Corrigendum: The cyl Genes Reveal the Biosynthetic and Evolutionary Origins of the Group B Streptococcus Hemolytic Lipid, Granadaene
- 1Department of Global Health, University of Washington, Seattle, WA, United States
- 2Center for Global Infectious Disease Research, Seattle Children’s Research Institute, Seattle, WA, United States
- 3Computational Biology Branch, National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD, United States
- 4Department of Organic Chemistry, University of Granada, Granada, Spain
- 5Department of Pediatrics, University of Washington School of Medicine, Seattle, WA, United States
- 6Department of Pathology, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa
Group B Streptococcus (GBS) is a β-hemolytic, Gram-positive bacterium that commonly colonizes the female lower genital tract and is associated with fetal injury, preterm birth, spontaneous abortion, and neonatal infections. A major factor promoting GBS virulence is the β-hemolysin/cytolysin, which is cytotoxic to several host cells. We recently showed that the ornithine rhamnolipid pigment, Granadaene, produced by the gene products of the cyl operon, is hemolytic. Here, we demonstrate that heterologous expression of the GBS cyl operon conferred hemolysis, pigmentation, and cytoxicity to Lactococcus lactis, a model non-hemolytic Gram-positive bacterium. Similarly, pigment purified from L. lactis is hemolytic, cytolytic, and identical in structure to Granadaene extracted from GBS, indicating the cyl operon is sufficient for Granadaene production in a heterologous host. Using a systematic survey of phyletic patterns and contextual associations of the cyl genes, we identify homologs of the cyl operon in physiologically diverse Gram-positive bacteria and propose undescribed functions of cyl gene products. Together, these findings bring greater understanding to the biosynthesis and evolutionary foundations of a key GBS virulence factor and suggest that such potentially toxic lipids may be encoded by other bacteria.
Introduction
Hemolytic activity is a key determinant of colonization and pathogenesis in Group B Streptococcus (GBS) (Nizet et al., 1996; Doran et al., 2002, 2003; Patras et al., 2013; Whidbey et al., 2013, 2015b; Boldenow et al., 2016), a Gram-positive bacterium that resides in the lower genital and/or gastrointestinal tract of approximately 18% of women and is a major cause of preterm birth and severe neonatal infections (Russell et al., 2017a, b; Seale et al., 2017a, b). Nearly 100 years ago, GBS isolates of human origin were first described as β-hemolytic (Brown, 1920; Ayers and Philip, 1922), which was invariantly linked to a pigmented phenotype (Nizet et al., 1996; Spellerberg et al., 2000; Pritzlaff et al., 2001). Recently, the GBS hemolysin and pigment were shown to be one and the same (Whidbey et al., 2013), initiating a shift in understanding this virulence factor. The GBS hemolytic pigment, also known as Granadaene (Rosa-Fraile et al., 2006), is a cell surface-associated (Platt, 1995) ornithine rhamnolipid consisting of a 12-alkene chain and is unrelated to other commonly studied Gram-positive pore-forming toxins, such as lysteriolysin O (Hamon et al., 2012) or alpha toxin (Seilie and Bubeck Wardenburg, 2017), which are proteinaceous in nature. Many gaps remain in our understanding of Granadaene, including the specifics of its biosynthesis in bacterial cells and reasons why GBS may have evolved to produce this potent toxin. Such insights would improve our understanding of this critical GBS virulence factor as well that of other similar, potentially toxic microbial lipids.
The cyl operon (cylX-K) is necessary for hemolytic pigment production in GBS (Spellerberg et al., 1999, 2000; Pritzlaff et al., 2001; Whidbey et al., 2013). We have previously postulated that enzymes encoded in the cyl operon use acetyl-CoA, malonyl Co-A, ornithine, and rhamnose as the building blocks for pigment biosynthesis in GBS (Whidbey et al., 2013). Here, for the first time, we show that heterologous expression of the GBS cyl operon is sufficient to confer the production of functional Granadaene in Lactococcus lactis, a generally recognized as safe (GRAS) Gram-positive bacterial strain (Cavanagh et al., 2015). Phyletic analysis revealed previously undescribed functional categories of cyl gene products, indicated that the cyl operon genes are present in a diverse range of Gram-positive bacteria, and suggested that pigment biosynthesis evolved in free-living bacteria, likely as a photoprotectant or as a defense mechanism against competing organisms. Collectively, these findings provide biosynthetic and evolutionary insight into a critical GBS virulence factor.
Results and Discussion
Heterologous Expression of the GBS cyl Operon in Lactococcus lactis Confers Hemolysis, Pigmentation, and Cytotoxicity
Our recent studies (Whidbey et al., 2013, 2015b; Gendrin et al., 2015; Boldenow et al., 2016) have revealed that the hemolytic pigment encoded by the GBS cyl operon genes is cytotoxic to many eukaryotic cells. Given that GBS colonizes the human host with other commensal microbes, we wondered whether horizontal gene transfer of this operon would enable production of the hemolytic lipid pigment [also known as Granadaene (Rosa-Fraile et al., 2006)] to non-hemolytic bacteria. To test this hypothesis, we expressed the cyl operon genes in a model Gram-positive bacterium, Lactococcus lactis. First, we cloned the 12-gene cyl operon (i.e., genes cylX-cylK) into a complementation vector pDC123 (Chaffin and Rubens, 1998) to generate the pcylX-K plasmid. Then, we complemented a non-hemolytic/non-pigmented GBS strain lacking the entire cyl operon [GBSΔcylX-K (Whidbey et al., 2013)] with the pcylX-K plasmid and confirmed that the plasmid restored hemolysis and pigmentation as observed on blood agar and Granada media (Supplementary Figure S1). Next, we transformed the pcylX-K plasmid into L. lactis to generate L. lactis pcylX-K. As a control, L. lactis was transformed with the empty plasmid vector (L. lactis pEmpty). Like hemolytic GBS, β-hemolysis and pigmentation were observed with L. lactis pcylX-K and not L. lactis pEmpty (Figures 1A,B). Consistent with previous observations with GBS (Whidbey et al., 2013), plasmids encoding only cylE (pcylE) or cylABE (pcylABE) were insufficient to confer hemolysis and pigmentation to L. lactis (Figures 1C,D). Also, the recombinant pcylX-K plasmid did not confer hemolysis to E. coli (Supplementary Figure S2), which we predict is due to the abundance of rare codons present in the cyl operon (Supplementary Table S1). Together, these results indicate that expression of the cyl operon is necessary and sufficient for hemolysis and pigmentation in GBS and even in L. lactis.
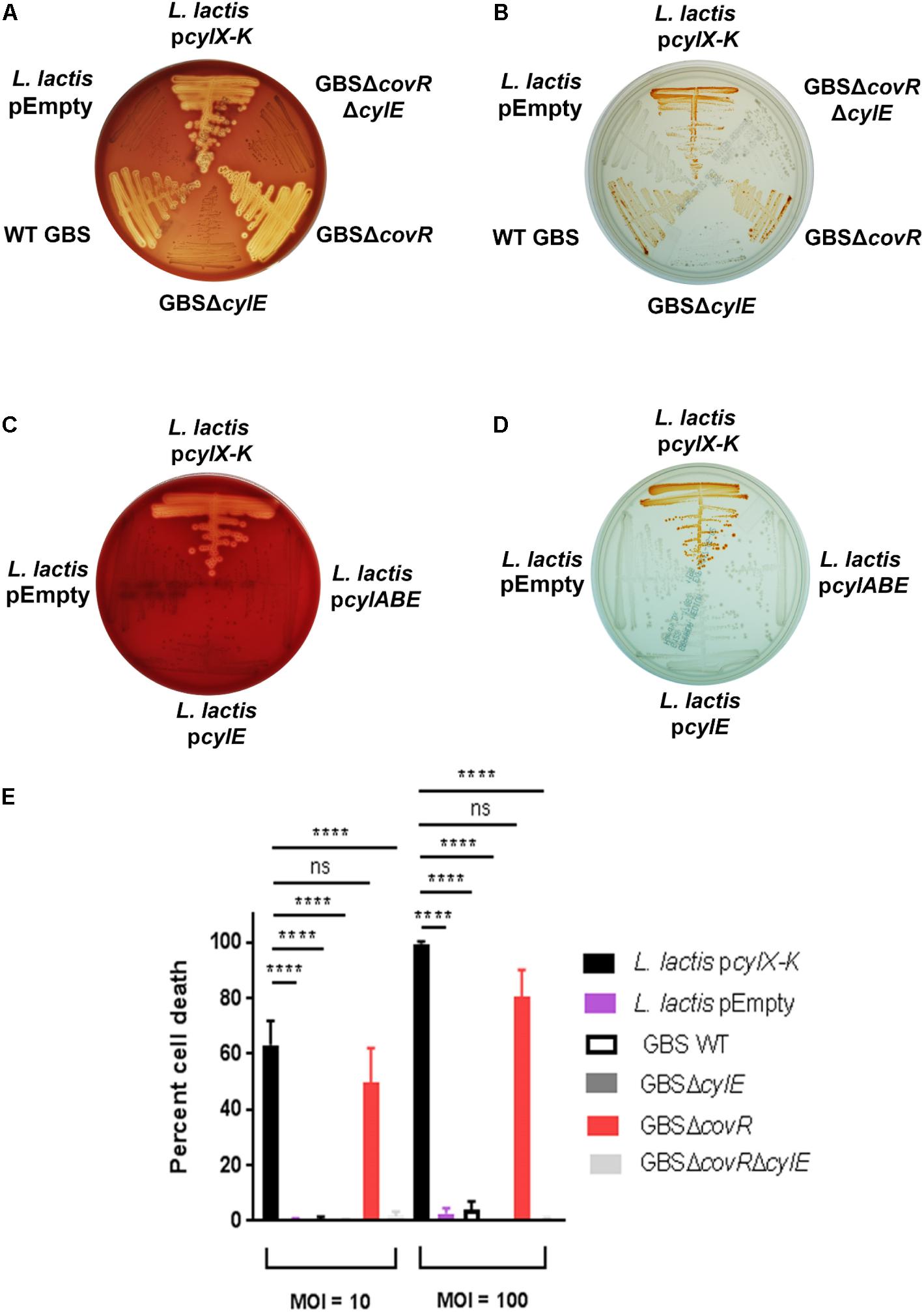
Figure 1. Complementation of L. lactis with pcylX-K, but not empty plasmid vector, pEmpty, conferred hemolysis (A) and pigmentation (B) similar to that observed for hemolytic GBS, including strains that lack the transcriptional repressor of the cyl operon, CovR (GBSΔcovR). Of note, GBS strains lacking CylE (i.e., GBSΔcylE, GBSΔcovRΔcylE) are neither hemolytic nor pigmented. Complementation of L. lactis with cylE alone or cylA, cylB, and cylE did not confer hemolysis (C) or pigmentation (D) to L. lactis. (E) L. lactis pcylX-K, L. lactis pEmpty, GBS WT, GBSΔcylE, GBSΔcovR, or GBSΔcovRΔcylE were incubated with human neutrophils for 4 h at an MOI of 10 and 100, and neutrophil death was measured by LDH release into cell supernatants relative to Triton X-100 and PBS-only controls. Mean and standard error from three independent experiments performed in triplicate are shown. One-way ANOVA with Tukey’s post-test was performed. ∗∗∗∗ indicates p < 0.0001, and ns indicates not significant, or p ≥ 0.05.
We next tested if the cyl operon expression by L. lactis conferred neutrophil cytotoxicity. In GBS, expression of the cyl operon is transcriptionally repressed by the CovR/S two-component system, and the absence of the repressor CovR results in increased expression of the hemolytic pigment, Granadaene (Lamy et al., 2004; Rajagopal et al., 2006; Lembo et al., 2010). GBS strains lacking a functional CovR (ΔcovR) cause significantly greater cytotoxicity in neutrophils compared to wildtype (WT) GBS and isogenic non-pigmented/non-hemolytic mutants due to enhanced production of Granadaene (Lamy et al., 2004; Jiang et al., 2005; Rajagopal et al., 2006; Boldenow et al., 2016). Furthermore, GBS strains lacking CovR have been identified and isolated from women in preterm labor (Whidbey et al., 2013) and from patients with other GBS infectious morbidities (Sendi et al., 2009; Lupo et al., 2014; Almeida et al., 2015, 2017; Whidbey et al., 2015a). We hypothesized that L. lactis pcylX-K, which lacks transcriptional repressors specific to the cyl operon, would induce neutrophil cytotoxicity similar to GBSΔcovR. To test this, we exposed primary human neutrophils to L. lactis pcylX-K at a multiplicity of infection (MOI) of 10 or 100 and measured cytotoxicity by lactate dehydrogenase (LDH) release in cell supernatants as described (Whidbey et al., 2013; Gendrin et al., 2015; Boldenow et al., 2016). WT GBS or hyper-hemolytic/pigmented GBSΔcovR were also included for comparison. As controls, we exposed neutrophils to L. lactis pEmpty, non-pigmented/non-hemolytic GBSΔcylE, and non-pigmented/non-hemolytic GBSΔcovRΔcylE. The results, shown in Figure 1E, demonstrate that the cell death caused by L. lactis pcylX-K was similar to that caused by the hyper-hemolytic strain GBSΔcovR. Importantly, cytotoxicity in cells exposed to L. lactis pcylX-K was greater than those exposed to L. lactis pEmpty. These results demonstrate that expression of the cyl operon in L. lactis is sufficient to confer neutrophil cytotoxicity like hyper-hemolytic/hyper-pigmented GBS.
Pigment Isolated From L. lactis pcylX-K Is Identical to Granadaene From GBS
We next tested if pigment produced by L. lactis pcylX-K was similar in structure to the GBS pigment, Granadaene. Accordingly, we extracted and purified pigment from L. lactis pcylX-K using methods described with GBS (Rosa-Fraile et al., 2006; Whidbey et al., 2013, 2015b; Gendrin et al., 2015; Boldenow et al., 2016) (see section “Materials and Methods”). As a control, we performed the same extraction and purification procedure on L. lactis pEmpty. In mass spectrometry studies, pigment purified from L. lactis pcylX-K demonstrated an M + H ion at m/z 677.3795 (Figure 2A). This exact mass is associated with an ion formula of C39H53N2O8 (expected mass of 677.3796), which is identical to that of Granadaene from GBS, as described (Rosa-Fraile et al., 2006; Whidbey et al., 2013). As expected, no peaks corresponding to the mass of Granadaene were observed in the extract from control L. lactis pEmpty. To gain further insight into the structure of pigment produced by L. lactis pcylX-K, we performed 1H NMR studies (Figure 2B). These analyses revealed a signal corresponding to a polyene structure (7.20–5.5 ppm), as well as signals corresponding to an ornithine (4.32, 2.77, 1.80, 1.59, and 1.64 ppm) and a rhamnose (4.63, 3.52, 3.43, 3.39, 3.16, and 1.09 ppm), which are identical to those described for natural Granadaene (Rosa-Fraile et al., 2006; Whidbey et al., 2013). The ornithine and rhamnose were also confirmed to be present in L. lactis pcylX-K pigment extract by 1D TOCSY. Importantly, none of these structures were present in the NMR analysis of L. lactis pEmpty extract. Of note, little is known about how Granadaene is tethered to the bacterial cell membrane, and while ornithine is present on the terminal end of extracted Granadaene from GBS and L. lactis pcylX-K, is it possible that solvents (namely trifluoracetic acid) may alter the terminal moiety during the extraction procedure. Future studies examining the structure of Granadaene embedded in intact bacterial membranes will help discern the nature of Granadaene’s terminal moiety prior to extraction. Nevertheless, our results demonstrate that the nominal formula and the structure of pigment produced by L. lactis pcylX-K are indistinguishable from those previously assigned to Granadaene (Figure 2B; Rosa-Fraile et al., 2006; Whidbey et al., 2013).
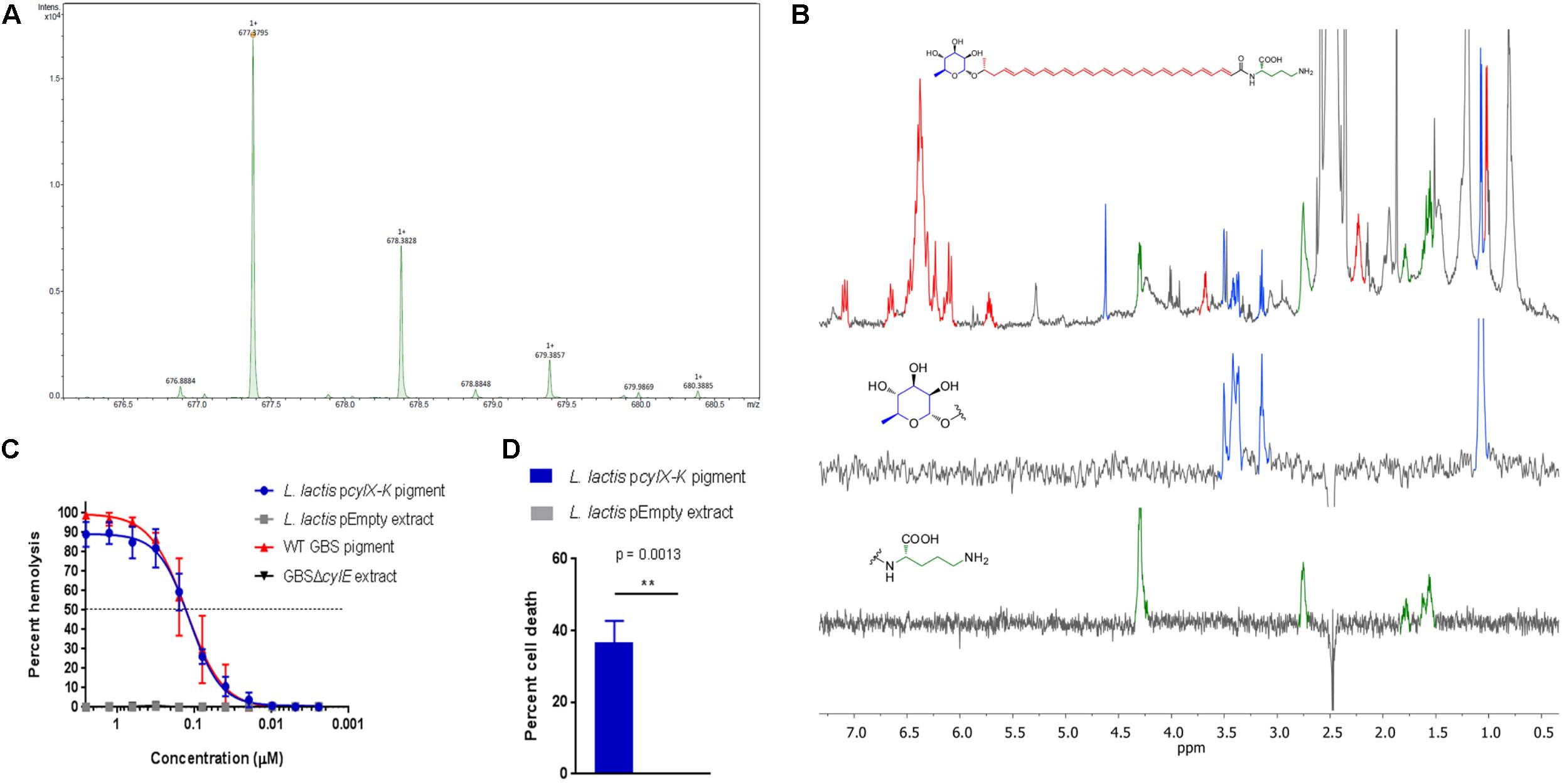
Figure 2. Pigment extracted and purified from L. lactis pcylX-K is identical to Granadaene extracted from WT GBS. (A) High resolution mass spectrometry was performed on pigment extracted from L. lactis pcylX-K and demonstrated an M + H ion at m/z 677.3795, which is associated with an ion formula of C39H53N2O8 (expected mass of 677.3796). (B) 1H NMR analyses on L. lactis pcylX-K pigment revealed a signal corresponding to a polyene structure (7.20–5.5 ppm), as well as signals corresponding to an ornithine (4.32, 2.77, 1.80, 1.59, and 1.64 ppm) and a rhamnose (4.63, 3.52, 3.43, 3.39, 3.16, and 1.09 ppm). (C) HPLC-purified pigment from L. lactis pcylX-K or WT GBS in DMSO + trifluoroacetic acid + starch (DTS) was added to human erythrocytes in twofold serial dilutions starting from 2.5 to 0.0024 μM for 1 h. As controls, equivalent amounts of extracts from L. lactis pEmpty and from non-pigmented/hemolytic GBSΔcylE were tested. Mean and standard error from three independent experiments performed in triplicate are shown. The EC50 for L. lactis pcylX-K pigment (0.128 μM, 95% CI: 0.108, 0.153) is no different than that of WT GBS pigment/Granadaene (0.127 μM, 95% CI: 0.100, 0.162). (D) Primary human neutrophils were incubated with 0.5 μM purified pigment from L. lactis pcylX-K for 4 h at 37°C. As a control, neutrophils were treated with an equivalent amount of L. lactis pEmpty extract. Neutrophil death was measured by LDH release into cell supernatants relative to Triton X-100 and PBS-only controls. Mean and standard error from three independent experiments performed in triplicate are shown. An unpaired t-test was performed, and p = 0.0013. ∗∗ indicates that p < 0.01.
To confirm that pigment purified from L. lactis pcylX-K possessed hemolytic and cytolytic activity analogous to Granadaene, we performed a hemolytic titer assay using the L. lactis pcylX-K pigment extract (Nizet et al., 1996; Whidbey et al., 2013). The results shown in Figure 2C indicate that purified pigment from L. lactis pcylX-K possessed hemolytic activity similar to that of Granadaene (Whidbey et al., 2013). Moreover, the EC50 (the effective concentration at which 50% of human red blood cells are lysed) of L. lactis pcylX-K pigment (0.128 μM, 95% CI: 0.108, 0.153) was no different than that of Granadaene (0.127 μM, 95% CI: 0.100, 0.162).
Finally, we sought to determine if L. lactis pcylX-K pigment is cytolytic to host cells like Granadaene. Purified Granadaene was previously reported to be cytolytic to host cells such as neutrophils (Boldenow et al., 2016). Human neutrophils isolated from fresh adult blood were treated with L. lactis pcylX-K pigment (0.5 μM) or equivalent amount of control extract from L. lactis pEmpty for 4 h, as described (Boldenow et al., 2016), and cytotoxicity was measured by LDH release in cell supernatants. As observed previously with Granadaene, L. lactis pcylX-K pigment caused significantly greater cell death than non-hemolytic control extracts (Figure 2D; Boldenow et al., 2016). Together, these data indicate that purified pigment from L. lactis pcylX-K possesses hemolytic and cytolytic activity that are no different than Granadaene. Furthermore, our findings confirm that the cyl operon encode proteins sufficient for production of this ornithine rhamnolipid in a heterologous Gram-positive bacterium.
Phyletic Analysis Suggests the cyl Operon Evolved Prior to the Diversification of Gram-Positive Bacteria
Our heterologous expression experiments suggest a possible advantage for lateral transfer of cyl genes between various bacteria. To understand the evolutionary origins of the ornithine rhamnolipid pigment, we conducted a systematic survey of the phyletic patterns and contextual associations of the cyl genes. We found that cyl genes are present in diverse Gram-positive phyla such as firmicutes and actinobacteria as well as the chloroflexi Thermosporothrix. A phylogenetic tree using the cylE acyl transferase gene, which is universally present in cyl operons, showed that the actinobacterial and firmicute proteins group within their respective bacterial clades, whereas the Thermosporothrix cylE groups outside the two principal clades (Figure 3A and Supplementary Figure S3). The Thermosporothrix operons seem to contain elements of both the actinobacterial and firmicute types of operon, supporting that it has retained features of an ancestral version (Figure 3A and Supplementary Figure S3). The presence of cyl operons in diverse firmicutes (spore formers and non-spore formers) and actinobacteria, and a largely vertical inheritance of the firmicute and actinobacterial versions, indicates that the pigment biosynthesis genes evolved prior to the diversification of Gram-positive bacteria.
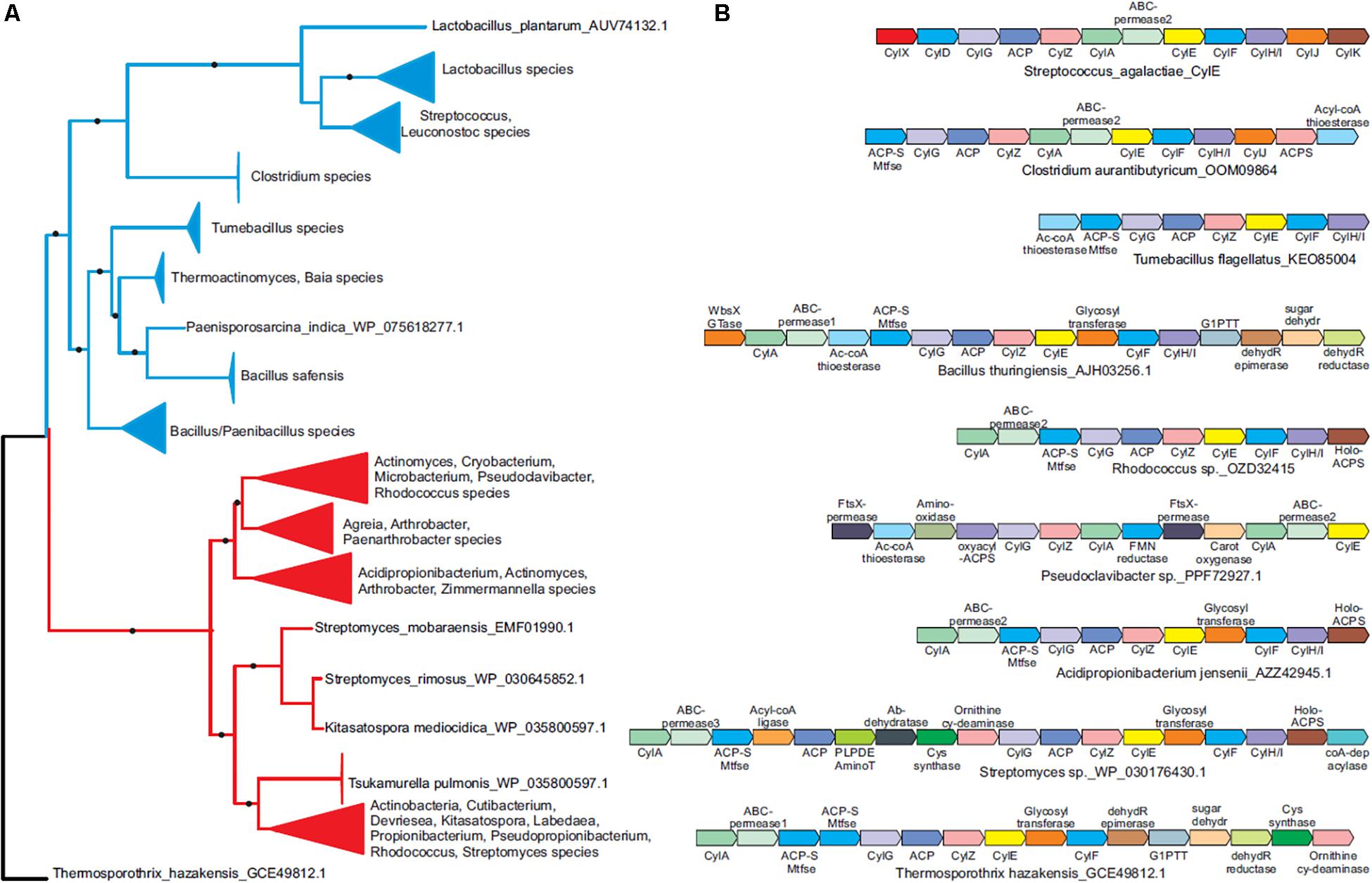
Figure 3. Phyletic analysis suggests the cyl operon evolved prior to the diversification of Gram-positive bacteria. (A) A phylogenetic tree of the CylE protein is shown. Clades with strong bootstrap support were collapsed into filled triangles for convenience (see Supplementary Figure S3 for full tree). Firmicute and actinobacterial branches are colored blue and red, respectively. (B) Representative cyl operons derived from the phylogenetic tree in (A) are shown. Genes are shown as boxed arrows, with the arrow head pointing to the 3′ gene. Operons are labeled using the accession number of the cylE gene in the operon. Proteins are denoted using their species names followed by their Genbank accession number or gene names.
We identified five functional groups of genes in homologous cyl operons (Table 1): (1) those involved in lipid biosynthesis, (2) ornithine biosynthesis, (3) acyl-lipid transfer to the ornithine, (4) sugar biosynthesis and transfer, and (5) pigment export. Of these, our data indicate a vertically inherited orthologous core of seven genes comprising of cylA, cylG, acpC, cylZ, cylE, cylF, and cylH. This likely comprises the most ancient core of the operon. Further, non-orthologous but biochemically functionally equivalent versions of four genes are observed, which encode the following: (1) CylK (Streptococcus, Lactococcus, Leuconostoc) and ACP synthase (actinobacteria, Clostridium and Paenisporosarcina species); (2) CylJ (Streptococci, certain Lactococci and Clostridium species) and one or two distinct glycosyltransferases (various firmicutes, actinobacteria and chloroflexi) involved in sugar conjugation; (3) CylD (Streptococci, Leuconostoc and Lactococci) and ACP-S-malonyltransferase (in various firmicutes, chloroflexi and actinobacteria); and 4) at least three distinct versions of ABC-transporter permease subunits have been alternatively recruited for pigment transport (Figure 3B and Table 1). Of note, the non-orthologous displacements are supported by change in operon position of the genes (Supplementary Figure S4). Taken together, this suggests a core of approximately eleven genes in an ancient operon prior to the divergence of actinobacteria and firmicutes, which encoded proteins belonging to one of the five functional groups involved in pigment synthesis (Table 1).
We observe that some cyl genes are exclusive to specific bacterial lineages, suggesting that they were acquired post-divergence. For instance, genes encoding CylJ and CylD are likely to have displaced those encoding the ancestral glycosyltransferase and ACP-S-malonyltransferase, respectively, in certain Streptococci (Figure 3B). These also include rhamnose biosynthesis genes, which are only seen in firmicutes and chloroflexi, as well as ornithine cyclodeaminase (OCD), PLP-dependent aminotransferases, acyl coA ligase and various redox enzymes, which are only found in actinobacteria and partly in chloroflexi. We predict that these genes evolved to increase the availability of the building blocks for pigment biosynthesis or add diversity to the functional groups on the pigment. For example, the presence of two glycosyltransferases in various Bacillus species in the cyl operons (Figure 3B and Supplementary Figure S3) could imply that these species possess a more complex sugar modification than rhamnose, and this warrants further study. In short, while there is little evidence to suggest extensive lateral transfer of cyl genes, our data indicate that individual genes were likely lost or displaced by non-orthologous genes across various species.
The Evolution of CylE and CylF Conferred Critical Functions in Granadaene Biosynthesis to the cyl Operon
One of the previously unresolved questions was the role of the lipoate-dependent aminomethyltransferase (CylF), as this is a universally conserved gene encoded by cyl operons. We predict that this is involved in ornithine biosynthesis. We infer this based on the presence of OCD-encoding genes in some actinobacterial cyl operons (Figure 3B and Table 1), which suggests in these organisms and, perhaps more generally in all organisms with the pigment, ornithine is synthesized by combining proline with ammonia. This provides a role for the aminomethylase CylF as this enzyme splits amino acids such as glycine to give rise to ammonia (Kikuchi et al., 2008). Given the need for ammonia in the ornithine synthesis by the OCD enzyme, we predict that CylF generates ammonia in the local milieu of the pigment biosynthesis complex, which is then used by OCD to make ornithine. Of the five basic components of the cyl operon, the fatty acid biosynthesis genes often appear together in other contexts. However, the accretion of the acetyltransferase CylE and CylF in an ancient bacterium were perhaps the key events in the fixation of the cyl genes for pigment biosynthesis.
Given the phyletic patterns, we hypothesize that the ancient cyl operon evolved in free-living bacteria, where we conceive possible biological roles that might not be mutually exclusive. First, the ornithine rhamnolipid might be involved in photoprotection and/or membrane stabilization, as with many microbial pigments (Taylor, 1984; Howardwilliams et al., 1989; Rajagopal et al., 1997; Slouf et al., 2012; Madden et al., 2019). Second, it might have evolved as a general defense against competing organisms in which the pigment disrupts their cell membrane. The latter is supported by the hemolytic and cytolytic activity of the Granadaene pigment in GBS (Whidbey et al., 2013, 2015b; Boldenow et al., 2016) and cyl-expressing L. lactis (Figures 1, 2). The presence of an identical or similar hemolytic pigment in a range of Gram-positive bacteria with cyl genes (Vanberg et al., 2007; Deptula et al., 2019), as well as our heterologous expression experiments, suggest that Granadaene can potentially function across a physiologically diverse group of organisms if they likely possess a Gram-positive-type cell wall structure. In fact, a recent study correlated pigmentation with hemolysis and cyl gene expression in Acidipropionibacterium species that have roles in cheese production and food preservation (Deptula et al., 2019). Accordingly, the hemolytic and cytotoxic properties of other pigmented, cyl operon-containing microbes found in soil environments or utilized for industrial application warrant further examination.
These findings broaden our understanding of the biosynthesis and evolutionary history of Granadaene in GBS, along with that of other homologous microbial lipid pigments. Although our phyletic analyses do not establish horizontal gene transfer of the entire cyl operon, our heterologous gene expression studies indicate that this remains a formal possibility, particularly in Gram-positive bacteria. Because GBS cohabitates the human lower genital and gastrointestinal tract with other commensal bacteria, the possibility of lateral transfer of the cyl operon to otherwise avirulent bacteria in the human niche should be explored further.
Materials and Methods
Ethics Statement
Written informed consent for donation of human blood was obtained from patients, per the Principles in the WMA Declaration of Helsinki and Department of Health and Human Services Belmont Report. The study was approved by the Seattle Children’s Research Institute Institutional Review Board (protocol #11117). Children under the age of 18 were not recruited for donation of human blood.
Bacterial Strains
The GBS strain A909 was used in all studies. A909 is a clinical isolate obtained from an infected human neonate and is classified as serotype Ia (Lancefield et al., 1975). The ΔcylE, ΔcovR, and ΔcovRΔcylE mutants were derived from A909 as described (Rajagopal et al., 2006; Lembo et al., 2010). Cultures of GBS were grown in tryptic soy broth (TSB; Difco Laboratories) at 37°C in 5% CO2, and cultures of Escherichia coli MC1016 were grown in lysogeny broth (LB; Difco Laboratories) at 37°C. Cultures of Lactococcus lactis were grown in TSB at 30°C in 5% CO2. Culture growth for all bacterial strains was measured at 600 nm, and bacterial strains were washed twice in PBS before being used in experiments. Photographs of bacterial strains on blood agar (Remel) or Granada medium (Hardy Diagnostics) were captured with an SLR camera (EOS Rebel XSi 12.2MP; Cannon) with an 18–55 mm zoom lens and processed using Photoshop CC (Adobe).
Heterologous Expression of the cyl Operon in Lactococcus lactis
The cyl operon was amplified using high fidelity PCR and primer pairs cyl1, cyl2, and cyl3 (IDT, see Supplementary Table S2) to obtain three overlapping DNA fragments. The PCR fragments were ligated to a PCR fragment representing the vector pDC123 (Chaffin and Rubens, 1998), referred to in the main text and hereafter as pEmpty) using Gibson Assembly. The recombinant plasmid, which contained a chloramphenicol resistance gene, was electroporated into Escherichia coli MC1061 and then isolated by midiprep (Qiagen). After the presence of the cyl operon within the recombinant plasmid was confirmed by PCR amplification, the recombinant plasmid was transformed into electrocompetent Lactococcus lactis. Upon the addition of the recombinant or empty plasmid, L. lactis was grown in the presence of 5 μg/mL chloramphenicol (Sigma-Aldrich).
Isolation and Purification of Pigment From GBS and L. lactis
Pigment was isolated from wildtype GBS and L. lactis pcylX-K as described, with minor modifications (Rosa-Fraile et al., 2006; Whidbey et al., 2013, 2015b; Gendrin et al., 2015; Boldenow et al., 2016). Wildtype GBS A909 or L. lactis pcylX-K were grown in 500 mL Granada medium (De La Rosa et al., 1992), and the bacterial pellet was washed twice with HPLC-grade water and twice with DMSO. Then, the pellet was resuspended in DMSO + 0.1% trifluoroacetic acid (TFA; Sigma-Aldrich) to extract the pigment. Extractions with DMSO + 0.1% TFA continued until no pigmentation was observed in the bacterial pellet. The pigment extract was pooled and precipitated overnight with NH4OH (Scientific Products). The precipitated pellet was washed twice with HPLC-grade water, and twice with DMSO. Pigment was then extracted using DMSO + 0.1% TFA as above, pooled, and then column-purified using high pressure liquid chromatography (HPLC, Shimadzu 10A system) with a Vydac 214TP C4 column (solvent: DMSO + 0.1% TFA, flow rate: 1 mL/min; detection: 489 nm). Fractions were collected between 10.5 min and 17.5 min of each run (when detection level was >100,000 μV, see Supplementary Figure S5). Purified fractions were pooled, precipitated with NH4OH (Scientific Products), washed three times with HPLC-grade water and then twice with DMSO, and lyophilized to dryness. Lyophilized pigment toxin was stored at −80°C, and working pigment toxin solutions were dissolved in DMSO + 0.1% TFA + 20% starch (DTS), as described (Whidbey et al., 2013). The purification and isolation procedure was also performed on corresponding GBS A909ΔcylE (non-pigmented/non-hemolytic isogenic mutant of wildtype A909) and L. lactis pEmpty (transformed with empty vector, non-pigmented/non-hemolytic) as described above. Extract from A909ΔcylE and L. lactis pEmpty were used as controls for pigment in all experiments, along with the DTS solvent. For NMR analysis, purified pigment from L. lactis pcylX-K or control extract from L. lactis pEmpty were resuspended in DMSO + 0.1% TFA. 1H, 13C, 1H-COSY NMR experiments were performed at 298K on a Varian 600 MHz NMR Spectrometer. Residual DMSO-d5 was used to calibrate chemical shifts. High resolution mass spectra (HRMS) of samples dilute in DMSO + 0.1% TFA were recorded on a mass spectrometer using via ESI.
Hemolytic Assays
Lyophilized pigment toxin or control extracts were dissolved in DTS to a final concentration of 200 μM and were incubated overnight at room temperature and protected from light before use. To perform the hemolytic titer, twofold serial dilutions of purified pigment in DTS from A909 WT or L. lactis pcylX-K were performed in PBS in a final volume of 100 μL. Twofold serial dilutions of control extracts (from GBSΔcylE or L. lactis pEmpty) in DTS were also performed in PBS as controls. The samples were incubated with 100 μL of EDTA-treated human red blood cells (0.5% in PBS) in 96-well plates for 1 h at 37°C. Then, the 96-well plate was centrifuged for 4 min at 300 × g to pellet the unlysed red blood cells, and supernatants were transferred to a replica 96-well plate. Hemoglobin release was measured by recording the absorbance at 420 nm of the supernatants (Spectramax i3x Plate Reader, Molecular Devices), and percent hemolysis was determined relative to red blood cells treated with positive control Triton X-100 (0.1%, Sigma-Aldrich) and negative control PBS only.
Isolation of Neutrophils From Adult Human Blood
As described (Boldenow et al., 2016), 5–15 mL of human adult blood was collected from independent healthy human adults into EDTA tubes (BD Bioscience). Immediately following collection, neutrophils were isolated using a MACSxpress neutrophil isolation kit, per the manufacturer’s instructions (Miltenyi Biotec). Cells were then pelleted, and any residual red blood cells (RBC) were removed by re-suspending the cell pellet in RBC lysis solution (0.15 nM NH4Cl, 1 mM NaHCO3) for 15 min at room temperature. Following RBC lysis, cells were washed with Roswell Park Memorial Institute 1640 tissue culture medium containing L-glutamine (Corning; hereafter referred to as RPMI-G). Neutrophil purity in the prepared cell suspension was assessed by examining the proportion of cells positive for the neutrophil cell markers CD15 (PerCP/Cy5.5, clone HI98, BD Biosciences) and CD16 (FITC, clone 3G8, BD Biosciences) by flow cytometry. Briefly, approximately 1 × 106 cells from the neutrophil purification preparation or 1 × 106 cells from whole blood (following two RBC lysis steps, as described above) were incubated with Fc receptor block (1:200, BD Biosciences) for 15 min at room temperature. Then, immunofluorescent antibodies were added to the cells at concentrations recommended by the manufacturer (1:10, CD15-PerCP/Cy5.5; 1:200, CD16-FITC), and cells incubated for 30 min at room temperature. Stained cells were washed twice in FACS (fluorescence-activated cell sorting) buffer (1 mM EDTA, 25 mM HEPES, 1% BSA (w/v) in PBS) and were analyzed immediately on an LSR II flow cytometer (BD Biosciences). Single-stained flurochrome-reactive AbC beads (Thermo Fisher) and unstained cells were used for compensation. Data were analyzed using FlowJo v. 10.1 (FlowJo, LLC).
Testing Bacterial Strains and Pigment Extracts for Neutrophil Cytotoxicity
Neutrophils were isolated as described above and were seeded in a U-bottom 96-well plate at approximately 4 × 105 cells/mL in 90 μL RPMI-G. L. lactis pcylX-K and L. lactis pEmpty were grown to mid-exponential growth phase (OD600 = 0.3), washed twice in sterile PBS, and added to neutrophils at a multiplicity of infection (MOI) of 10 and 100. MOIs were confirmed by dilution plating. Additionally, pigment in DTS from L. lactis pcylX-K or WT GBS was added to neutrophils at a final concentration of 0.5 μM. As positive and negative controls for all assays, neutrophils were incubated in 0.1% Triton X-100 (Sigma Aldrich) or sterile PBS, respectively. After incubating for 4 h at 37°C, cells were analyzed for cytotoxicity by the presence of cytoplasmic lactate dehydrogenase (LDH) in cell supernatants using the colorimetric LDH kit (Clontech), per the manufacturer’s instructions. Percent cytotoxicity was calculated by normalizing to PBS-treated cells (0% cell death) and Triton X-100-treated cells (100% cell death), as described (Whidbey et al., 2013, 2015b; Boldenow et al., 2016).
Phyletic Analysis
Phyletic pattern searches were done using the PSI-BLAST program (Altschul et al., 1997) against a locally constructed compressed non-redundant database (NR50) of the National Center for Biotechnology Information (NCBI). Multiple sequence alignments were built using the Kalign program (Lassmann et al., 2009). Contextual information from prokaryotic gene neighborhoods was retrieved by a Perl custom script that extracts the upstream and downstream genes of the query gene. Phylogenetic analysis was conducted using an approximately maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (Price et al., 2010)1. Network analysis was performed using the R language with the igraph package (Csardi and Nepusz, 2006).
Statistical Analysis
A value of p < 0.05 was considered significant. Non-linear regression analysis was used to interpolate EC50 concentrations. In LDH release assays, an unpaired t-test or one-way ANOVA with Tukey’s post-test was employed as appropriate to analyze differences between treatment groups, unless otherwise noted. GraphPad Prism (version 7.03) was used to compute all statistical tests.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by the Seattle Children’s Research Institute Institutional Review Board (protocol #11117). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BA, CW, LI, LA, JC, and LR designed the experiments and analyzed the results. BA, CW, LI, PH-F, PQ, and AH performed the experiments. BA, LI, LA, HJ, and LR wrote the manuscript.
Funding
This work was supported by funding from the National Institutes of Health grants R01AI112619, R01AI133976, R01AI100989, R21AI125907, and T32AI007509 to LR. The NIH training grant T32AI07509 (PI: Lee Ann Campbell) supported CW. LI and LA were supported by intramural funds from NLM, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Connie Hughes for administrative support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03123/full#supplementary-material
Footnotes
References
Almeida, A., Villain, A., Joubrel, C., Touak, G., Sauvage, E., Rosinski-Chupin, I., et al. (2015). Whole-Genome comparison uncovers genomic mutations between group B streptococci sampled from infected newborns and their mothers. J. Bacteriol. 197, 3354–3366. doi: 10.1128/JB.00429-15
Almeida, A., Rosinski-Chupin, I., Plainvert, C., Douarre, P. E., Borrego, M. J., Poyart, C., et al. (2017). Parallel evolution of group B Streptococcus hypervirulent clonal complex 17 unveils new pathoadaptive mutations. mSystems 2:e00074-17.. doi: 10.1128/mSystems.00074-17
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped Blast and Psi-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ayers, S. H., and Philip, R. (1922). Differentiation of hemolytic streptococci from human and bovine sources by the hydrolysis of sodium hippurate. J. Infect. Dis.30, 388–399. doi: 10.1093/infdis/30.4.388
Boldenow, E., Gendrin, C., Ngo, L., Bierle, C., Vornhagen, J., Coleman, M., et al. (2016). Group B Streptococcus circumvents neutrophils and neutrophil extracellular traps during amniotic cavity invasion and preterm labor. Sci. Immunol. 1:eaah4576. doi: 10.1126/sciimmunol.aah4576
Brown, J. H. (1920). The cultural differentiation of beta hemolytic streptococci of human and bovine origin. J. Exp. Med. 31, 35–47. doi: 10.1084/jem.31.1.35
Cavanagh, D., Fitzgerald, G. F., and Mcauliffe, O. (2015). From field to fermentation: the origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol. 47, 45–61. doi: 10.1016/j.fm.2014.11.001
Chaffin, D. O., and Rubens, C. E. (1998). Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219, 91–99. doi: 10.1016/s0378-1119(98)00396-5
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Complex Syst. 1695, 1–9.
De La Rosa, M., Perez, M., Carazo, C., Pareja, L., Peis, J. I., and Hernandez, F. (1992). New granada medium for detection and identification of group B streptococci. J. Clin. Microbiol. 30, 1019–1021.
Deptula, P., Loivamaa, I., Smolander, O. P., Laine, P., Roberts, R. J., Piironen, V., et al. (2019). Red-brown pigmentation of Acidipropionibacterium jensenii Is tied to haemolytic activity and cyl-Like gene cluster. Microorganisms 7:E512. doi: 10.3390/microorganisms7110512
Doran, K. S., Chang, J. C., Benoit, V. M., Eckmann, L., and Nizet, V. (2002). Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis/ 185, 196–203. doi: 10.1086/338475
Doran, K. S., Liu, G. Y., and Nizet, V. (2003). Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J. Clin. Invest. 112, 736–744. doi: 10.1172/jci17335
Forquin, M. P., Tazi, A., Rosa-Fraile, M., Poyart, C., Trieu-Cuot, P., and Dramsi, S. (2007). The putative glycosyltransferase-encoding gene cylJ and the group B Streptococcus (Gbs)-specific gene cylK modulate hemolysin production and virulence of Gbs. Infect. Immun. 75, 2063–2066. doi: 10.1128/iai.01565-06
Gendrin, C., Vornhagen, J., Ngo, L., Whidbey, C., Boldenow, E., Santana-Ufret, V., et al. (2015). Mast cell degranulation by a hemolytic lipid toxin decreases Gbs colonization and infection. Sci. Adv. 1:e1400225. doi: 10.1126/sciadv.1400225
Gottschalk, B., Broker, G., Kuhn, M., Aymanns, S., Gleich-Theurer, U., and Spellerberg, B. (2006). Transport of multidrug resistance substrates by the Streptococcus agalactiae hemolysin transporter. J. Bacteriol. 188, 5984–5992. doi: 10.1128/jb.00768-05
Hamon, M. A., Ribet, D., Stavru, F., and Cossart, P. (2012). Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20, 360–368. doi: 10.1016/j.tim.2012.04.006
Howardwilliams, C., Pridmore, R., Downes, M. T., and Vincent, W. F. (1989). Microbial biomass, photosynthesis and chlorophyll-a related pigments in the ponds of the mcmurdo ice shelf, antarctica. Antarct.Sci. 1, 125–131. doi: 10.1017/s0954102089000192
Jiang, S. M., Cieslewicz, M. J., Kasper, D. L., and Wessels, M. R. (2005). Regulation of virulence by a two-component system in group B Streptococcus. J. Bacteriol. 187, 1105–1113. doi: 10.1128/jb.187.3.1105-1113.2005
Kikuchi, G., Motokawa, Y., Yoshida, T., and Hiraga, K. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84, 246–263. doi: 10.2183/pjab.84.246
Lamy, M. C., Zouine, M., Fert, J., Vergassola, M., Couve, E., Pellegrini, E., et al. (2004). CovS/CovR of group B Streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 54, 1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x
Lancefield, R. C., Mccarty, M., and Everly, W. N. (1975). Multiple mouse-protective antibodies directed against group B streptococci. Special reference to antibodies effective against protein antigens. J. Exp. Med. 142, 165–179. doi: 10.1084/jem.142.1.165
Lassmann, T., Frings, O., and Sonnhammer, E. L. (2009). Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865. doi: 10.1093/nar/gkn1006
Lembo, A., Gurney, M. A., Burnside, K., Banerjee, A., De Los Reyes, M., Connelly, J. E., et al. (2010). Regulation of CovR expression in Group B Streptococcus impacts blood-brain barrier penetration. Mol. Microbiol. 77, 431–443. doi: 10.1111/j.1365-2958.2010.07215.x
Lupo, A., Ruppen, C., Hemphill, A., Spellerberg, B., and Sendi, P. (2014). Phenotypic and molecular characterization of hyperpigmented group B Streptococci. Int. J. Med. Microbiol. 304, 717–724. doi: 10.1016/j.ijmm.2014.05.003
Madden, K. S., Jokhoo, H. R. E., Conradi, F. D., Knowles, J. P., Mullineaux, C. W., and Whiting, A. (2019). Using Nature’s polyenes as templates: studies of synthetic xanthomonadin analogues and realising their potential as antioxidants. Org. Biomol. Chem. 17, 3752–3759. doi: 10.1039/c9ob00275h
Nizet, V., Gibson, R. L., Chi, E. Y., Framson, P. E., Hulse, M., and Rubens, C. E. (1996). Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect. Immun. 64, 3818–3826.
Patras, K. A., Wang, N. Y., Fletcher, E. M., Cavaco, C. K., Jimenez, A., Garg, M., et al. (2013). Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol. 15, 1154–1167. doi: 10.1111/cmi.12105
Platt, M. W. (1995). In vivo hemolytic activity of group B Streptococcus is dependent on erythrocyte-bacteria contact and independent of a carrier molecule. Curr. Microbiol. 31, 5–9. doi: 10.1007/bf00294625
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5:e9490. doi: 10.1371/journal.pone.0009490
Pritzlaff, C. A., Chang, J. C., Kuo, S. P., Tamura, G. S., Rubens, C. E., and Nizet, V. (2001). Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol. Microbiol. 39, 236–247.
Rajagopal, L., Sundari, C. S., Balasubramanian, D., and Sonti, R. V. (1997). The bacterial pigment xanthomonadin offers protection against photodamage. Febs Lett. 415, 125–128. doi: 10.1016/s0014-5793(97)01109-5
Rajagopal, L., Vo, A., Silvestroni, A., and Rubens, C. E. (2006). Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62, 941–957. doi: 10.1111/j.1365-2958.2006.05431.x
Rosa-Fraile, M., Rodriguez-Granger, J., Haidour-Benamin, A., Cuerva, J. M., and Sampedro, A. (2006). Granadaene: proposed structure of the group B Streptococcus polyenic pigment. Appl. Environ Microbiol. 72, 6367–6370. doi: 10.1128/aem.00756-06
Russell, N. J., Seale, A. C., O’driscoll, M., O’sullivan, C., Bianchi-Jassir, F., Gonzalez-Guarin, J., et al. (2017a). Maternal colonization with group B Streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin. Infect. Dis. 65, S100–S111.
Russell, N. J., Seale, A. C., O’sullivan, C., Le Doare, K., Heath, P. T., Lawn, J. E., et al. (2017b). Risk of early-onset neonatal Group B Streptococcal Disease with maternal colonization worldwide: systematic review and meta-analyses. Clin. Infect. Dis. 65, S152–S159. doi: 10.1093/cid/cix655
Seale, A. C., Bianchi-Jassir, F., Russell, N. J., Kohli-Lynch, M., Tann, C. J., Hall, J., et al. (2017a). Estimates of the burden of group B Streptococcal Disease worldwide for pregnant women. Stillbirths, and Children. Clin. Infect. Dis. 65, S200–S219. doi: 10.1093/cid/cix664
Seale, A. C., Blencowe, H., Bianchi-Jassir, F., Embleton, N., Bassat, Q., Ordi, J., et al. (2017b). Stillbirth with group B Streptococcus Disease worldwide: systematic review and meta-analyses. Clin. Infect. Dis. 65, S125–S132.
Seilie, E. S., and Bubeck Wardenburg, J. (2017). Staphylococcus aureus pore-forming toxins: The interface of pathogen and host complexity. Semin. Cell Dev. Biol. 72, 101–116. doi: 10.1016/j.semcdb.2017.04.003
Sendi, P., Johansson, L., Dahesh, S., Van-Sorge, N. M., Darenberg, J., Norgren, M., et al. (2009). Bacterial phenotype variants in group B streptococcal toxic shock syndrome. Emerg Infect. Dis. 15, 223–232. doi: 10.3201/eid1502.080990
Slouf, V., Chabera, P., Olsen, J. D., Martin, E. C., Qian, P., Hunter, C. N., et al. (2012). Photoprotection in a purple phototrophic bacterium mediated by oxygen-dependent alteration of carotenoid excited-state properties. Proc. Natl. Acad. Sci US.A. 109, 8570–8575. doi: 10.1073/pnas.1201413109
Spellerberg, B., Martin, S., Brandt, C., and Lutticken, R. (2000). The cyl genes of Streptococcus agalactiae are involved in the production of pigment. Fems. Microbiol. Lett. 188, 125–128. doi: 10.1016/s0378-1097(00)00224-x
Spellerberg, B., Pohl, B., Haase, G., Martin, S., Weber-Heynemann, J., and Lutticken, R. (1999). Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by Iss1 transposition. J. Bacteriol. 181, 3212–3219.
Vanberg, C., Lutnaes, B. F., Langsrud, T., Nes, I. F., and Holo, H. (2007). Propionibacterium jensenii produces the polyene pigment granadaene and has hemolytic properties similar to those of Streptococcus agalactiae. Appl.Environ.Microbiol. 73, 5501–5506. doi: 10.1128/aem.00545-07
Whidbey, C., Burnside, K., Martinez, R. M., Gendrin, C., Vornhagen, J., Frando, A., et al. (2015a). A Hyperhemolytic/Hyperpigmented Group B Streptococcus strain with a CovR mutation isolated from an adolescent patient with sore throat. Clin. Res. Infect. Dis. 2:1018.
Whidbey, C., Vornhagen, J., Gendrin, C., Boldenow, E., Samson, J. M., Doering, K., et al. (2015b). A streptococcal lipid toxin induces membrane permeabilization and pyroptosis leading to fetal injury. Embo. Mol. Med. 7, 488–505. doi: 10.15252/emmm.201404883
Keywords: Group B Streptococcus, bacterial toxin, microbial evolution, virulence factor, Gram-positive bacteria
Citation: Armistead B, Whidbey C, Iyer LM, Herrero-Foncubierta P, Quach P, Haidour A, Aravind L, Cuerva JM, Jaspan HB and Rajagopal L (2020) The cyl Genes Reveal the Biosynthetic and Evolutionary Origins of the Group B Streptococcus Hemolytic Lipid, Granadaene. Front. Microbiol. 10:3123. doi: 10.3389/fmicb.2019.03123
Received: 15 November 2019; Accepted: 24 December 2019;
Published: 21 January 2020.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Shaynoor Dramsi, Institut Pasteur, FranceManuel Rosa-Fraile, Hospital Virgen Nieves, Spain
Copyright © 2020 Armistead, Whidbey, Iyer, Herrero-Foncubierta, Quach, Haidour, Aravind, Cuerva, Jaspan and Rajagopal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lakshmi Rajagopal, lakshmi.rajagopal@seattlechildrens.org
†Present address: Christopher Whidbey, Seattle University, Seattle, WA, United States
 Blair Armistead
Blair Armistead Christopher Whidbey
Christopher Whidbey Lakshminarayan M. Iyer
Lakshminarayan M. Iyer Pilar Herrero-Foncubierta4
Pilar Herrero-Foncubierta4 Phoenicia Quach
Phoenicia Quach Ali Haidour
Ali Haidour L. Aravind
L. Aravind Juan Manuel Cuerva
Juan Manuel Cuerva Heather B. Jaspan
Heather B. Jaspan Lakshmi Rajagopal
Lakshmi Rajagopal