotics, prebiotics and competitive exclusion for prophylaxis against bacterial disease. Anim. Health Res. Rev. 9, 217–225. doi: 10.1017/S1466252308001540
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Celasco, G., Moro, L., Aiello, C., Mangano, K., Milasi, A., Quattrocchi, C., et al. (2014). Calcium butyrate: anti-inflammatory effect on experimental colitis in rats and antitumor properties. Biomed. Reports 2, 559–563. doi: 10.3892/br.2014.273
Chen, C.-H. (2002). Generalized association plots: information visualization via iteratively generated correlation matrices. Statist. Sinica 12, 7–29.
Chinivasagam, H. N., Redding, M., Runge, G., and Blackall, P. J. (2010). Presence and incidence of food-borne pathogens in Australian chicken litter. Br. Poult. Sci. 51, 311–318. doi: 10.1080/00071668.2010.499424
Choi, J. H., Kim, G. B., and Cha, C. J. (2014). Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult. Sci. 93, 1942–1950. doi: 10.3382/ps.2014-03974
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–145. doi: 10.1093/nar/gkn879
Coufal, C. D., Chavez, C., Niemeyer, P. R., and Carey, J. B. (2006). Measurement of broiler litter production rates and nutrient content using recycled litter. Poult. Sci. 85, 398–403. doi: 10.1093/ps/85.3.398
Cox, N. A., Richardson, L. J., Maurer, J. J., Berrang, M. E., Fedorka-Cray, P. J., Buhr, R. J., et al. (2012). Evidence for horizontal and vertical transmission in Campylobacter passage from hen to her progeny. J. Food Prot. 75, 1896–1902. doi: 10.4315/0362-028.JFP-11-322
Cressman, M. D., Yu, Z., Nelson, M. C., Moeller, S. J., Lilburn, M. S., and Zerby, H. N. (2010). Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl. Environ. Microbiol. 76, 6572–6582. doi: 10.1128/AEM.00180-10
Danzeisen, J. L., Calvert, A. J., Noll, S. L., McComb, B., Sherwood, J. S., Logue, C. M., et al. (2013). Succession of the turkey gastrointestinal bacterial microbiome related to weight gain. PeerJ 1:e237. doi: 10.7717/peerj.237
Danzeisen, J. L., Kim, H. B., Isaacson, R. E., Tu, Z. J., and Johnson, T. J. (2011). Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE 6:e27949. doi: 10.1371/journal.pone.0027949
Fonseca, B. B., Beletti, M. E., Melo, R. T., Mendonça, E. P., Vieira, C. U., Levenhagen, M. A., et al. (2011). Transfer, viability and colonisation of Campylobacter jejuni in the chicken vitellus and in embryos. Br. Poult. Sci. 52, 279–286. doi: 10.1080/00071668.2011.577056
Garrido, M. N., Skjervheim, M., Oppegaard, H., and Sørum, H. (2004). Acidified litter benefits the intestinal flora balance of broiler chickens. Appl. Environ. Microbiol. 70, 5208–5213. doi: 10.1128/AEM.70.9.5208-5213.2004
Gong, J., Forster, R. J., Yu, H., Chambers, J. R., Wheatcroft, R., Sabour, P. M., et al. (2002). Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 41, 171–179. doi: 10.1111/j.1574-6941.2002.tb00978.x
Gong, J., Si, W., Forster, R. J., Huang, R., Yu, H., Yin, Y., et al. (2007). 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 59, 147–157. doi: 10.1111/j.1574-6941.2006.00193.x
Gong, J., Yu, H., Liu, T., Gill, J. J., Chambers, J. R., Wheatcroft, R., et al. (2008). Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 104, 1372–1382. doi: 10.1111/j.1365-2672.2007.03699.x
Hammons, S., Oh, P. L., Martínez, I., Clark, K., Schlegel, V. L., Sitorius, E., et al. (2010). A small variation in diet influences the Lactobacillus strain composition in the crop of broiler chickens. Syst. Appl. Microbiol. 33, 275–281. doi: 10.1016/j.syapm.2010.04.003
Heyrman, J., and Vos, P. D. (2015). Oceanobacillus, in Bergey's Manual of Syst. Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons Ltd.
Hiett, K. L., Cox, N. A., and Rothrock, M. J. Jr. (2013). Polymerase chain reaction detection of naturally occurring Campylobacter in commercial broiler chicken embryos. Poult. Sci. 92, 1134–1137. doi: 10.3382/ps.2012-02812
Jia, W., Slominski, B. A., Bruce, H. L., Blank, G., Crow, G., and Jones, O. (2009). Effects of diet type and enzyme addition on growth performance and gut health of broiler chickens during subclinical Clostridium perfringens challenge. Poult. Sci. 88, 132–140. doi: 10.3382/ps.2008-00204
Kassem, I. I., Sanad, Y., Gangaiah, D., Lilburn, M., Lejeune, J., and Rajashekara, G. (2010). Use of bioluminescence imaging to monitor Campylobacter survival in chicken litter. J. Appl. Microbiol. 109, 1988–1997. doi: 10.1111/j.1365-2672.2010.04828.x
Kerr, A. K., Farrar, A. M., Waddell, L. A., Wilkins, W., Wilhelm, B. J., Bucher, O., et al. (2013). A systematic review-meta-analysis and meta-regression on the effect of selected competitive exclusion products on Salmonella spp. prevalence and concentration in broiler chickens. Prev. Vet. Med. 111, 112–125. doi: 10.1016/j.prevetmed.2013.04.005
Kim, M., Morrison, M., and Yu, Z. (2011). Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 84, 81–87. doi: 10.1016/j.mimet.2010.10.020
Kim, M., and Yu, Z. (2014). Variations in 16S rRNA-based microbiome profiling between pyrosequencing runs and between pyrosequencing facilities. J. Microbiol. 52, 355–365. doi: 10.1007/s12275-014-3443-3
Lee, K. W., Lillehoj, H. S., Lee, S. H., Jang, S. I., Ritter, G. D., Bautista, D. A., et al. (2011). Impact of fresh or used litter on the posthatch immune system of commercial broilers. Avian Dis. 55, 539–544. doi: 10.1637/9695-022511-Reg.1
Li, Y.-F., Chen, P.-H., and Yu, Z. (2014). Spatial and temporal variations of microbial community in a mixed plug-flow loop reactor fed with dairy manure. Microbial Biotechnol. 7, 332–346. doi: 10.1111/1751-7915.12125
Li, Y.-F., Nelson, M., Chen, P.-H., Graf, J., Li, Y., and Yu, Z. (2015). Comparison of the microbial communities in solid-state anaerobic digestion (SS-AD) reactors operated at mesophilic and thermophilic temperatures. Appl. Microbiol. Biotechnol. 99, 969–980. doi: 10.1007/s00253-014-6036-5
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Lovanh, N., Cook, K. L., Rothrock, M. J., Miles, D. M., and Sistani, K. (2007). Spatial shifts in microbial population structure within poultry litter associated with physicochemical properties. Poult. Sci. 86, 1840–1849. doi: 10.1093/ps/86.9.1840
Lu, J., Idris, U., Harmon, B., Hofacre, C., Maurer, J. J., and Lee, M. D. (2003a). Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69, 6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003
Lu, J., Sanchez, S., Hofacre, C., Maurer, J. J., Harmon, B. G., and Lee, M. D. (2003b). Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69, 901–908. doi: 10.1128/aem.69.2.901-908.2003
Lund, M., Bjerrum, L., and Pedersen, K. (2010). Quantification of Faecalibacterium prausnitzii- and Subdoligranulum variabile-like bacteria in the cecum of chickens by real-time PCR. Poult. Sci. 89, 1217–1224. doi: 10.3382/ps.2010-00653
Omeira, N., Barbour, E. K., Nehme, P. A., Hamadeh, S. K., Zurayk, R., and Bashour, I. (2006). Microbiological and chemical properties of litter from different chicken types and production systems. Sci. Total Environ. 367, 156–162. doi: 10.1016/j.scitotenv.2006.02.019
Pan, D., and Yu, Z. (2014). Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 5, 108–119. doi: 10.4161/gmic.26945
Peinado, M. J., Ruiz, R., Echávarri, A., and Rubio, L. A. (2012). Garlic derivative propyl propane thiosulfonate is effective against broiler enteropathogens in vivo. Poult. Sci. 91, 2148–2157. doi: 10.3382/ps.2012-02280
Ridley, A. M., Morris, V. K., Cawthraw, S. A., Ellis-Iversen, J., Harris, J. A., Kennedy, E. M., et al. (2011). Longitudinal molecular epidemiological study of thermophilic campylobacters on one conventional broiler chicken farm. Appl. Environ. Microbiol. 77, 98–107. doi: 10.1128/AEM.01388-10
Rodriguez, M. L., Rebole, A., Velasco, S., Ortiz, L. T., Trevino, J., and Alzueta, C. (2012). Wheat- and barley-based diets with or without additives influence broiler chicken performance, nutrient digestibility and intestinal microflora. J. Sci. Food Agric. 92, 184–190. doi: 10.1002/jsfa.4561
Roll, V. F., Dai Prá, M. A., and Roll, A. P. (2011). Research on Salmonella in broiler litter reused for up to 14 consecutive flocks. Poult. Sci. 90, 2257–2262. doi: 10.3382/ps.2011-01583
Santos, F. B., Sheldon, B. W., Santos, A. A. Jr., and Ferket, P. R. (2008). Influence of housing system, grain type, and particle size on Salmonella colonization and shedding of broilers fed triticale or corn-soybean meal diets. Poult. Sci. 87, 405–420. doi: 10.3382/ps.2006-00417
Shanmugasundaram, R., Lilburn, M. S., and Selvaraj, R. K. (2012). Effect of recycled litter on immune cells in the cecal tonsils of chickens. Poult. Sci. 91, 95–100. doi: 10.3382/ps.2011-01800
Si, W., Ni, X., Gong, J., Yu, H., Tsao, R., Han, Y., et al. (2009). Antimicrobial activity of essential oils and structurally related synthetic food additives towards Clostridium perfringens. J. Appl. Microbiol. 106, 213–220. doi: 10.1111/j.1365-2672.2008.03994.x
Snel, J., Heinen, P. P., Blok, H. J., Carman, R. J., Duncan, A. J., Allen, P. C., et al. (1995). Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int. J. Syst. Bacteriol. 45, 780–782. doi: 10.1099/00207713-45-4-780
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humaran, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Spees, A. M., Lopez, C. A., Kingsbury, D. D., Winter, S. E., and Baumler, A. J. (2013). Colonization resistance: battle of the bugs or Menage a Trois with the host? PLoS Pathog. 9:e1003730. doi: 10.1371/journal.ppat.1003730
Stanley, D., Denman, S. E., Hughes, R. J., Geier, M. S., Crowley, T. M., Chen, H., et al. (2012). Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 96, 1361–1369. doi: 10.1007/s00253-011-3847-5
Stanley, D., Geier, M. S., Denman, S. E., Haring, V. R., Crowley, T. M., Hughes, R. J., et al. (2013). Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet. Microbiol. 164, 85–92. doi: 10.1016/j.vetmic.2013.01.030
Thompson, C. L., Mikaelyan, A., and Brune, A. (2013). Immune-modulating gut symbionts are not “Candidatus Arthromitus”. Mucosal Immunol. 6, 200–201. doi: 10.1038/mi.2012.91
Thompson, C. L., Vier, R., Mikaelyan, A., Wienemann, T., and Brune, A. (2012). ‘Candidatus Arthromitus’ revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ. Microbiol. 14, 1454–1465. doi: 10.1111/j.1462-2920.2012.02731.x
Torok, V. A., Hughes, R. J., Mikkelsen, L. L., Perez-Maldonado, R., Balding, K., MacAlpine, R., et al. (2011). Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl. Environ. Microbiol. 77, 5868–5878. doi: 10.1128/aem.00165-11
Torok, V. A., Hughes, R. J., Ophel-Keller, K., Ali, M., and Macalpine, R. (2009). Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult. Sci. 88, 2474–2481. doi: 10.3382/ps.2008-00381
van der Wielen, P. W., Keuzenkamp, D. A., Lipman, L. J., van Knapen, F., and Biesterveld, S. (2002). Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44, 286–293. doi: 10.1007/s00248-002-2015-y
Van Immerseel, F., Ducatelle, R., De Vos, M., Boon, N., Van De Wiele, T., Verbeke, K., et al. (2010). Butyric acid-producing anaerobic bacteria as a novel probiotic treatment approach for inflammatory bowel disease. J. Med. Microbiol. 59, 141–143. doi: 10.1099/jmm.0.017541-0
Ventosa, A. (2015). Salinicoccus, in Bergey's Manual of Syst. Archaea and Bacteria. Hoboken, NJ: John Wiley Sons Ltd.
Wade, W. G. (2015). Dialister, in Bergey's Manual of Syst. Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons Ltd.
Wagner, R. D. (2006). Efficacy and food safety considerations of poultry competitive exclusion products. Mol. Nutr. Food Res. 50, 1061–1071. doi: 10.1002/mnfr.200600058
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wei, S., Gutek, A., Lilburn, M., and Yu, Z. (2013a). Abundance of pathogens in the gut and litter of broiler chickens as affected by bacitracin and litter management. Vet. Microbiol. 166, 595–601. doi: 10.1016/j.vetmic.2013.06.006
Wei, S., Morrison, M., and Yu, Z. (2013b). Bacterial census of poultry intestinal microbiome. Poult. Sci. 92, 671–683. doi: 10.3382/ps.2012-02822
Wise, M. G., and Siragusa, G. R. (2007). Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102, 1138–1149. doi: 10.1111/j.1365-2672.2006.03153.x
Wu, H.-M., Tien, Y.-J., and Chen, C.-H. (2010). GAP: a graphical environment for matrix visualization and cluster analysis. Comput. Stat. Data Analysis 54, 767–778. doi: 10.1016/j.csda.2008.09.029
Yeoman, C. J., Chia, N., Jeraldo, P., Sipos, M., Goldenfeld, N. D., and White, B. A. (2012). The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 13, 89–99. doi: 10.1017/S1466252312000138
Yin, Y., Lei, F., Zhu, L., Li, S., Wu, Z., Zhang, R., et al. (2010). Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 4, 367–376. doi: 10.1038/ismej.2009.128
Yu, Z., and Morrison, M. (2004a). Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70, 4800–4806. doi: 10.1128/AEM.70.8.4800-4806.2004
Keywords: bacteria, broiler chickens, gastrointestinal microbiota, litter, poultry, pyrosequencing
Citation: Wang L, Lilburn M and Yu Z (2016) Intestinal Microbiota of Broiler Chickens As Affected by Litter Management Regimens. Front. Microbiol. 7:593. doi: 10.3389/fmicb.2016.00593
Received: 16 February 2016; Accepted: 11 April 2016;
Published: 18 May 2016.
Edited by:
Mike Taylor, The University of Auckland, New ZealandReviewed by:
David William Waite, Ministry for Primary Industries, New ZealandAmelia Camarinha-Silva, University Hohenheim, Germany
Copyright © 2016 Wang, Lilburn and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongtang Yu, yu.226@osu.edu
 Lingling Wang1
Lingling Wang1 Zhongtang Yu
Zhongtang Yu