- 1College of Food Science and Engineering, Qingdao Agricultural University, Qingdao, China
- 2Key Laboratory of Agro-products Quality and Safety Control in Storage and Transport Process, Ministry of Agriculture and Rural Affairs/Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, Beijing, China
As the most carcinogenic, toxic, and economically costly mycotoxins, aflatoxin B1 (AFB1) is primarily biosynthesized by Aspergillus flavus and Aspergillus parasiticus. Aflatoxin biosynthesis is related to oxidative stress and functions as a second line of defense from excessive reactive oxygen species. Here, we find that ethanol can inhibit fungal growth and AFB1 production by A. flavus in a dose-dependent manner. Then, the ethanol’s molecular mechanism of action on AFB1 biosynthesis was revealed using a comparative transcriptomic analysis. RNA-Seq data indicated that all the genes except for aflC in the aflatoxin gene cluster were down-regulated by 3.5% ethanol. The drastic repression of aflatoxin structural genes including the complete inhibition of aflK and aflLa may be correlated with the down-regulation of the transcription regulator genes aflR and aflS in the cluster. This may be due to the repression of several global regulator genes and the subsequent overexpression of some oxidative stress-related genes. The suppression of several key aflatoxin genes including aflR, aflD, aflM, and aflP may also be associated with the decreased expression of the global regulator gene veA. In particular, ethanol exposure caused the decreased expression of stress response transcription factor srrA and the overexpression of bZIP transcription factor ap-1, C2H2 transcription factors msnA and mtfA, together with the enhanced levels of anti-oxidant enzymatic genes including Cat, Cat1, Cat2, CatA, and Cu, Zn superoxide dismutase gene sod1. Taken together, these RNA-Seq data strongly suggest that ethanol inhibits AFB1 biosynthesis by A. flavus via enhancing fungal oxidative stress response. In conclusion, this study served to reveal the anti-aflatoxigenic mechanisms of ethanol in A. flavus and to provide solid evidence for its use in controlling AFB1 contamination.
Introduction
Aspergillus flavus is a saprophytic fungus being often found in mildewed grains, grain products, and other moldy organic matter, and causes the wastage of several important agricultural crops (Wild and Gong, 2010; Liang et al., 2015). In addition, this fungus is an opportunistic human and animal pathogen causing aspergillosis diseases (Amaike and Keller, 2010). It is more important to notice that this fungus can produce aflatoxins (AFs), the most potent natural carcinogen and toxic compounds ever characterized (Da Rocha et al., 2014). In 1993, AFs are classified as a Class 1 carcinogen by the (International Agency for Research on Cancer [IARC], 1993, 2002), and were estimated to induce up to 28% of the total global cases of hepatocellular carcinoma (HCC) (Wu, 2014; Liu et al., 2017). AFs are mainly produced by A. flavus and Aspergillus parasiticus, and the former is the predominant aflatoxigenic species of contaminated foods and feeds in China (Xing et al., 2017). The most common AF-contaminated food and feed are aflatoxin B1 (AFB1), B2, G1, and G2 (Bennett and Klich, 2003). Among AFs, AFB1 is the most potent natural carcinogen and toxic compound known (Squire, 1981; Marin et al., 2013). Therefore, it is urgent to develop simple, economical, and effective ways to control A. flavus and subsequent AF contamination in food and feed, especially during storage and processing.
As we all know, ethanol is an inhibitor of the growth of bacteria and fungi (Ma et al., 2019). Previous studies showed that the accumulation of ethanol inhibited yeast cell growth and viability, affected the integrity of the cell membrane, and inactivated cellular enzymes, resulting in cell death during fermentation (Gibson et al., 2007; Kim et al., 2016; Ma et al., 2019). Ma et al. (2019) indicated that ethanol stress induced an obvious suppression of Aspergillus oryzae growth and conidia formation, and the inhibitory effect increased with ethanol concentration. As a general cell toxic substance, ethanol disturbed many cellular processes, such as irregular nuclei, the aggregation of scattered vacuoles, the increase of unsaturated fatty acid, and the overexpression of related fatty acid desaturases (Ma et al., 2019).
Transcriptional sequencing (RNA-Seq) has been widely applied to study lots of eukaryotic transcriptomes because of high sensitivity, low false-positive rates, and broad expression range coverage (Wilhelm et al., 2008; Wang et al., 2009; Lin et al., 2013; Lv et al., 2018). For A. flavus, this technology has been used to explore the mechanism of action of water activity (aw) and temperature on fungal growth and AF production (Yu et al., 2011; Zhang et al., 2014; Bai et al., 2015). Moreover, it also has been used to decipher the inhibitory mechanism of 5-azacytidine (5-AC) (Lin et al., 2013), 2-phenylethanol (Chang et al., 2015), eugenol (Lv et al., 2018), gallic acid (Zhao et al., 2018), and cinnamaldehyde (Wang et al., 2019) on A. flavus growth and AF formation. The objective of this study was to determine transcriptomic changes in A. flavus treated with ethanol and untreated samples using RNA-Seq technology. In particular, ethanol’s molecular mechanism of action on AF biosynthesis was elucidated. This study may pave a way for further understanding the inhibitory mechanism of action of ethanol on AF formation at the transcriptomic level.
Materials and Methods
Chemicals, Fungal Strain, and Growth Conditions
Ethanol (100% purity) was purchased from Beijing Chemical Works (Beijing, China). Chromatographic grade methanol and acetonitrile were purchased from Thermo Fisher Scientific (Waltham, MA, United States). The AFB1 standard was purchased from Sigma-Aldrich (Sigma-Aldrich Chemicals, St. Louis, MO, United States).
The A. flavus strain NRRL3357 was obtained from Dr. Wenbing Yin, Institute of Microbiology, Chinese Academy of Sciences, and was maintained in the dark on potato dextrose agar (PDA, purchased from Hopebio, Qingdao, China) at 4°C. Conidia suspension of 1 × 107 conidia/ml was prepared by surface washing PDA culture with 0.1% Tween-80 solution.
In order to investigate the effect of ethanol on A. flavus growth, after filtering with 0.22 μm filters, ethanol was added into the autoclaved PDA medium to obtain the final concentrations of 2, 2.5, 3, 3.5, 4, 5, and 6%. As the control group, PDA plates without ethanol were prepared. Then, 5 μl of 103–107 conidia/ml suspension was inoculated on PDA medium and incubated at 28°C for 7 days. A requisite amount of the ethanol was added to the autoclaved yeast extract sucrose (YES, purchased from Hopebio, Qingdao, China) broth to obtain the final concentrations of 1, 2, 2.5, 3, 3.5, and 4%. Then, 100 μl of 107 conidia/ml suspension was added to 100 ml of YES broth containing different concentrations of ethanol. The control cultures were treated similarly but without ethanol. After incubation at 28°C and 180 rpm/min in the dark for 7 days, fungal mycelia were collected. Each treatment was conducted in triplicate.
Determination of Mycelia Weights and AF Production
The dry weights of fugal mycelia were determined according to the method described by Yamazaki et al. (2007). AFB1 levels were determined according to the method described by Liang et al. (2015). It was extracted with acetonitrile:water (84:16) mixture from 10 ml of culture broth and purified using a ToxinFast immunoaffinity column (Huaan Magnech Biotech, Beijing, China). AFB1 was quantified using an HPLC system with a fluorescence detector (Agilent 1220 Infinity II System, Santa Clara, CA, United States) and a post-column derivation system (Huaan Magnech Biotech), and a TC-C18 column (250 mm × 4.6 mm, 5 μm particle size; Agilent, Santa Clara, CA, United States). The mean recovery of AFB1 (1–100 ng/ml) was 95.3% ± 7.5%, and the lowest detection limit was 1 ng/ml.
Preparation of cDNA Libraries, RNA Sequencing, and Data Analysis
RNA extraction, cDNA libraries preparation, and data analysis were conducted according to the methods described by Lv et al. (2018). An Illumina® HiSeq 4000TM system (San Diego, CA, United States) was used to sequence the cDNA libraries. The RNA-seq data have been deposited in the NCBI Sequence Read Archive with accession code SRP217458.
The EST sequencing, rRNA sequencing, and assembling were performed using the programs TopHat v2.0.12 (Trapnell et al., 2009), Bowtie2 (Langmead et al., 2009), and Cufflinks, respectively. The transcription levels of genes were normalized using the FPKM values (Trapnell et al., 2010). The differential expression of genes was analyzed using DEseq software (Anders and Huber, 2010). The significant differentially expressed genes were identified as log2Ratio ≥ 1 and q < 0.05 between these compared samples (Zhao et al., 2018).
Quantitative Reverse Transcription QRT-PCR Analysis of AF Biosynthesis Genes
All genes in the AF biosynthesis cluster were analyzed using QRT-PCR according to the methods described by Lv et al. (2018).
Results
Inhibitory Effect of Ethanol on Fungal Growth and AFB1 Production by A. flavus
As shown in Figure 1, some significant morphological changes of mycelial colonies were observed in A. flavus treated with ethanol compared with the control. The diameters of A. flavus colonies appeared much smaller than the control after treatment with 2–6% ethanol in a dose-dependent manner, and the mycelia growth was completely inhibited by 6% ethanol when the initial concentration was ≤104 conidia/ml (Figure 1A). In YES broth, as shown in Figure 1B, the dry mycelia weights of A. flavus appeared much lower in 3.5–4.0% ethanol application compared to the control. AFB1 production was significantly inhibited by 3.0–4.0% ethanol with the inhibition rate up to 99.8%. Interestingly, the mycelia weight was higher in 2.0–2.5% ethanol application compared to the control, but the AFB1 level was obviously decreased. Taken together, these findings suggested that ethanol significantly inhibited fungal growth and AFB1 production by A. flavus. Moreover, the suppressive effect increased with the rising levels of ethanol.
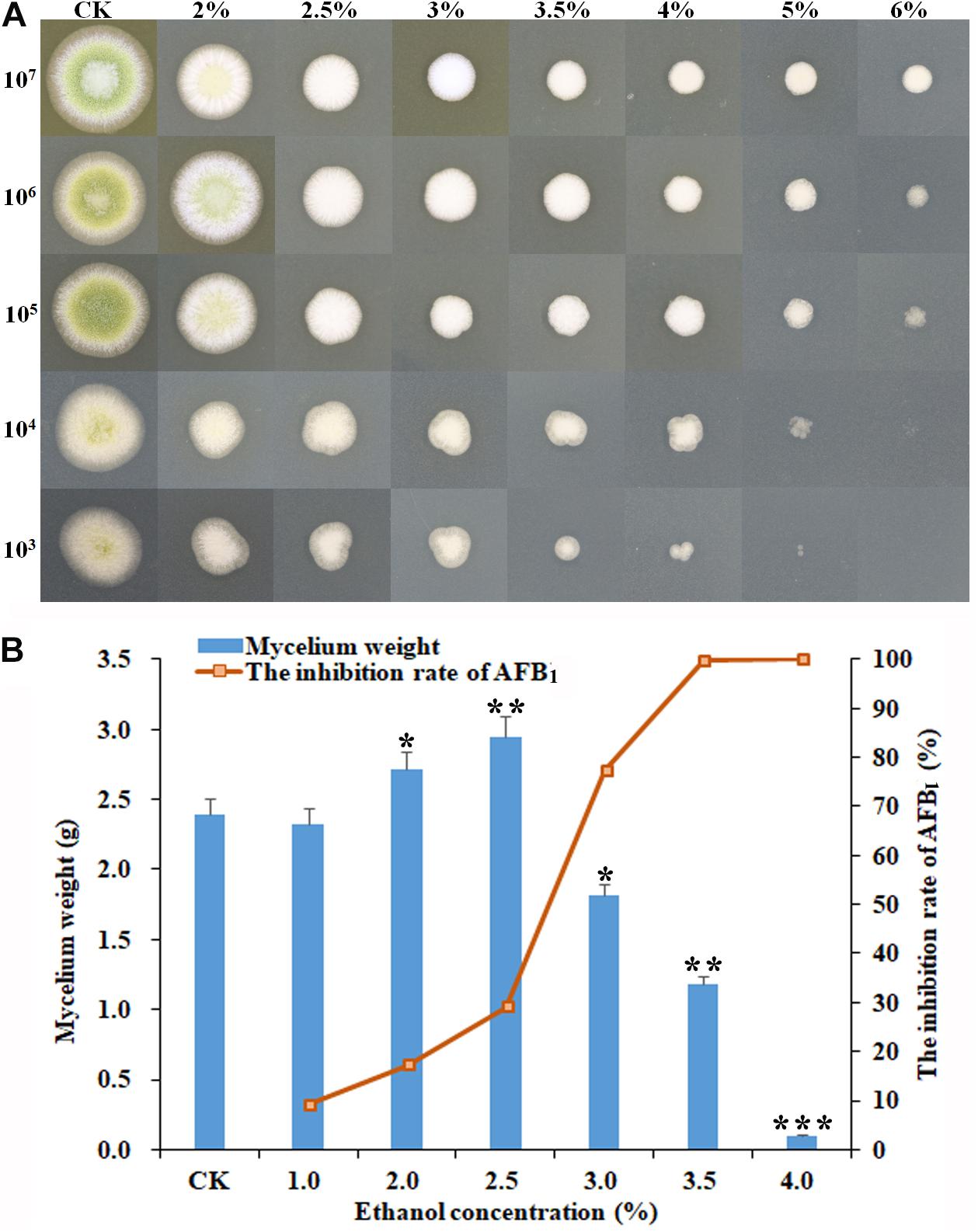
Figure 1. Effect of ethanol on the mycelial growth and AFB1 production of A. flavus NRRL3357. (A) The inhibitory effect of ethanol at different concentrations (from 0 to 6%) on mycelial colonies on PDA plates by inoculating the serial dilutions of A. flavus conidia (from 107 to 103) at 48 h post-treatment. (B) The mycelia biomass of A. flavus and the inhibition rate of AFB1 in YES broth at 120 h post-treatment. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Overall Transcriptional Response Profile of A. flavus to Ethanol
To decipher the potential inhibitory mechanism of ethanol on A. flavus growth and AFB1 biosynthesis, a transcriptome analysis was carried out. Via RNA-seq, averagely 47.81 million, 46.01 million, and 49.49 million raw reads were generated from control, 2.5 and 3.5% of ethanol treatment samples, respectively. After filtering, 46.30 million, 44.85 million, and 47.34 million clean reads were obtained, and 96.09, 93.99, and 94.32% of total clean reads from control, 2.5 and 3.5% ethanol group were aligned to reference sequences. Based on the FPKM values with FDR ≤ 0.05 and Log2Ratio ≥ 1 or ≤ −1, 2240 and 2434 differentially expression genes (DEGs) were down-regulated and up-regulated under 2.5% ethanol treatment compared with control. Under 3.5% ethanol treatment, 2636 and 3105 DEGs were down-regulated and up-regulated compared with control, respectively. Compared with 2.5% ethanol, 973 and 1547 DEGs were down-regulated and up-regulated under 3.5% ethanol treatment, respectively.
Functional and Pathway Analysis of DEGs
The DEGs between the ethanol treatment and the control provided an important clue to decipher the molecular mechanism of action of ethanol on fungal growth and AFB1 production. The functions, metabolic pathways and interactions of these DEGs were analyzed using GO and KEGG enrichment analysis. Figure 2A showed the top 30 enriched functional categories of 2240 down-regulated DEGs in A. flavus treated with 2.5% ethanol. Therein, cellular protein metabolic process, organonitrogen compound metabolic process, organonitrogen compound biosynthetic process, etc. were obvious enrichment terms in the biological process. Adenyl nucleotide binding, adenyl ribonucleotide binding, ATP binding, etc. were the main terms in molecular function. For the up-regulated DEGs in the 2.5% ethanol group (Figure 2B), carbohydrate metabolic process, phosphorus metabolic process, phosphate-containing compound metabolic, etc. were the predominant terms belonging to the biological process. The significant enrichment terms in the molecular function were hydrogen ion transmembrane transporter activity, monovalent inorganic cation transmembrane transporter activity, cation transmembrane transporter activity, etc. For the down-regulated DEGs in the 3.5% ethanol group (Figure 2C), cellular protein metabolic process, organonitrogen compound metabolic process, and organonitrogen biosynthetic process were the most abundant in the biological process. Structural constituent of ribosome, structural molecule activity, and RNA binding were the most abundant in the molecular function. For the up-regulated DEGs in this group (Figure 2D), carbohydrate metabolic process, single-organism catabolic process, and single-organism carbohydrate metabolic process were the main terms belonging to the biological process. Hydrolase activity, cofactor binding, FMN binding, etc. were the main enrichment terms in molecular function.
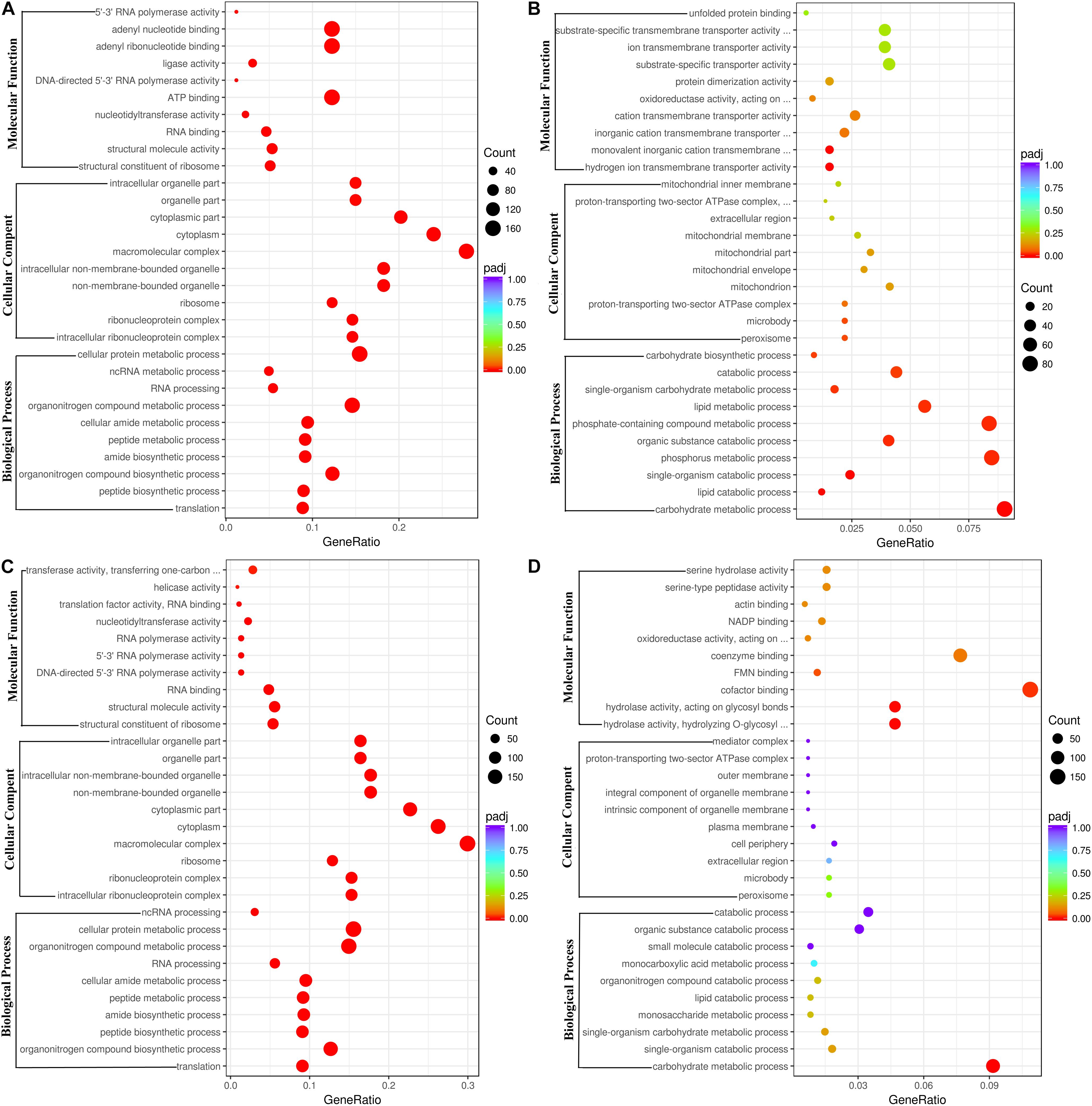
Figure 2. Go functional classification of down-regulated (A,C) and up-regulated (B,D) DEGs. (A,B) The ordinate means with 2.5% ethanol treatment. (C,D) The ordinate means with 3.5% ethanol treatment. The size of the plot represents the number of DEGs in one GO term; the color of the plot close to red represents more significant enrichment.
In A. flavus treated with 2.5% ethanol, the top 20 enriched KEGG pathway were shown in Figures 3A,B. For the down-regulated DEGs, the most abundant genes (83 DEGs) were enriched in ribosome (afv03010), and 54, 50, and 50 DEGs were enriched in RNA transport (afv03013), ribosome biogenesis (afv03008), and spliceosome (afv03040), respectively. For the up-regulated DEGs (Figure 3B), the most abundant genes (48 DEGs) were enriched in carbon metabolism (afv01200), and 33, 28, 26, and 26 DEGs were enriched in oxidative phosphorylation (afv00190), autophagy-yeast (afv04138), glycolysis/gluconeogenesis (afv00010), and protein processing in endoplasmic reticulum (afv04141), respectively. For the 3.5% ethanol group, the most abundant down-regulated DEGs (Figure 3C) were enriched in ribosome (afv03010, 97 DEGs), and 63, 63, and 52 DEGs were enriched in spliceosome (afv03008), RNA transport (afv03013), and ribosome biogenesis in eukaryotes (afv03008), respectively. The most abundant up-regulated DEGs (Figure 3D) were enriched in biosynthesis of secondary metabolites (afv01110, 136 DEGs), and 96, 58, and 32 DEGs were enriched in biosynthesis of antibiotics (afv01130), carbon metabolism (afv01200), and glycolysis/gluconeogenesis (afv00010), respectively.
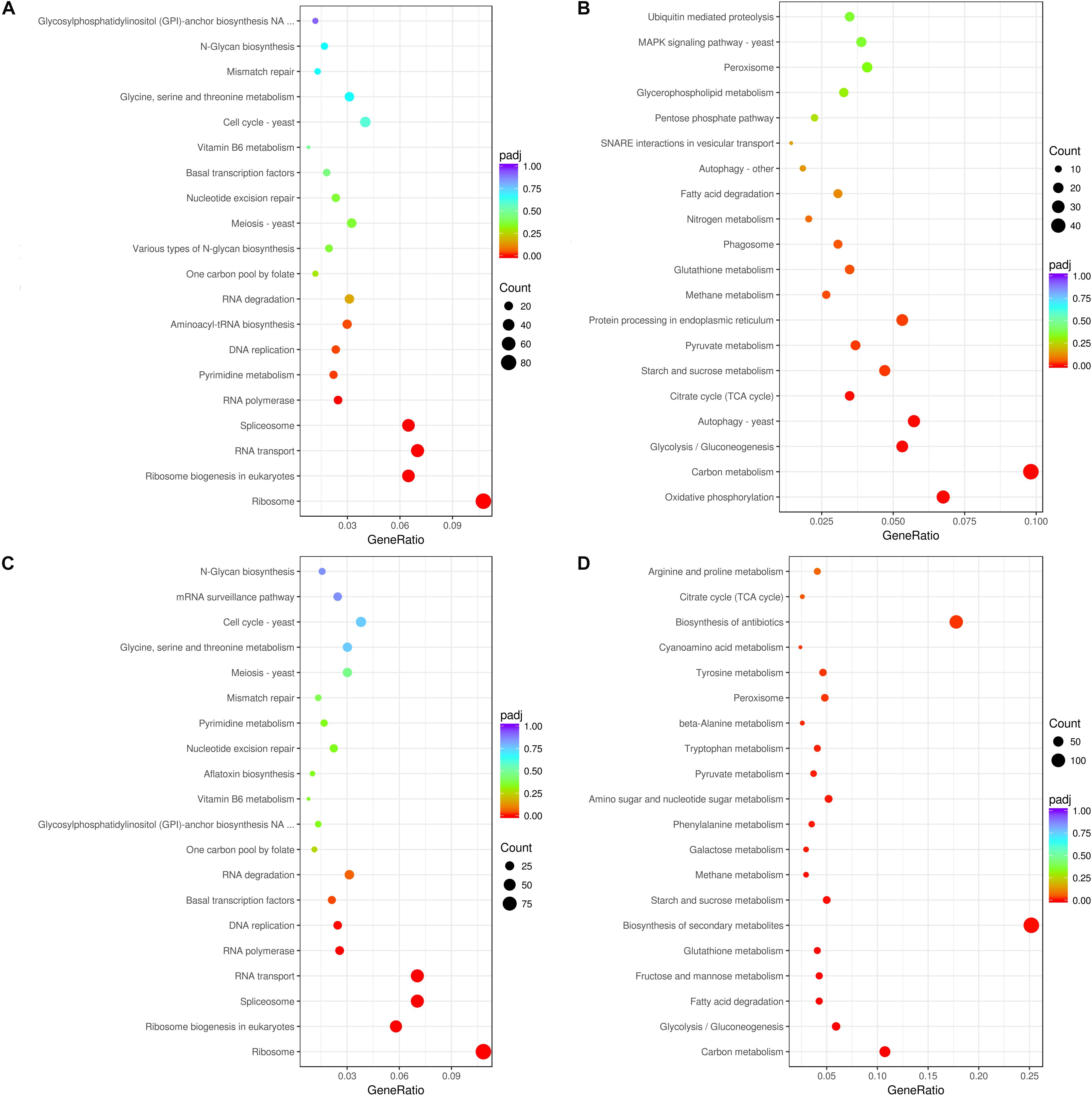
Figure 3. KEGG enrichment of down-regulated (A,C) and up-regulated (B,D) DEGs. (A,B) The ordinate means with 2.5% ethanol treatment. (C,D) The ordinate means with 3.5% ethanol treatment. The size of the plot represents the number of DEGs in one GO term; the color of the plot close to red represents more significant enrichment.
Expression Analysis of Pigment (#10), Aflatrem (#15), Aflatoxin (#54), and Cyclopiazonic Acid (#55) Biosynthesis Genes in Response to Ethanol
As shown in Table 1, in pathway #10, AFLA_016120 encoding an O-methyltransferase family protein and AFLA_016130 were down-regulated by 2.5% ethanol, but all three genes in this pathway were up-regulated by 3.5% ethanol. In pathway #15, the expression levels of most genes were very low except for AFLA_045450. In pathway #55, AFLA_139470 encoding a FAD-dependent oxidoreductase, AFLA_139480 encoding a tryptophan dimethylallyl transferase, and AFLA_139480 encoding a hybrid PKS/NRPS enzyme were down-regulated by 2.5% ethanol, while AFLA_139460 coding a MFS multidrug transporter was up-regulated. Under 3.5% ethanol treatment, four genes in pathway #55 were all down-regulated. In AF pathway #54, aflLa (a similar hypothetical gene of aflL), and aflG were up-regulated by 2.5% ethanol, while aflYd and aflYb (aflYa-e are genes in sugar cluster and the last letters indicate the sequence of genes in the cluster) were down-regulated. The expression changes of other genes in pathway #54 were slight after 2.5% ethanol treatment. Interestingly, all of AF cluster genes were down-regulated by 3.5% ethanol except for aflC. The two key regulator genes aflR and aflS were both down-regulated by 3.5% ethanol compared to the control with log2FC values −1.31 and −1.73, respectively. For the structural genes, the expression of aflK and aflLa was completely inhibited, and aflV, aflP, aflO, aflL, and aflM were markedly down-regulated with log2FC values ≤ −10, and aflY, aflX, aflW, aflQ, aflI, aflG, aflN, aflMa, aflE, and aflJ were down-regulated with log2FC values ≤ −5. It is worth mentioning that aflY(a–d) genes belong to the sugar cluster and most of them appear to be more down-regulated when 2.5% ethanol was applied. However, the aflYa gene encoding NADH oxidase was significantly down-regulated by 3.5% ethanol, while the other four genes did not change significantly.
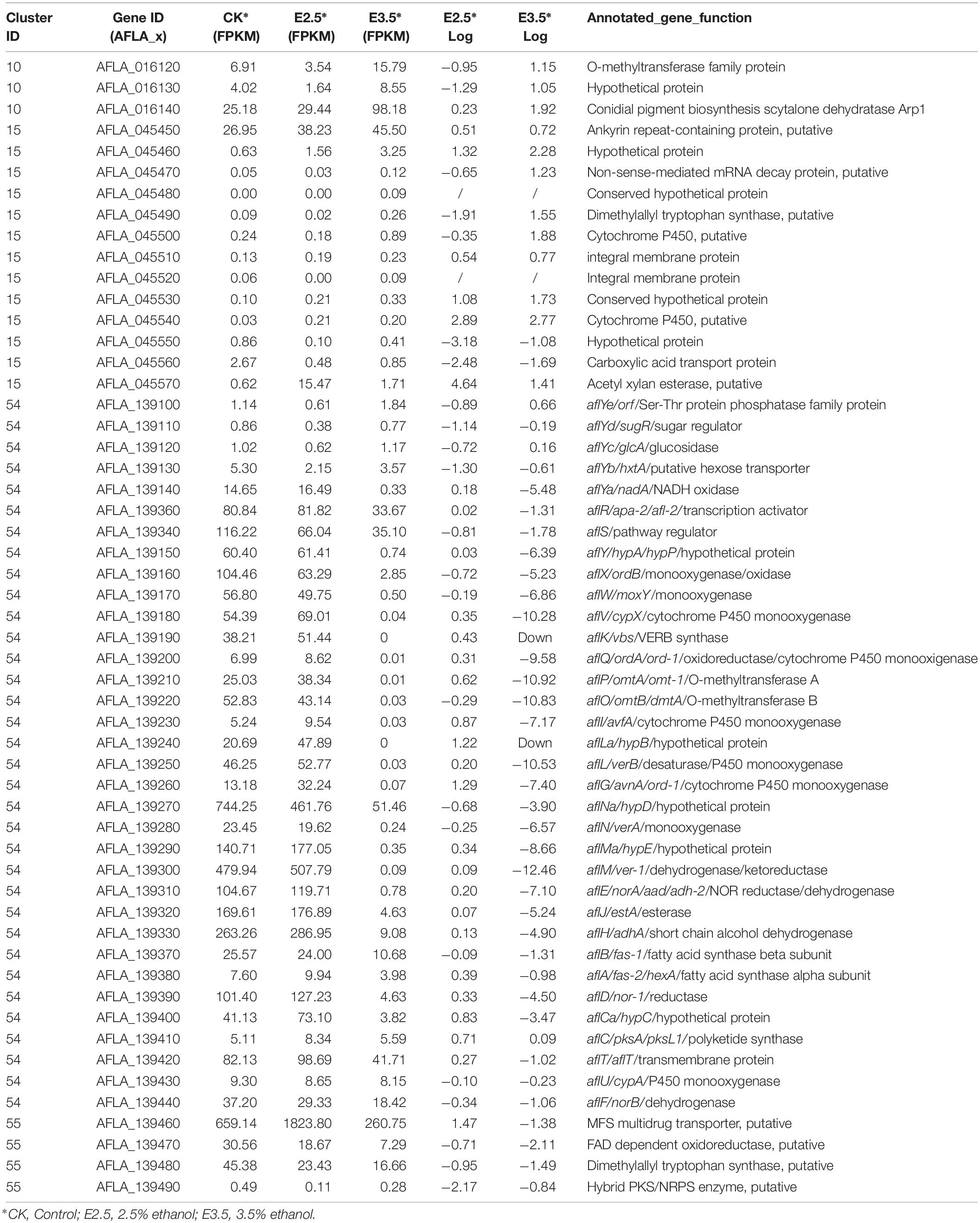
Table 1. Transcriptional activity of genes in the biosynthesis of conidial pigment (#10), aflatrem (#15), aflatoxin (#54), and cyclopiazonic acid (#55).
The RNA-seq results were confirmed by analyzing the expression of AF cluster genes in A. flavus treated with 3.5% ethanol using qRT-PCR method. As shown in Figure 4, the expression mode of these genes was consistent with the RNA-seq data.
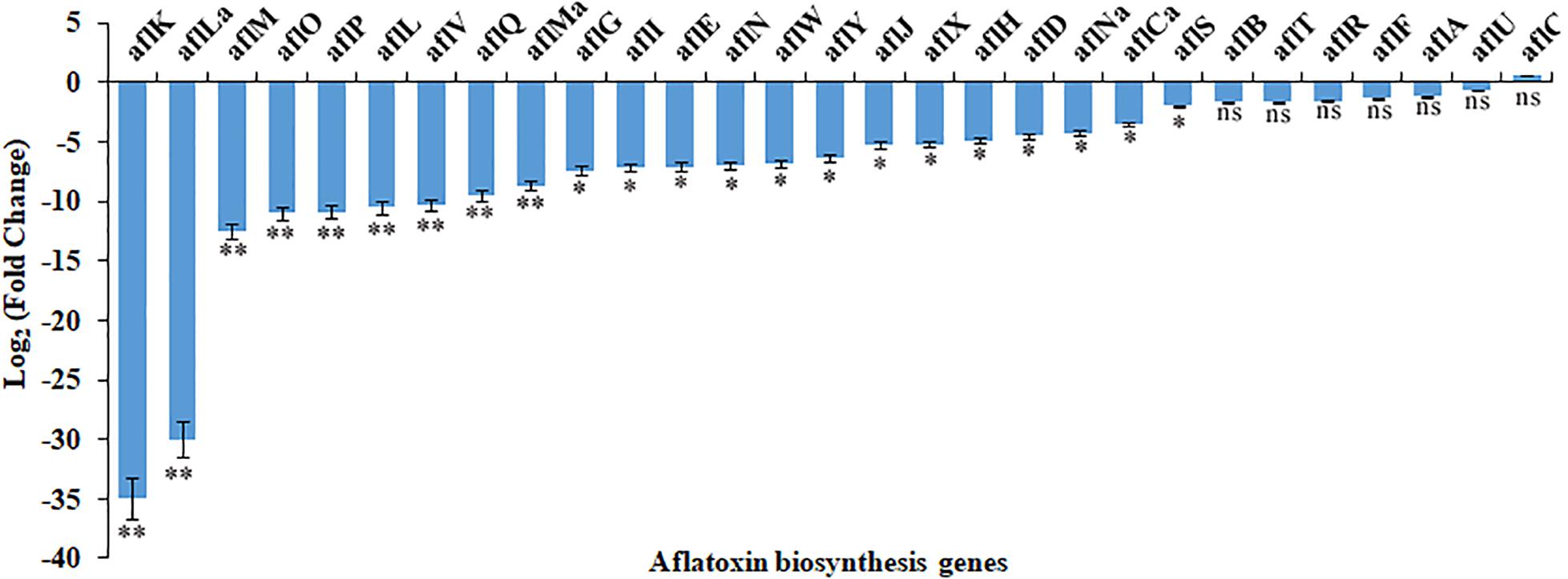
Figure 4. The differential expression of genes in aflatoxin biosynthesis cluster in response to 3.5% ethanol. ns, not significant; ∗p < 0.05; ∗∗p < 0.01.
Genes Involved in the Development
The transcription levels of genes involved in development are shown in Supplementary Table S1. From the expression profile data, we found that some genes involved in conidiophores development including FlbA, FlbC, FlbD, and HymA were down-regulated by 2.5 and 3.5% ethanol. For the velvet complex, VeA was up-regulated by 2.5% ethanol, but was down-regulated by 3.5% ethanol. FluG (AFLA_039530) and VosA were down-regulated by 2.5 and 3.5% ethanol. However, LaeA did not show a significant differential expression with ethanol treatment. AbaA controlling phialide differentiation, development regulator Mod-A (AFLA_009340), and conidial hydrophobin RodB were down-regulated by 2.5 and 3.5% ethanol. The BrlA mediating conidiophores was up-regulated by 3.5% ethanol.
Genes Involved in Fungal Oxidative Stress
The expression levels of genes involved in oxidative stress response are shown in Supplementary Table S2. The catalase/peroxidase/superoxide dismutase genes were all significantly modulated by ethanol. The expression of Cat1, Cat2, CatA, and sod1 were up-regulated by 2.5 and 3.5% ethanol while mnSOD was down-regulated. The transcriptional levels of Cat were down-regulated by 2.5% ethanol, but were up-regulated by 3.5% ethanol. The bZIP transcription factor ap-1 and two C2H2 transcription factors msnA and mtfA were up-regulated by 2.5 and 3.5% ethanol. However, the stress response transcription factor srrA was down-regulated by 2.5 and 3.5% ethanol. The MAP kinase sakA gene was obviously down-regulated by 2.5 and 3.5% ethanol. The transcriptional level of fatty acid oxygenase ppoA was down-regulated by 2.5 and 3.5% ethanol, but ppoC was up-regulated. Meantime, ppoB was expressed at a very low level. The expression of GPCRs gprC, gprH, gprM, gprR, and gprS was down-regulated by 2.5 and 3.5% ethanol, while that of gprD and gprG was up-regulated. The transcriptional level of gprK was down-regulated by 2.5% ethanol, but was up-regulated by 3.5% ethanol.
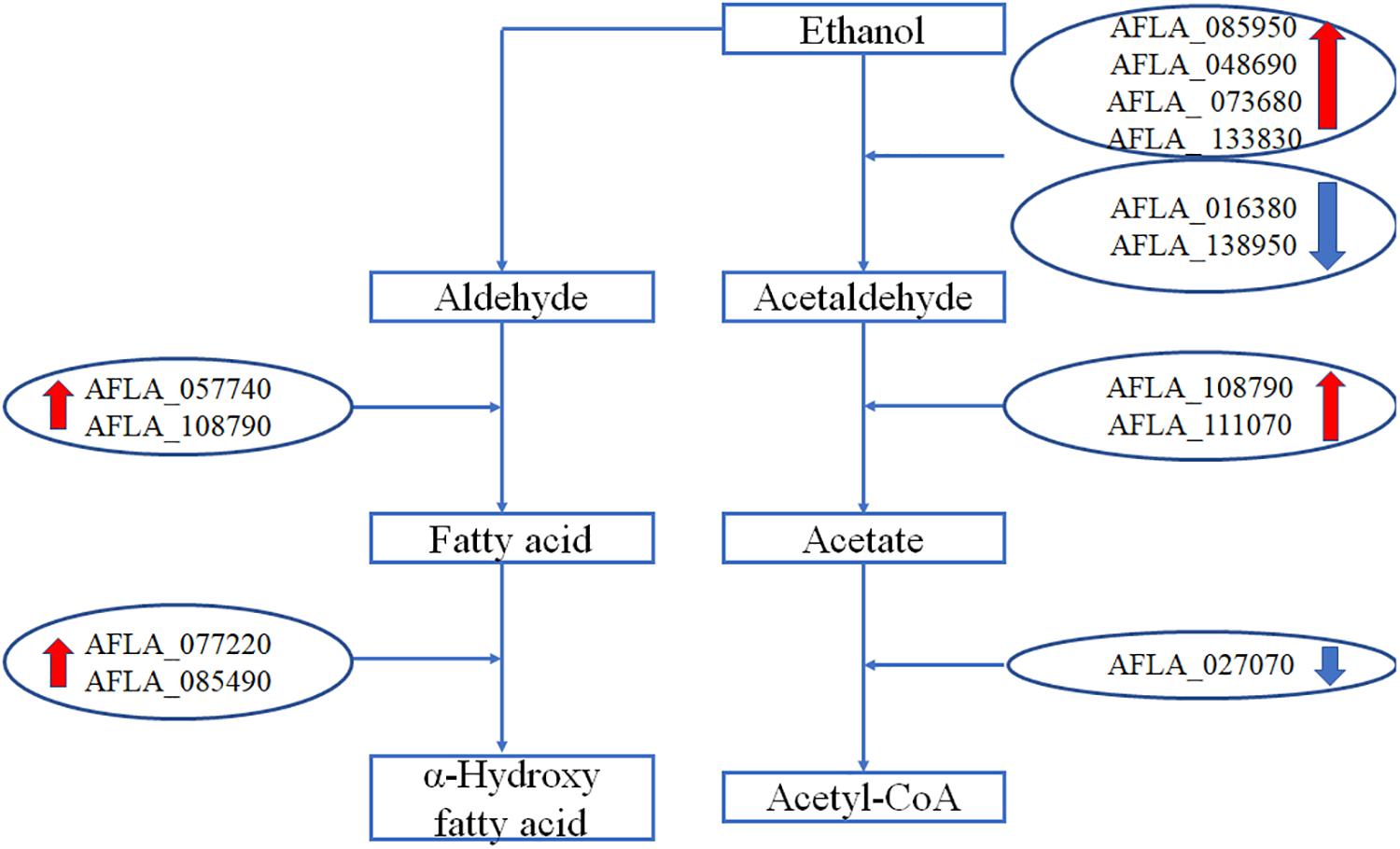
Genes Involved in Metabolism of Ethanol
The expression levels of genes involved in metabolism of ethanol are shown in Figure 5. After treatment with 3.5% ethanol, most of the genes involved in the metabolism of ethanol were up-regulated except for the two alcohol dehydrogenase genes, AFLA_016380 and AFLA_138950, involved in the process converting ethanol to acetaldehyde and the acetate and CoA ligase gene AFLA_027070 involved in the conversion of acetate to acetyl-CoA. The four alcohol dehydrogenase genes AFLA_085950, AFLA_048690, AFLA_073680, and AFLA_0133830 were up-regulated by 3.5% ethanol with Log2FC values of 2.94, 1.48, 2.82, and 1.54, respectively. The two aldehyde dehydrogenase AldA genes were up-regulated by 3.5% ethanol with Log2FC values of 2.33 and 1.69, respectively. The NADPH flavin oxidoreductase gene AFLA_077220 and P450 family fatty acid hydroxylase AFLA_085490 involved in the conversion of fatty acid to α-hydroxy fatty acid were up-regulated by 3.5% ethanol with Log2FC of 1.65 and 1.86, respectively.
Discussion
AF biosynthesis needs more than 23 enzymatic reactions (Cleveland et al., 2009). In A. flavus, the genes encoding these enzymes are located in an AF pathway gene cluster and are regulated by AFLR and AFLS (Bhatnagar et al., 2003; Cleveland et al., 2009). In our RNA-Seq data, the transcriptional level changes of the AF cluster genes were stronger in A. flavus treated with 3.5% ethanol compared to 2.5% ethanol. Of 30 AF cluster genes, the expression of 27 genes was significantly down-regulated by 3.5% ethanol except for aflA, aflC, and aflU. It is important to notice that the two key regulator genes aflR and aflS were both down-regulated by 3.5% ethanol, together with the down-regulation of the structural genes in the cluster. The gene aflK, encoding a versicolorin (VERB) synthase involved in conversion of versiconal (VAL) to VERB (McGuire et al., 1996; Silva and Townsend, 1996; Silva et al., 1996), was completely inhibited. This conversion is a critical step in AF biosynthesis because it closes the bifuran ring of AFs, which is a prerequisite for binding to DNA and gives AFs the mode of action as a mutagen (Yu et al., 2004). In addition, the expression of aflLa/hypB, a hypothetical protein gene, was also completely inhibited by 3.5% ethanol. Similarly, Lin et al. (2013) found that aflLa/hypB was completely inhibited by 5-azacytidine (5-AC), an inactivator of DNA methyltransferase. It was reported that aflLa/hypB might be involved in the second oxidation step converting O-methylsterigmatocystin (OMST) to a 7-membered ring lactone, the precursor for AFB1 formation (Ehrlich, 2009). Our previous study indicated that aflLa/hypB was one of the target genes for rapid identification of atoxigenic strains (Wei et al., 2014). These findings suggested that 3.5% ethanol inhibited AF biosynthesis by down-regulating the transcriptional levels of transcriptional factor aflR, the cofactor aflS, and subsequently most of the structural genes.
As a general cell toxic substance, ethanol affects the integrity of the cell membrane, inactivates cellular enzymes, and destroys protein structure, leading to the inhibition of fungal growth, viability, and conidia formation (Ma et al., 2019). In addition, ethanol triggered internal cellular perturbations like irregular nuclei and the aggregation of scattered vacuoles in fungal cells. The abovementioned disorders of cellular functions in turn could lead to the reduction of AFs biosynthesis. Moreover, ethanol also influenced the transcription levels of some global regulator factors. The velvet family proteins VeA, VelB, and LaeA of A. flavus form a heterotrimeric velvet complex to coordinate sexual development and biosynthesis of several secondary metabolites in the dark (Bayram et al., 2008; Chang et al., 2013). The coordinating and balanced interactions among the velvet family proteins together with FluG play a key role in maintaining programmed AFs biosynthesis and conidiation and sclerotial production (Chang et al., 2013). After treatment with 3.5% ethanol, the expression of veA and fluG was significantly down-regulated with Log2FC −2.97 and −4.03, respectively. The down-regulation of veA suppressed the expression of several key AFs genes including aflR, aflD, aflM, and aflP and resulted in the inhibition of AF biosynthesis (Duran et al., 2007).
The oxidative stress was recognized as a prerequisite for AFs formation in A. flavus and A. parasiticus (Reverberi et al., 2008; Zhang et al., 2015; Lv et al., 2018; Guan et al., 2019). In the meantime, AFs biosynthesis is thought to protect the fungus against oxidative stress (Wang et al., 2019). Several previous studies have indicated that some AFs inhibitors can regulate the stress response system of fungi (Reverberi et al., 2005; Grintzalis et al., 2014; Sun et al., 2015; Caceres et al., 2017). After treatment with 3.5% ethanol, all catalase genes including Cat, Cat1, Cat2, CatA, and Cu, Zn superoxide dismutase gene sod1 were up-regulated, while only Mn superoxide dismutase gene mnSOD was down-regulated. Similarly, piperine exposure significantly induced decreased expression of veA together with the overexpression of several bZIP transcription factors genes like atfA, atfB, and ap-1 and genes encoding catalase such as catA, cat2, and superoxide dismutase like sod1 in A. flavus (Caceres et al., 2017). Moreover, this gene response was coupled with an obvious increase of catalase enzymatic activity (Caceres et al., 2017). Cinnamaldehyde exposure resulted in the up-regulation of several transcription factors genes like srrA, msnA, and atfB and genes encoding catalase like cat, cat1, catA, and superoxide dismutase including sod1 and mnSOD (Wang et al., 2019).
The transcriptional levels of genes involved in the antioxidant system were modulated by the upstream transcription factors including ap-1, atfA, atfB, msnA, mtfA, and PacC (Hong et al., 2013). As a redox-state sensor protein, the functions of Ap-1 are highly conserved in yeast, fungi, and mammals (Toone et al., 2001; Caceres et al., 2017). In fungi, the N- and C-terminal cysteine-rich domains of Ap-1-like protein might act as a sensor target of reactive oxygen species (ROS) like H2O2 (Sies, 2014). In A. parasiticus, the deletion of ApyapA causes the increase of AFs biosynthesis, oxidative stress, premature conidiogenesis, and an earlier transcription of AFs cluster genes like aflR and aflE (Reverberi et al., 2008; Caceres et al., 2017). The bZIP transcription factor SrrA, an ortholog of Saccharomyces cerevisiae Skn7 and Saccharomyces pombe Prr1, mediates cellular response to environmental stimuli (Hagiwara et al., 2007; Vargas-Perez et al., 2007). In A. parasiticus, Hong et al. (2013) identified a recognition site of SrrA in promoters of the antioxidant genes cat1 and mnsod, and AFs biosynthetic genes aflB (fas-1) and aflM (ver-1). Moreover, the adjacent binding sites of SrrA and AP-1 in the promoter suggest that they can interact and are involved in the transcriptional regulation of AFs genes (Hong et al., 2013). In the present study, an up-regulation of ap-1 and a down-regulation of srrA were observed upon 3.5% ethanol addition. MsnA is a C2H2 zinc finger transcription factor and can respond to some cellular stress such as oxidative stress, carbon starvation, heat shock, and osmotic stress (Martinez-Pastor et al., 1996; Hong et al., 2013). In A. flavus and A. parasiticus, disruption of msnA led to increased AFs biosynthesis and the production of conidia, ROS, and kojic acid, although fungal growth was inhibited (Chang et al., 2011). In addition, msnA deletion down-regulated transcription levels of genes encoding antioxidant enzymes, which protect fungus against ROS (Hong et al., 2013). Our previous studies revealed that eugenol and cinnamaldehyde up-regulated the expression of msnA and inhibited AFs biosynthesis (Lv et al., 2018; Wang et al., 2019). A similar finding, the up-regulation of msnA in A. flavus treated with 3.5% ethanol, was obtained in the present study. MtfA is another C2H2 zinc finger transcription factor, which was originally identified in Aspergillus nidulans and was involved in sterigmatocystin (ST) regulation (Ramamoorthy et al., 2013). The disruption and overexpression of mtfA both induced the decreased production of ST (Zhuang et al., 2016). In A. flavus, overexpression of mtfA dramatically reduced AFB1 production accompanied by a drastic reduction of aflR expression compared to the WT strain while deletion of mtfA did not significantly influenced AFB1 production (Zhuang et al., 2016). Caceres et al. (2016) indicated that eugenol up-regulated the expression of mtfA and inhibited AFB1 production. Similarly, the transcription level of mtfA was up-regulated by 3.5% ethanol in the present study.
It is important to point out that the transcriptional status is very fluctuating depending on transcription rate and half-life of the mRNA, which may be very short compared to the more accumulative and stable concentration of the AF produced. This means that the transcription may not be directly correlated with the amount of AF produced at each time point. Therefore, the following mechanism of action of ethanol on the inhibition of AFs proposed in this study is based on the RNA-seq data on the 7th day.
Based on the abovementioned results, we proposed a hypothetical mechanism of action of ethanol on the inhibition of AFs (Figure 6). Taken together, the enhanced transcription levels of the stress response system, such as bZIP transcription factor ap-1, C2H2 transcription factors msnA and mtfA, the down-regulation of stress response transcription factor srrA, and the overexpression of genes encoding for antioxidant system including catalase genes and superoxide dismutase gene in A. flavus treated with ethanol, significantly down-regulate the expression of AF biosynthesis genes and in turn result in the inhibition of AFs production.
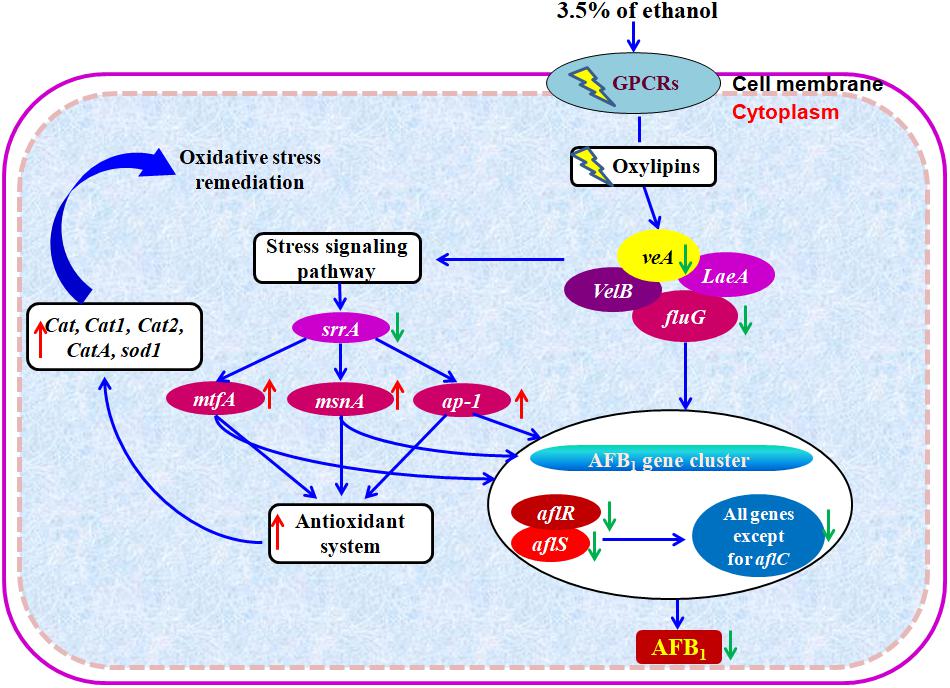
Figure 6. Hypothetical mechanism of action of ethanol on AFB1 biosynthesis. Down- and up-regulation of gene on ethanol addition is expressed using green and red arrows, respectively.
Conclusion
In the present study, we reveal the transcription modulation mechanism behind ethanol’s AFB1-repressing action using an RNA-Seq. The RNA data indicated that (1) with ethanol treatment, AFB1 cluster genes were dramatically down-regulated following the up-regulation of their specific regulators aflS/aflR; (2) ethanol’s mechanism of action involved the down-regulation of the global regulator veA and fluG; (3) ethanol’s transcription modulation mechanism involved the decreased expression of stress response transcription factor srrA together with overexpression of bZIP transcription factor ap-1 and C2H2 transcription factors msnA and mtfA; (4) ethanol induced enhanced levels of anti-oxidant enzymatic genes including Cat, Cat1, Cat2, CatA, and Cu, Zn superoxide dismutase gene sod1. In conclusion, these results strongly suggest that ethanol inhibits AFB1 biosynthesis by A. flavus via enhancing fungal oxidative stress response.
Data Availability Statement
The datasets generated for this study can be found in the https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA558521.
Author Contributions
FX and QY conceived and designed the experiments. YR, JJ, and MZ performed the experiments. YR and FX analyzed the data and wrote the manuscript.
Funding
We gratefully acknowledge the financial support of the National Key Research and Development Program of China (2016YFD0400105), the National Natural Science Foundation of China (31571938), and the Fundamental Research Funds for Central Non-profit Scientific Institution (S2016JC02). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02946/full#supplementary-material
References
Amaike, S., and Keller, N. P. (2010). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Bai, Y., Wang, S., Zhong, H., Yang, Q., Zhang, F., Zhuang, Z., et al. (2015). Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature. Sci. Rep. 5:14582. doi: 10.1038/srep14582
Bayram, O., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Bennett, J. W., and Klich, M. (2003). Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. doi: 10.1128/cmr.16.3.497-516.2003
Bhatnagar, D., Ehrlich, K. C., and Cleveland, T. E. (2003). Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61, 83–93. doi: 10.1007/s00253-002-1199-x
Caceres, I., El Khoury, R., Bailly, S., Oswald, I. P., Puel, O., and Bailly, J. D. (2017). Piperine inhibits aflatoxin B1 production in Aspergillus flavus by modulating fungal oxidative stress response. Fungal Genet. Biol. 107, 77–85. doi: 10.1016/j.fgb.2017.08.005
Caceres, I., Khoury, R. E., Medina, A., Lippi, Y., Naylies, C., Atoui, A., et al. (2016). Deciphering the anti-aflatoxinogenic properties of eugenol using a large-scale q-PCR approach. Toxins 8:123. doi: 10.3390/toxins8050123
Chang, P. K., Hu, S. S., Sarreal, S. B., and Li, R. W. (2015). Suppression of aflatoxin biosynthesis in Aspergillus flavus by 2-phenylethanol is associated with stimulated growth and decreased degradation of branched-chain amino acids. Toxins 7, 3887–3902. doi: 10.3390/toxins7103887
Chang, P. K., Scharfenstein, L. L., Li, P., and Ehrlich, K. C. (2013). Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 58–59, 71–79. doi: 10.1016/j.fgb.2013.08.009
Chang, P. K., Scharfenstein, L. L., Luo, M., Mahoney, N., Molyneux, R. J., Yu, J., et al. (2011). Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 3, 82–104. doi: 10.3390/toxins3010082
Cleveland, T. T., Yu, J., Fedorova, N., Bhatnagar, D., Payne, G. A., Nierman, W. C., et al. (2009). Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27, 151–157. doi: 10.1016/j.tibtech.2008.11.008
Da Rocha, M. E. B., Freire, F. D. C. O., Maia, F. E. F., Guedes, M. I. F., and Rondina, D. (2014). Mycotoxins and their effects on human and animal health. Food Control 36, 159–165. doi: 10.1016/j.foodcont.2013.08.021
Duran, R. M., Cary, J. W., and Calvo, A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus, is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Ehrlich, K. C. (2009). Predicted roles of the uncharacterized clustered genes in aflatoxin biosynthesis. Toxins 1, 37–58. doi: 10.3390/toxins1010037
Gibson, B. R., Lawrence, S. J., Leclaire, J. P., Powell, C. D., and Smart, K. A. (2007). Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 31, 535–569. doi: 10.1111/j.1574-6976.2007.00076.x
Grintzalis, K., Vernardis, S. I., Klapa, M. I., and Georgiou, C. D. (2014). Role of oxidative stress in sclerotial differentiation and aflatoxin B1 biosynthesis in Aspergillus flavus. Appl. Environ. Microbiol. 80, 5561–5571. doi: 10.1128/AEM.01282-14
Guan, X., Zhao, Y., Liu, X., Shang, B., Xing, F., Zhou, L., et al. (2019). The bZIP transcription factor Afap1 mediates the oxidative stress response and aflatoxin biosynthesis in Aspergillus flavus. El factor de transcripción bZIP Afap1 afecta al estrés oxidativo y la biosíntesis de aflatoxinas en Aspergillus flavus. Rev. Argent. Microbiol. 51, 292–301. doi: 10.1016/j.ram.2018.07.003
Hagiwara, D., Asano, Y., Marui, J., Furukawa, K., Kanamaru, K., Kato, M., et al. (2007). SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1003–1014. doi: 10.1271/bbb.60665
Hong, S. Y., Roze, L. V., Wee, J., and Linz, J. E. (2013). Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in Aspergilli. Microbiologyopen 2, 144–160. doi: 10.1002/mbo3.63
International Agency for Research on Cancer [IARC] (1993). Some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monogr. Eval. Carcinog. Risks Hum. 56, 245–395.
International Agency for Research on Cancer [IARC] (2002). Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monogr. Eval. Carcinog. Risks Hum. 82, 1–556.
Kim, S., Kim, J., Song, J. H., Jung, Y. H., Choi, I. S., Choi, W., et al. (2016). Elucidation of ethanol tolerance mechanisms in Saccharomyces cerevisiae by global metabolite profiling. Biotechnol. J. 11, 1221–1229. doi: 10.1002/biot.201500613
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Liang, D., Xing, F., Selvaraj, J. N., Liu, X., Wang, L., Hua, H., et al. (2015). Inhibitory effect of cinnamaldehyde, citral, and eugenol on aflatoxin biosynthetic gene expression and aflatoxin B1 biosynthesis in Aspergillus flavus. J. Food Sci. 80, M2917–M2924. doi: 10.1111/1750-3841.13144
Lin, J. Q., Zhao, X. X., Zhi, Q. Q., Zhao, M., and He, Z. M. (2013). Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 56, 78–86. doi: 10.1016/j.fgb.2013.04.007
Liu, X., Guan, X., Xing, F., Lv, C., Dai, X., and Liu, Y. (2017). Effect of water activity and temperature on the growth of Aspergillus flavus, the expression of aflaoxin biosynthetic genes and aflatoxin production in shelled peanuts. Food Control 82, 325–332. doi: 10.1016/j.foodcont.2017.07.012
Lv, C., Wang, P., Ma, L., Zheng, M., Liu, Y., and Xing, F. (2018). Large-scale comparative analysis of eugenol-induced/repressed genes expression in Aspergillus flavus using RNA-seq. Front. Microbiol. 9:1116. doi: 10.3389/fmicb.2018.01116
Ma, L., Fu, L., Hu, Z., Li, Y., Zheng, X., Zhang, Z., et al. (2019). Modulation of fatty acid composition of Aspergillus oryzae in response to ethanol stress. Microorganisms 7:158. doi: 10.3390/microorganisms7060158
Marin, S., Ramos, A. J., Cano-Sancho, G., and Sanchis, V. (2013). Mycotoxins: occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 60, 218–237. doi: 10.1016/j.fct.2013.07.047
Martinez-Pastor, M. T., Marchler, G., Schuller, C., Marchler-Bauer, A., Ruis, H., and Estruch, F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235. doi: 10.1089/dna.1996.15.423
McGuire, S. M., Silva, J. C., Casillas, E. G., and Townsend, C. A. (1996). Purification and characterization of versicolorin B synthase from Aspergillus parasiticus. Biochemistry 35, 11470–11486. doi: 10.1021/bi960924s
Ramamoorthy, V., Dhingra, S., Kincaid, A., Shantappa, S., Feng, X., and Calvo, A. M. (2013). The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS One 8:e74122. doi: 10.1371/journal.pone.0074122
Reverberi, M., Fabbri, A. A., Zjalic, S., Ricelli, A., Punelli, F., and Fanelli, C. (2005). Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl. Microbiol. Biotechnol. 69, 207–215. doi: 10.1007/s00253-005-1979-1
Reverberi, M., Zialic, S., Ricelli, A., Punelli, F., Camera, E., Fabbri, C., et al. (2008). Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role for the ApyapA gene. Eukaryot. Cell 7, 988–1000. doi: 10.1128/EC.00228-07
Sies, H. (2014). Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 289, 8735–8741. doi: 10.1074/jbc.R113.544635
Silva, J. C., Minto, R. E., Barry, C. E., Holland, K. A., and Townsend, C. A. (1996). Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus: expansion of the aflatoxin B1 biosynthetic cluster. J. Biol. Chem. 271, 13600–13608. doi: 10.1074/jbc.271.23.13600
Silva, J. C., and Townsend, C. A. (1996). Heterologous expression, isolation, and characterization of versicolorin B synthase from Aspergillus parasiticus. J. Biol. Chem. 272, 804–813. doi: 10.1074/jbc.272.2.804
Squire, R. A. (1981). Ranking animal carcinogens: a proposed regulatory approach. Science 214, 877–880. doi: 10.1126/science.7302565
Sun, Q., Shang, B., Wang, L., Lu, Z., and Liu, Y. (2015). Cinnamaldehyde inhibits fungal growth and aflatoxin B1 biosynthesis by modulating the oxidative stress response of Aspergillus flavus. Appl. Microbiol. Biotechnol. 100, 1355–1364. doi: 10.1007/s00253-015-7159-z
Toone, W. M., Morgan, B. A., and Jones, N. (2001). Redox control of AP-1-like factors in yeast and beyond. Oncogene 20, 2336–2346. doi: 10.1038/sj.onc.1204384
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vargas-Perez, I., Sanchez, O., Kawasaki, L., Georgellis, D., and Aguirre, J. (2007). Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 6, 1570–1583. doi: 10.1128/EC.00085-07
Wang, P., Ma, L., Jin, J., Zheng, M., Pan, L., Zhao, Y., et al. (2019). The anti-aflatoxigenic mechanism of cinnamaldehyde in Aspergillus flavus. Sci. Rep. 9, 10499. doi: 10.1038/s41598-019-47003-z
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wei, D., Zhou, L., Selvaraj, J. N., Zhang, C., Xing, F., Zhao, Y., et al. (2014). Molecular characterization of atoxigenic Aspergillus flavus isolates collected in China. J. Microbiol. 52, 559–565. doi: 10.1007/s12275-014-3629-8
Wild, C. P., and Gong, Y. Y. (2010). Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis 31, 71–82. doi: 10.1093/carcin/bgp264
Wilhelm, B. T., Marguerat, S., Watt, S., Schubert, F., Wood, V., Goodhead, I., et al. (2008). Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243. doi: 10.1038/nature07002
Xing, F., Liu, X., Wang, L., Selvaraj, J. N., Jin, N., Wang, Y., et al. (2017). Distribution and variation of fungi and major mycotoxins in pre- and post-nature drying maize in North China plain. Food Control 80, 244–251. doi: 10.1016/j.foodcont.2017.03.055
Yamazaki, H., Yamazaki, D., Takaya, N., Takagi, M., Ohta, A., and Horiuchi, H. (2007). A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr. Genet. 51, 89–98. doi: 10.1007/s00294-006-0109-7
Yu, J., Chang, P. K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004
Yu, J., Fedorova, N. D., Montalbano, B. G., Bhatnagar, D., Cleveland, T. E., Bennett, J. W., et al. (2011). Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 322, 145–149. doi: 10.1111/j.1574-6968.2011.02345.x
Zhang, F., Guo, Z., Zhong, H., Wang, S., Yang, W., Liu, Y., et al. (2014). RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 6, 3187–3207. doi: 10.3390/toxins6113187
Zhang, Z., Qin, G., Li, B., and Tian, S. (2015). Effect of cinnamic acid for controlling gray mold on table grape and its possible mechanisms of action. Curr. Microbiol. 71, 396–402. doi: 10.1007/s00284-015-0863-1
Zhao, X., Zhi, Q. Q., Li, J. Y., Keller, N. P., and He, Z. M. (2018). The antioxidant gallic acid inhibits aflatoxin formation in Aspergillus flavus by modulating transcription factors FarB and CreA. Toxins 10:270. doi: 10.3390/toxins10070270
Keywords: aflatoxin B1, Aspergillus flavus, transcriptome, RNA-seq, oxidative stress, ethanol
Citation: Ren Y, Jin J, Zheng M, Yang Q and Xing F (2020) Ethanol Inhibits Aflatoxin B1 Biosynthesis in Aspergillus flavus by Up-Regulating Oxidative Stress-Related Genes. Front. Microbiol. 10:2946. doi: 10.3389/fmicb.2019.02946
Received: 07 August 2019; Accepted: 06 December 2019;
Published: 17 January 2020.
Edited by:
Antonio Francesco Logrieco, Italian National Research Council (CNR), ItalyReviewed by:
Antonia Gallo, Italian National Research Council (CNR), ItalyRolf Geisen, Max Rubner Institut Karlsruhe, Germany
Copyright © 2020 Ren, Jin, Zheng, Yang and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingli Yang, rice407@163.com; Fuguo Xing, xingfuguo@caas.cn; fgxing@163.com
 Yaoyao Ren1
Yaoyao Ren1 Qingli Yang
Qingli Yang Fuguo Xing
Fuguo Xing