- 1Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
- 2Institute of Engineering Biology and Health, Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals, College of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou, China
WhiB3 is a conserved cytoplasmic redox sensor which is required in the infection and lipid anabolism of Mycobacterium tuberculosis. The response of WhiB3 to environmental nutrient and its regulatory cascades are crucial during the persistent infection, while little is known about the relationship between WhiB3 and emergence of nutrient stress in this process. Here, we found that nitrogen regulator GlnR directly interacted with the WhiB3 promoter region and activated its transcription in response to nitrogen availability. In whiB3 promoter region, the typical GlnR-box was also identified. Moreover, GlnR controlled cell resistance to redox stress and SL-1 lipid anabolism by directly activating whiB3 expression. These results demonstrated that GlnR regulated redox sensor WhiB3 at the transcriptional level and mediated the interplay among nitrogen metabolism, redox sensing, and lipid anabolism.
Importance
The regulatory network of mycobacteria has delicately evolved to adapt to the extreme nutrient deficiencies and various stress including acidic stress and redox stress during infection. Identification the roles of critical members that participate in this network is crucial to the development of novel clinical therapies. Here, we found that WhiB3, a redox-sensing regulator, was under direct transcription activation of nitrogen regulator GlnR during nitrogen starvation. In addition, GlnR controlled cell resistance to redox/acidic stress and SL-1 lipid anabolism through WhiB3, which suggested that the GlnR-mediated WhiB3 regulatory pathway might influence mycobacterial infection. These findings provide novel insight into the tight connection of intracellular redox status, potential virulence, and nitrogen metabolism.
Introduction
Tuberculosis is one of the major global health emergency, whose pathogenesis is closely related to its cell wall lipid (Dye et al., 1999). The enzymes and regulators involved in the lipid synthesis and regulation are typically ideal drug targets due to their uniqueness (Brennan and Nikaido, 1995; Kolattukudy et al., 1997; Barry et al., 1998; Daffe and Draper, 1998), the regulatory protein WhiB3 was one of which has been well characterized associating with redox sensing and lipid anabolism in Mycobacterium tuberculosis during infection (Singh et al., 2009).
WhiB3 is a member of the WhiB subfamily with the common characteristic of small molecular weight and high cysteine content (Flardh et al., 1999). WhiB protein is initially found in Actinobacteria (Chater, 1972) and predicted to be important transcriptional factor participated in pathogenesis, cell division, and other stresses sensing. M. tuberculosis genome contains seven annotated whiB genes (Alam et al., 2009), elucidation of their functions and regulatory network in mycobacteria is beneficial for understanding the pathogen biology and finding out appropriate countermeasure. Previous studies had shown that M. tuberculosis WhiB3 controlled virulence in two animal models (Steyn et al., 2002), and the WhiB orthologs were implicated in varieties of pathways including sporulation, cell division (Gomez and Bishai, 2000), oxidative stress (Kim et al., 2005), pathogenesis, and antibiotic resistance (Morris et al., 2005). During infection, M. tuberculosis whiB3 expression was significantly induced (Rohde et al., 2007). The whiB3 deficiency strain exhibited a pathology defect, altered colony rugosity, and growth properties (Steyn et al., 2002; Singh et al., 2007). WhiB3 was also involved in maintaining intracellular redox homeostasis through regulating the fatty acids metabolisms via a redox switching mechanism (Singh et al., 2009; Cumming et al., 2017). M. tuberculosis WhiB3 was identified as a regulator of virulence lipid anabolism and distinctively modulated the propionate assimilation into complex virulence lipids including polyacyltrehalose (PAT), sulfolipid (SL-1), phthiocerol dimycocerosate (PDIM), and storage lipid triacylglycerol (TAG) under defined oxidizing/reducing conditions.
Mycobacterium smegmatis is a fast-growing non-pathogenic species generally used as a model for slow-growing pathogenic M. tuberculosis. In M. smegmatis, the WhiB subfamily protein WhmD (homologous to WhiB2 in M. tuberculosis) was an essential gene necessary in proper septation and cell division (Raghunand and Bishai, 2006). Moreover, during nutrient starvation, WhiB2, WhiB3, and WhiB4 participated in the development of both mono-nucleoided small resting cells and log-phase-sized resting cells with similar temporal expression pattern (Wu et al., 2016). Differing from the well-studied function of WhiB3, the regulatory factors and mechanisms acted on WhiB3 is still little known except that its DNA binding ability is regulated by a thiol-disulfide redox switch reversibly (Singh et al., 2009), it is a downstream target of RelA (Primm et al., 2000; Rifat et al., 2009), and it interacts with σ factor RpoV (Steyn et al., 2002).
In the present study, we investigated a notable role of nitrogen regulator GlnR in regulating whiB3 transcription responding to nitrogen availability of M. smegmatis. We found out that GlnR directly bound with whiB3 promoter region and activated whiB3 transcription during nitrogen starvation. Moreover, GlnR controlled the cell resistance to redox stress and SL-1 lipid anabolism by directly activating the expression of whiB3. These results elucidated the mechanism of GlnR-mediated interplay between nitrogen metabolism and stress sensing, which possibly contributed toward understanding pathogen biology of mycobacteria.
Materials and Methods
Strains, Plasmids, and Growth Conditions
Strains and plasmids used in this study were listed in Supplementary Table S1. E. coli DH5α (TransGen Biotech) was used for cloning and cultured in LB media supplemented with 50 μg/ml kanamycin. M. smegmatis MC2 155 WT, ΔglnR and complemented strains were cultured in LB media supplemented with 0.05% tween 80 or Sauton’s medium. Nitrogen free Sauton’s medium (0.05% (w/v) MgSO4, 0.05% (w/v) KH2PO4, 0.2% (w/v) citric acid, 0.2% (v/v) glycerol, 0.005% (w/v) ferric citrate, 0.015% (v/v) Tyloxapol, 0.0001% (v/v) ZnSO4) supplemented with 1 mM (nitrogen-limited, NL) or 30 mM (nitrogen-rich, NXS) (NH4)2SO4 was used for the assessment of transcription level (Jenkins et al., 2013).
Overproduction and Purification of GlnR and WhiB3 Proteins
The overproduction and purification of His-tag GlnR protein was performed as previously described (Xu et al., 2017). The whiB3 gene was amplified from M. smegmatis with primers listed in Supplementary Table S2. The purified PCR product using the PCR purification kit (TransGen Biotech) was cloned into pET-28a (+), and generated the recombinant plasmid pET-whiB3. The single colony was grown overnight in 5 mL LB medium containing 50 μg/ml kanamycin at 37°C. Then transferred into 100 mL LB medium until the OD600 reached 0.6 and induced with 0.5 mM IPTG. The His-WhiB3 protein was purified from soluble fraction performed as described (Singh et al., 2009; You et al., 2014). Apo-WhiB3 protein was prepared as described previously (Singh et al., 2009), the purified apo-WhiB3 protein was then used for in vitro EMSA experiments.
Electrophoretic Mobility Shift Assays (EMSAs)
The upstream 350 bp region of whiB3 gene containing the putative GlnR-box was amplified with primers listed in Supplementary Table S2. PCR products were labeled with biotin using the universal biotinylated primer (5’-AGCCAGTGGCGATAAG-3’). The EMSA probes were purified using PCR purification kit (TransGen Biotech) and analyzed by agarose gel electrophoresis. The probes concentration was determined with microplate reader (BioTek, United States). EMSA assays were carried out with the Chemiluminescent EMSA Kit (Beyotime Biotechnology, China). After incubation at 25°C for 20 min, samples were loaded and separated on 6% non-denaturing PAGE gel in ice-cold 0.5% Tris-borate-EDTA at 100 V and determined by BeyoECL Plus.
RNA Preparation and RT-PCR
Mycobacterium smegmatis MC2 155 and its mutant strains were activated in LB medium (containing 0.05% tween 80) for 36–48 h at 37°C and then transferred into NXS and NL Sauton’s medium. Mid-exponential cells of M. smegmatis were collected by centrifugation (12000 rpm, 5 min, 4°C). Total RNA was obtained with the RNAprep Pure Cell/Bacteria kit (Tiangen Biotech, Beijing, China). The quality of RNA was analyzed by electrophoresis. The RNA concentrations were determined with microplate reader (BioTek). 1 μg RNA was used as template for cDNA synthesis with the PrimeScript reverse transcription (RT) regent kit (TaKaRa, Japan). The genomic DNA was removed before reverse transcription by DNase digestion for 5 min at 42°C. RT-PCR experiments were conducted with SYBR Premix Ex Taq GC kit (TaKaRa, Japan), using the primers listed in Supplementary Table S2. PCR assays were carried out as described previously (You et al., 2017).
Construction of whiB3MU Strain
A whiB3MU mutant was generated by electroporation with recombined plasmid pPR27 (Pelicic et al., 1997) which consisted of the mutated GlnR-box (consistent with the Figure 1C) and kanamycin resistance. The recombination cassette was obtained through homologous recombination and confirmed by PCR analysis and DNA sequencing. After the above cloning steps, the recombination cassette was transferred into the suicide plasmid pPR27 to generate final plasmid pPR27-whiB3MU and then introduced into M. smegmatis strains by electroporation. The selected mutants were further confirmed by PCR analysis and DNA sequencing.
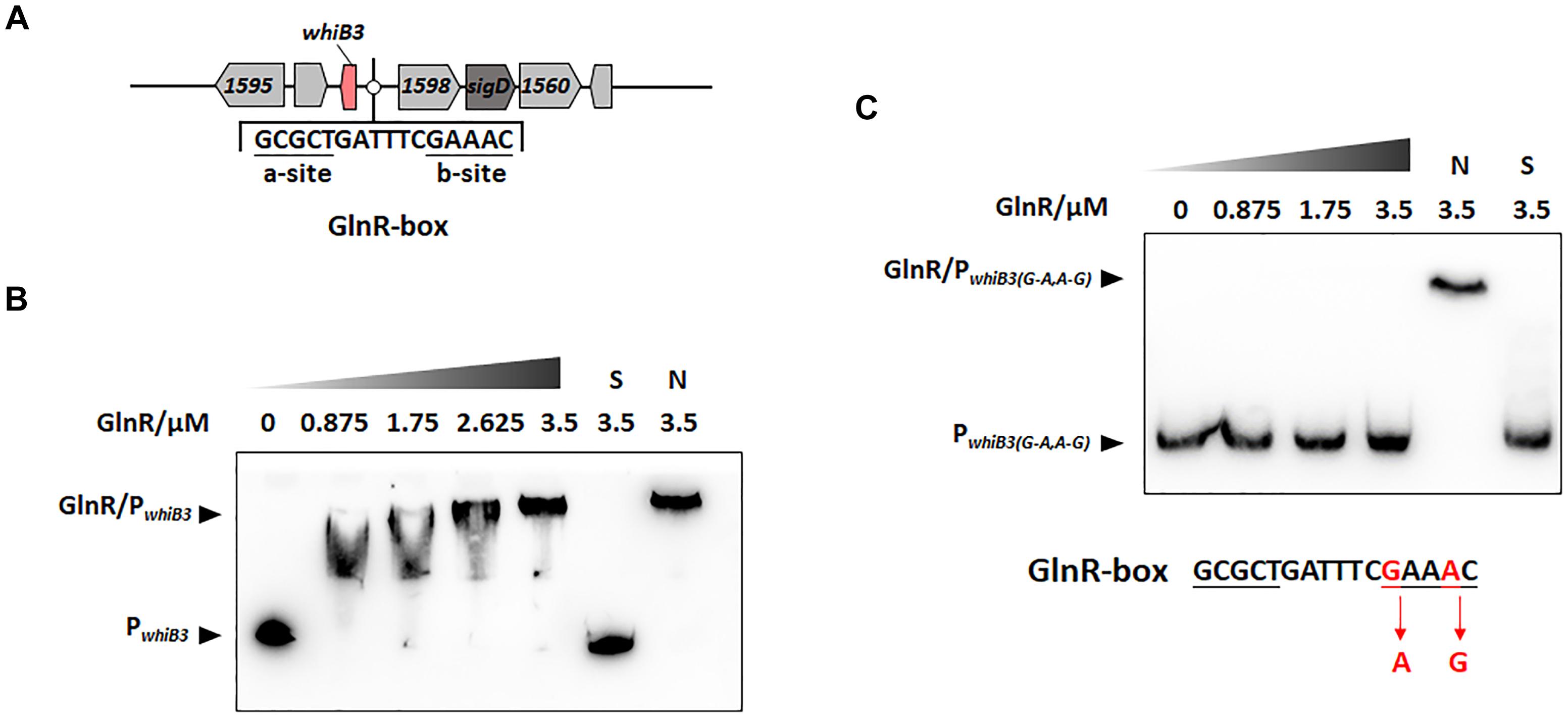
Figure 1. GlnR binding with the upstream promoter region of whiB3 in M. smegmatis. (A) GlnR-binding site (GlnR-box) in whiB3 promoter region. (B) EMSA of purified GlnR with the whiB3 promoter region. 10 nM DNA probes were incubated with a gradient concentration of purified GlnR. (C) EMSAs of purified GlnR protein with mutated whiB3 promoter regions.
Overexpression of WhiB3 in M. smegmatis
With M. smegmatis genomic DNA as template, whib3 was amplified with the primers listed in Supplementary Table S2. Purified PCR products were digested with BamHI and HindIII, then inserted into the corresponding sites of integrative plasmid pMV261 previously digested with the same enzymes. By electroporation transformation (Manganelli et al., 2001), the plasmid was introduced into M. smegmatis strains (wild type and ΔglnR). The final overexpression strains were screened using kanamycin resistance and confirmed by PCR with primers PMV-F and PMV-R (Supplementary Table S2).
CFU Colony Count
Mycobacterium smegmatis WT, ΔglnR, ΔglnR::glnR and ΔglnR::whiB3 strains were inoculated in 5 mL LB medium with Tween for approximately 48 h. Then transferred into NL Sauton’s medium with different pH and different H2O2 concentrations at 37°C for 24 h. Cultures unexposed to acid or H2O2 were used as controls. The exposed/unexposed cultures were then diluted for bacterial viability at the indicated time points.
Construction of the whiB3-lacZ+ Reporters
DNA fragments containing the lacZ gene from Escherichia coli (E. coli str. K-12 substr. MG1655) were amplified by PCR assays using lacZ-F-O and H-HindIII-lacZ-R primers (Supplementary Table S2). The promoter of whiB3 gene was amplified using the H-KpnI-1597P-F and 1597P-R-O primers (Supplementary Table S2). The two pairs of primers contained an overlap sequence. Using the above PCR products as template, H-KpnI-1597P-F and H-HindIII-lacZ-R as primers, lacZ gene and whiB3 promoter were linked together. The PCR fragment was then cloned into plasmid pMV261 (Stover et al., 1991). The recombinant plasmid was introduced into M. smegmatis strains by transformation of electroporation (Manganelli et al., 2001). The whiB3-lacZ+ reporter with kanamycin resistance was selected for subsequent experiments.
In vitroβ-Galactosidase Assays
In vitroβ-Galactosidase activity was analyzed using previous methods with some modifications. The strains contain lacZ were cultured in LB medium supplemented with 0.05% tween 80 and Sauton’s medium until the exponential phase. 10 μL cells were added into 990 μL Z buffer (40 mM NaH2PO4, 6 mM Na2HPO4, 10 mM KCl, 50 mM β-mercaptoethanol, 1 mM MgSO4). The mixtures were incubated for 15 min at room temperature. 200 μL substrate O-nitrophenyl-β-D-galactosidase (ONPG, 4 mg/mL in 100 mM KH2PO4, pH 7.0) was added, and the reaction was stopped with the addition of 0.2 mL 2.5 mM Na2CO3. Optical density of the solution at 420 nm was then measured. β-galactosidase activity in modified Miller units was calculated as following: (OD420 × 1000)/(t ×V × OD600), in which “V” represents the volume of culture used in milliliters and “t” represents the incubation time in minutes.
Polar Lipid Extraction and TLC Analysis
Lipids extraction, fractionation and analysis were performed as previously described (Jackson et al., 1999). The M. smegmatis WT and WhiB3 overexpression strain (OwhiB3) were activated in 5 mL LB medium with Tween for approximately 48 h. Then transferred into NL Sauton’s medium for 24 h. The bacteria were centrifuged at 12,000 × g for 10 min for several times to collect 50 mg bacteria (wet weight). Bacteria were extracted first with CHCl3/CH3OH (1:2, v/v), and then with CHCl3/CH3OH (2:1, v/v). After centrifugation, polar lipids were extracted in the supernatant. The dried lipid extracts were analyzed using chloroform: ethanol: water (90:10:1) by TLC. Glycolipid spots were visualized by spraying anthrone (0.2% in sulfuric acid), and followed with charring at 115°C. The band intensities were quantified by densitometry with Image J software using the Substract Background option.
Results
Nitrogen Response Regulator GlnR Directly Activated whiB3 Gene in M. smegmatis
The DNA binding sites of GlnR (GlnR-box) in actinobacteria (Yao et al., 2014) and Mycobacteria (Xu et al., 2017; Liu et al., 2018) were identified in previous studies, the putative GlnR-box was identified in the upstream region of whiB3 gene in M. smegmatis, consisting of a-site and b-site separated with six nucleotides (a-site-n6-b-site) (Figure 1A). To investigate whether GlnR could directly bind to the whiB3 upstream region, EMSA was performed. A 200-fold excess of unlabeled specific probes (S) and non-specific competitor DNA (sperm DNA) (N) were used as controls. The results in Figure 1B showed obvious shift bands following incubation with purified His-tag GlnR, suggesting that GlnR bound to the promoter region of whiB3. A mutation of GlnR-box was constructed (G mutated to A, A mutated to G in the conserved site of b-site shown in red, Figure 1C) and then subjected to EMSA assay. EMSA revealed that the mutation inhibited GlnR binding with whiB3 promoter region (Figure 1C). The result indicated that these nucleotides in the b-site are essential for binding of GlnR to the whiB3 promoter region.
We then examined whether GlnR has regulatory effect on whiB3 using M. smegmatis glnR-deletion strain (ΔglnR) and glnR complemented strain (ΔglnR::glnR) constructed as described previously (Xu et al., 2017; Liu et al., 2018). During growth in NL medium, the transcription level of whiB3 in M. smegmatis wild type (WT), ΔglnR and complemented strains were detected. As shown in Figure 2A, glnR deficiency caused a 70% decrease in whiB3 transcription compared with the WT strain, and was restored in the glnR complemented strain. Thus, GlnR directly activated the transcription of whiB3 gene in M. smegmatis.
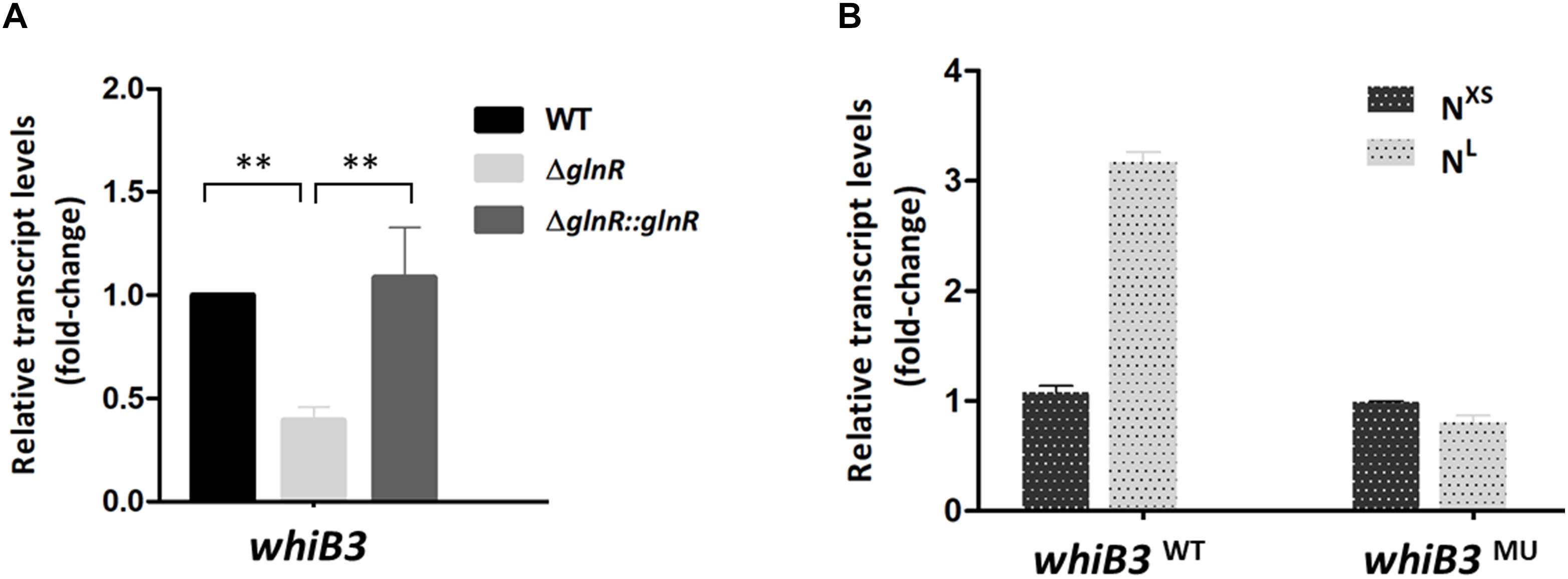
Figure 2. The whiB3 expression is directly controlled by GlnR. (A) The whiB3 transcription levels in Mycobacterium smegmatis WT, ΔglnR and ΔglnR::glnR strains grown in NL medium till the middle exponential phase. Fold change represented the expression level compared to the whiB3 expression in WT strain. (B) The whiB3 transcription levels in M. smegmatis whiB3WT and whiB3MU strains grown in NXS/NL condition. Fold change represented the whiB3 transcription level compared to whiB3WT strains grown in the NXS condition. The error bars showed standard deviations from three independent experiments. ∗∗P < 0.01.
Intracellular whiB3 Expression Is Influenced by Nitrogen Availability
The transcription response of whiB3 gene to nitrogen signals was then investigated. Using homologous replacement, we constructed a mutant strain (whiB3MU) with a mutant GlnR-box (GCGCTGATTTCAAAGC) in the whiB3 promoter region as used in the EMSA experiment above (Figure 1C). To confirm GlnR is necessary in the whiB3 expression changes influenced by nitrogen, whiB3 with wild type GlnR-box (whiB3WT) was used as control. The two strains were grown in NXS or NL media. As shown in Figure 2B, in the whiB3WT strain, the nitrogen starvation resulted in a 3-fold higher whiB3 expression compared to the level in NXS media. While in the whiB3MU strain, whiB3 transcription showed almost no response to nitrogen signals. EMSA result also revealed that GlnR did not bind with the mutant GlnR-box in vitro (Figure 1C). Taken together, these observations demonstrated that whiB3 expression was influenced by GlnR-mediated nitrogen availability in M. smegmatis.
WhiB3 Is Responsible for Response to Acidic and Redox Stress
In Mycobacterium tuberculosis, WhiB3 is reported necessary for maintaining redox homeostasis (Singh et al., 2009). Here we constructed a chromosomal whiB3-lacZ+ reporter to monitor the whiB3 transcription changes to pH and redox stress in M. smegmatis. The whiB3-lacZ+ reporter strain was confirmed by PCR and SDS-PAGE analysis, then screened by β-galactosidase activity, leading to the final identification of whiB3-lacZ+-4 strain (Supplementary Figure S1). The β-galactosidase activity was used to reflect whiB3 expression level accordingly. The reporter strain was cultured in Sauton’s medium for 24 h and transferred to fresh Sauton’s medium with different pH or H2O2 concentration prior to exponential phase. The strains transferred directly to Sauton’s medium with pH 7.0 or containing 0 mM H2O2 were used as controls, respectively. β-galactosidase activity in the M. smegmatis reporter strain was measured under different pH and H2O2 concentration, it achieved ∼5.5-fold higher at pH 4.5 than the control strain, and 10-fold higher when grown in 1.5 mM H2O2 concentration (Figures 3A,C). Meanwhile, the RT-PCR results displayed a relatively consistent trend of whiB3 expression in response to different environment stress (Figures 3B,D). According to these data, we concluded that oxidative stress and acidic (low pH) conditions induced WhiB3 expression in M. smegmatis.
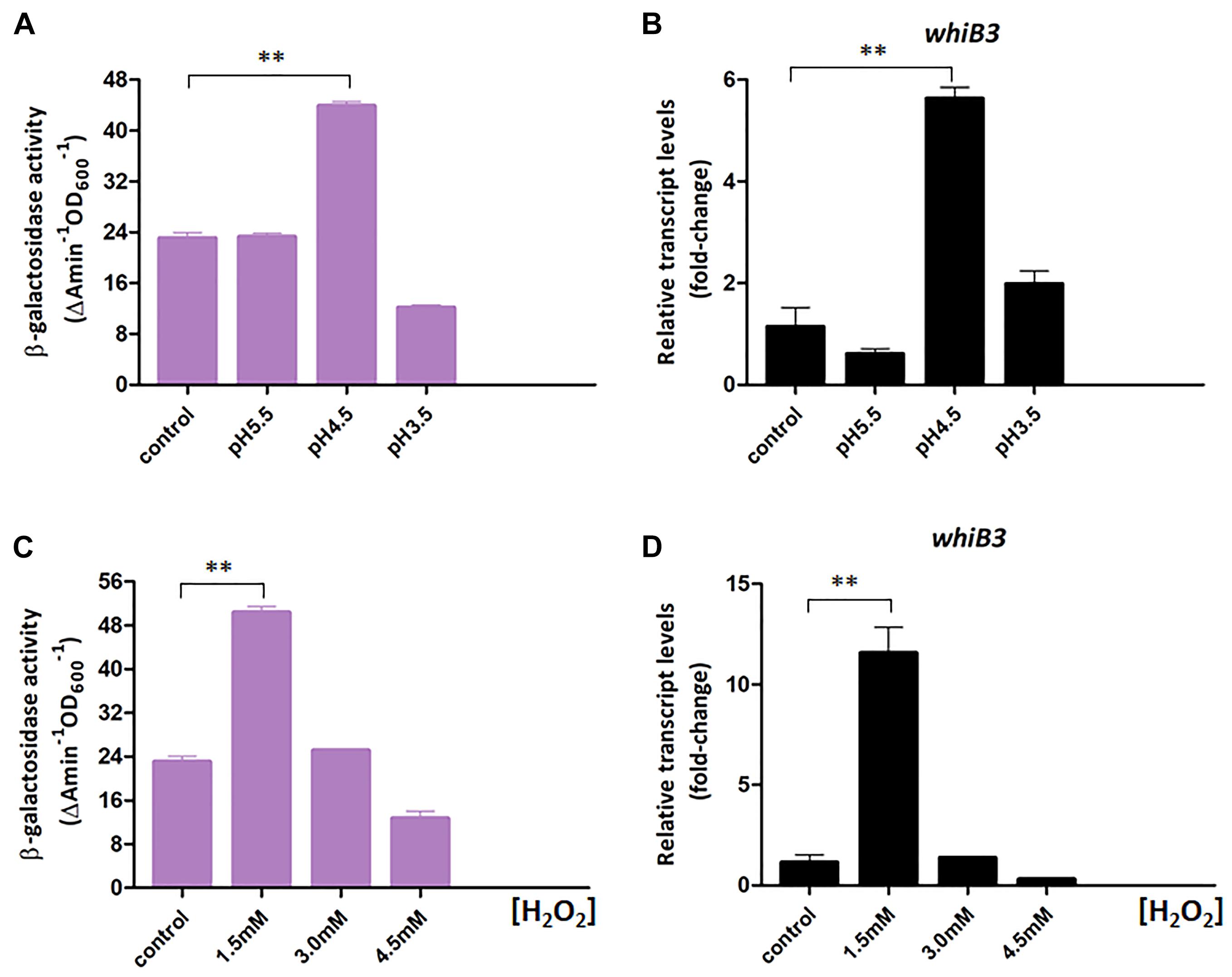
Figure 3. WhiB3 is responsible for response to environmental stress. (A) The β-galactosidase activity of M. smegmatis WT strain under different pH conditions. (B) The expression of whiB3 under different pH conditions. (C) The β-galactosidase activity of M. smegmatis WT strain under different H2O2 concentrations. (D) The expression of whiB3 under different H2O2 concentrations. Error bars represented standard deviations of three independent experiments. ∗∗P < 0.01.
M. smegmatis Responses to Environmental Stress Are Influenced by Lack of GlnR
The GlnR-mediated WhiB3 was responsible for the response to redox and acidic stress, raising a possibility that GlnR activation of M. smegmatis grown in NL condition might exert an impact on the response to redox and acidic stress. We thus investigated whether GlnR influenced M. smegmatis growth under stress exposures via comparing the survival of WT, ΔglnR, and ΔglnR::glnR cells in different pH and H2O2 concentrations. The M. smegmatis WT, ΔglnR and ΔglnR::glnR strains were first activated in LB medium and then transferred into NL Sauton’s medium with different pH or different H2O2 concentration, respectively. After 24 h incubation, the CFU was determined. As shown in Figures 4A,B, ΔglnR strain showed higher sensitivity to redox and acidic stress, the cell viability has a ∼2.4-fold decrease at pH 4.5 and ∼2.5-fold decrease in 3 mM H2O2. The resistance was substantially restored in the ΔglnR-complemented strain. When grown under pH 4.5 or 3 mM H2O2 condition, respectively, the most obvious differences in response to redox and acidic stress were observed (Figures 4A,B). The growth defect of ΔglnR strain under redox and acidic stress indicated that the phenotype might due to decrease of whiB3 expression in ΔglnR strain. Next, we examined the cell survival of ΔglnR strain overexpressing WhiB3 (ΔglnR::whiB3) under pH 4.5 or 3 mM H2O2 condition. As shown in Figures 4C,D, the peroxide and acidic stress sensitivity of ΔglnR was substantially restored after overexpression of whib3, suggesting that the peroxide and acidity resistance was mainly contributed by GlnR-mediated activation of whiB3.
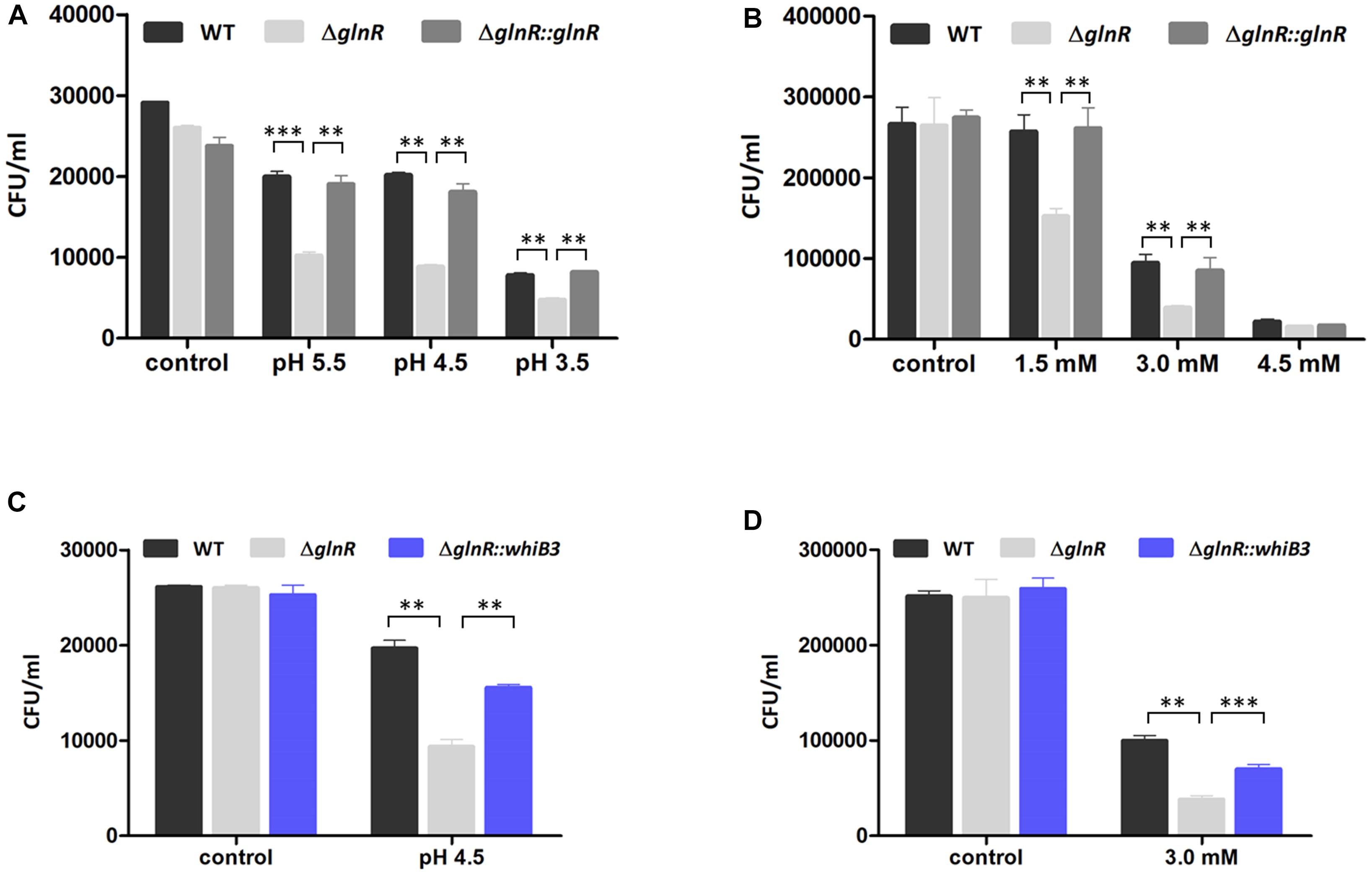
Figure 4. GlnR influenced the resistance of M. smegmatis to acidic and oxidative stress. (A) Effect of different pH condition on survival of WT, ΔglnR and ΔglnR-complemented strains grown in NL Sauton’s medium. (B) Effect of different H2O2 concentration on survival of M. smegmatis strains grown in NL Sauton’s medium. (C) Survival of WT, ΔglnR and ΔglnR::whib3 strains grown in NL Sauton’s medium with pH 4.5. (D) Survival of WT, ΔglnR and ΔglnR::whib3 strains grown in NL Sauton’s medium with 3 mM H2O2. Error bars showed standard deviations from three independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
The Transcription of Polyketide Synthase (MSMEG_4727, pks5) Is Influenced by GlnR-Mediated Nitrogen Availability
WhiB3 was identified as a physiological regulator of virulence lipid anabolism (Singh et al., 2009; Cumming et al., 2017). In M. tuberculosis, WhiB3 directly regulated the expression of polyketide biosynthetic gene pks2 (necessary for SL-1 production), pks3 (necessary for PAT/DAT production), ppsA, mas, fbpA (necessary for TDM production) fadD26 or fadD28 (necessary for PDIM production), hence influenced the production of complex lipids (Singh et al., 2009). The MSMEG_4727 gene (pks5) in M. smegmatis was identified to be homologous to M. tuberculosis pks2 with a 65% identity according to the KEGG database. To validate whether pks5 was the target gene of WhiB3 in M. smegmatis, the EMSA assay was performed with apo-WhiB3 protein treated with diamide as described previously (Singh et al., 2009) under anaerobic conditions. As shown in Figure 5A, the result revealed a direct binding interaction, as DNA probes clearly shifted following incubation with WhiB3.
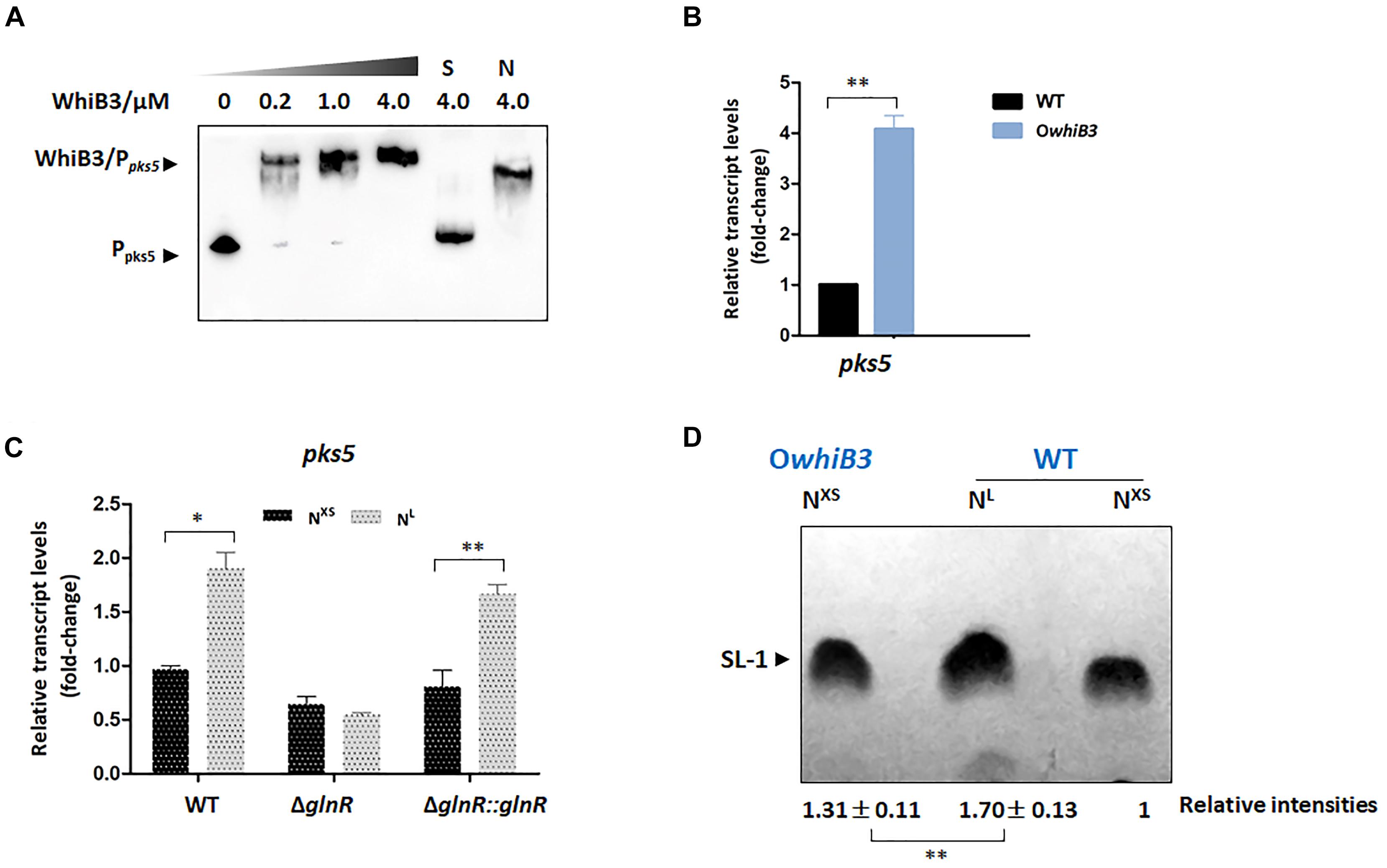
Figure 5. The biosynthesis of SL-1 lipid is regulated by GlnR-mediated WhiB3. (A) EMSA of apo-WhiB3 protein with pks5 promoter region. A concentration gradient of apo-WhiB3 was incubated with 10 nM DNA probes. (B) The pks5 transcript levels in M. smegmatis WT and OwhiB3 strains grown in NL medium till the middle exponential phase. Fold changes represented the expression level compared to the pks5 expression of WT. (C) The pks5 transcript levels in M. smegmatis WT, ΔglnR and ΔglnR::glnR strains grown in NXS/NL Sauton’s medium. Fold changes represented the pks5 transcript levels compared to the pks5 expression in NXS condition. (D) M. smegmatis WT and OwhiB3 strains grown in NXS/NL Sauton’s medium. In each lane, silica TLC plates were loaded with equal amount of polar lipids and developed in chloroform: ethanol: water (90:10:1). The band intensities were quantified by densitometry using Image J software. Relative band intensities were shown. Error bars showed standard deviations from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
To investigate the regulatory effect of WhiB3 on pks5, we tried to construct the M. smegmatis ΔwhiB3 strains, unfortunately, we were unable to construct the deletion strain. The WT strain and WhiB3 overexpression strain (OwhiB3) were then used for the RT-PCR experiments. The data showed that pks5 transcript level was markedly increased (4-fold) in OwhiB3 strain (Figure 5B), indicating that WhiB3 directly activated the transcription of pks5. The impact of nitrogen availability on pks5 transcription was then analyzed. We compared pks5 transcription level under defined NXS and NL conditions. The result showed that transcript level of pks5 was approximately 2-fold higher under the NL condition compared with the NXS condition, while in the ΔglnR mutant, no obvious changes in pks5 transcription level were observed in response to nitrogen starvation. The glnR complementation partially restored the NL activation effect as observed in WT strain (Figure 5C). These results indicated that GlnR activated pks5 transcription during nitrogen starvation.
GlnR Activates the Synthesis of SL-1 Lipid Through WhiB3
The SL-1 lipid was informed to play a crucial role in organization and pathogenesis of M. tuberculosis cell envelopes and pks2 was demonstrated necessary for its biosynthesis (Jackson et al., 2007). We chose to evaluate the effect of GlnR and nitrogen availability on the SL-1 production using thin layer chromatography (TLC) assay and quantified the band intensities using Image J software. As shown in Figure 5D, production of SL-1 had a 31% increase in M. smegmatis OwhiB3 over WT strain when grown in the same condition, which confirmed that WhiB3 was a positive regulator for SL-1 production. Next, we analyzed the effect of GlnR-mediated nitrogen availability on SL-1 production. In M. smegmatis WT strain, the production of SL-1 showed an obvious 70% improvement in response to nitrogen starvation (Figure 5D). Since GlnR was activated during M. smegmatis adaptation in nitrogen starvation, the implications of these findings strongly suggested that GlnR activated the synthesis of SL-1 lipid through WhiB3, which exerted new influence of nitrogen metabolism on lipid anabolism in M. smegmatis.
Discussion
The WhiB-like protein family is reported to occupy an important place in actinobacteria pathogenesis and biology. The known function of WhiB in mycobacteria is as a sensor and regulator: sensing fluctuations of the intracellular redox state to maintain redox balance, and regulating the production of inflammatory lipids including PAT, DAT, SL-1, PDIM, and TAG via a redox-dependent switching mechanism (Rohde et al., 2007; Singh et al., 2007, 2009; Cumming et al., 2017). Both of which are associated with normal cellular metabolism, especially the extensively studied carbon metabolism. However, the mechanisms associated with nitrogen metabolism have not yet been established.
Nitrogen is one of the essential elements required for bacterial growth. Bacteria must adapt to nitrogen limitation for survival. In M. smegmatis, the transcriptional response to nitrogen limitation is thought to be regulated by OmpR-family protein GlnR (Amon et al., 2008, 2009, 2010). At present, knowledge of the genetic response to nitrogen starvation and nitrogen metabolism in mycobacteria is still limited (Malm et al., 2009; Jessberger et al., 2013). In this article, we provided unique insight into the regulation mechanism of nitrogen regulator GlnR on WhiB3 in M. smegmatis and proved that GlnR bound directly to whiB3 promoter region, exerting regulatory effect on whiB3 transcription as a strong positive regulator. Furthermore, GlnR was demonstrated essential in the survival of M. smegmatis under acidic and oxidative stress.
WhiB3 was reported as a regulator of complex virulence lipid production including SL-1, PAT, PDIM, and TAG. SL-1 was suggested to play a key role in the virulence of M. tuberculosis. SL-I prevented fusion of phagosome and lysosome in macrophages, inhibited mitochondrial oxidative phosphorylation, modulated the cytokine secretion and oxidative responses of human neutrophils and monocytes (Jackson et al., 2007). The pks2 expression was necessary for the synthesis of the hydroxyphthioceranic and phthioceranic acids found in SL-1 (Graham and Clark-Curtiss, 1999). In M. smegmatis, pks5, homologous to M. tuberculosis pks2, was verified as target gene of WhiB3 and its transcription was strongly activated by WhiB3. Consequently, the SL-1 synthesis was under control of WhiB3 and its regulatory factor GlnR. It is noteworthy that M. smegmatis acquire more SL-1 lipid during nitrogen starvation, revealing positive regulatory effect of GlnR on SL-1 production. It could be speculated that transcription of other WhiB3 targets such as fbpA (necessary for TDM production), pks3 (necessary for PAT/DAT production), ppsA, mas, fadD26 or fadD28 (necessary for PDIM production) might also be regulated by GlnR-mediated nitrogen signals, which hence influenced the production of complex lipids.
Recent studies have shown that GlnR regulates short-chain fatty acid assimilation (Liu et al., 2018), post-translational modifications (Xu et al., 2017) and mycobacterial biofilm development (Yang et al., 2018) in mycobacteria. These facts further supported a global role of GlnR expanding beyond nitrogen assimilation and widely influencing mycobacterial adaptation. Moreover, our observation here confirmed that GlnR regulate redox sensing and lipid anabolism via transcriptional control of WhiB3. These findings suggest a tight connection between intracellular redox status, virulence and nitrogen metabolism (Figure 6). Due to the high homology of WhiB3 between M. smegmatis and M. tuberculosis, our research might serve as an enrichment of pathogen biology and the foundation for new therapeutic discovery efforts.
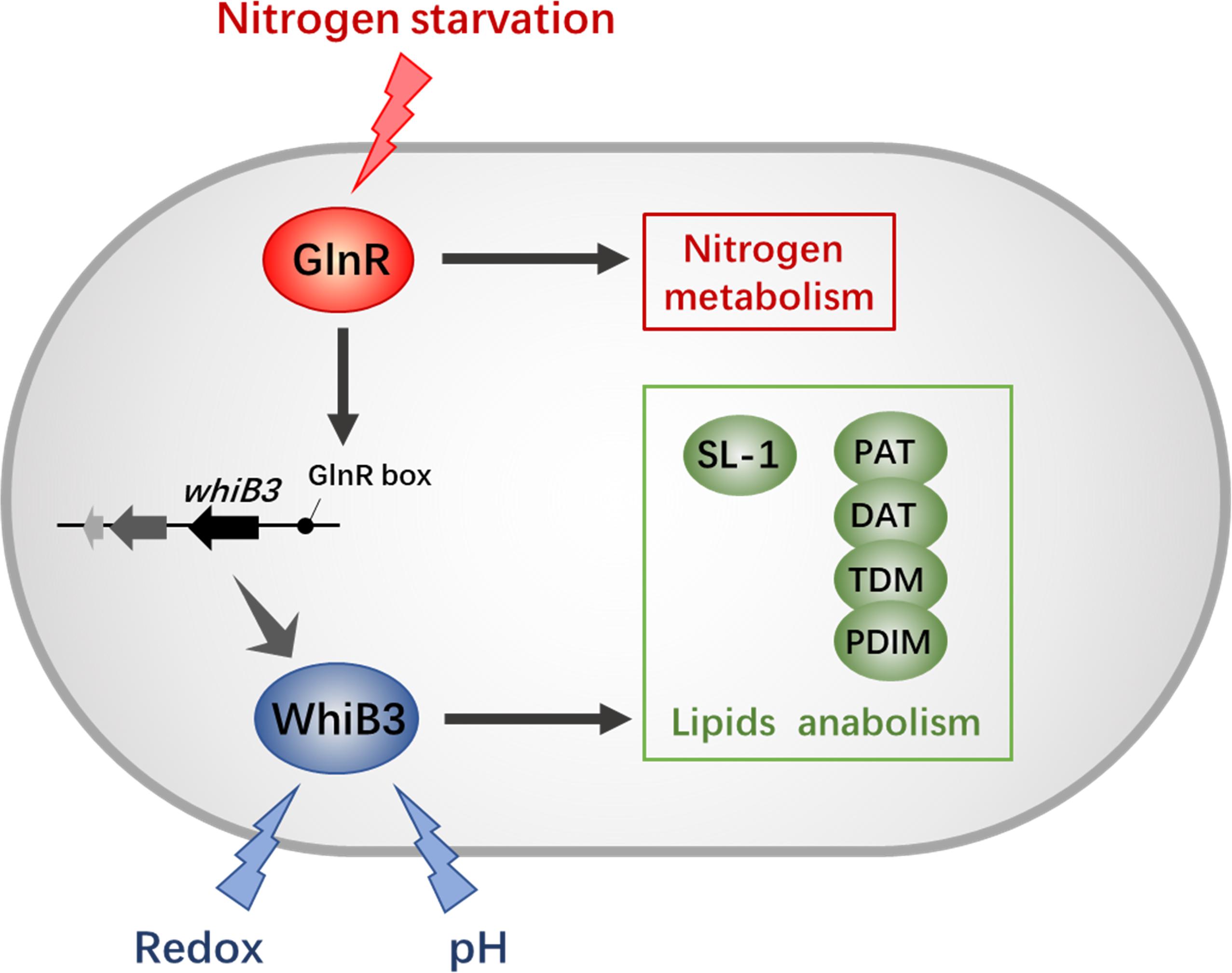
Figure 6. The GlnR-mediated network of redox sensing and lipid anabolism. The black solid lines with arrows indicate positive transcriptional regulation; arrows indicate the positive control. The GlnR box indicates a GlnR-binding motif.
Author Contributions
Ba-CY and DY designed the research. DY, YX, and Bi-CY performed the experiments. DY, YX, Bi-CY, and Ba-CY contributed to data analysis and manuscript writing. All authors contributed to manuscript revision and approved the submission.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31730004, 31700058, and 21575089), the China Postdoctoral Science Foundation (2017M610232), and the Fundamental Research Funds for the Central Universities (222201714025).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00074/full#supplementary-material
References
Alam, M. S., Garg, S. K., and Agrawal, P. (2009). Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 276, 76–93. doi: 10.1111/j.1742-4658.2008.06755.x
Amon, J., Brau, T., Grimrath, A., Hanbler, E., Hasselt, K., Holler, M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. doi: 10.1128/JB.00855-08
Amon, J., Titgemeyer, F., and Burkovski, A. (2009). A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 17, 20–29. doi: 10.1159/000159195
Amon, J., Titgemeyer, F., and Burkovski, A. (2010). Common patterns - unique features: nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol. Rev. 34, 588–605. doi: 10.1111/j.1574-6976.2010.00216.x
Barry, C. E. III, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A., et al. (1998). Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37, 143–179. doi: 10.1016/S0163-7827(98)00008-3
Brennan, P. J., and Nikaido, H. (1995). The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63. doi: 10.1146/annurev.bi.64.070195.000333
Chater, K. F. (1972). A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 72, 9–28. doi: 10.1099/00221287-72-1-9
Cumming, B. M., Rahman, M. A., Lamprecht, D. A., Rohde, K. H., Saini, V., Adamson, J. H., et al. (2017). Mycobacterium tuberculosis arrests host cycle at the G(1)/S transition to establish long term infection. PLoS Pathog. 13:e1006389. doi: 10.1371/journal.ppat.1006389
Daffe, M., and Draper, P. (1998). The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39, 131–203. doi: 10.1016/S0065-2911(08)60016-8
Dye, C., Scheele, S., Dolin, P., Pathania, V., and Raviglione, R. C. (1999). Global burden of tuberculosis - estimated incidence, prevalence, and mortality by country. WHO global surveillance and monitoring project. JAMA 282, 677–686. doi: 10.1001/jama.282.7.677
Flardh, K., Findlay, K. C., and Chater, K. F. (1999). Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145(Pt 9), 2229–2243. doi: 10.1099/00221287-145-9-2229
Gomez, J. E., and Bishai, W. R. (2000). whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl. Acad. Sci. U.S.A. 97, 8554–8559. doi: 10.1073/pnas.140225297
Graham, J. E., and Clark-Curtiss, J. E. (1999). Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U.S.A. 96, 11554–11559. doi: 10.1073/pnas.96.20.11554
Jackson, M., Raynaud, C., Laneelle, M. A., Guilhot, C., Laurent-Winter, C., Ensergueix, D., et al. (1999). Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31, 1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x
Jackson, M., Stadthagen, G., and Gicquel, B. (2007). Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis 87, 78–86. doi: 10.1016/j.tube.2006.05.003
Jenkins, V. A., Barton, G. R., Robertson, B. D., and Williams, K. J. (2013). Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 14:301. doi: 10.1186/1471-2164-14-301
Jessberger, N., Lu, Y., Amon, J., Titgemeyer, F., Sonnewald, S., Reid, S., et al. (2013). Nitrogen starvation-induced transcriptome alterations and influence of transcription regulator mutants in Mycobacterium smegmatis. BMC Res. Notes 6:482. doi: 10.1186/1756-0500-6-482
Kim, T. H., Park, J. S., Kim, H. J., Kim, Y., Kim, P., and Lee, H. S. (2005). The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337, 757–764. doi: 10.1016/j.bbrc.2005.09.115
Kolattukudy, P. E., Fernandes, N. D., Azad, A. K., Fitzmaurice, A. M., and Sirakova, T. D. (1997). Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24, 263–270. doi: 10.1046/j.1365-2958.1997.3361705.x
Liu, X. X., Shen, M. J., Liu, W. B., and Ye, B. C. (2018). GlnR-mediated regulation of short-chain fatty acid assimilation in Mycobacterium smegmatis. Front. Microbiol. 9:1311. doi: 10.3389/fmicb.2018.01311
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339. doi: 10.1099/mic.0.023275-0
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., and Smith, I. (2001). The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437. doi: 10.1046/j.1365-2958.2001.02525.x
Morris, R. P., Nguyen, L., Gatfield, J., Visconti, K., Nguyen, K., Schnappinger, D., et al. (2005). Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 12200–12205. doi: 10.1073/pnas.0505446102
Pelicic, V., Jackson, M., Reyrat, J. M., Jacobs, W. R. Jr., Gicquel, B., and Guilhot, C. (1997). Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10955–10960. doi: 10.1073/pnas.94.20.10955
Primm, T. P., Andersen, S. J., Mizrahi, V., Avarbock, D., Rubin, H., and Barry, C. E. (2000). The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000
Raghunand, T. R., and Bishai, W. R. (2006). Mapping essential domains of Mycobacterium smegmatis WhmD: insights into WhiB structure and function. J. Bacteriol. 188, 6966–6976. doi: 10.1128/JB.00384-06
Rifat, D., Bishai, W. R., and Karakousis, P. C. (2009). Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 200, 1126–1135. doi: 10.1086/605700
Rohde, K. H., Abramovitch, R. B., and Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2, 352–364. doi: 10.1016/j.chom.2007.09.006
Singh, A., Crossman, D. K., Mai, D., Guidry, L., Voskuil, M. I., Renfrow, M. B., et al. (2009). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545. doi: 10.1371/journal.ppat.1000545
Singh, A., Guidry, L., Narasimhulu, K. V., Mai, D., Trombley, J., Redding, K. E., et al. (2007). Mycobacterium tuberculosis WhiB3 responds to O-2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U.S.A. 104, 11562–11567. doi: 10.1073/pnas.0700490104
Steyn, A. J. C., Collins, D. M., Hondalus, M. K., Jacobs, W. R., Kawakami, R. P., and Bloom, B. R. (2002). Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. U.S.A. 99, 3147–3152. doi: 10.1073/pnas.052705399
Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., et al. (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. doi: 10.1038/351456a0
Wu, M. L., Gengenbacher, M., Chung, J. C., Chen, S. L., Mollenkopf, H. J., Kaufmann, S. H., et al. (2016). Developmental transcriptome of resting cell formation in Mycobacterium smegmatis. BMC Genomics 17:837. doi: 10.1186/s12864-016-3190-4
Xu, Y., You, D., and Ye, B. C. (2017). Nitrogen regulator GlnR directly controls transcription of genes encoding lysine deacetylases in actinobacteria. Microbiology 163, 1702–1710. doi: 10.1099/mic.0.000553
Yang, Y., Richards, J. P., Gundrum, J., and Ojha, A. K. (2018). GlnR activation induces peroxide resistance in mycobacterial biofilms. Front. Microbiol. 9:1428. doi: 10.3389/fmicb.2018.01428
Yao, L. L., Liao, C. H., Huang, G., Zhou, Y., Rigali, S., Zhang, B. C., et al. (2014). GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 98, 7935–7948. doi: 10.1007/s00253-014-5878-1
You, D., Wang, M. M., and Ye, B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103, 845–859. doi: 10.1111/mmi.13595
You, D., Yao, L. L., Huang, D., Escalante-Semerena, J. C., and Ye, B. C. (2014). Acetyl coenzyme a synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-Type N-acetyltransferase (GNAT) domain in Saccharopolyspora erythraea. J. Bacteriol. 196, 3169–3178. doi: 10.1128/JB.01961-14
Keywords: Mycobacterium, GlnR, WhiB3, nitrogen metabolism, SL-1, redox stress
Citation: You D, Xu Y, Yin B-C and Ye B-C (2019) Nitrogen Regulator GlnR Controls Redox Sensing and Lipids Anabolism by Directly Activating the whiB3 in Mycobacterium smegmatis. Front. Microbiol. 10:74. doi: 10.3389/fmicb.2019.00074
Received: 02 October 2018; Accepted: 15 January 2019;
Published: 29 January 2019.
Edited by:
Chris Greening, Monash University, AustraliaReviewed by:
Harold J. Schreier, University of Maryland – Baltimore County, United StatesAdrie J.C. Steyn, University of Alabama at Birmingham, United States
Copyright © 2019 You, Xu, Yin and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bang-Ce Ye, YmN5ZUBlY3VzdC5lZHUuY24=
 Di You1
Di You1 Bang-Ce Ye
Bang-Ce Ye