- 1State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
- 3Novogene Bioinformatics Technology Co. Ltd, Beijing, China
The genus Legionella comprises over 60 species, which are important human pathogens. Secretion systems in Legionella pneumophila have been studied extensively because of the essential role of protein secretion in bacterial infection. However, there are few reports describing the secretion systems in non-L. pneumophila species. In this study, we analyzed the distribution of secretion systems in L. pneumophila and 18 species of non-L. pneumophila based on whole genome sequences. A total of 74 whole genome sequences from 19 species of Legionella were analyzed. Type II and IVB secretion systems were detected in all Legionella strains, but the type I secretion systems was restricted to L. pneumophila. The type IVA secretion system was randomly distributed among different species. Furthermore, we found the type VI secretion system in three non-L. pneumophila strains (Legionella cherrii DSM 19213, Legionella dumoffii Tex-KL, and Legionella gormanii ATCC 33297). In population structure analysis, L. pneumophila formed a conservative cluster and was located at the terminal of the evolutionary tree. At the same time, L. pneumophila, especially eight clone groups (named MCGG1–MCGG8), showed higher intracellular growth ability than non-L. pneumophila species. These results suggest that L. pneumophila has acquired additional secretion systems during evolution, resulting in increased pathogenicity.
Introduction
Legionella is the causative agent of Legionnaires’ disease in humans (Fields et al., 2002). It is a Gram-negative facultative intracellular parasite of fresh water amoebae, monocytes and alveolar macrophages (Rogers et al., 1994). Legionella is an inhabitant of natural and man-made fresh water environments, where it is present either in a biofilm-associated state or in various protozoa where it can reside and proliferate as an intracellular pathogen (Rogers et al., 1994). Infection rates have increased as a result of industrial settings and the use of technical devices such as air conditioning systems, whirlpools and spas, showers, dental unit water lines and other medical devices, vegetable misters and fountains, which harbor Legionella species, combined with an increase in the population of elderly and immuno-compromised individuals (Diederen, 2008).
Transmission of bacteria from the environment to humans occurs via inhalation or aspiration of Legionella-containing aerosols (Breiman et al., 1990; Blatt et al., 1993). The genus Legionella comprises more than 60 species, among which Legionella pneumophila is the major causative agent of human infections, although other species have also been reported to cause disease (Fields et al., 2002; Diederen, 2008). In humans, Legionella causes a potentially fatal form of pneumonia, known as legionellosis, as well as a milder form, known as Pontiac fever.
Legionella invades, replicates, and survives in human macrophages, mainly by being surrounded by a membrane-bound vacuole that prevents lysosomal degradation (Fields et al., 2002). The mechanism of pathogenicity for Legionella, especially the non-L. pneumophila species, remains to be elucidated. Protein secretion is a universal process of fundamental importance for various aspects of cell physiology, including the infection of a host organism by a bacterial pathogen. Many Gram-negative pathogens achieve host infection by exporting virulence-associated proteins across one or two cell membranes to the site of action using a wide variety of secretory pathways. These protein secretion systems, which are considered to be important virulence factors in L. pneumophila, have been studied extensively. L. pneumophila genomes encode multiple protein secretion systems, including the putative type I Lss secretion machinery, type II PilD-dependent Lsp, type IVA Lvh, and type IVB Icm/Dot secretion pathways (De Buck et al., 2007). L. pneumophila use these specialized secretion systems to transport effectors and toxins to the extracellular environment or directly into the host cell to modify host physiology and promote interactions (Tseng et al., 2009). Type II and type IVB secretion systems (T4BSS) have been reported to play roles in L. pneumophila pathology, containing intracellular infection of protozoans and persistence in the lungs of A/J mice (Liles et al., 1998; Segal and Shuman, 1998; Vogel et al., 1998; Rossier and Cianciotto, 2001; Rossier et al., 2004; De Buck et al., 2007). In this study, we analyzed the genomic sequences of 53 L. pneumophila strains and 21 non-L. pneumophila strains. The distribution and gene cluster structure of the secretion systems were analyzed, in addition to the population structure and intracellular growth ability to ascertain the relationship between secretion systems and pathogenicity during the evolution of Legionella.
Materials and Methods
Sequenced Strains and Whole Genome Sequence Analysis
A total of 74 whole genomes of Legionella were analyzed in this study. Among them, six isolates were newly sequenced in this study, 44 isolates were sequenced in our previous study (Qin et al., 2016), and the remaining 24 were obtained from the NCBI database. The six newly sequenced strains are L. bozemanii strain ATCC 33217, L. feeleii strain ATCC 35072, L. jordanis strain ATCC 33623, L. micdadei strain ATCC 33218, L. oakridgensis strain ATCC 33761 and L. gormanii strain ATCC 33297. Of the 74 strains, 53 were L. pneumophila, comprising 41 serogroup 1 and 12 of other serogroups. The strain ATCC35289# was passed by three generations of strain ATCC35289. The 41 L. pneumophila serogroup 1 strains belonged to 26 different sequence-based typing (SBT) types with six SBT groups and three singles were formed among them, showing representative genotypes. As shown in our previous study, the L. pneumophila strains belonged to nine minimum core genome groups (MCGG1–MCGG9) (Qin et al., 2016). Another 21 strains belonged to other 19 Legionella species. Metadata (species, serogroups, dates and countries of isolation and accession numbers) for the strains included in the genome comparisons are presented in Supplementary Table S1.
DNA Sequencing and Assembly
The genomes were sequenced in Novogene Bioinformatics Technology Co. Ltd, Beijing, China. Bacterial strains were sequenced using an Illumina Hiseq2000 (Illumina Inc., San Diego, CA, USA) with a multiplexed protocol. Paired-end 90 nt-long reads of 500 bp sequencing libraries were generated for six strains. Raw data was processed in a four-step process involving removal of reads with ambiguous bases (5 bp), 20 bp of low quality reads (≤Q20), adapter contamination, and duplicated reads. In total, 100 libraries (500 bp) were obtained with clean paired-end read data. Assembly was performed using SOAPdenovo v1.05 (Li et al., 2008).
Genes Prediction and Function Annotation
Genes were predicted using Glimmer v3.02 (Delcher et al., 1999). This software predicts start sites and coding regions effectively and achieves good interpolation of hidden Markov models to minimize the frequency of false positive predictions. Function annotation was based on protein sequence analysis and completed by comparing BLAST v2.2.251 results in M8 format to the Kyoto Encyclopedia of Genes and Genomes (KEGG) v59 (Kanehisa et al., 2006), Cluster of Orthologous Groups of proteins (COG) v20090331 (Tatusov et al., 2003; Kanehisa et al., 2004), SwissProt v2011_10_19 (Magrane and Consortium, 2011), NR v2012-02-29, and Gene Ontology (GO) v1.419 databases (Ashburner et al., 2000). Genes were aligned with database sequences to obtain the annotation corresponding to homologs, with the highest quality alignment chosen for the gene annotation.
Identification of Secretion System Components
We used the ‘tblastn’ algorithm in BLAST (2.2.25 release) (Altschul et al., 1990) to search the L. pneumophila genomes for homologs of secretion system components. Perl scripts were developed to select hits with sequence identity >30%, and alignment extended would be implemented, if there’s star or end codon at alignment block boundary areas of 3X bp within 100 bp. Regions of the final alignment with gene coverage homology >40% were considered to be secretion systems components.
Phylogenetic Analysis
Core-pan analysis of 74 genomes was done and the single-copy orthologous genes were identified. A phylogenetic tree based on concatenated sequences of single-copy orthologous genes was constructed using the maximum-likelihood method with TreeBeST software (Nandi et al., 2010). A phylogenetic tree based on the core genome SNPs was constructed using the neighbor-joining or minimum evolution algorithms in MEGA (version 5.1) (Tamura et al., 2007). Bootstraps were performed with 1,000 replicates. The MEGA program was also used to calculate the p-distance (p = nd/n) within and between population groups, where nd is the number of sites with differences and n is the total number of sites.
Intracellular Growth Assay
The Legionella strains were revived from lyophilized isolate. The bacteria were streaked onto buffered charcoal yeast extract (BCYE) agar plates, and one typical colony of each strain was picked up and inoculated onto buffered yeast extract (BYE) broth at 35°C until they reached early stationary phase. Approximately 2 × 109 bacteria were pelleted, resuspended and diluted (1:1,000) in Roswell Park Memorial Institute (RPMI) 1640 tissue culture medium. The bacteria were then added to J774 cells or guinea pig peritoneal macrophages (2 × 105 per well) in 24-well dishes to give a multiplicity of infection (MOI) of approximately 10. The infected cells were incubated at 37°C under 5% CO2-air for 1.5 h and then washed three times with PBS to remove extracellular bacteria. To measure bacterial internalization, 1 ml of sterile distilled water was added to the wells to release intracellular bacteria from the host cells. The colony-forming units (CFUs) were determined by plating dilutions on BCYE agar plates. To each of the wells, 0.5 ml of fresh tissue culture medium was added, and the intracellular and extracellular bacteria in each well were combined at 24-h intervals. The total number of CFUs was determined by plating the dilutions onto BCYE agar plates.
The J774 cell monolayers were prepared on cover slips by the same procedures as described above. The cells were infected with L. pneumophila philadelphia-1 and the other isolated strains. After 48 h of infection, the infected J774 cells were stained by Gimenez staining and observed under a light microscope.
Nucleotide Sequence Accession Numbers
This Whole Genome Shotgun project has been deposited at GenBank under the Bioproject ID PRJNA281151, with accession numbers LBAW00000000, LBHK00000000, LBAX00000000, LCUB00000000, LCUA00000000, and LBAY00000000.
Results and Discussion
In this study, we analyzed the distribution of L. pneumophila and non-L. pneumophila secretion system protein types I, II, III, IV and VI by searching the NR, KEGG, COG, POG, GO, PHI, VFDB, ARDB, CAZy databases by using the Legionella pan-genome (Figure 1). Genes encoding types II and IV secretion system homologs were found in all the strains analyzed, while genes encoding types I and III secretion genes were absent from some non-L. pneumophila strains. Additionally, type VI secreted system genes were found in six non-L. pneumophila strains.
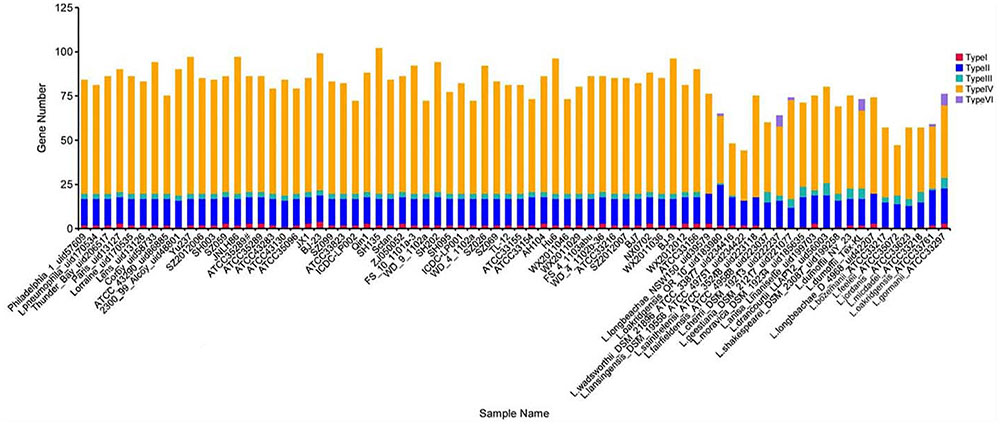
FIGURE 1. Number of TnSS functional genes in 74 Legionella genomes. Here, the TnSS contained secretion system protein types I, II, III, IV and VI, collected from the NR, KEGG, COG, POG, GO, PHI, VFDB, ARDB, CAZy databases.
However, searches conducted using identified secretion system gene cluster sequences in L. pneumophila strain Philadelphia 1 revealed genes encoding types I, II, IVA, and IVB secretion system homologs in other genomes, although there were several notable differences. Type II and IVB secretion systems were detected in all Legionella strains, but the type I secretion systems (T1SS) was restricted to L. pneumophila. The type IVA secretion system (T4ASS) was randomly distributed among different species. Furthermore, we found the type VI secretion system in three non-L. pneumophila strains.
Type I Secretion Systems
The T1SS allows secretion of substrates into the extracellular space in a one-step process, without a stable periplasmic intermediate. A T1SS consists of three proteins: an inner-membrane ATPase, known as the ABC (ATP-binding cassette) transporter, a membrane fusion protein spanning the periplasmic space and an outer-membrane protein (Gerlach and Hensel, 2007). In L. pneumophila, a putative T1SS designated Lss has been identified. Analysis of the lss gene distribution in various Legionella strains by Southern blotting suggested that a complete lss gene cluster is restricted to L. pneumophila, while the lssD gene was not detected in non-L. pneumophila species (Jacobi and Heuner, 2003). The role of T1SS in L. pneumophila virulence has recently been identified and repeats-in-toxin protein RtxA is secreted through an LssB-LssD-TolC-dependent mechanism (Fuche et al., 2015). In this study, all L. pneumophila strains contained the lssXYZABD locus encoding a putative T1SS. In contrast, the lssXYZABD locus was not found in non-L. pneumophila species (Figure 2), thus indicating that the lssXYZABD secretion system is restricted and conserved in L. pneumophila.
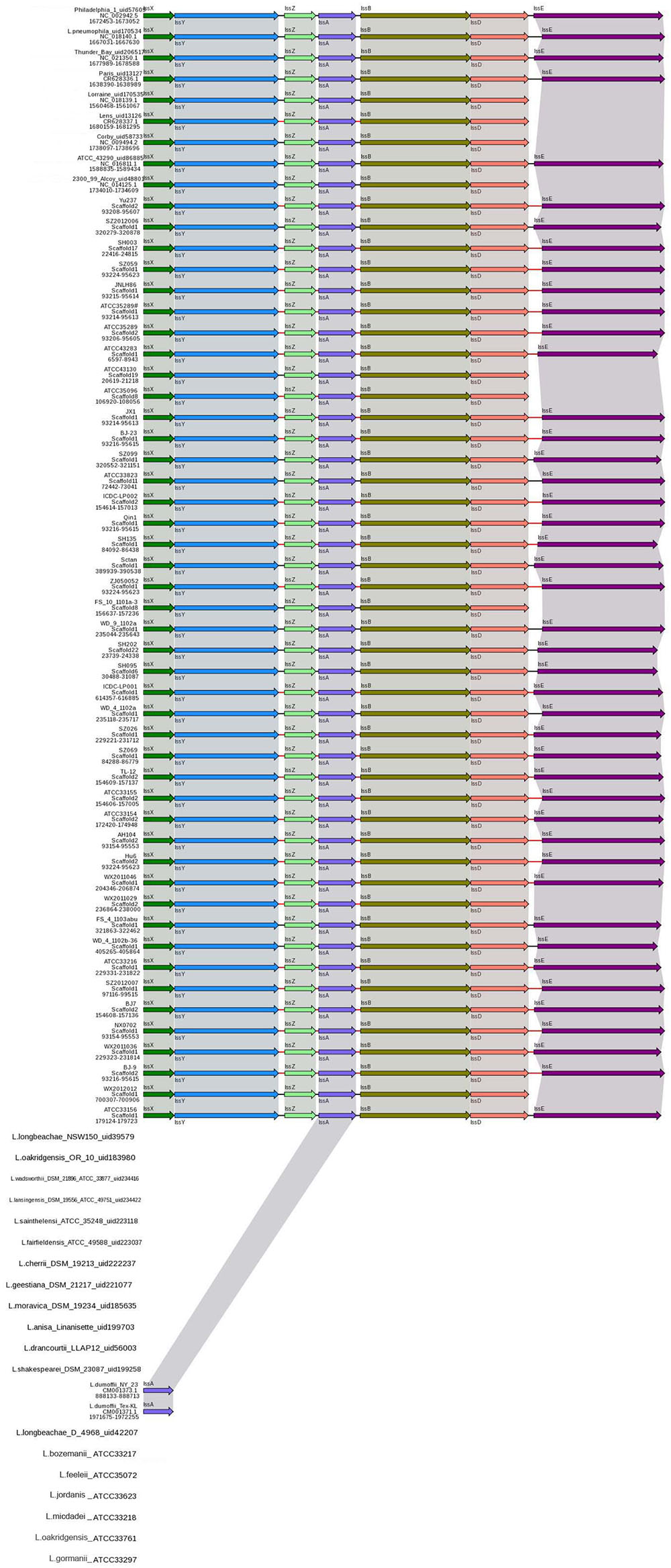
FIGURE 2. Structure and distribution of T1SS in 74 Legionella genomes. The box arrows represent open reading frames (ORFs); black lines represent forward scaffolds; red lines represent reverse scaffolds; and genes linked by the same dotted line are homologous.
Type II Secretion Systems
To date, L. pneumophila is the only intracellular pathogen known to possess a functional type II secretion system (T2SS) (Cianciotto, 2005). The T2SS system has been shown to promote L. pneumophila growth in low temperature environments and is required for biofilm establishment, sliding motility, intracellular infection of protozoans and mammalian macrophages, and persistence in the lungs of A/J mice (Rossier et al., 2004). In a previous study, T2SS genes were identified in several non-L. pneumophila species and it was hypothesized that the T2SS occurs throughout the genus Legionella (Rossier et al., 2004); this speculation was confirmed by our results. In this study, all analyzed strains, regardless of the species, harbored T2SS genes, containing pilD gene and Lsp system (Figure 3).
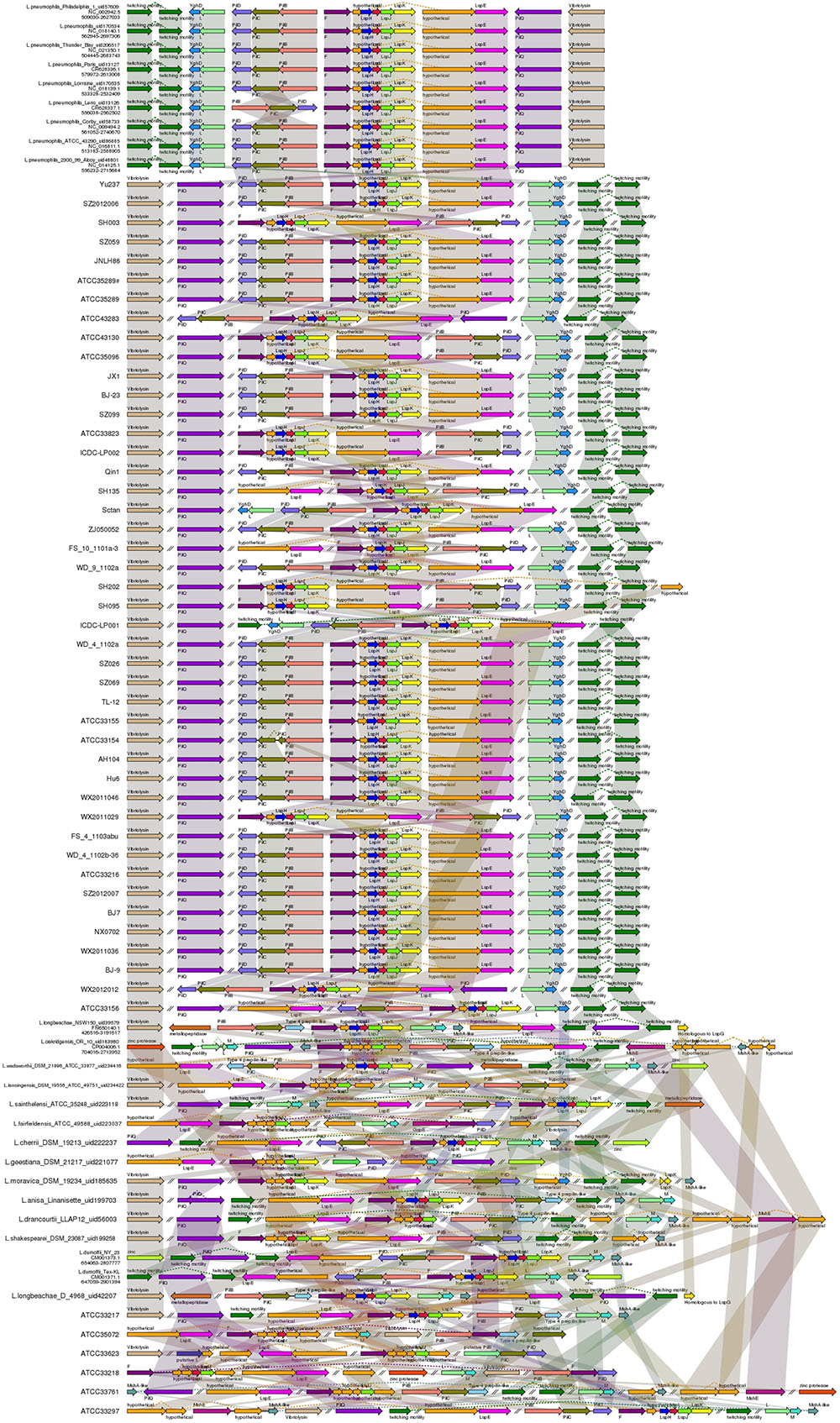
FIGURE 3. Structure and distribution of T2SS in 74 Legionella genomes. The box arrows represent ORFs; black lines represent forward scaffolds; red lines represent reverse scaffolds; genes linked by the same dotted line are homologous; and the double forward slash indicates that the genes are located in different scaffolds.
Type III Secretion System
The type III secretion systems (T3SS) are found in Gram-negative bacteria such as Salmonella typhimurium, Yersinia pestis and Pseudomonas syringae, which interact with both plant and animal hosts (Tseng et al., 2009). In this study, flagellum-encoding genes were found in all L. pneumophila and 14 of 21 non-L. pneumophila strains. Flagellum-encoding genes were not found in seven non-L. pneumophila strains; there were L. longbeachae NSW150, L. wadsworthii DSM 21896 (ATCC 33877), L. lansingensis DSM 19556 (ATCC 49751), L. sainthelensi ATCC 35248, L. longbeachae D4968, L. oakridgensis OR 10 and L. oakridgensis ATCC 33761. However, no non-flagellar T3SSs were found.
Type IV Secretion Systems
The type IV secretion systems (T4SSs) have been grouped into two subclasses; type IVA, which is similar to the Agrobacterium tumefaciens Vir system, and type IVB, which is similar to the Tra/Trb bacterial conjugation systems (De Buck et al., 2007). The type IVA Lvh and type IVB Icm/Dot secretion systems are present in L. pneumophila.
The gene cluster encoding T4ASS, designated Lvh, is located in a an elevated G+C content region of plasmid-like elements, which also contain genes encoding mobility factors and enzymes, such as phage integrases and transposases (Doleans-Jordheim et al., 2006; Glockner et al., 2008). Furthermore, Kozak et al. (2010) hypothesized that these plasmid-like elements can exist in either integrated or excised forms with widespread mobility of the lvh region in the genus Legionella, thus contributing to interspecies exchange of genetic information; our analyses support this view. In this study, the Lvh genes were found in 40 L. pneumophila and seven non-L. pneumophila strains. The order and sequence of T4ASS Lvh genes are highly conserved among these strains and interestingly, strain SZ026 was found to possess two copies of the Lvh secretion system genes (Figure 4).
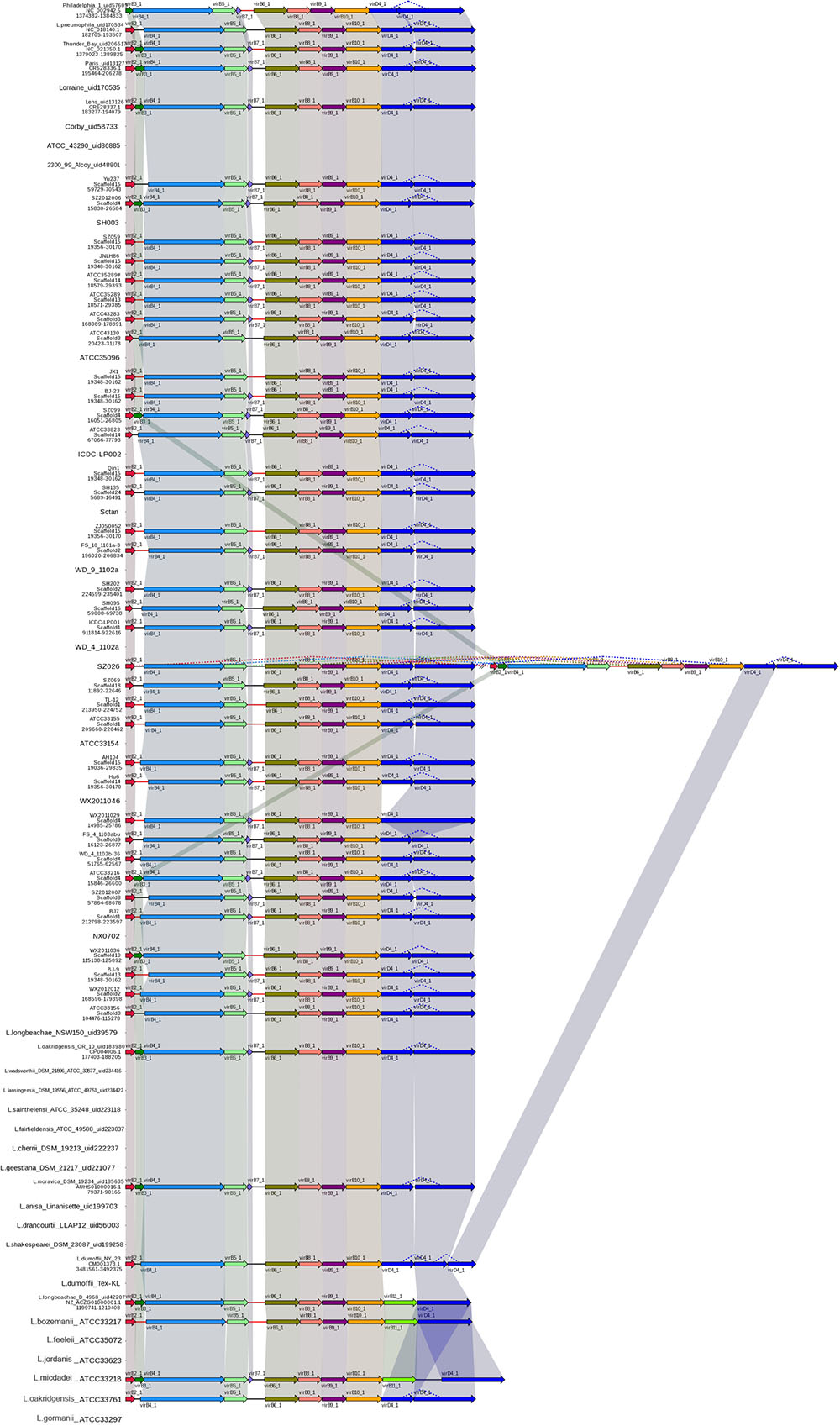
FIGURE 4. Structure and distribution of T4ASS in 74 Legionella genomes. The box arrows represent ORFs; black lines represent forward scaffolds; red lines represent reverse scaffolds; genes linked by the same dotted line are homologous; and the double forward slash indicates that the genes are located in different scaffolds.
In L. pneumophila, the T4BSS, designated the Icm/Dot secretion system, is encoded by two separate pathogenicity regions on the chromosome. The first region contains 17 genes (icmTSRQ-PONMLKEGCDJBF), while the second region carries icmXWV and dotABCD (Nagai and Kubori, 2011). Our results reveal that the gene organization of both regions is highly conserved in all L. pneumophila strains, with relative diversity observed in non-L. pneumophila species (Figure 5). In region I, all non-L. pneumophila strains lack icmR and other genes were observed in three non-L. pneumophila strains (L. longbeachae strain NSW150, L. cherrii DSM 19213 and L. dumoffii strain NY 23). Furthermore, the icmF-icmH (dotU) genes of region I are separated from the rest of the gene cluster in L. wadsworthii DSM 21896 (ATCC 33877), L. sainthelensi ATCC 35248, L. geestiana DSM 21217, L. anisa Linanisette, L. drancourtii LLAP12, L. shakespearei DSM 23087, L. longbeachae D-4968, L. bozemanii ATCC 33217, L. feeleii ATCC 35072, L. gormanii ATCC 33297 (Figure 5A). In region II, all strains contained all the genes, except icmV in L. micdadei strain ATCC 33218; relative diversity was observed among the neighboring Icm/Dot genes (Figure 5B). However, in both Icm/Dot regions, the order and orientation of the genes were conserved throughout the genus; this is coincident with the previous findings (Burstein et al., 2016).
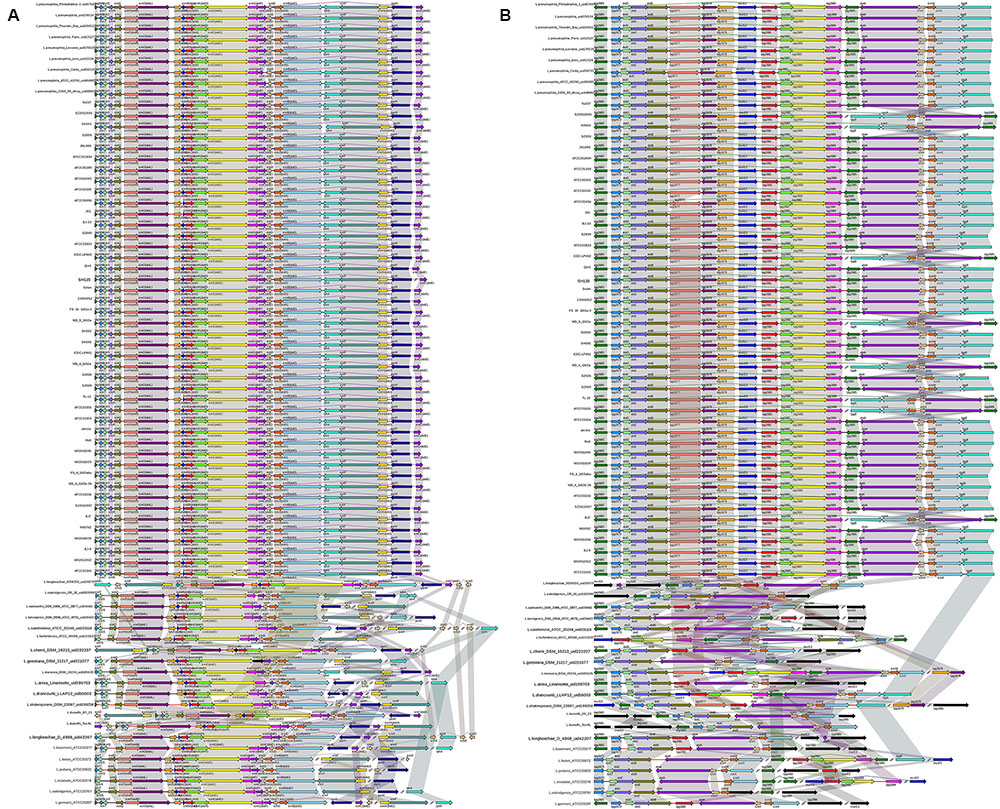
FIGURE 5. Structure and distribution of T4BSS region 1 (A) and region 2 (B) in 74 Legionella genomes. The box arrows represent ORFs; black lines represent forward scaffolds; red lines represent reverse scaffolds; genes linked by the same dotted line are homologous; and the double forward slash indicates that the genes are located in different scaffolds.
Type VI Secretion System
The recently characterized type VI secretion system (T6SS) appears to constitute a phage-tail-spike-like injectisome that has the potential to introduce effector proteins directly into the cytoplasm of host cells. Genes encoding T6SS components are detected in more than a 25% of sequenced bacterial genomes. The T6SS is required for virulence in human, animal and plant pathogens, for efficient root colonization, and may contribute to environmental adaptations, such as biofilm formation, adhesion and cytotoxicity to host cells (Bingle et al., 2008; Cascales, 2008; Filloux et al., 2008; Liu et al., 2008; Shrivastava and Mande, 2008; Wu et al., 2008; Liu et al., 2015). The T6SS in Legionella has not been reported previously. In this study, type VI secreted system genes were identified in six strains by searching the NR, KEGG, COG, POG, GO, PHI, VFDB, ARDB, CAZy databases. However, the gene identified in three strains (L. oakridgensis OR 10, L. geestiana DSM 21217 and L. oakridgensis ATCC 33761) was icmF, which is a type IV secreted system gene in Legionella, and also one of the VI secreted system genes in Escherichia coli (de Pace et al., 2011). Another three strains, Legionella cherrii DSM 19213, Legionella dumoffii Tex-KL, Legionella gormanii ATCC 33297, harbored a gene cluster previously identified as T6SS. This gene cluster contains six genes with conserved order (Figure 6). This is the first report of the identification and function of T6SS in Legionella. It is worth noting that only one of two L. dumoffii strains harbors T6SS, suggesting that the presence of T6SS is strain but not species-specific.
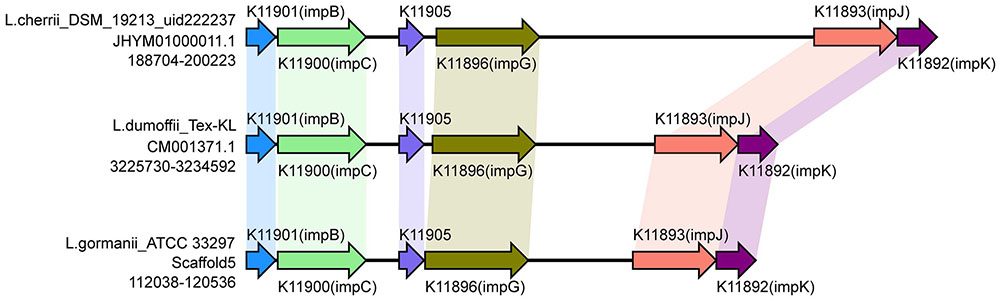
FIGURE 6. Structure and distribution of T6SS in 74 Legionella genomes. The box arrows represent ORFs; black lines represent forward scaffolds; red lines represent reverse scaffolds; genes linked by the same dotted line are homologous; and the double forward slash indicates that the genes are located in different scaffolds.
Population Structure and Intracellular Growth Ability
We used a maximum-likelihood tree to explore the relationships among our strains, using 300 single-copy orthologous genes identified in 74 Legionella genomes (Figure 7). Most of the branches in this tree had high supporting bootstrap values, suggesting stable genomic differences among different Legionella species. Fifty L. pneumophila were clustered as a monophyl, which is consistent with a previous classification system (Qin et al., 2016). L. moravica and L. shakespearei were more closely related to L. pneumophila than to other species, suggesting that these three species had the most recent common ancestor. Among these strains, some strains such as L. longbeachae and L. sainthelensi strains, and L. bozemanii and L. anisa strains are more closely related than others. An additional tree constructed using the Rickettsia strain CbuG_Q212 as an outgroup (Figure 8). While Legionella geestiana diverged first from the rest of the species, in this tree, the L. pneumophila cluster is more recently derived. This is also supported by the short branches within the L. pneumophila cluster. These results suggest that L. geestiana may be the first Legionella species generated, and that L. pneumophila has evolved from other species, most likely L. moravica and L. shakespearei.
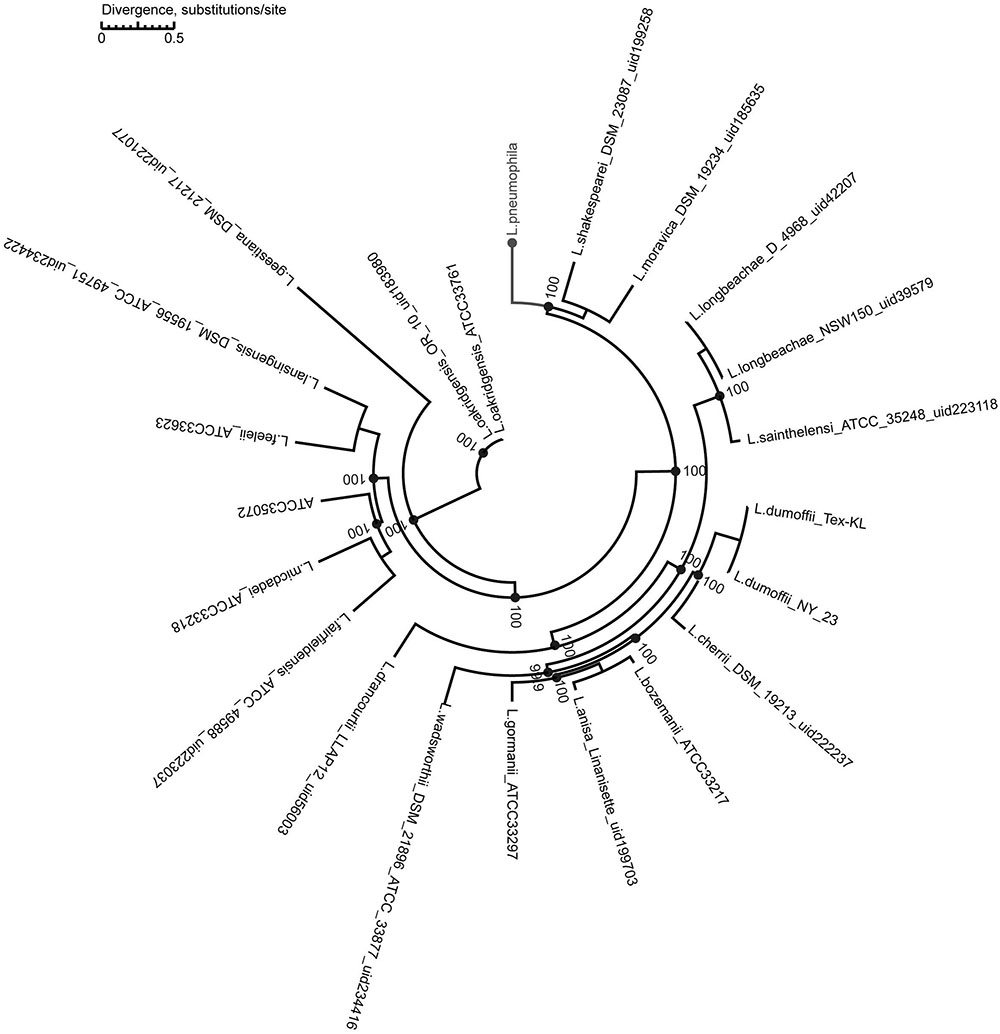
FIGURE 7. Maximum-likelihood tree of 74 Legionella strains. This tree was constructed using 300 single-copy orthologous genes identified in 74 Legionella genomes.
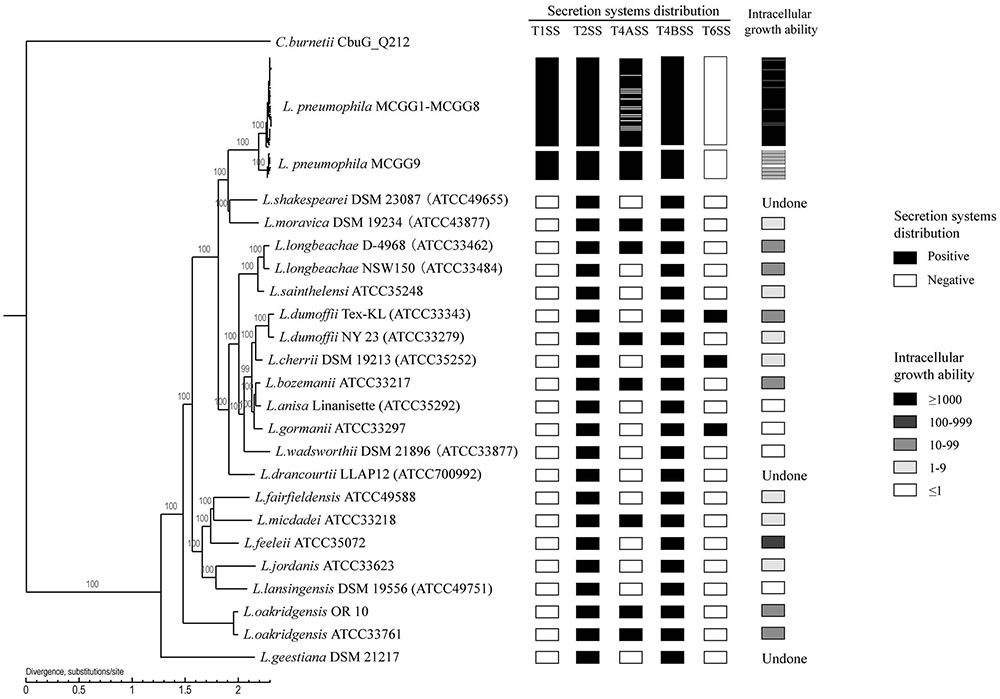
FIGURE 8. Rooted phylogenetic tree of 74 Legionella strains using 300 single-copy orthologous genes identified in 74 Legionella genomes. Rickettsia strain CbuG_Q212 was used as an outgroup. L. pneumophila MCGG1–MCGG8 and L. pneumophila MCGG9 are reduced to a single cluster each. Secretion system distribution and intracellular growth ability are shown in the left columns. The intracellular growth ability is represented by the fold-increase in bacterial number on day 3 of infection compared with the initial number of bacteria. The two L. oakridgensis strains, OR 10 and ATCC 33761, are the same isolate. The genome of OR 10 was obtained from the NCBI database and that of ATCC 33761 was newly sequenced in this study.
We also tested the intracellular growth ability of L. pneumophila and some non-L. pneumophila strains. MCGG1–MCGG8 L. pneumophila showed significantly higher intracellular growth ability than MCGG9 L. pneumophila strains and non-L. pneumophila strains. There was no difference in the distribution of secretion systems between MCGG1–MCGG8 and MCGG9 L. pneumophila strains; therefore, it can be speculated that the difference in intracellular growth ability is caused by other factors. The differences in distribution of T1SS and the rearrangement and separation of T4BSS region II between L. pneumophila and non-L. pneumophila strains might account for the difference in intracellular growth ability; however, further studies are required to confirm this speculation.
Conclusion
Secretion systems are widely distributed in genus Legionella. In this study, all the Legionella strains analyzed harbor complete T2SS and T4BSS gene clusters; however, T1SS is restricted to L. pneumophila. T4ASSs exist in some of the L. pneumophila and non-L. pneumophila species. Three non-L. pneumophila strains harbor T6SS. Population structure analysis revealed that L. pneumophila forms a conservative cluster and located on the terminal of the evolutionary tree. L. pneumophila also showed higher intracellular growth ability than non-L. pneumophila species. The limitation of this study is only 20 species were analyzed, as well as there are more than 60 species of Legionella. However, these results suggest that L. pneumophila has acquired additional secretion systems during evolution, resulting in increased pathogenicity.
Author Contributions
TQ and HZ designed experiments. TQ and HR performed experiments. TQ, HZ, and WL analyzed data. TQ and WL contributed reagents, materials, and analysis tools. TQ and HZ wrote this manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants the Science Foundation for the State Key Laboratory for Infectious Disease Prevention and Control from China (Grant No. 2015SKLID508) and the National Science Foundation of China (Grant No. 81671985).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00388/full#supplementary-material
TABLE S1 | Metadata for the strains included in the genome comparisons in this study.
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology: the gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Blatt, S. P., Parkinson, M. D., Pace, E., Hoffman, P., Dolan, D., Lauderdale, P., et al. (1993). Nosocomial Legionnaires’ disease: aspiration as a primary mode of disease acquisition. Am. J. Med. 95, 16–22. doi: 10.1016/0002-9343(93)90227-G
Breiman, R. F., Cozen, W., Fields, B. S., Mastro, T. D., Carr, S. J., Spika, J. S., et al. (1990). Role of air sampling in investigation of an outbreak of legionnaires’ disease associated with exposure to aerosols from an evaporative condenser. J. Infect. Dis. 161, 1257–1261. doi: 10.1093/infdis/161.6.1257
Burstein, D., Amaro, F., Zusman, T., Lifshitz, Z., Cohen, O., Gilbert, J. A., et al. (2016). Uncovering the Legionella genus effector repertoire – strength in diversity and numbers. Nat. Genet. 48, 167–175. doi: 10.1038/ng.3481
Cascales, E. (2008). The type VI secretion toolkit. EMBO Rep. 9, 735–741. doi: 10.1038/embor.2008.131
Cianciotto, N. P. (2005). Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588. doi: 10.1016/j.tim.2005.09.005
De Buck, E., Anné, J., and Lammertyn, E. (2007). The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology 153, 3948–3953. doi: 10.1099/mic.0.2007/012039-0
de Pace, F., Boldrin de Paivazzz, J., Nakazato, G., Lancellotti, M., Sircili, M. P., Guedes Stehling, E., et al. (2011). Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology 157, 2954–2962. doi: 10.1099/mic.0.050005-0
Delcher, A. L., Harmon, D., Kasif, S., White, O., and Salzberg, S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641. doi: 10.1093/nar/27.23.4636
Diederen, B. M. (2008). Legionella spp. and Legionnaires’disease. J. Infect. 56, 1–12. doi: 10.1016/j.jinf.2007.09.010
Doleans-Jordheim, A., Akermi, M., Ginevra, C., Cazalet, C., Kay, E., Schneider, D., et al. (2006). Growth-phase-dependent mobility of the lvh-encoding region in Legionella pneumophila strain Paris. Microbiology 152, 3561–3568. doi: 10.1099/mic.0.29227-0
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires’disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. doi: 10.1128/CMR.15.3.506-526.2002
Filloux, A., Hachani, A., and Bleves, S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583. doi: 10.1099/mic.0.2008/016840-0
Fuche, F., Vianney, A., Andrea, C., Doublet, P., and Gilbert, C. (2015). Functional type 1 secretion system involved in Legionella pneumophila virulence. J. Bacteriol. 197, 563–571. doi: 10.1128/JB.02164-14
Gerlach, R. G., and Hensel, M. (2007). Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297, 401–415. doi: 10.1016/j.ijmm.2007.03.017
Glockner, G., Albert-Weissenberger, C., Weinmann, E., Jacobi, S., Schunder, E., Steinert, M., et al. (2008). Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428. doi: 10.1016/j.ijmm.2007.07.012
Jacobi, S., and Heuner, K. (2003). Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 293, 349–358. doi: 10.1078/1438-4221-00276
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., et al. (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357. doi: 10.1093/nar/gkj102
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., et al. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044. doi: 10.1128/JB.01272-09
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Liles, M. R., Viswanathan, V. K., and Cianciotto, N. P. (1998). Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66, 1776–1782.
Liu, H., Coulthurst, S. J., Pritchard, L., Hedley, P. E., Ravensdale, M., Humphris, S., et al. (2008). Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. doi: 10.1371/journal.ppat.1000093
Liu, L., Hao, S., Lan, R., Wang, G., Xiao, D., Sun, H., et al. (2015). The type VI secretion system modulates flagellar gene expression and secretion in Citrobacter freundii and contributes to the adhesion and cytotoxicity to host cells. Infect. Immun. 83, 2596–2604. doi: 10.1128/IAI.03071-14
Magrane, M., and Consortium, U. (2011). UniProt knowledgebase: a hub of integrated protein data. Database 2011:bar009. doi: 10.1093/database/bar009
Nagai, H., and Kubori, T. (2011). Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2:136. doi: 10.3389/fmicb.2011.00136
Nandi, T., Ong, C., Singh, A. P., Boddey, J., Atkins, T., Sarkar-Tyson, M., et al. (2010). A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 6:e1000845. doi: 10.1371/journal.ppat.1000845
Qin, T., Zhang, W., Liu, W., Zhou, H., Ren, H., Shao, Z., et al. (2016). Population structure and minimum core genome typing of Legionella pneumophila. Sci. Rep. 6:21356. doi: 10.1038/srep21356
Rogers, J., Dowsett, A. B., Dennis, P. J., Lee, J. V., and Keevil, C. W. (1994). Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60, 1585–1592.
Rossier, O., and Cianciotto, N. P. (2001). Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69, 2092–2098. doi: 10.1128/IAI.69.4.2092-2098.2001
Rossier, O., Starkenburg, S. R., and Cianciotto, N. P. (2004). Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect. Immun. 72, 310–321. doi: 10.1128/IAI.72.1.310-321.2004
Segal, G., and Shuman, H. A. (1998). How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6, 253–255.
Shrivastava, S., and Mande, S. S. (2008). Identification and functional characterization of gene components of Type VI secretion system in bacterial genomes. PLoS ONE 3:e2955. doi: 10.1371/journal.pone.0002955
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Tseng, T. T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol. 9:S2. doi: 10.1186/1471-2180-9-S1-S2
Vogel, J. P., Andrews, H. L., Wong, S. K., and Isberg, R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876. doi: 10.1126/science.279.5352.873
Keywords: secretion systems, Legionella, L. pneumophila, whole genome sequence, population structure, pathogenicity evolution
Citation: Qin T, Zhou H, Ren H and Liu W (2017) Distribution of Secretion Systems in the Genus Legionella and Its Correlation with Pathogenicity. Front. Microbiol. 8:388. doi: 10.3389/fmicb.2017.00388
Received: 28 November 2016; Accepted: 24 February 2017;
Published: 14 March 2017.
Edited by:
Leonard Peruski, US Centers for Disease Control and Prevention, USAReviewed by:
Shaofu Qiu, Academy of Military Medical Sciences, ChinaJunko Amemura-Maekawa, National Institute of Infectious Diseases, Japan
Copyright © 2017 Qin, Zhou, Ren and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian Qin, qintian@icdc.cn
 Tian Qin1,2*
Tian Qin1,2* Haijian Zhou
Haijian Zhou