- 1Department of Epidemiology and Statistics, North China University of Science and Technology, Tangshan, China
- 2Department of Laboratory Medicine, The Fifth Affiliated Hospital of Shijiazhuang, North China University of Science and Technology, Shijiazhuang, China
The emergence of drug-resistant tuberculosis (TB) is involved in ineffective treatment of TB, especially multidrug resistant/extensively resistant TB (MDR/XDR-TB), leading to acquired resistance and transmission of drug-resistant strains. Second-line drugs (SLD), including both fluoroquinolones and injectable drugs, were commonly proved to be the effective drugs for treatment of drug-resistant TB. The purpose of this study was to investigate the prevalence of SLD-resistant strains and its specific mutations in drug-resistant Mycobacterium tuberculosis clinical isolates, and to acknowledge the transmission pattern of SLD resistance strains in Hebei. The genes gyrA, gyrB, rrs, eis promoter and tlyA of 257 drug-resistant clinical isolates were sequenced to identify mutations that could be responsible for resistance against fluoroquinolones and second-line injectable drugs. Each isolate was genotyped by Spoligotyping and 15-loci MIRU-VNTR. Our results indicated that 48.2% isolates were resistant to at least one of five SLD. Of them, 37.7% isolates were resistant to fluoroquinolones and 24.5% isolates were resistant to second-line injectable drugs. Mutations in genes gyrA, gyrB, rrs, eis promoter and tlyA were detected in 73 (75.3%), 7 (7.2%), 24 (38.1%), 5 (7.9%), and 3 (4.8%) isolates, respectively. The most prevalent mutations were the D94G (23.7%) in gyrA gene and the A1401G (33.3%) in rrs gene. A combination of gyrA, rrs and eis promoter can act as a valuable predicator for predicting XDR phenotype. These results highlight the development of rapid diagnosis are the effective manners for the control of SLD-TB or XDR-TB.
Introduction
Today, tuberculosis (TB) remains a major threat worldwide than any other single infectious disease. Millions of people continue to fall sick with TB each year. Moreover, the increasing rates of drug-resistant TB (DR-TB) worldwide and the emergence of multidrug/extensively-drug resistant TB (MDR/XDR-TB), leading to a high mortality as well as the financial burden. Regarding cases, estimates of the burden of DR-TB have focused on MDR-TB, there were 457,560 people (range: 396,060–523,980) developed MDR-TB (World Health Organization [WHO], 2018). Among cases of MDR-TB, 8.5% (95%CI: 6.2–11.0) were estimated to have XDR-TB (World Health Organization [WHO], 2018).
MDR-TB patients ordinarily require 18 months of treatment with recommendatory second-line anti-TB drugs (SLD) that primarily include fluoroquinolones (FQ) and second-line injectable drugs (SLID). Similar to other anti-TB agents, the main mechanisms of Mycobacterium tuberculosis (M. tuberculosis) resistant to SLD rely on spontaneous chromosomal mutations. Associated-mutations in the fluroquinolone resistance-determining region (QRDR) of DNA gyrase-coding genes gyrA and gyrB turned out to be responsible for FQ resistance (FQr) (Chien et al., 2016). The genetic determinants of SLID resistance (SLIDr) are more complicated and suggested that being involved in three well-known genes. Cross-resistance between kanamycin (KAN), amikacin (AMK) and capreomycin (CAP) is thought to be associated with mutations in the16S rRNA gene rrs, specifically at position A1401G (Alangaden et al., 1998; Oudghiri et al., 2018). Mutations in promoter region of eis gene, encoding an aminoglycoside acetyltransferase, caused low-level KAN resistance (Bauskenieks et al., 2015; Ngo et al., 2018). CAP resistance has been correlated with mutations in tlyA gene, which encodes a putative 2′-O-methyltransferase (TlyA) (Freihofer et al., 2016; Witek et al., 2017).
Currently, little is known about mutation profiles of SLD resistance (SLDr) in clinical M. tuberculosis isolates in our area, the objectives of this study were to compare the sequencing data of SLDr associated-genes (FQ for gyrA and gyrB, SLID for rrs region 1400, eis promoter and tlyA) with the phenotypic results by the traditional proportion method in 275 drug-resistant clinical M. tuberculosis isolates collected in Hebei, and to analyze the M. tuberculosis genetic diversity of SLDr-TB (covering FQr-TB and SLIDr-TB), and to elucidate transmission pattern by using Spoligotyping and 15-loci MIRU-VNTR typing.
Materials and Methods
M. tuberculosis Isolates
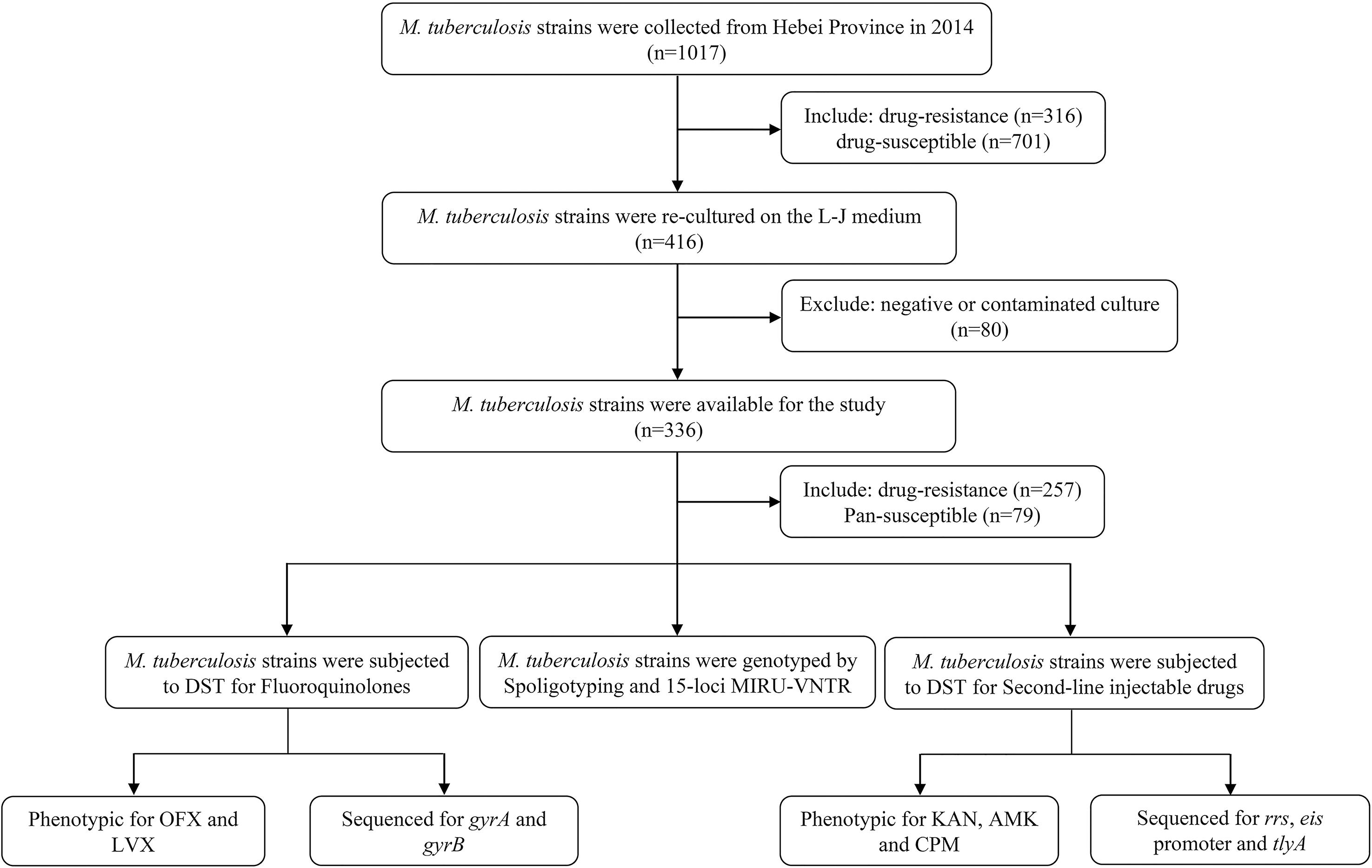
For the present study, we successfully recovered 257 clinical M. tuberculosis isolates. Isolates were obtained from sputum samples provided by confirmed pulmonary patients (167 males and 90 females; age range: 14–83 years; median age: 41 years; 146 new patients and 111 retreated patients) attending eight hospitals in Hebei over 1-year period (From January to December in 2014). One hundred pan-susceptible M. tuberculosis isolates served as negative control. Figure 1 shows the selection process of clinical M. tuberculosis isolates originated from Hebei Province.
Drug Susceptibility Testing
Drug susceptibility testing against SLD was performed using conventional proportion method on Lowenstein-Jensen (L-J) medium (Celnovte-bio, Henan, China) cultured for 4 weeks (World Health Organization [WHO], 1997), with the following break-out values: ofloxacin (OFX) 2 μg/mL; levofloxacin (LVX) 2 μg/mL; KAN 30 μg/mL; AMK 30 μg/mL, and CAP 40 μg/mL. Each DST was used M. tuberculosis H37Rv (ATC27294) as a reference control.
DNA Isolation, Amplification, and Sequencing
Genomic DNA extractions were re-suspended a loopful of bacilli in 200 uL of fast lysis buffer (Qiagen, Valencia, CA, United States) and heat inactivated at 80°C for 10 min, followed by centrifugation at 13,000 g for 5 min. All genes were chosen on the basis of documented association with resistance to SLD and included gyrA and gyrB for FQ and rrs, eis promoter, and tlyA for SLID. Primers and amplicon sizes are presented in the Supplementary Table S1. Each 20 μL PCR mixture was prepared as follows: 10 μL 2× Taq Master Mix (CWBOIO, Beijing, China), 1 μL of the forward and reverse 10 μM primers, 1 μL genomic DNA, and final 7 μL distilled H2O complement. PCR program for amplification were 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, 30 s at 72°C, and a final extension 72°C for 10 min.
Sequencing services were provided by Tsingke Biological Technology company (TsingKe, Beijing, China). All the mutations were identified and aligned with the homologous sequences of the reference M. tuberculosis H37Rv strain (GenBank accession number NC_000962) using the BLASTN algorithm1.
Genotyping
Spoligotyping was used to identify Beijing family as described previously by Kamerbeek et al. (1997). The direct repeat region was amplified with the primer pairs included DRa (5′-GGTTTTGGGTCTGACGAC-3′) and DRb (5′-CCGAGAGGGGACGGAAAC-3′). All PCR products were hybridized to a set of 43 oligonucleotide probes corresponding to each spacer that were covalently bound to a membrane. Spoligotypes in binary formats were matched with Spoldb.4.0 database2, and applied the published rules for definition of Beijing family (hybridization to at least three of the spacers 35–43 in direct repeat region and absence of hybridization to spacers 1–34).
MIRU-VNTR typing method was based on 15-loci set, including Mtub04, ETRC, ETRD, MIRU40, MIRU10, MIRU16, Mtub21, QUB11b, ETRA, Mtub30, MIRU26, ETRE, Mtub39, QUB26, QUB4156, as previously reported (Supply et al., 2001; Li et al., 2016). A phylogenetic tree was built using UPGMA algorithm at the web site MIRU-VNTRplus3.
Definitions
Based on the following definitions, we utilize these terms throughout the rest of this article. (i) Drug-resistant (DR): defined as resistance to at least one first- or second-line drug; (ii) Fluoroquinolones (FQ): defined as resistance to at least one OFX or LVX; (iii) Second-line injectable drugs (SLID): defined as resistance to at least one AMK, KAN, and CAP; (iv) Second-line anti-TB drugs (SLD): defined as resistance to at least one fluoroquinolones or second-line injectable drugs; (v) Multidrug-resistant (MDR): defined as resistance to at least two of the most effective anti-TB drugs, isoniazid and rifampin (World Health Organization [WHO], 2018); (vi) Pre-extensively drug-resistant (Pre-XDR): defined as MDR resistance to either fluoroquinolones or second-line injectable drugs; (vii) Extensively drug-resistant (XDR): defined as MDR-TB plus resistance to a fluoroquinolone and at least one second-line injectable drug (World Health Organization [WHO], 2018).
Data Analysis
To assess the associations between variables (DST, mutations, and genotyping), the χ2 test, odds ratio (OR) and 95% confidence interval (95%CI) were calculated. And the Fishers’ exact was used if any expected counts are less than 5. A P-value of < 0.05 was considered statistically significant. All statistical data analyzed with SPSS version 22.0 (IBM SPSS, Chicago, IL, United States).
Resolution of Discrepant Results
When the results between DST and DNA sequencing were not consistent, two methods were used for repeated testing. If the duplicate result conflicted with the original data, a third round of testing was accepted as a final value.
Results
Strains Selection and Drug Resistance Patterns
Initially, a total of 316 DR isolates were re-cultured and finally only 257 eligible isolates were included in this study. The selection procedure of strains is shown in Figure 1. A total 257 eligible isolates, 170 (170/257, 66.1%) isolates were resistant to isoniazid; 152 (152/257, 59.1%) to rifampicin; 74 (74/257, 28.8%) to ethambutol; and 176 (176/257, 56.8%) to streptomycin.
Among 257 DR isolates, 48.2% (124/257) were resistant to at least one of five SLD and the remaining 51.8% (133/257) were susceptible to all SLD. Of them, 37.7% (97/257) were FQr isolates and 24.5% (63/257) were SLIDr isolates. A total of 118 isolates (45.9%, 118/257) identified as MDR, of which 46 isolates (17.9%, 46/257) were Pre-XDR and 10.9% (28/257) were XDR. At the same time, there were 32.7% (84/257), 21.7% (56/257), 20.2% (52/257), 7.0% (18/257), and 6.2% (16/257) of 257 DR isolates were found to be resistant to OFX, LVX, KAN, AMK and CAP, respectively. Detailed profiles of 275 DR isolates against five SLD were summarized in Table 1.
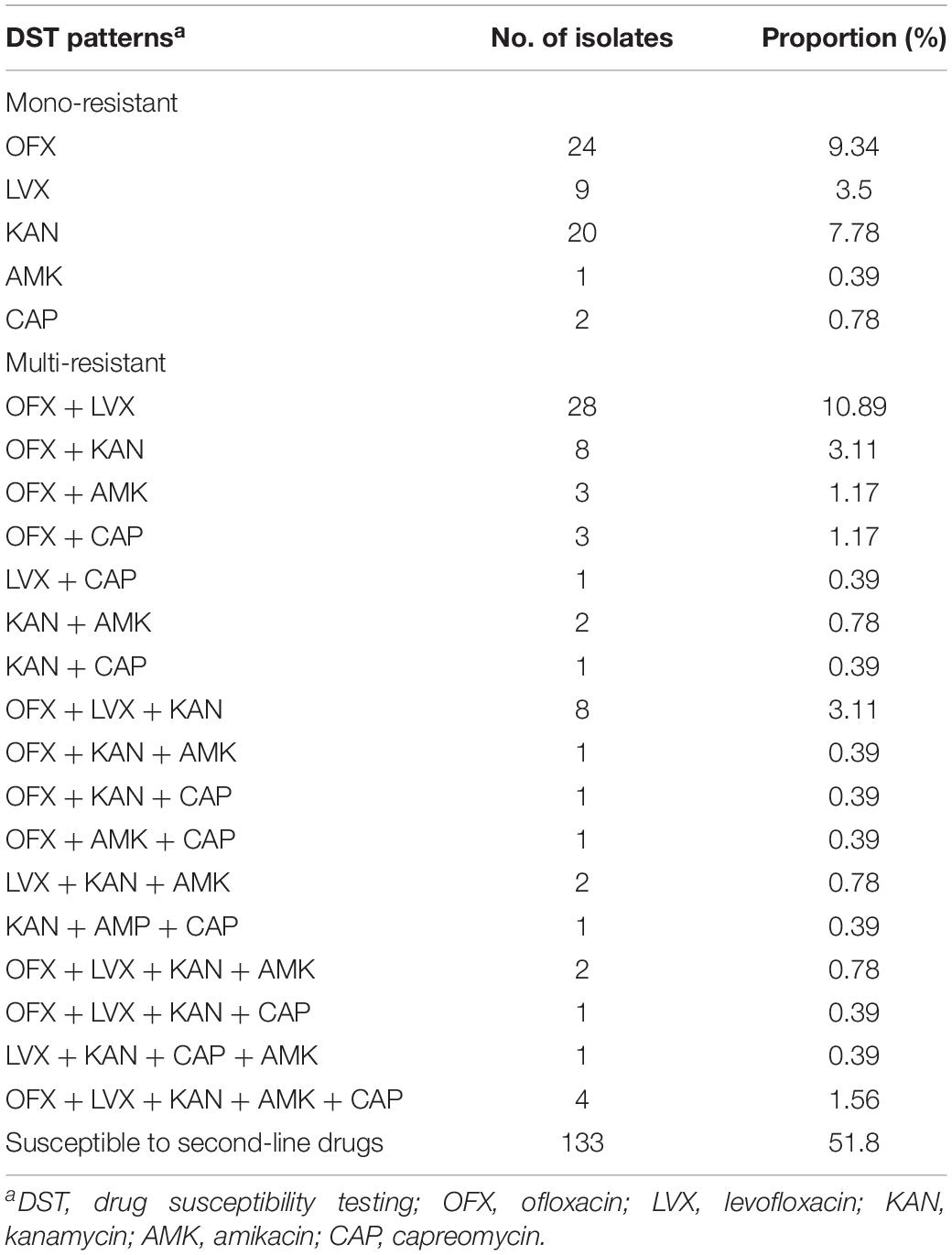
Table 1. Profiles of drug resistance to second-line drugs among the studied drug-resistant isolates.
Out of 84 OFXr isolates, 43 isolates (44.3%, 43/97) were simultaneously resistant to LVX (Figure 2A). Among 52 KANr isolates, 6 isolates were resistant to both AMK and CAP, the cross-resistance rate between KAN, AMK and CAP was 9.5% (6/64) (Figure 2B). Compared with FQr, there was significant difference in SLIDr (37.7 vs. 24.5%, 1.9 OR, 95%CI [1.3, 2.7], P = 0.001), MDR (37.7 vs. 45.9%, 2.6 OR, 95%CI [1.8, 3.8], P < 0.001), Pre-XDR (37.7 vs. 17.9%, 0.4 OR, 95%CI [0.2, 0.5], P < 0.001) and XDR (37.7 vs. 10.9%, 0.2 OR, 95%CI [0.1, 0.3], P < 0.001) (Figure 3). Detailed distribution of drug-resistant strains resistant to OFX, LVX, KAN, AMK and CAP as shown in Supplementary Figure S1.
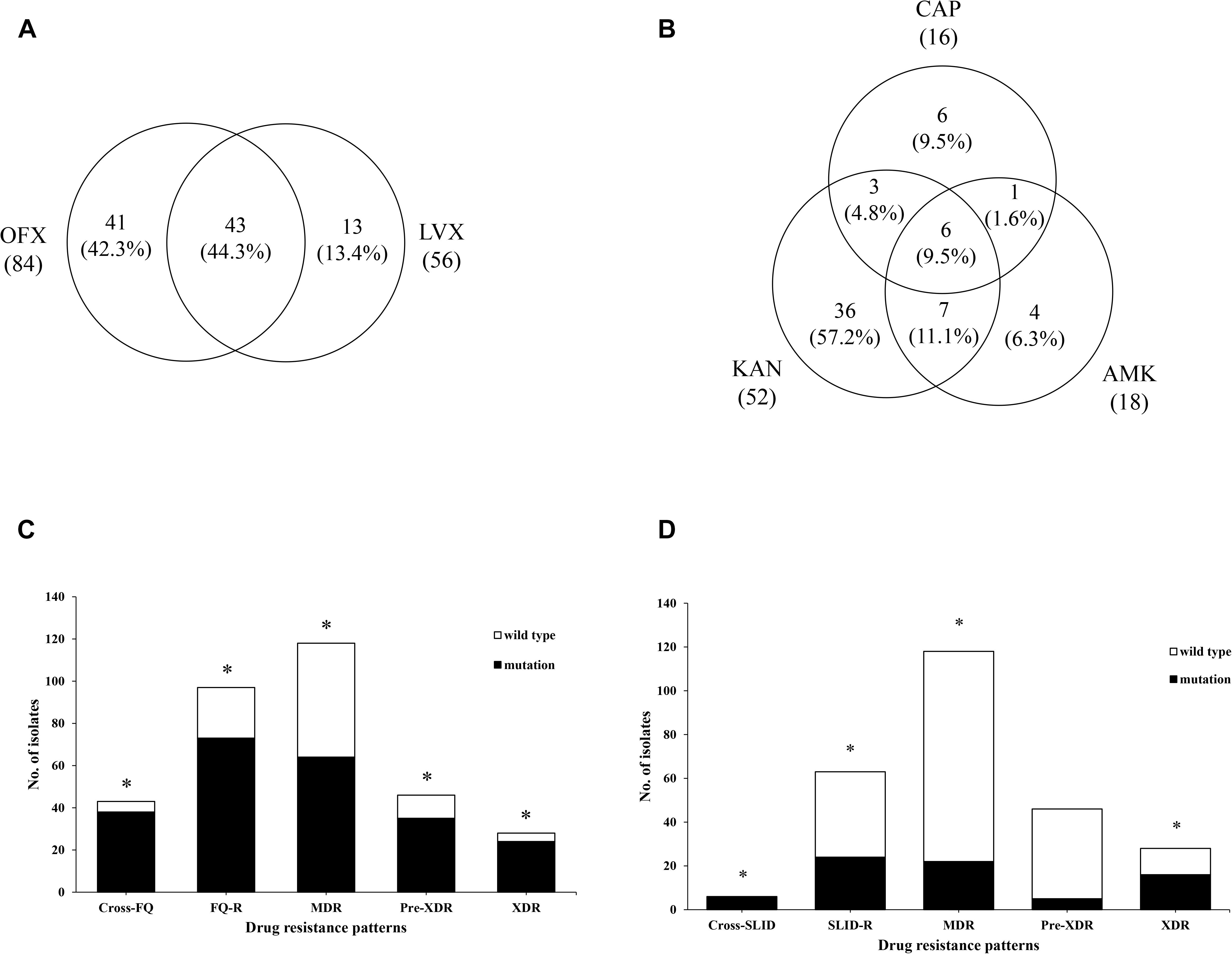
Figure 2. Analysis of phenotypic and genetic characterizations in different drug-resistance patterns. (A) Venn diagram of isolates identified from FQ group. The left circle shows any resistance to OFX, the right circle shows any resistance to LVX, the overlapping region shows isolates resistant to both OFX and LVX. (B) Venn diagram of isolates identified from SLID group. Each circles show any resistance to KAN, AMK, and CAP, respectively. The overlapping regions show isolates cross-resistant to the corresponding drugs. (C) The gyrA mutations were significantly associated with cross-resistance of FQ (4.13 OR, 95%CI [1.39, 12.23], P = 0.009), resistance of FQ (45.63 OR, 95%CI [20.73,100.42], P < 0.001), MDR (7.49 OR, 95%CI [4.09, 13.70], P < 0.001), Pre-XDR (10.81OR, 95%CI [5.10, 22.88], P < 0.001), and XDR (17.29 OR, 95%CI [5.76, 51.89], P < 0.001). (D) The rrs mutations were significantly associated with cross-resistance of SLID (3.17 OR, 95%CI [2.16, 4.64], P = 0.002), resistance of SLID (9.23 OR, 95%CI [9.60, 88.98], P < 0.001), MDR (5.08 OR, 95%CI [1.98, 13.00], P < 0.001), and XDR (24.11 OR, 95%CI [9.35, 62.20], P < 0.001). *Indicates the difference is statistically significant (P < 0.05).
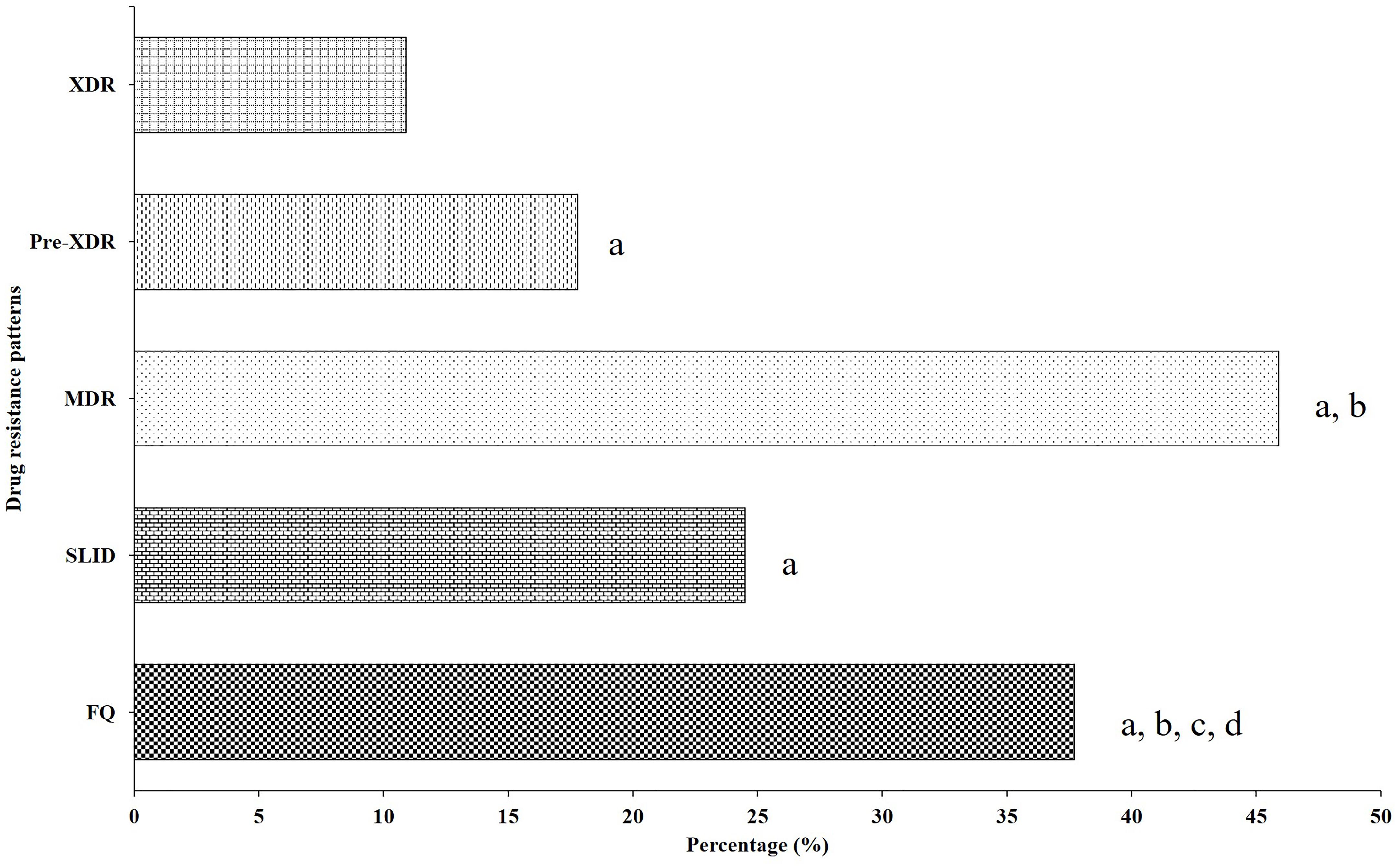
Figure 3. Distribution of drug-resistant strains resistant to different second-line drug group. aCompared with XDR group, there was a significantly lower than FQ (0.20 OR, 95%CI [0.13, 0.32], P < 0.001), SLID (0.38 OR, 95%CI [0.23, 0.61], P < 0.001), Pre-XDR (0.56 OR, 95%CI [0.34, 0.93], P = 0.024) and MDR (0.14 OR, 95%CI [0.09, 0.23], P < 0.001). bCompared with Pre-XDR group, there was significantly lower than FQ (0.36 OR, 95%CI [0.24, 0.54], P < 0.001), and MDR (0.26 OR, 95%CI [0.17, 0.38], P < 0.001). cCompared with MDR group, there was significantly higher than FQ (2.61 OR, 95%CI [1.80, 3.81], P < 0.001). dCompared with SLID group, there was significantly lower than FQ (1.87 OR, 95%CI [1.27, 2.73], P = 0.001).
Mutations in gyrA and gyrB
It is known to that the main molecular mechanism of FQr was caused by mutations in the QRDR of DNA gyrase, which composed of GyrA and GyrB subunits, encoded by gyrA and gyrB genes, respectively. DNA sequencing results of these two genes from FQr, FQs and pan-susceptible isolates were summarized in Table 2.
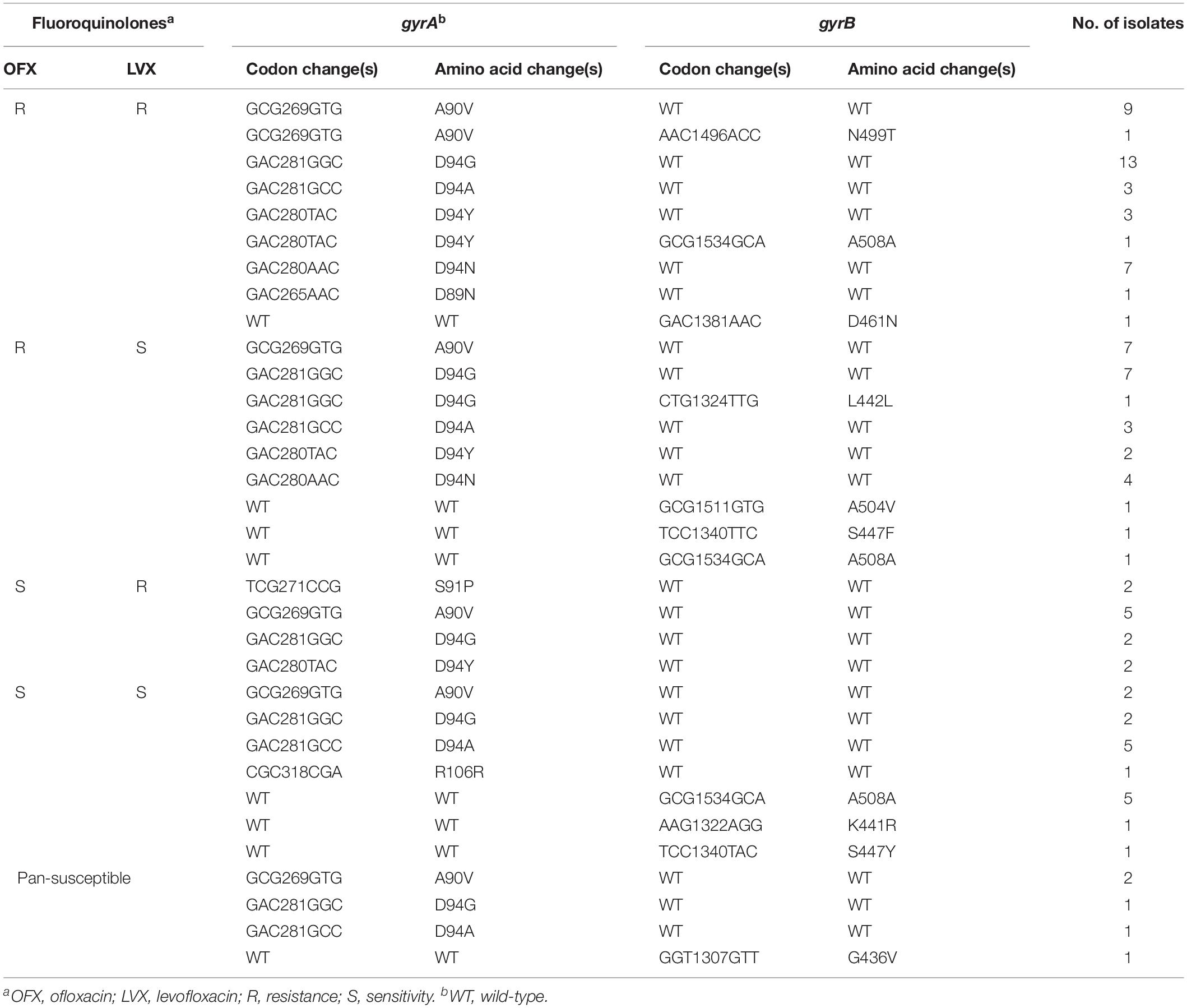
Table 2. Analysis of the gyrA and gyrB mutations in resistant, sensitive to FQ and to pan-susceptible M. tuberculosis isolates.
Of 97 FQr isolates, 73 isolates (73/97, 75.3%) carried mutations within gyrA gene, including codons 89, 90, 91, and 94. The most predominant mutation occurred at codon 94, with four different amino acid changes, D94G (23/97, 23.7%), D94A (6/97, 6.2%), D94Y (8/97, 8.2%), and D94N (11/97, 11.3%), which accounted for 49.5% (48/97) of FQr isolates. The A90V was the next most prevalent mutation, 22.7% (22/97) of isolates harbored this mutation. Besides, a single mutation in or near the QRDR of gyrB gene associated with FQr was detected in 4 isolates, including D461N (1/97, 1.0%), A504V (1/97, 1.0%), S447F (1/97, 1.0%) and A508A (1/97, 1.0%). Double mutations in both gyrA and gyrB were observed in three isolates, one with A90V and N499T mutation, one with D94Y and A508A mutation and one with D94G and L442L.
Of 160 FQs isolates, 17 isolates (17/160, 10.6%) displayed mutations within these two target fragments (gyrA and gyrB). Among them, the mutations within gyrA included A90V (2/160, 1.3%), D94G (2/160, 1.3%), D94A (5/160, 3.1%), and R106R (1/160, 0.6%) and mutations within gyrB were observed in 7 isolates with A508A (5/160, 3.1%), K441R (1/160, 0.6%), and S447Y (1/160, 0.6%).
Among 79 pan-susceptible isolates, 3 isolates each had mutations at codon 90 (A-V),94 (D-G), or 94 (D-A) in gyrA gene. Mutation at codon 436 (G-V) of gyrB gene was observed only one isolate.
Mutations in rrs, eis Promoter and tlyA
To determine the molecular basis of resistance to SLID, the rrs, eis promoter, and tlyA region were sequenced both in SLIDr, SLIDs, and pan-susceptible strains. Table 3 showed the mutations in the 1400 region of rrs, eis promoter and tlyA as well as the corresponding resistance phenotypes.
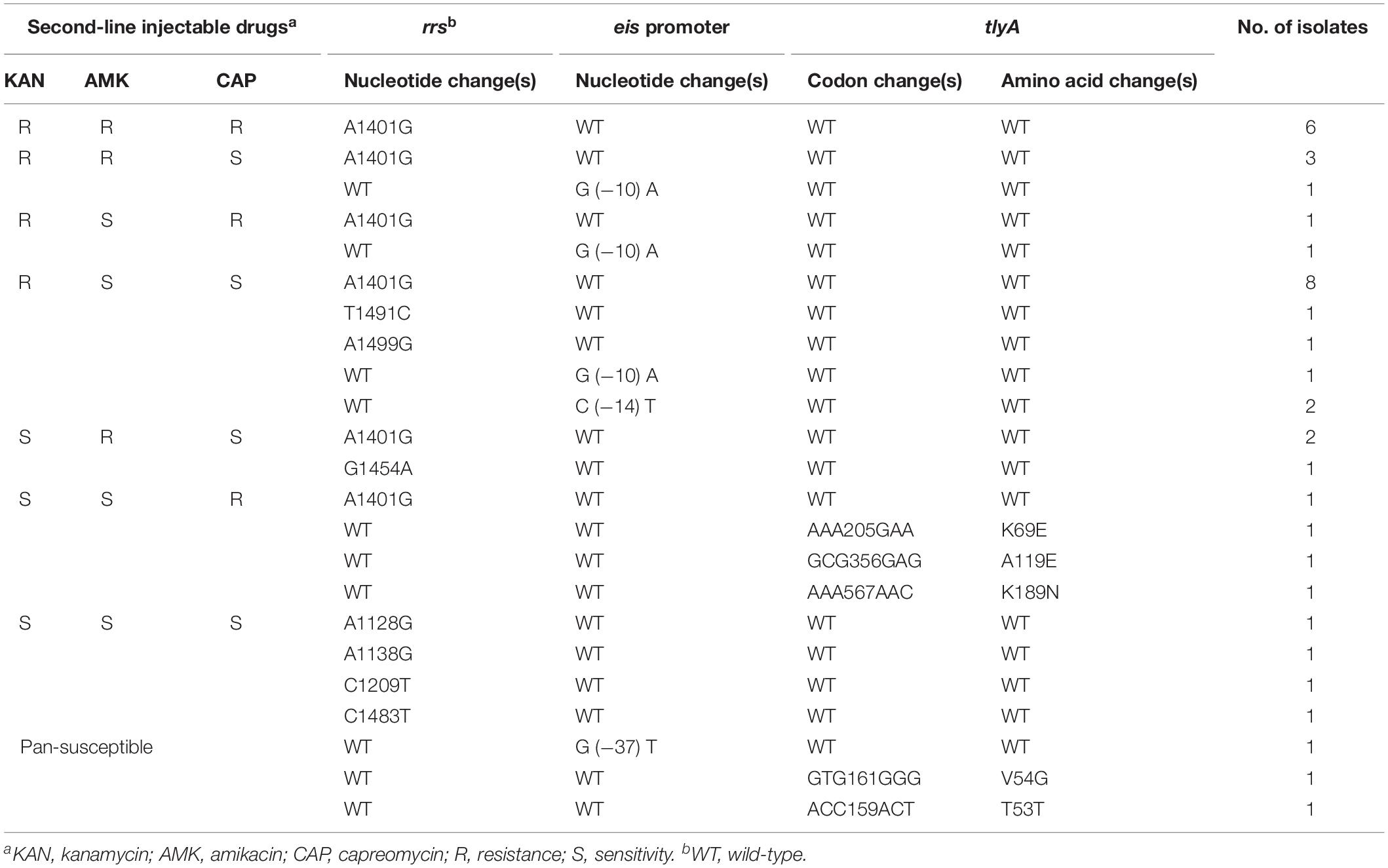
Table 3. Analysis of rrs, eis promoter and tlyA mutations in resistant, sensitive to SLID and to pan-susceptible M. tuberculosis isolates.
Of 63 SLIDr isolates, the most frequent mutation was the change from A to G at position 1401 of rrs gene, observed in 33.3% (21/63) of isolates. Three additional isolates displayed mutations at T1491C, G1454A, and A1499G in rrs. Mutations within the promoter region of eis included G (−10) A (3/63, 4.8%) and C (−14) T (2/63, 3.2%). Nevertheless, mutations in tlyA conferring amino acid substitutions K69E (1/63, 1.6%), A119E (1/63, 1.6%), and K189N (1/63, 1.6%), solely appeared in CAPr isolates.
Of 193 SLIDs isolates, one isolate each of A1128G, A1138G, C1209T, and C1483T, all occurred in rrs. Furthermore, for eis promoter, a change from G to T at −37 was detected only one isolate.
Among 79 pan-susceptible isolates, none of mutations were tested by us, except that, two clinical isolates with mutations V54G and T53T in tlyA.
Association Between Gene Mutations and Phenotypes
Among the 97 FQr isolates with gyrA mutations, 63.9% (62/97) were resistant to OFX and 50.5% (49/97) were resistant to LVX. As seen in Figure 2C, strongly evidence showed that gyrA mutations were associated with the cross-resistance of FQ (4.1 OR, 95%CI [1.4, 12.2], P = 0.009), FQ (45.6 OR, 95%CI [20.7, 100.4], P < 0.001), MDR (7.5 OR, 95%CI [4.1, 13.7], P < 0.001), Pre-XDR (10.8 OR, 95%CI [5.1, 22.9], P < 0.001), and XDR (17.3 OR, 95%CI [5.8, 51.9], P < 0.001). On the contrary, there was no significant correlation between FQr isolates with gyrB mutations (1.7 OR, 95%CI [0.6, 5.0], P = 0.331) (Table 4).
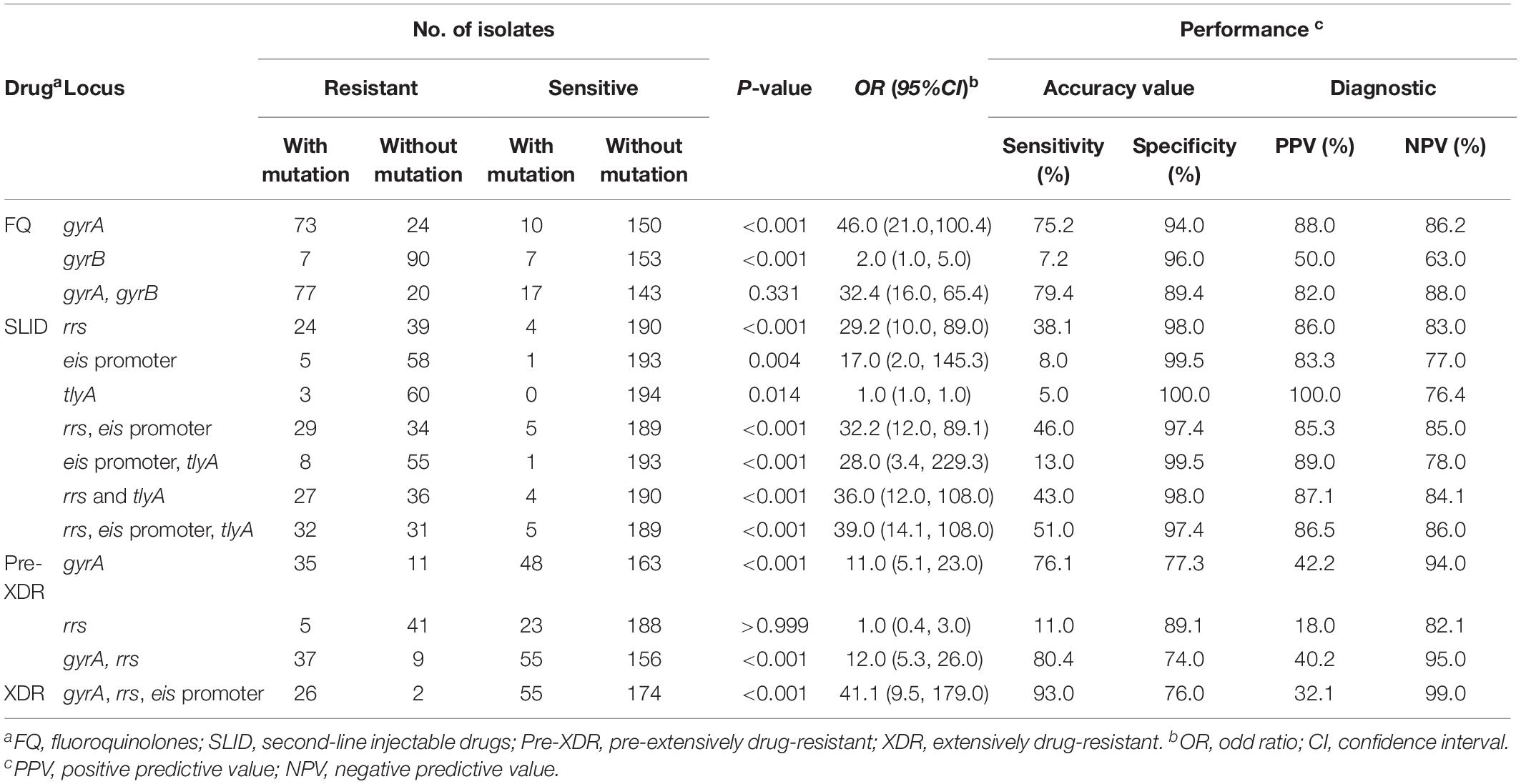
Table 4. Evaluation of phenotypic resistance of second-line anti-tuberculosis strains by mutations in second-line drug-resistant genes.
On the other side, we observed that 28.6% (18/63), 17.5% (11/63), and 12.7% (8/63) of resistant isolates with an A1401G mutation in rrs were resistant to KAN, AMK, and CAP, respectively. Also, we found rrs mutations were significantly associated with the cross-resistance of SLID (3.2 OR, 95%CI [2.2, 4.6], P = 0.002), SLID (29.2 OR, 95%CI [9.6, 89.0], P < 0.001), MDR (5.1 OR, 95%CI [2.0, 13.0], P < 0.001), and XDR (24.1 OR, 95%CI [9.4, 62.2], P < 0.001) (Figure 2D). Moreover, the isolates with eis promoter or tlyA gene mutations were related to the drug resistance to SLID (16.6 OR, 95%CI [1.9, 145.3], P = 0.004; 1.0 OR, 95%CI [0.9, 1.0], P = 0.014, respectively) (Table 4). Table 4 showed the sensitivity and specificity for evaluating phenotypes. For FQr, mutations in gyrA and gyrB had a combined sensitivity of 79.4% (95%CI [69.7, 86.7]), and specificity of 89.4% (95%CI [83.3, 93.5]). For SLIDr, rrs mutations were predicted phenotypes with 38.1% (95%CI [26.4, 51.2]) sensitivity, and 97.9% (95%CI [94.5, 99.3]) specificity. When adding the eis promoter and tlyA mutations, a combined sensitivity of 50.8% (95%CI [38.0, 63.5]), specificity of 97.4% (95%CI [93.8, 99.0]). The individual gyrA mutations for assessing Pre-XDR had sensitivity, and specificity values of 76.1% (95%CI [60.9, 86.9]), and 77.3% (95%CI [70.9, 82.6]), respectively, but adding the rrs mutations increased the values of sensitivity and for Pre-XDR detection to 80.4% (95%CI [65.6, 90.1]), and 73.9% (95%CI [67.4, 79.6]), respectively. A combined mutation of gyrA, rrs and eis promoter achieved the best evaluation for XDR with the sensitivity, and specificity values of 92.9% (95%CI [75.0, 98.8]), and 73.9% (95%CI [67.4, 79.6]), respectively.
Genotyping
Three major diverse families were identified among the 124 SLDr isolates, including Beijing, T and H family. Beijing family was the largest sub-lineage with 112 isolates. Nine isolates belonged to the T family, 6 isolates were T1 sublineage, 2 isolates were T2, and 1 isolate was T3. The remaining 3 isolates were identified as H3 sub-lineage.
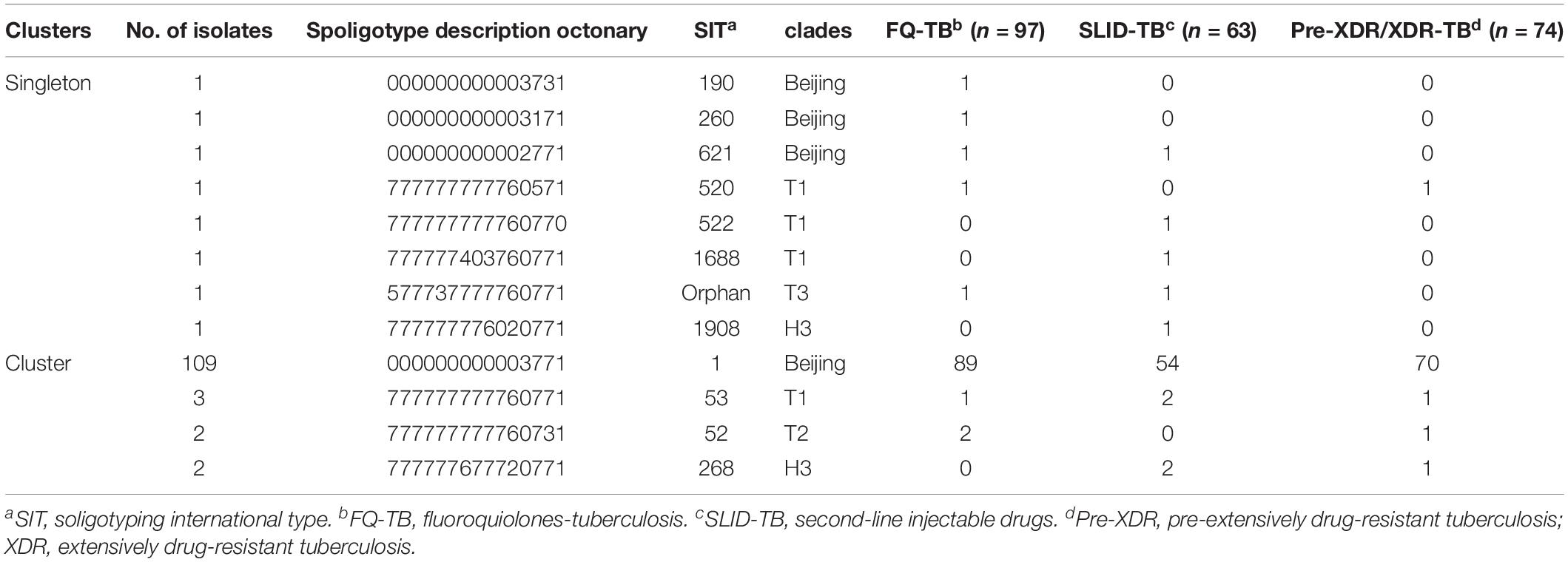
Comparison of Spoligotyping results with three drug resistance patterns (Table 5), 116 isolates were grouped in 4 clusters and 8 isolates had singletons. Of the singletons, 7 isolates were distributed in clades, Beijing (SIT190, 260, and 621), T1 (SIT522 and 1688), T3 (Orphan) and H3 (SIT1908) with FQ-TB or SLID-TB. Only one Pre-XDR isolate was defined as T1 family (SIT520). The predominance of Beijing family (SIT1) contained 109 isolates clone in FQ-TB (71.8%, 89/124), SLID-TB (43.5%, 54/124) and Pre-XDR/XDR-TB group (56.5%, 70/124). Other three clusters, T1 (SIT53), T2 (SIT52) and H3 (SIT268), 2–3 isolates each, were found in Pre-XDR/XDR-TB and/or additional FQ or SLID. No significant associations were found between the genotypes and specific types of resistance (Supplementary Table S2). Meanwhile, no significant correlation was observed between any mutation and a Spoligotype family (Supplementary Table S3 and Supplementary Figure S2).
A total of 124 SLDr isolates were genotyped by 15-loci MIRU-VNTR, revealed that the strains were divided into 113 genotypes, of which 107 were identified as unique patterns and 17 isolates were categorized to cluster patterns (Figure 4). Six small clusters ranging from 2 to 4 isolates were observed in all genotypes. The cumulative clustering rate and recent transmission rate rely on the MIRU-VNTR typing was 13.7% (17/124) and 4.8% (6/124), respectively.
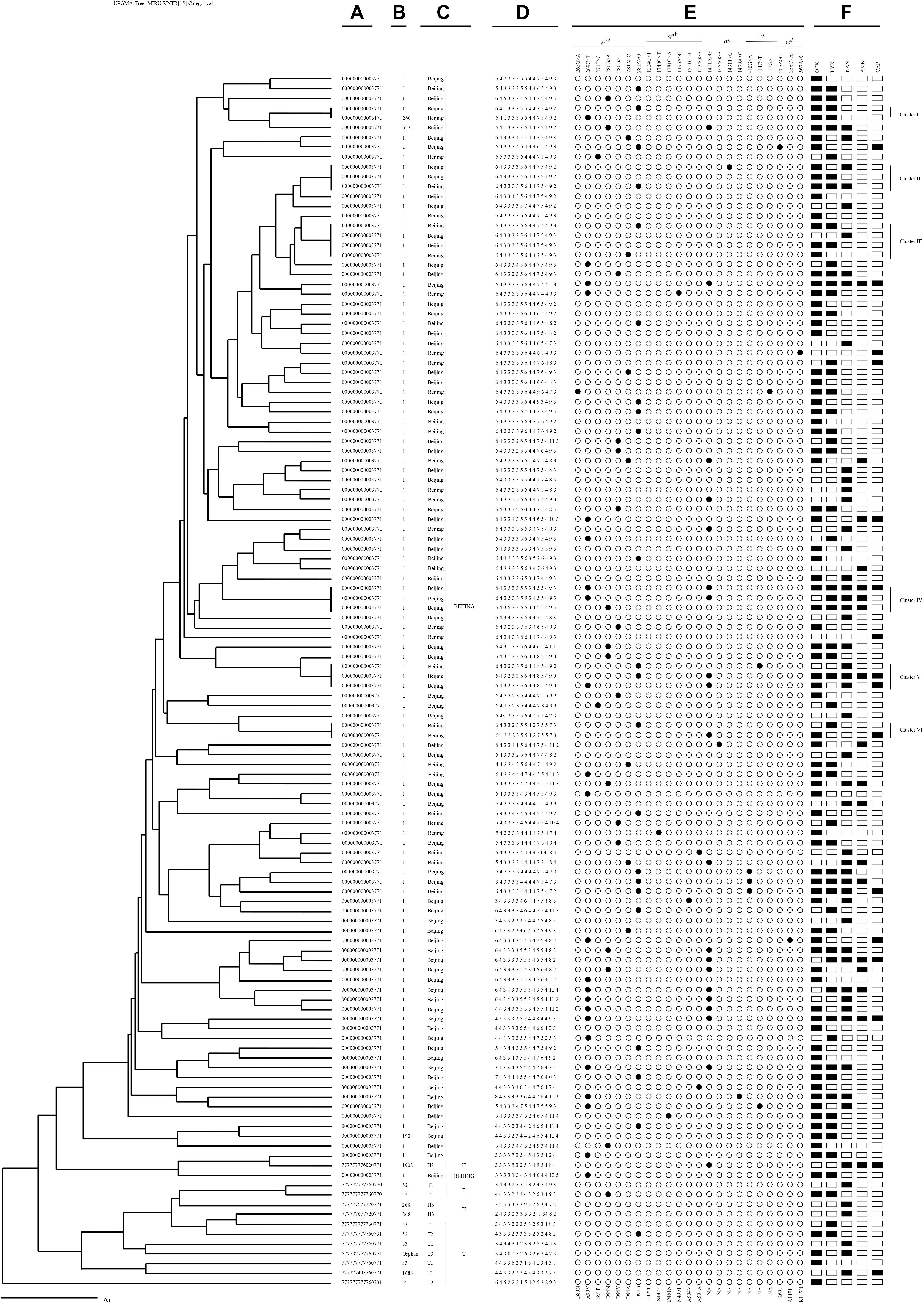
Figure 4. An UPGMA-tree based on 15-loci MIRU-VNTR of the clinical 124 M. tuberculosis isolates resistant to second-line drugs. From left to right: (A) Spoligotype octonary; (B) Spoligotype international type (SIT); (C) Spoligotype clade; (D) 15-loci MIRU-VNTR profile; (E) mutational pattern; (F) second-line drugs resistance pattern.
Among six clusters with SLDr strains, two Pre-XDR isolates with a gyrA (A90V or D94G) mutation occurred exclusively in Cluster I. Cluster II contained three isolates, two Pre-XDR isolates presented at gyrA mutation (D94G), one of them accompanied with a mutation at rrs (T1491C), the rest of FQ had no mutation. Cluster III grouped in 4 isolates, the Pre-XDR included 2 isolates with a mutation at gyrA (D94G or D94A), one was FQ and another was SLID without any mutation. Three isolates of cluster IV had three different polymorphisms at either gyrA (A90V or D94N) or rrs (A1401G) locus. Cluster V comprised 3 isolates, the XDR had gyrA (A90V or D94G) and rrs (A1401G) mutations sharing with 2 isolates, whereas one Pre-XDR isolate with gyrA (D94G) and eis promoter C (−14) T. Cluster VI had one XDR isolate with a mutation at rrs (A1401G) and FQ with a mutation at gyrA (D94G).
Discussion
To our best knowledge, the present study is the first to provide a description of mutations associated with SLDr in M. tuberculosis strains from Hebei. Nevertheless, we examined the phylogenetic diversity in SLDr strains through genotyping profiles. The present findings revealed that 48.2% (124/257) were resistant to at least one five SLD. Previously, several studies reported the resistance rate of SLD strains has been estimated to range from 25.7 to 51.9% (Bakuła et al., 2016; Brossier et al., 2017; Hu et al., 2017). According to a recently published World Health Organization (WHO) report, among cases of MDR-TB in 2017, 8.5% (95%CI, 6.2–11.0%) were estimated to have XDR-TB (World Health Organization [WHO], 2018). We reported the frequency of XDR-TB is 10.9%.
FQr isolates is primarily attributed to mutations in the QRDR of gyrA. According to a systematic review, gyrA mutations were reported in codons 88–94 appeared to account for roughly 60.0–90.0% of FQr globally (Avalos et al., 2015). In our present study, mutations at gyrA codon 89, 90, 91, and 94 were observed in 75.3% of resistant isolates. The frequency of mutations conferring FQr is similar to Shanghai (76.0%), although it is lower than those in Russia (83.0%), India (81.0%) and Thailand (92.3%), and higher than those in Morocco (30.0%) and New York (67.0%) (Sullivan et al., 1895; Mokrousov et al., 2008; Zhu et al., 2012; Disratthakit et al., 2016; Singhal et al., 2016; Chaoui et al., 2018), indicating that mutations in gyrA tend to differ by geographic region. In line with previous studies (Singhal et al., 2016; Pang et al., 2017; Chawla et al., 2018), substitutions at codon 94 were the most frequent mutation among FQr isolates. This phenomenon may be explained by the fact that codon 94, which aims at the water-magnesium ion bridge with a conserved C3/C4 keto acid moiety of quinolones, plays an important role in stabilizing the quinolone molecule in the quinolone binding pocket, an amino acid substitution at this position will exaggerate the deleterious effect of the binding between most quinolones and DNA gyrase (Aldred et al., 2016; Blower et al., 2016; Disratthakit et al., 2016).
Interestingly, significant evidence has demonstrated a link between gyrA mutations and FQ, MDR, Pre-XDR and XDR, suggesting that mutations at gyrA might be act as a candidate diagnostic marker for FQ, MDR and a possible indicator of Pre-XDR-TB or XDR-TB. In Mycobacterium, the interactive effect between rifampicin- and FQ-resistant mutations was influenced by epistasis and produced a varying degree of loss in fitness (Borrell et al., 2013). We hypothesized that the progression of MDR to Pre-XDR or XDR may be caused by the positive epistasis between gyrA mutations and mutations in drug resistant gene conferring rifampicin. Certainly, further research is required to confirm this hypothesis.
Unlike the high frequency of gyrA mutations, mutations in gyrB gene are seldom commonly associated with FQr in M. tuberculosis isolates. A gyrB mutation (L442L, S447F, N499T, A504V, or A508A) was identified in 7 FQr isolates. As far as we known, a L442L, S447F, N499T, A504V mutation has not been previously reported. Due to the small numbers of isolates with gyrB mutations, no such mutation was significant independently associated with FQr. Additionally, we found that both FQr and FQs strains exhibited an A508A mutation in gyrB, making it difficult to acknowledge the contribution to phenotypic resistance. Several publications reported double mutations in gyrA, gyrB or both gyrA and gyrB, which ranged from 1.0 to 3.0% (Avalos et al., 2015; Chien et al., 2016; Kateete et al., 2019). Double mutations in both gyrA and gyrB were found in FQr isolates but the absence of susceptible isolates, suggesting that although rare, it can be as the highly specific predictor of FQr.
Previous reports indicated the A1401G in rrs effectively identify the phenotypic resistance to KAN, AMK and CAP (Malinga et al., 2016; Varghese and Al-Hajoj, 2017). In our study, 36.5, 63.2, and 50.0% of sensitivities with an A1401G mutation in rrs were resistant to KAN, AMK, and CAP, respectively (Supplementary Tables S6–S8). Compared to DST test, it seems that a mutation at A1401G provided a better maker for AMK and CAP than for KAN. However, the canonical A1401G rrs mutation explains only around 56.0% of KANr, while the mutations G10A and C14T in the promoter of eis, explain another 33.0% of low-level KANr as estimated by one review (Georghiou et al., 2012). Our study supports these finding, as 5 KANr isolates were found to have mutations in eis promoter, including three of G (−10) A and two of C (−14) T. Mutations in tlyA is also known to responsible for resistance to CAP. In fact, we observed three isolates were resistance to CAP alone, but KAN-sensitive and AMK-sensitive isolates had no mutations in tlyA gene. Consequently, tlyA should be included in the molecular analysis of CAP-resistance.
In our study, rrs mutations were significantly associated with MDR-TB (5.1 OR, 95%CI [2.0, 13.0], P < 0.001), and XDR-TB (24.1 OR, 95%CI [9.4, 62.2], P < 0.001), suggesting the detection of rrs mutations provide the valuable information to support the initiation of effective treatment regimens for MDR-TB or even XDR-TB cases. Specifically, as previously reported (Zhang et al., 2014; Ogari et al., 2019), we observed a significant cross-resistance between KAN, AMK and CAP associated with rrs gene. Therefore, rrs might serve as a marker to predict the cross-resistance among SLID agents.
The traditional phenotypic DSTs lead serious delays to the detection of resistance just because of the extremely slow growth of M. tuberculosis. However, several commercial diagnostic tests such as GeneXpert MTB/RIF, Genotype MTBDRplus/MTBDRsl assays, have been developed to rapidly detect both first- and second-line drug resistance in M. tuberculosis by scanning the associated mutations (Xie et al., 2017; Kim et al., 2019). Our performance showed that analysis of the associated mutations was recommended to provide a good sensitivity for rapid verification of FQr (79.4%), SLIDr (50.8%), Pre-XDR (80.4%), and XDR (92.9%). In addition, sensitivities and specificities for predicting phenotypic resistance to five SLD in M. tuberculosis isolates as shown in Supplementary Tables S4–S8.
Spoligotyping result showed that the predominance of Beijing family (SIT1), which accounted for the largest cluster (71.8%) among SLDr strains in this study. No significant associations were found between the genotypes and specific types of resistance. But a high frequency of Beijing family strains among FQr isolates was previously reported from Vietnam and Russia (Mokrousov et al., 2008; Duong et al., 2009). One possibility is that the Beijing family strains have a high level of intrinsic resistance to FQ, providing an opportunity to tolerate low level of FQ and subsequently generate gyrA mutations, or that these mutations confer an advantage under the absence of antibiotics pressure (Mokrousov et al., 2008). An observation revealed that eis mutations were considered to be associated with the Beijing clades (Casali et al., 2014); however, this is not applicable for all geographical settings. Generally, we found no significant correlation between any mutation and a spoligotype family, which is accordance with reports from China (Zhang et al., 2013, 2014).
In this study, an UPGMA-tree was generated from 15-loci MIRU-VNTR among 124 SLDr isolates, which was slightly similar with our previous study on the acquired resistance of MDR-TB in this region (Li et al., 2019). The highly different genotypic patterns and drug resistance profiles suggest that acquisition of resistance is also an important cause for the emergence of SLDr in Hebei Province. MIRU-VNTR data revealed a moderate level of genotypic diversity, which implicated composite mechanisms of resistance, including transmitted and acquired resistance, as a potential cause for the emergence of SLDr strains. Transmission of clustered SLDr strains with identical mutations may indicated the acquisition of drug resistance typically confers a reduction in fitness cost, and mutations may further contribute to the spreading of Pre-XDR-TB or XDR-TB. Therefore, the strict DOTS is the necessary principle to reduce the incidence of acquired strains. In addition, 28.6% of XDR strains were clustered, and it can be inferred that there exists cloning transmission between a small number of XDR strains.
The main limitation of this study was only ability to perform at critical concentrations for DST but not established MICs. This discrepancy of the critical-concentration method used for DST, such that up to 5% of wild-type M. tuberculosis strains are classified as drug resistant. While our study is also limited by the number of strains, further studies continue to collect larger additional strains to depict acquired resistance in the community and household and build transmission chain.
Conclusion
In summary, this study showed that 48.2% of isolates were resistance to at least one of five SLD, and most of which were resistant to OFX, LVX, and KAN. The majority of FQr and SLIDr strains were associated with gyrA mutation at D94G and rrs mutation at A1401G, respectively. Mutations in gyrA, rrs, and eis promoter seem to be the causative biomarker for the screening of resistance to SLD, even XDR-TB itself. No correlation was found between any mutation and a spoligotype family. Furthermore, acquired resistance is one of the critical factors driving the SLDr strains in Hebei Province. These results highlight the use of appropriate treatment regimens and the development of early rapid diagnosis are the effective manners for the control of SLD-TB or XDR-TB patients.
Data Availability
All the data analyzed throughout this research are included in this published article.
Ethics Statement
Approval for this study was obtained from the Medical Ethics Committee of Scientific Research Project of the Fifth Hospital of Shijiazhuang. Written informed consent was obtained from each participant according to the Federal and Institutional Guidelines.
Author Contributions
QL performed the DST and DNA extraction, analyzed the data, and wrote the first draft of the manuscript. HG, ZZ, and YT performed the DST and spoligotyping. TL performed the MIRU-VNTR typing. YW collected the samples and supported the study. JL and YL contributed to the conception and design of the study. ED designed the study, collected the samples, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Program of Mega Infectious Disease of 12th Five Year Plan (Grant No. 2015ZX10004801) and the Scientific Research Fund Project of Health Department from Hebei Province (Grant No. 20130650).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the staffs from the Laboratory of Shijiazhuang Fifth Hospital for supplying strains and DNA samples. We wish to thank the staffs of the Chinese Centre for Disease Control and Prevention for their excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01838/full#supplementary-material
Abbreviations
AMK, amikacin; CAP, capreomycin; CI, confidence interval; DOTS, directly observed treatment short course; DR-TB, drug-resistant tuberculosis; DST, drug susceptibility testing; FQ, fluoroquinolones; KAN, kanamycin; L-J, Lowenstein-Jensen; LVX, levofloxacin; MDR-TB, multidrug-resistant tuberculosis; MIC, minimum inhibitory concentration; MIRU-VNTR, mycobacterial interspersed repetitive unit–variable number of tandem repeats; M. tuberculosis, Mycobacterium tuberculosis; NPV, negative predictive value; OFX, ofloxacin; OR, odd ratio; PCR, polymerase chain reaction; PPV, positive predictive value; Pre-XDR, pre-extensively drug-resistant tuberculosis; QRDR, fluoroquinolone resistance-determining region; SIT, spoligotyping international type; SLD, second-line anti-tuberculosis drugs; SLID, second-line injectable drugs; SPSS, statistical package for the social sciences; TB, tuberculosis; XDR-TB, extensively drug-resistant tuberculosis; UPGMA, unweighted pair group method with arithmetic mean; WHO, world health organization.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/BLAST
- ^ http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/index.jsp
- ^ http://www.miru-vntrplus.org
References
Alangaden, G. J., Kreiswirth, B. N., Aouad, A., Khetarpal, M., Igno, F. R., Moghazeh, S. L., et al. (1998). Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42, 1295–1297. doi: 10.1128/aac.42.5.1295
Aldred, K. J., Blower, T. R., Kerns, R. J., Berger, J. M., and Osheroff, N. (2016). Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: enhancing drug activity against wild-type and resistant gyrase. Proc. Natl. Acad. Sci. U.S.A. 113, E839–E846. doi: 10.1073/pnas.1525055113
Avalos, E., Catanzaro, D., Catanzaro, A., Ganiats, T., Brodine, S., Alcaraz, J., et al. (2015). Frequency and geographic distribution of gyrA and gyrB mutations associate with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One 1:e0120470. doi: 10.1371/journal.pone.0120470
Bakuła, Z., Napiórkowska, A., Kamiński, M., Augustynowicz-Kopeć, E., Zwolska, Z., Bielecki, J., et al. (2016). Second-line anti-tuberculosis drug resistance and its genetic determinants in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J. Microbiol. Immunol. 49, 439–444. doi: 10.1016/j.jmii.2015.04.003
Bauskenieks, M., Pole, I., Skenders, G., Jansone, I., Broka, L., Nodieva, A., et al. (2015). Genotypic and phenotypic characteristics of aminoglycoside-resistant Mycobacterium tuberculosis isolates in latvia. Diagn. Microbiol. Infect. Dis. 81, 177–182. doi: 10.1016/j.diagmicrobio.2014.12.004
Blower, T. R., Williamson, B. H., Kerns, R. J., and Berger, J. M. (2016). Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 113, 1706–1713. doi: 10.1073/pnas.1525047113
Borrell, S., Teo, Y., Giardina, F., Streicher, E. M., Klopper, M., Feldmann, J., et al. (2013). Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol. Med. Public Health. 2013, 65–74. doi: 10.1093/emph/eot003
Brossier, F., Pham, A., Bernard, C., Aubry, A., Jarlier, V., Veziris, N., et al. (2017). Molecular investigation of resistance to second-line injectable drugs in multidrug-resistant clinical isolates of Mycobacterium tuberculosis in France. Antimicrob. Agents Chemother. 61:e1299-16. doi: 10.1128/AAC.01299-16
Casali, N., Nikolayevskyy, V., Balabanova, Y., and Harris, S. R. (2014). Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46, 279–286. doi: 10.1038/ng.2878
Chaoui, I., Oudghiri, A., and El Mzibri, M. (2018). Characterization of gyrA and gyrB mutations associated with fluoroquinolone resistance in Mycobacterium tuberculosis isolates from morocco. J. Glob. Antimicrob. Resist. 12, 171–174. doi: 10.1016/j.jgar.2017.10.003
Chawla, K., Kumar, A., Shenoy, V. P., Chakrabarty, S., and Satyamoorthy, K. (2018). Genotypic detection of fluoroquinolone resistance in drug-resistant Mycobacterium tuberculosis at a tertiary care centre in south Coastal Karnataka. India. J. Glob. Antimicrob. Resis. 13, 250–253. doi: 10.1016/j.jgar.2018.01.023
Chien, J. Y., Chiu, W. Y., Chien, S. T., Chiang, C. J., Yu, C. J., and Hsueh, P. R. (2016). Mutations in gyrA and gyrB among fluoroquinolone- and multidrug resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 60, 2090–2096. doi: 10.1128/AAC.01049-15
Disratthakit, A., Prammananan, T., Tribuddharat, C., Thaipisuttikul, I., Doi, N., Leechawengwongs, M., et al. (2016). Role of gyrB mutations in pre-extensively and extensively drug resistant tuberculosis in thai clinical isolates. Antimicrob. Agents Chemother. 60, 5189–5197. doi: 10.1128/AAC.00539-16
Duong, D. A., Nguyen, T. H., Nguyen, T. N., Dai, V. H., Dang, T. M., Vo, S. K., et al. (2009). Beijing genotype of Mycobacterium tuberculosis is significantly associated with high-level fluoroquinolone resistance in Vietnam. Antimicrob. Agents Chemother. 53, 4835–4839. doi: 10.1128/AAC.00541-09
Freihofer, P., Akbergenov, R., Teo, Y., Juskeviciene, R., Andersson, D. I., and Böttger, E. C. (2016). Nonmutational compensation of the fitness cost of antibiotic resistance in mycobacteria by overexpression of tlyA rRNA methylase. RNA 22, 1–5. doi: 10.1261/rna.057257.116
Georghiou, S. B., Magana, M., Garfein, R. S., Catanzaro, D. G., Catanzaro, A., and Rodwell, T. C. (2012). Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275. doi: 10.1371/journal.pone.0033275
Hu, Y., Xu, L., He, Y. L., Pang, Y., Lu, N., Liu, L., et al. (2017). Prevalence and molecular characterization of second-line drugs resistance among multidrug-resistant Mycobacterium tuberculosis isolates in southwest of China. Biomed. Res. Int. 11, 1–9. doi: 10.1155/2017/4563826
Kamerbeek, J., Schouls, L., Kolk, A., van Agterveld, M., van Soolingen, D., Kuijper, S., et al. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914.
Kateete, D. P., Kamulegeya, R., Kigozi, E., Katabazi, F. A., Lukoye, D., Sebit, S. I., et al. (2019). Frequency and patterns of second-line resistance conferring mutations among MDR-TB isolates resistant to a second-line drug from eSwatini, Somalia and Uganda (2014-2016). BMC Pulm. Med. 19:124. doi: 10.1186/s12890-019-0891-x
Kim, S., Kim, Y., Chang, Y., Hirgo, W. K., Chang, C. L., Shim, T. S., et al. (2019). Comparison of quantamatrix multiplexed assay platform and GenoType MTBDR assay Using smear-positive sputum specimens from patients with multidrug- resistant/extensively drug-resistant tuberculosis in South Korea. Front. Microbiol. 10:1075. doi: 10.3389/fmicb.2019.01075
Li, Q. L., Wang, Y. L., Li, Y. N., Gao, H. X., Zhang, Z., Feng, F. M., et al. (2019). Characterization of drug resistance-associated mutations among clinical multidrug-resistant Mycobacterium tuberculosis isolates from hebei province, China. J. Glob. Antimicrob. Resist. doi: 10.1016/j.jgar.2019.03.012 [Epub ahead of print].
Li, Y., Cao, X., Li, S., Wang, H., Wei, J., Liu, P., et al. (2016). Characterization of Mycobacterium tuberculosis isolates from hebei, China: genotypes and drug susceptibility phenotypes. BMC Infect. Dis. 16:107. doi: 10.1186/s12879-016-1441-2
Malinga, L., Brand, J., Olorunju, S., Stoltz, A., and van der Walt, M. (2016). Molecular analysis of genetic mutations among cross-resistant second-line injectable drugs reveals a new resistant mutation in Mycobacterium tuberculosis. Diagn. Microbiol. Infect. Dis. 85, 433–437. doi: 10.1016/j.diagmicrobio.2016.05.010
Mokrousov, I., Otten, T., Manicheva, O., Potapova, Y., Vishnevsky, B., Narvskaya, O., et al. (2008). Molecular characterization of ofloxacin resistant Mycobacterium tuberculosis strains from Russia. Antimicrob. Agents Chemother. 52, 2937–2939. doi: 10.1128/AAC.00036-08
Ngo, H. X., Green, K. D., Gajadeera, C. S., Willby, M. J., Holbrook, S. Y. L., Hou, C., et al. (2018). Potent 1,2,4-Triazino[5,6b]indole-3-thioether inhibitors of the kanamycin resistance enzyme Eis from Mycobacterium tuberculosis. ACS Infect. Dis. 4, 1030–1040. doi: 10.1021/acsinfecdis.8b00074
Ogari, C. O., Nyamache, A. K., Nonoh, J., and Amukoye, E. (2019). Prevalence and detection of drug resistant mutations in Mycobacterium tuberculosis among drug naïve patients in Nairobi. Kenya. BMC Infect. Dis. 19:279. doi: 10.1186/s12879-019-3911-9
Oudghiri, A., Karimi, H., Chetioui, F., Zakham, F., Bourkadi, J. E., Elmessaoudi, M. D., et al. (2018). Molecular characterization of mutations associated with resistance to second-line tuberculosis drug among multidrug-resistant tuberculosis patients from high prevalence tuberculosis city in Morocco. BMC Infect. Dis. 18:98. doi: 10.1186/s12879-018-3009-9
Pang, Y., Zong, Z., Huo, F., Jing, W., Ma, Y., Dong, L., et al. (2017). Drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing. China. Antimicrob. Agents Chemother. 61, 1–17. doi: 10.1128/AAC.00900-17
Singhal, R., Reynolds, P. R., Marola, J. L., Epperson, L. E., Arora, J., Sarin, R., et al. (2016). Sequence analysis of fluoroquinolone resistance-associated genes gyrA and gyrB in clinical Mycobacterium tuberculosis isolates from patients suspected of having multidrug-resistant tuberculosis in New Delhi. India. J. Clin. Microbiol. 54, 2298–2305. doi: 10.1128/JCM.00670-16
Sullivan, E. A., Kreiswirth, B. N., Palumbo, L., Kapur, V., Musser, J. M., Ebrahimzadeh, A., et al. (1895). Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet 345, 1148–1150. doi: 10.1016/S0140-6736(95)90980-X
Supply, P., Lesjean, S., Savine, E., Kremer, K., van Soolingen, D., and Locht, C. (2001). Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39, 3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001
Varghese, B., and Al-Hajoj, S. (2017). First insight into the fluoroquinolone and aminoglycoside resistance of multidrug-resistant in Saudi Arabia. Am. J. Trop. Med. Hyg. 96, 1066–1070. doi: 10.4269/ajtmh.16-0579
Witek, M. A., Kuiper, E. G., Minten, E., Crispell, E. K., and Conn, G. L. (2017). A novel motif for S-Adenosyl-l-methionine binding by the ribosomal RNA methyltransferase TlyA from Mycobacterium tuberculosis. J. Biol. Chem. 292, 1977–1987. doi: 10.1074/jbc.M116.752659
World Health Organization [WHO] (1997). Global Working Group on Antituberculosis Drug Resistance Surveillance. Guideline for Surveillance of Drug Resistance in Tuberculosis. Paris: IUATLD.
World Health Organization [WHO] (2018). Global Tuberculosis Report 2018.WHO/CDS/TB/2016.20. Geneva: World Health Organization.
Xie, Y. L., Chakravorty, S., Armstrong, D. T., Hall, S. L., Via, L. E., Song, T., et al. (2017). Evaluation of a rapid molecular drug susceptibility test for tuberculosis. N. Engl. J. Med. 377, 1043–1054. doi: 10.1056/NEJMoa1614915
Zhang, Z., Liu, M., Wang, Y., Pang, Y., Kam, K. M., and Zhao, Y. (2014). Molecular and phenotypic characterization of multidrug-resistant Mycobacterium tuberculosis isolates resistant to kanamycin, amikacin, and capreomycin in China. Eur. J. Clin. Microbiol. 33, 1859–1866. doi: 10.1007/s10096-014-2144-5
Zhang, Z. J., Lu, J., Wang, Y. F., Pang, Y., and Zhao, Y. L. (2013). Prevalence and molecular characterization of fluoroquinolone resistant Mycobacterium tuberculosis isolates in China. Antimicrob. Agents Chemother. 58, 364–369. doi: 10.1128/AAC.01228-13
Keywords: second-line anti-tuberculosis drugs, Mycobacterium tuberculosis, associated-resistance mutation, acquired resistance, genotype
Citation: Li Q, Gao H, Zhang Z, Tian Y, Liu T, Wang Y, Lu J, Liu Y and Dai E (2019) Mutation and Transmission Profiles of Second-Line Drug Resistance in Clinical Isolates of Drug-Resistant Mycobacterium tuberculosis From Hebei Province, China. Front. Microbiol. 10:1838. doi: 10.3389/fmicb.2019.01838
Received: 10 June 2019; Accepted: 25 July 2019;
Published: 07 August 2019.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Ibrahim Bitar, Charles University, CzechiaIris Spiliopoulou, University of Patras, Greece
Copyright © 2019 Li, Gao, Zhang, Tian, Liu, Wang, Lu, Liu and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erhei Dai, daieh2008@126.com
 Qianlin Li
Qianlin Li Huixia Gao2
Huixia Gao2