- 1Department of Genetics, Genomics, and Informatics, University of Tennessee Health Science Center, Memphis, TN, United States
- 2Centers for Disease Control and Prevention, National Institute for Occupational Safety and Health, Morgantown, WV, United States
- 3Department of Preventive Medicine, University of Tennessee Health Science Center, Memphis, TN, United States
Introduction: Gulf War Illness is a chronic multisymptomatic disorder that affects as many as 25-35% of the military personnel who were sent to the Persian Gulf war in 1991. The illness has many debilitating symptoms, including cognitive problems, gastrointestinal symptoms, and musculoskeletal pain. Those so afflicted have been sick for more than 30 years and, therefore, it has become imperative to understand the etiology of Gulf War Illness and then produce treatments to ease the symptoms. We hypothesized that the length of the disease was reflected in epigenetic modification of possibly several genes related to the symptoms.
Methods: We subjected male and female mice from 11 BXD strains to combined corticosterone and the sarin surrogate, diisopropylfluorophosphate, to emulate the physiological stress of war and the potential exposures to organophosphate pesticides and nerve agent in theater. Three hundred days after treatment, we used Methyl-CpG-binding domain sequencing (MBD-seq) to assay genome-wide methylation.
Results: The analysis revealed 20 methylated genes, notably Eif2b5, that regulates myelin production.
Discussion: Loss of myelin with accompanying musculoskeletal pain is a major symptom of Gulf War Illness. Our work demonstrates multiple genes were methylated by exposure to organophosphates and glucocorticoids. These genes point to biochemical mechanisms that may be targets for therapeutic intervention.
Introduction
In 1991, a 42-nation coalition led by the United States, initiated combat against Iraq because of its invasion of Kuwait (Engelhardt, 2019). Nearly one million personnel were involved and of those who participated in combat actions, between 25 and 35 percent became sick with a multi-symptom malaise now called Gulf War illness (GWI). Symptoms include gastrointestinal, respiratory, fatigue and cognitive problems (Fappiano and Baraniuk, 2020). One of the hypotheses concerning the cause of the syndrome is exposure to organophosphates (OP), including sarin and chlorpyrifos, combined with increased circulating glucocorticoids as might be expected in the stress of combat. Moreover, for many of the afflicted veterans, the symptoms have persisted for more than 30 years since the conflict ended (Petry et al., 2024). Earlier research from our group presented evidence for genetic contributions to individual differences in susceptibility to the acute effects of exposure (Jones et al., 2020; Xu et al., 2020; Gao et al., 2020). In that series of experiments, we identified two candidate genes underlying individual differences in susceptibility to the exposures experienced by the troops in theater. That many of the afflicted veterans experience symptoms more than 30 years after cessation of the conflict poses a different, but important feature of GWI. The question is what is the reason these individuals are sick for so long? One highly likely possibility is the exposure modified the expression potential of one or more genes. Accordingly, we performed small studies to examine the possible methylation of genes, thus reducing their expression potential. The first of these involved the acute effects of a sarin surrogate coupled with a glucocorticoid (Ashbrook et al., 2018) in a single inbred mouse strain, C57BL/6J, one of the foundation strains for the genetic reference family of recombinant inbred mouse strains, the BXD group (Peirce et al., 2004). The second study involved the two foundation strains for the BXD group, C57BL/6J and DBA/2J (Mozhui et al., 2023). The basis for the present study was the observation in two previous studies that one of the targets for GWI exposure profile in our animal model was myelin (Ashbrook et al., 2018; Xu et al., 2020). Importantly, one of the pathophysiological chronic signs in afflicted GWI veterans is diminished myelin with associated muscular pain (Heaton et al., 2007; Chao et al., 2015; Belgrad et al., 2019). Accordingly, the major hypothesis that we proposed is that chronic GWI has major impact on myelination and the genes that regulate oligodendrocytes. In this study, we performed the same treatments in our genetic reference population of mice, exposure to corticosterone followed by treatment with a sarin surrogate but harvesting the hippocampus for analysis of genome-wide methylation 43 weeks after the sarin surrogate treatment, to model the years-long persistent nature of the disorder in ill veterans. Here we report the results from a study that expanded the number of strains and time after treatment.
Methods
Mice and treatment
Male and female mice from11 BXD strains age 60–65 days of age were the subjects for this study. All subjects received corticosterone in their drinking water (20 mg% w/v) for 7 days. On the eighth day, the animals received 4.0 mg kg−1 i.p. diisopropylfluorophosphate (DFP), a sarin surrogate. These are the same doses of corticosterone and DFP reported in our previous work on the acute effects of these treatments (Jones et al., 2020) and developed by O’Callaghan and colleagues (O'Callaghan et al., 2015). Over the next 43 weeks and for the first 12 weeks on alternate weeks, the animals received corticosterone as above in their drinking water. On the 42nd week, the animals received corticosterone in their drinking water. These repeated corticosterone treatments were done to prime and maintain the neuroinflammatory effect of the original corticosterone/DFP treatment. Glucocorticoids are regarded as having anti-inflammatory effects; however, they can also show pro-inflammatory effects, (Frank et al., 2010; Bolshakov et al., 2021). One week following the last corticosterone treatment, the mice were euthanized, and the hippocampus harvested. The DNA was extracted from the tissues and prepared for Methyl-CpG-binding domain sequencing (MBD-seq) to identify genome-wide methylation and RNA extracted for analysis. The interval between DFP treatment and tissue harvest was based on mouse-human age comparison to evaluate mice at an equivalent human age in the early 50s based on Jackson Laboratory website.
Design summary
Week 1, days 1–7, Corticosterone in the drinking water.
Week 2, day 1 (8 days since corticosterone treatment) i.p. injection of 4 mg/kg DFP.
Week 3, days 2–7 no corticosterone in the drinking water.
Weeks 4–12 Corticosterone in drinking water on alternate weeks beginning with week 4
Weeks 13–41 No treatments.
Week 42 Corticosterone in drinking water.
Week 43 days 1, euthanize mice, harvest tissue.
Mouse n = 5 per strain, sex, and treatment.
Tissue harvest and sample preparation
Genomic DNA was extracted from the hippocampus using the Quick-DNA/RNA Miniprep Plus kit (Zymo Research, Irvine, CA, United States) and checked for purity and quantity using a NanoDrop spectrophotometer (ThermoFisher Scientific, Waltham, MA, United States), and a Qubit™ fluorometer and the dsDNA BR (Broad Range) Assay kit (Invitrogen). Affinity based CpG enrichment was done using the Invitrogen MethylMiner Methylated DNA Enrichment Kit (ThermoFisher Scientific, Waltham, MA, United States), which relies on the methyl-CpG binding domain protein 2 (MBD2) protein to capture DNA fragments containing methyl-CpGs. MBD2 preferentially binds to methylated CpGs, and this depletes the DNA sample of DNA regions without CpGs, and enriches for methyl-CpGs (Aberg et al., 2018; Aberg et al., 2020) First, 1 µg of DNA in 110 μL low TE (tris-EDTA) buffer was sheared to∼150 bp fragments using a Covaris S2 ultrasonicator (Covaris, Woburn, MA, United States). Sonication settings were the same as described by Sandoval-Sierra (Sandoval-Sierra et al., 2020) with cycle/burst of 1 for 10 cycles of 60 s, duty cycle of 10%, and intensity of 5.0. DNA fragment size and quality were assessed using the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, United States). MBD-capture reaction was done according to the standard manufacturer’s protocol, followed by a single step elution with 2 M NaCl solution. The enriched DNA was then reconcentrated by ethanol precipitation, and the final concentration of methylated-CpG enriched DNA ranged from 0.17 to 2.1 ηg per μl (0.87 ± 0.39).
Sequencing and initial data processing
Sequencing was performed to 40 million reads per sample (150 paired-end) on Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States). Mus musculus (mouse) reference genome (GRCm38) and gene model annotation files were downloaded from the Ensembl genome browser (https://useast.ensembl.org/). Indices of the reference genome were built using STAR v2.5.0a (Dobin et al., 2013) and paired-end clean reads aligned to the reference genome.
Following alignment, the bam files were loaded to the MEDIPS R package (version 1.52.0) (Lienhard et al., 2014). The parameters for the MEDIPS.createSet function were: uniq = 1, extend = 150, window_size = 150, shift = 0, and the genome used was BSgenome.Mmusculus.UCSC.mm10. DNA methylation quantification was based on number of read counts within a 150 bp non-overlapping bins. The local CpG density (the coupling factor or CF) was computed using the MEDIPS.couplingVector function, and the read counts were normalized to the CF using the function MEDIPS.meth. We excluded all bins with CF = 0 (i.e., no CpG in the 150 bp bin), and this resulted in 10,647,424 bins. The raw read counts were the further processed using the EdgeR R package (version 3.42.4) (Robinson et al., 2010). Prior to statistical tests, we further filtered out bins with counts per million (CPM) less than 1 per library. This retained 432,694 DNA methlation regions that were normalized by the library size using the normLibSizes function. These 150 bp bin read counts were converted to logRPKM values. We then used the ChIPseeker (Yu et al., 2015) and AnnotationDbi (ref: see below for the website) R packages to annotate the bins for relevant gene information and genomic features (e.g., promoters, UTRs, introns, intergenic, etc.) (Pagès et al., 2024).
Statistical analyses
We performed multivariate regression for an epigenome-wide association study (EWAS). The primary goal was to identify the main effect of treatment in a genetically complex population. For initial quality check, we applied a principal component analysis (PCA), and the top PCs were used to detect possible outliers, and verify strain identify and sex. Background strain has a strong effect on DNA methylation and these are captured by the top PCs, and this gives us a variable to adjust for background genotype during EWAS for the main effect of treatment (Sandoval-Sierra et al., 2020). For the initial EWAS, we used the following regression model: glm(yi ∼ treatment + sex + body weight + PC1 + PC2 + PC3), where yi is methylation level at CpG region 1–432,694. Following the initial identification of CpGs associated with the main effect of treatment, we then examine the strain differences as a post hoc analysis.
Results
Effect of CORT + DFP on DNA methylation
To define site specific changes induced by the long-term exposure to CORT + DFP, we performed an epigenome-wide association study (EWAS) to compare between the treatment and control groups. For the first test (EWAS 1), we examined the main effect of treatment, with sex and body weight as cofactor, and adjusted for the top 3 PCs as proxies of genetic heterogeneity and other sources of variance. At a nominal unadjusted p = 0.0001, 34 CpG regions were altered in methylation by treatment (Data S1). The strongest main effect of treatment was on the promoter methylation of the Eif2b5 gene (Figure 1a). This region showed no difference between the sexes and was significantly decreased in methylation by CORT + DFP (Figure 1b). Analysis of variance showed a significant treatment effect (F1,65 = 25.62, p < .001). The main effect for strain and strain X treatment interaction were not significant (F10,65 = 1.55, p < .2; F10,65 = 1.02, p < .4, respectively). The top 20 CpG regions associated with the CORT + DFP treatment are presented in Table 1. Many of the CpG regions were in intergenic sites, but also included CpGs located in the introns or exons of Ctif, Cdh6, etc. We followed up the main EWAS with a regression that include the sex-by-treatment interaction (EWAS 2). The addition of this additional parameter reduced the associations for the main effect of treatment (Figure 1c). However, as a sensitivity test, the association with treatment for most of the CpG regions shown in Table 1 remained consistent at p < 0.001. This also revealed CpGs that had evidence of sex-by-treatment interactions. The strongest interaction effect was for a CpG region in the intron of Nell1 (alternatively, a promoter of the Nell1os). While this region shows no difference by treatment when all samples are pooled, but when stratified by sex, females show a reduction in methylation, while males show an increase in methylation with treatment (Figure 1d). An intergenic region on chromosome X showed a contrasting effect on treatment between the two sexes (Figure 1e). The top 20 CpG regions with treatment-sex interaction are in Table 1. None of these were associated with treatment based on EWAS 1.
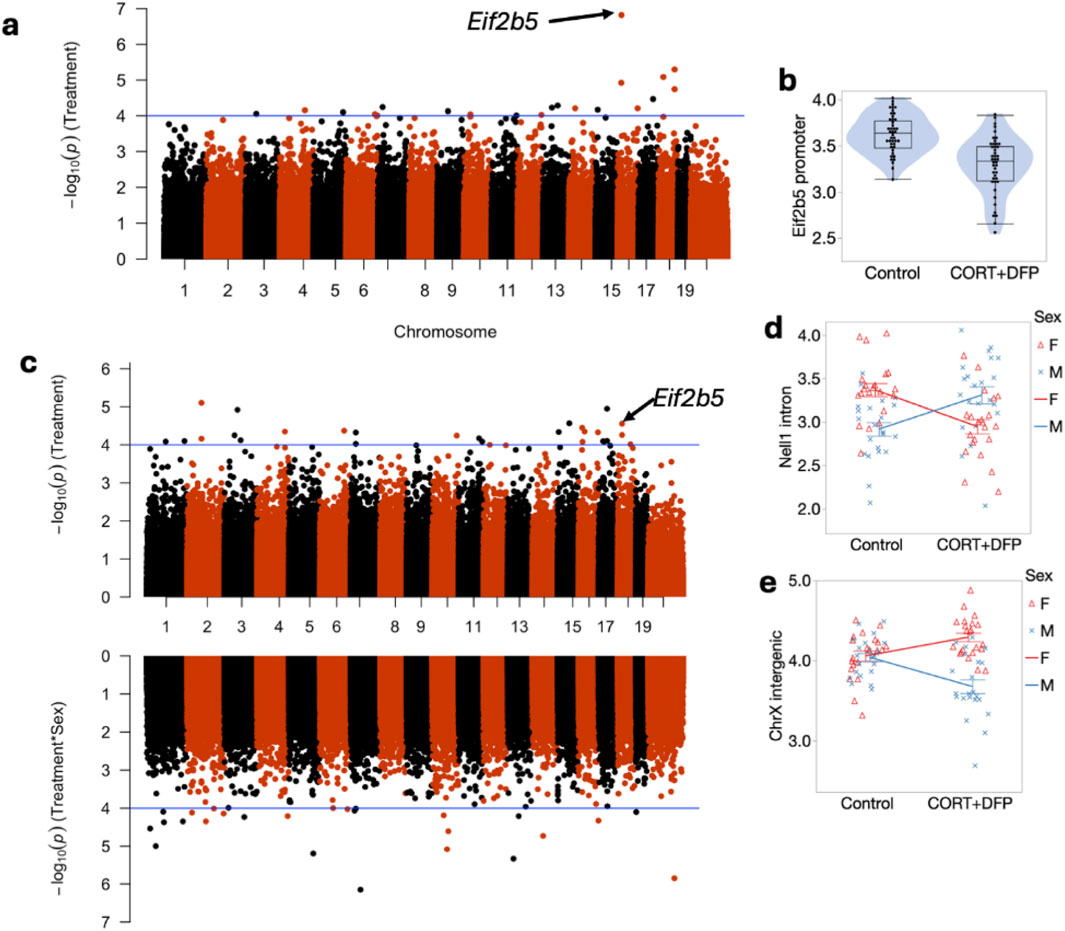
Figure 1. Manhattan plot of differentially methylated CpG regions. (a) main effect of CORT + DFP; (b) Effect of CORT + DFP on expression of the promoter region of Eif2b5; (c) Manhattan plot following addition of sex X treatment; (d) sex differences in methylation of Eif2b5; (e) sex differences in treatment in an intergenic region on X chromosome.
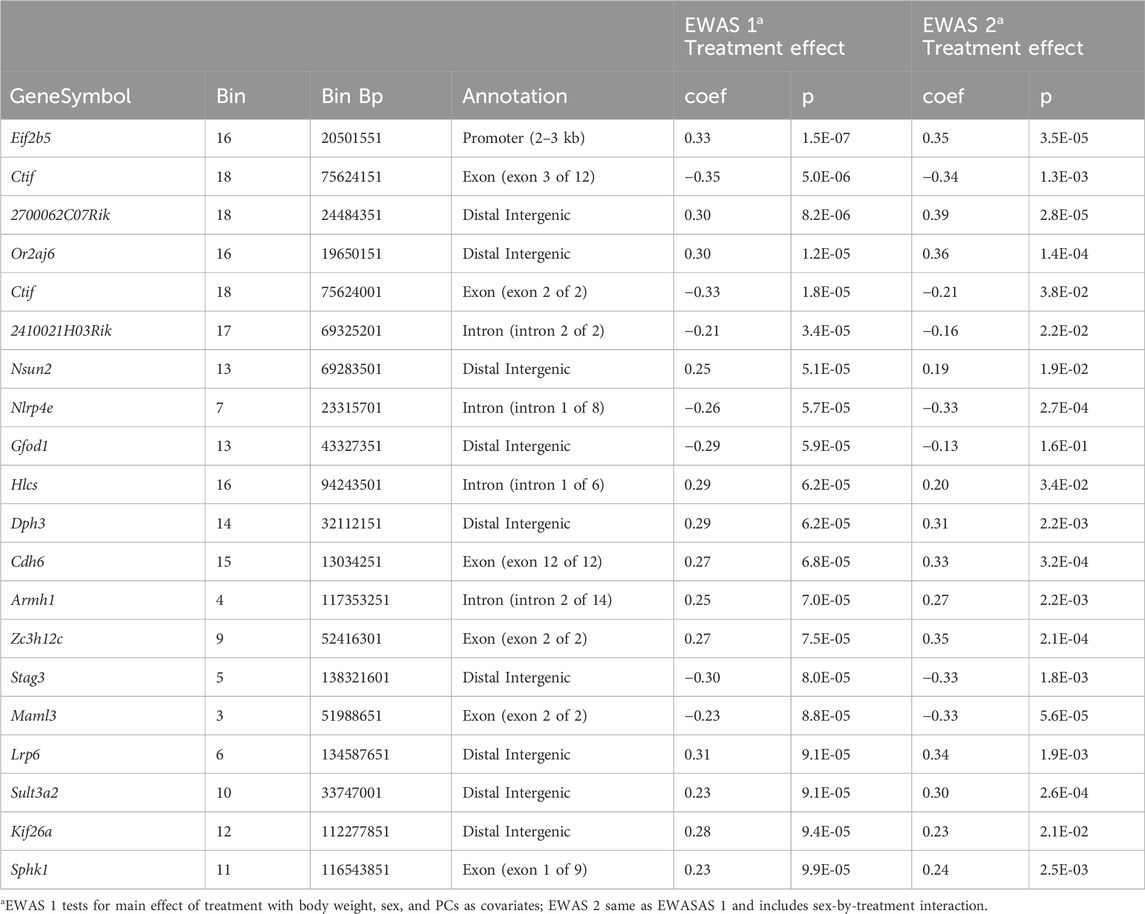
Table 1. Methylated genes identified for Main effect of CORT + DFP treatment and sex by treatment interaction.
Relating to strain variation
Figure 1 Presents the mapping and analysis of the effects of CORT + DFP main effect and for CORT + DFP X sex interaction. Compared to control, CORT + DFP resulted in lower overall methylation and the sex by treatment interaction showed males to evince increased methylation and females to show decreased methylation overall.
The strain distribution of effect is presented in Figure 2. As illustrated in Figure 2, many of the CpGs show significant heterogeneity by genetic background, and the top hits from the EWAS shown in Figure 1a are the ones that are robust to such heterogeneity.
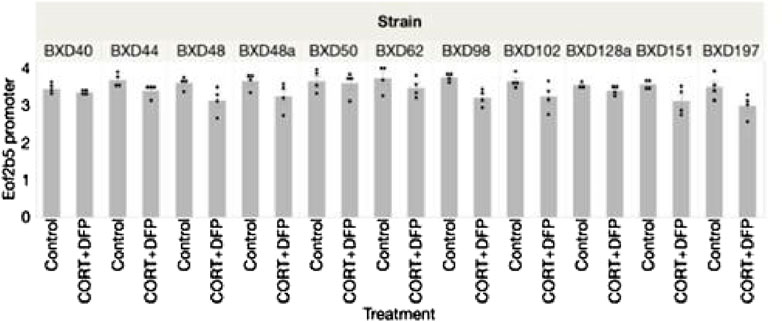
Figure 2. Strain distribution of methylation of Eif2b5 43 weeks following treatment with corticosterone and DFP.
Analysis of the MBD-seq results showed significant associations between CORT + DFP for the following genes: Eif2b5, Or2aj6, Nsun, Nlrp4e, and Cdh6. The strongest signal was for Eif2b5 (Figure 1a).
We then searched for genes that regulate Eif2b5 using the mapping software available on Genenetwork.org. using the Genome-wide Efficient Mixed Model Association (GEMMA) method. GEMMA is software implementing Linear Mixed Model Association for genome-wide association studies (GWAS). Gene mapping in the BXD family of mice is analogous to GWAS in humans except for replication of genotypes. Several BXD mouse strains are closely related genetically, therefore the need to account for population stratification.
Figure 3 illustrates quantitative trait loci analysis of Eif2b5 expression. We observed significant associations between the expression phenotype and markers on chromosomes 1 and 7. The area under the QTL curve on chromosome 7 is in a gene-impoverished region; however, the QTL curve on chromosome 1 contains several genes among which is Mapkapk2 (Mk2).
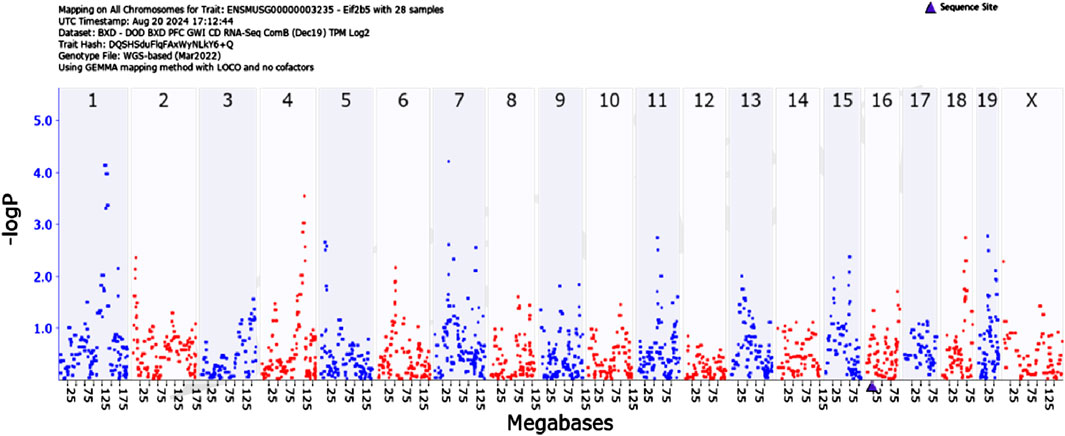
Figure 3. GEMMA quantitative trait analysis of Eif2b5 gene expression. Note the significant association on Chromosome 1. Search for candidate genes in the interval reveals Mapkapk2 (Mk2).
Figure 4 illustrates quantitative trait loci analysis of Mapkapk2 expression in hippocampus. The peak, indicating high association between expression and a marker near the coding region (blue triangle on the X-axis) shows that this gene is cis-regulated.
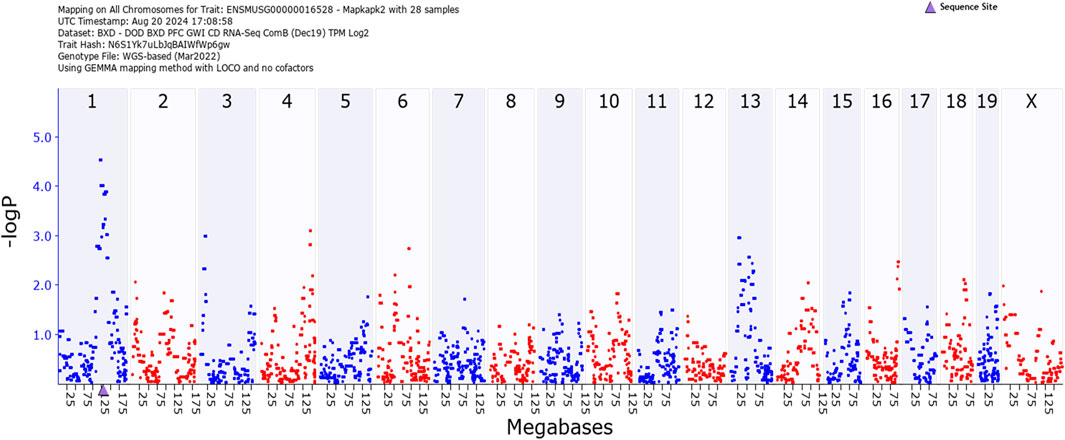
Figure 4. GEMMA quantitative trait analysis of Mapkapk2 gene expression. Note the significant association on Chromosome 1. The peak is near the coding region (blue triangle on the X-axis). This indicates that the gene is cis-regulated.
What about corticosterone?
In our previous study on the genetic basis of susceptibility to develop GWI, we showed that corticosterone exacerbated the effect of DFP on the expression of Il1b (Jones et al., 2020). We also presented data showing strain (genetic) differences in corticosterone consumption. We then asserted that because analysis of covariance showed no influence of corticosterone intake, that differences in corticosterone ingestion had no effect on Il1b expression. Similarly, here we observed differences in corticosterone intake among the 11 strains (Figure 5).
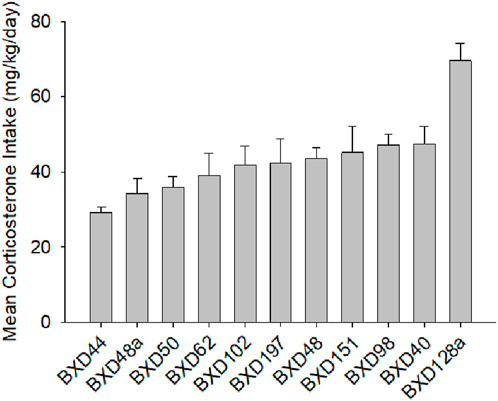
Figure 5. Mean daily consumption of corticosterone, males and females combined. Corticosterone intake represented here includes the pre DFP period and the weekly intakes for the 5 periods following DFP and the consumption on week 42. The data presented here are for both sexes the correlation for the sexes was r = 0.66, p < 0.01 one-tailed based on previous observation, so we combined the data for a single figure.
The variation that we observed among the strains in corticosterone consumption did not correlate with differences in methylation of Eif2b5.
Discussion
In this study, we focused on the hippocampus as those afflicted with GWI reported difficulties with memory (Li et al., 2011; Chao, 2023) and dysfunction of blood flow in this structure has been reported to be impaired with impairment increasing with age (Li et al., 2011). Also, we extend the findings of our previous small study involving just the parental strains of the BXD group and for 84 instead of 300 days (Mozhui et al., 2023). The small study was for proof of concept, and we identified four genes that had been differentially methylated by strain. The genes are Ttll7, which regulates glutamate as a neurotransmitter, Akr1c14, associated with the immune microenvironment, Slc44a4, a high-affinity, sodium-dependent choline transporter found in peripheral tissues and a choline supplier to other cells for membrane maintenance. Rusc2 is associated with cognition. The present study expanded the number of BXD strains to 11 and extended the time from exposure to DFP to 300 days. The time frame approximates the developmental age of Gulf War veterans who are now in their early to late 50s. The current study expands the number of methylated candidate genes, including Eif2b5, Or2aj6, Nsun, Nlrp4e, Cdh6. Eif2b5 is associated with myelination and GWI includes neurological problems involving deficits in myelin. Or2aj6 participates in olfactory processing. Some Gulf War Veterans report loss of olfaction and this is often a prodromal sign of neurodegeneration (Chao, 2024), Nsun2 is a methyltransferase and is associated with Dubowitz syndrome, an autosomal recessive disease that produces intellectual delay and microcephaly. At present there is no known association between GWI symptoms and this gene, Nlrp4e regulates actin cap formation during oögenesis and interferon 1 via inhibition of tank binding kinase 1. It has function in viral immunity and programmed cell death. Its involvement in GWI is presently unknown but seems to be a candidate for further study because of its role in neuroinflammation; Cdh6, cahedrin 6, is associated with gliomas. Its expression is positively associated with malignancy and negatively associated with prognosis. This is an important player in GWI as those so afflicted have increased risk for glioma. Of particular interest is our finding that Eif2b5 is regulated by Mapkapk2, a gene that participates in cellular stress response, including production of the proinflammatory cytokines, Tnfa, l1b, and IL6, among several other cellular processes. The sex X treatment interaction for Eif2b5 was not unexpected. Previously, we showed a sex by treatment interaction for the acute effects of CORT + DFP (Jones et al., 2020) with female mice showing less neuroinflammation indices than males. In humans, the reverse has been observed with females showing more severe symptoms of GWI than their male counterpoints (Heboyan et al., 2019). As many of those afflicted with GWI are still experiencing debilitating symptoms are aging, the emphasis needs to be placed on treatments. Indeed, relatively few of those Gulf War veterans afflicted have shown recovery (Van Riper et al., 2017). One of the primary symptoms of chronic GWI is peripheral pain, originating from compromised central nervous system myelin (Abbink et al., 2019). Vanishing white matter (VWM) disease is a fatal disorder but apparently not related to the white matter compromise seen in GWI. This disease, triggered by stress, is seen primarily in children (Wong et al., 2019). A recent study [31] reported that a small molecule compound, 2BAct, reversed the loss of myelin in a mouse model of VWM. This compound stimulates Eif2b5 expression. This or similar compounds may have promise for treating the chronic debilitating symptoms of GWI.
The value of our approach to show the genetic basis for individual differences in susceptibility to the acute and chronic manifestations of Gulf War Illness, shows the way to understanding the basic mechanisms of the diseases per se. Accordingly, this approach will point to the means for treatment or even prevention of the disease.
Conclusion
Loss of myelin with associated musculoskeletal pain is a major and debilitating symptom of chronic GWI. It is likely that the loss of myelin relates to cognitive and other neurological symptoms as well. Here, we have shown a promising gene-based and biochemical mechanism that underlies chronic symptoms of GWI and the possibility of treating these symptoms. Because the GWI population is aging, time is the important factor in prosecuting the discovery of therapies.
Data availability statement
The original contributions presented in the study are publicly available. These data can be found here: https://info.genenetwork.org/infofile/source.php?GN_AccesionId=1063.
Ethics statement
The animal study was approved by University of Tennessee Health Science Center Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BJ: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Writing – original draft, Writing – review and editing. JO: Conceptualization, Methodology, Writing – review and editing, Writing – original draft. DA: Formal Analysis, Writing – review and editing, Writing – original draft. LL: Data curation, Writing – review and editing, Writing – original draft. PP: Formal Analysis, Writing – review and editing, Writing – original draft. WZ: Data curation, Investigation, Supervision, Writing – review and editing. KM: Data curation, Formal Analysis, Visualization, Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Major funding by the National Institute of Environmental Health Sciences, National Institutes of Health.
Acknowledgments
The authors thank Daming Zhuang for expert technical assistance Major support by USPHS grant R02ES031656.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1553410/full#supplementary-material
References
Abbink, T. E. M., Wisse, L. E., Jaku, E., Thiecke, M. J., Voltolini-González, D., Fritsen, H., et al. (2019). Vanishing white matter: deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 6, 1407–1422. doi:10.1002/acn3.50826
Aberg, K. A., Chan, R. F., and van den Oord, EJCG (2020). MBD-seq - realities of a misunderstood method for high-quality methylome-wide association studies. Epigenetics 15, 431–438. doi:10.1080/15592294.2019.1695339
Aberg, K. A., Chan, R. F., Xie, L., Shabalin, A. A., and van den Oord, EJCG (2018). Methyl-CpG-binding domain sequencing: MBD-seq. Methods Mol. Biol. 1708, 171–189. doi:10.1007/978-1-4939-7481-8_10
Ashbrook, D. G., Hing, B., Michalovicz, L. T., Kelly, K. A., Miller, J. V., de Vega, W. C., et al. (2018). Epigenetic impacts of stress priming of the neuroinflammatory response to sarin surrogate in mice: a model of Gulf War illness. J. Neuroinflammation 15, 86. doi:10.1186/s12974-018-1113-9
Belgrad, J., Dutta, D. J., Bromley-Coolidge, S., Kelly, K. A., Michalovicz, L. T., Sullivan, K. A., et al. (2019). Oligodendrocyte involvement in gulf war illness. Glia 67, 2107–2124. doi:10.1002/glia.23668
Bolshakov, A. P., Tret'yakova, L. V., Kvichansky, A. A., and Gulyaeva, N. V. (2021). Glucocorticoids: dr. Jekyll and mr. Hyde of hippocampal neuroinflammation. Biochem. (Mosc). 86, 156–167. doi:10.1134/S0006297921020048
Chao, L. L. (2023). Examining the current health of Gulf War veterans with the veterans affairs frailty index. Front. Neurosci. 7, 1245811. doi:10.3389/fnins.2023.1245811
Chao, L. L. (2024). Olfactory and cognitive decrements in 1991 Gulf War veterans with gulf war illness/chronic multisymptom illness. Environ. Health 23, 14. doi:10.1186/s12940-024-01058-2
Chao, L. L., Zhang, Y., and Buckley, S. (2015). Effects of low-level sarin and cyclosarin exposure on white matter integrity in gulf war veterans. Neurotoxicology 48, 239–248. doi:10.1016/j.neuro.2015.04.005
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
Engelhardt, J. P. (2019). U. S. Army strategic studies Institute, special report: desert shield and desert storm: a chronology and troop list for the 1990-1991 Persian Gulf crisis. Middle East Studies CarlisLe, PA: Department of National Security and Strategy U.S. Army War College.
Fappiano, C. M., and Baraniuk, J. N. (2020). Gulf war illness symptom severity and onset: a cross-sectional survey. Mil. Med. 14 (185), e1120–e1127. doi:10.1093/milmed/usz471
Frank, M. G., Miguel, Z. D., Watkins, L. R., and Maier, S. F. (2010). Prior exposure to glucocorticoids sensitizes the neuroinflammatory and peripheral inflammatory responses to E. coli lipopolysaccharide. Brain Behav. Immun. 24, 19–30. doi:10.1016/j.bbi.2009.07.008
Gao, J., Xu, F., Starlard-Davenport, A., Miller, D. B., O'Callaghan, J. P., Jones, B. C., et al. (2020). Exploring the role of chemokine receptor 6 (Ccr6) in the BXD mouse model of gulf war illness. Front. Neurosci. 14, 818. doi:10.3389/fnins.2020.00818
Heaton, K. J., Palumbo, C. L., Proctor, S. P., Killiany, R. J., Yurgelun-Todd, D. A., and White, R. F. (2007). Quantitative magnetic resonance brain imaging in US army veterans of the 1991 gulf war potentially exposed to sarin and cyclosarin. Neurotoxicology 28, 761–769. doi:10.1016/j.neuro.2007.03.006
Heboyan, V., Krengel, M. H., Sullivan, K., Iobst, S., Klimas, N., Wilson, C., et al. (2019). Sex differences in gulf war illness: a reanalysis of data from the cdc air force study using cdc and modified Kansas case definitions. J. Occup. Environ. Med. 61, 610–616. doi:10.1097/JOM.0000000000001620
Jones, B. C., Miller, D. B., Lu, L., Zhao, W., Ashbrook, D. G., Xu, F., et al. (2020). Modeling the genetic basis of individual differences in susceptibility to gulf war illness. Brain Sci. 10 (3), 143. doi:10.3390/brainsci10030143
Li, X., Spence, J. S., Buhner, D. M., Hart, J., Cullum, C. M., Biggs, M. M., et al. (2011). Hippocampal dysfunction in Gulf War veterans: investigation with ASL perfusion MR imaging and physostigmine challenge. Radiology 261, 218–225. doi:10.1148/radiol.11101715
Lienhard, M., Grimm, C., Morkel, M., Herwig, R., and Chavez, L. (2014). MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286. doi:10.1093/bioinformatics/btt650
Mozhui, K., O’Callaghan, J. P., Ashbrook, D. G., Prins, P., Zhao, W., Lu, L., et al. (2023). Epigenetic analysis in a murine genetic model of Gulf War illness. Front. Toxicol. 5, 1162749. doi:10.3389/ftox.2023.1162749
O'Callaghan, J. P., Kelly, K. A., Locker, A. R., Miller, D. B., and Lasley, S. M. (2015). Corticosterone primes the neuroinflammatory response to DFP in mice: potential animal model of Gulf War Illness. J. Neurochem. 133, 708–721. doi:10.1111/jnc.13088
Pagès, H., Carlson, M., Falcon, S., and Li, N. (2024). AnnotationDbi: manipulation of SQLite-based annotations in bioconductor. R. package version 1.66.0. Available online at: https://bioconductor.org/packages/AnnotationDbi.
Peirce, J. L., Lu, L., Gu, J., Silver, L. M., and Williams, R. W. (2004). A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5, 7. doi:10.1186/1471-2156-5-7
Petry, S. E., Thompson, A. D., Hauser, E. R., Lynch, S. M., Boyle, S. H., Upchurch, J., et al. (2024). Characterizing deficit accumulation among gulf war era veterans. J. Frailty Aging. 13, 300–306. doi:10.14283/jfa.2024.44
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Sandoval-Sierra, J. V., Helbing, A. H. B., Williams, E. G., Ashbrook, D. G., Roy, S., Williams, R. W., et al. (2020). Body weight and high-fat diet are associated with epigenetic aging in female members of the BXD murine family. Aging Cell 9, e13207. doi:10.1111/acel.13207
Van Riper, S. M., Alexander, A. L., Koltyn, K. F., Stegner, A. J., Ellingson, L. D., Destiche, D. J., et al. (2017). Cerebral white matter structure is disrupted in Gulf War Veterans with chronic musculoskeletal pain. Pain 12, 2364–2375. doi:10.1097/j.pain.0000000000001038
Wong, Y. L., LeBon, L., Basso, A. M., Kohlhaas, K. L., Nikkel, A. L., Robb, H. M., et al. (2019). EIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife 8, e42940. doi:10.7554/eLife.42940
Xu, F., Ashbrook, D. G., Gao, J., Starlard-Davenport, A., Zhao, W., Miller, D. B., et al. (2020). Genome-wide transcriptome architecture in a mouse model of Gulf War Illness. Brain Behav. Immun. 89, 209–223. doi:10.1016/j.bbi.2020.06.018
Keywords: MBD-seq, Forward genetcs, QTL (loci of quantitative traits), BXD mice, neuroinflammation
Citation: Jones BC, O’Callaghan JP, Ashbrook DG, Lu L, Prins P, Zhao W and Mozhui K (2025) Epigenetic study of the long-term effects of gulf War illness. Front. Genet. 16:1553410. doi: 10.3389/fgene.2025.1553410
Received: 30 December 2024; Accepted: 08 May 2025;
Published: 18 June 2025.
Edited by:
Festenstein Richard, Imperial College London, United KingdomReviewed by:
Sanjay Kumar Singh Patel, Hemwati Nandan Bahuguna Garhwal University, IndiaPeter J. Bayley, United States Department of Veterans Affairs, United States
Israel Ramirez-Sanchez, National Polytechnic Institute (IPN), Mexico
Copyright © 2025 Jones, O’Callaghan, Ashbrook, Lu, Prins, Zhao and Mozhui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. C. Jones, YmpvbmUxMjlAdXRoc2MuZWR1
 B. C. Jones
B. C. Jones J. P. O’Callaghan
J. P. O’Callaghan D. G. Ashbrook
D. G. Ashbrook L. Lu
L. Lu P. Prins
P. Prins W. Zhao
W. Zhao K. Mozhui
K. Mozhui