Abstract
Over the last decade, strong evidence is emerging from multiple disciplines of scientific inquiry ranging from behavioral psychology to clinical medicine that many published scientific studies are not reproducible. The best-known retrospective analyses from psychology (Rahal and Open Science Collaboration, 2015) and cancer biology (Begley and Ellis, 2012) report staggeringly low replication rates of 40 and 20%, respectively. This has led to the phenomenon of a “reproducibility crisis” where a significant amount of funding, research effort and even public policy has come to depend on non-reproducible research (Goodman et al., 2016). In this perspective article, I outline how the blockchain can be used to power a decentralized peer-review system for digital health applications. This is followed by a discussion of how such a system impacts the broader issue of scientific reproducibility, and how existing blockchain protocols can help alleviate the barriers to reproducibility.
Introduction
In 2005, Dr. John Ioannidis of Stanford University published a report in PLoS Medicine titled “Why Most Published Research Findings Are False,” which eventually became a cornerstone of discussion regarding interpretation of statistical significance testing and the value it provides to the claims of a report (Ioannidis, 2005). In 2011, the Center of Open Sciences launched a project to replicate 100 different studies that were published in 2008 from the field of psychology (Rahal and Open Science Collaboration, 2015). The results of this initiative were published in 2015, and demonstrated that even though 97% of the original results claimed statistically significance, this was only recapitulated in 36% of the replication attempts (Rahal and Open Science Collaboration, 2015). This theme of non-replication was also brought to light by Dr. Lee Ellis from MD Anderson who wrote a similar article in 2012 on cancer biology titled “Raise standards for preclinical cancer research” reporting that only 10% of published preclinical studies could be validated to enter clinical trials (Begley and Ellis, 2012). Clinical data is a particularly sensitive instrument in the sense that it is prone to perturbations and noise introduced by a variety of sources ranging from investigator bias or errors related to experimental design and statistical analysis of the collected data. To that end, digital medicine provides robust statistical tools that subject the current research processes to intensive scrutiny, and allow for earlier interventions into the data-collection process. In addition, next generation digital health technologies, particularly distributed ledgers can offer a solution (in the form of audit trails and data quality control through smart contracts) needed to address reproducibility problems and the peer-review process – given sufficient funding and regulatory support. However, this cannot be accomplished by replicating the current research processes and just transforming them from paper to digital form. Rather, a complete re-thinking and re-engineering of the peer-review process with a focus on reproducibility using blockchain protocols is needed (Goodman et al., 2016). Here, I present a digital medicine use-case where machine learning powers a clinical application, and discuss how a new type of blockchain-based peer-review system that can help reduce the burden of reproducibility.
In a recent study, Tomašev et al. (2019) reported the use of a machine learning approach called recurrent neural networks to identify impending acute kidney injury, 1 or 2 days in advance of the diagnosis made using the standard of care clinical tests. The authors applied this methodology to data collected from more than 700,000 adults treated in hospitals and outpatient clinics run by the United States Department of Veterans Affairs and published their results. However, the codebase used to develop the trained recurrent neural network was not made open-source due to use of proprietary libraries and licensing. Instead, the authors provided extensive data from testing their machine learning model against training data in the supplementary information (Tomašev et al., 2019). This data provided insights into the framework behind the neural network, the training phases, results from supervised learning, prediction capabilities in the final model, and the kind of test data used for training (Tomašev et al., 2019). Ultimately, the data was used as a surrogate for sharing the code behind the network, and the quality control of validation was on the shoulders of the authors.
Increased use of machine learning models for clinical decision support has brought about a paradigm shift in handling the immense amounts of data generated by quantitative and qualitative measurements of physiological parameters. The two areas in evidence-based medicine that may benefit heavily from the application of machine learning techniques are diagnosis and outcome prediction (Kononenko, 2001; Darcy et al., 2016; Beam and Kohane, 2018; Zhang, 2019). A common end-point for machine learning algorithms in diagnostic medicine is to build a classifier: a predictive function that can map variables from an input stream onto discrete output categories. Classifiers can aid a highly skilled worker in the decision-making processes, for instance triaging a data-set containing new actionable observations (Kononenko, 2001; Beam and Kohane, 2018). However, the development and validation (peer-review) of machine learning classifiers for clinical applications require a few special considerations in order to help increase the chances of eventually having patient contact and improving patient outcomes (Darcy et al., 2016; Beam and Kohane, 2018; Zhang, 2019). The opportunities for monetization of classifiers built on top of private libraries are off-set by the cost of “black-boxing” parameters that are hidden and therefore not replicated and verified rigorously. We need to approach this problem in the context of burden of reproducibility – a standardized mechanism for peer-review of digital medicine apps and proprietary machine learning tools is needed. More precisely, one that involves authors of the study, provides a public interface for testing the tools in a secure environment with new data, and reports the final recommendations back to a public forum.
A constant pressure to publish high-impact work, selective reporting of results, poor use of statistics [including p-hacking (Head et al., 2015)], and unsophisticated protocols can all contribute to reducing overall replication potential of an experiment. Researchers can also be hampered by the technical difficulties involved in replicating a complex experiment that requires multiple difficult lab techniques, poorly described methods and incompletely reported setbacks from data (Head et al., 2015). Funding agencies and publishers are stepping in to reduce these problems. Funding agencies such as the National Science Foundation (NSF) and the National Institutes of Health (NIH) have changed grant requirements and have awarded grants to design classes aimed at improving statistical literacy (Stodden et al., 2014). Journals are designing policies that help address inadequate documentation and supporting technologies that facilitate data-sharing (Stodden et al., 2014). In addition, many high-impact journals are raising the data-standards by requesting authors to deposit more experimental data, and making it publicly available. Presently, the use of commercial machine learning applications is nascent in clinical medicine, private libraries can accelerate research and development by providing monetization opportunities. Any code built upon private libraries is not submitted to open-source repositories due to licensing dependencies, and this leads to a built-in assumption: if the authors of a given study provide sufficient indirect data and insight into the black-box model, the results from the study will be reproducible. There is a conscious effort from the entire scientific enterprise to increase data transparency, but the burden of reproducibility weighs heavily on the authors of a study. The broader question remains: Can we create a standardized peer-review approach where the authors of a study (in digital medicine) get involved in demonstrating the generalizability of their models?
Reproducibility crisis has become well recognized by researchers in many fields of science (Chalmers et al., 2014; Chan et al., 2014; Glasziou et al., 2014; Ioannidis et al., 2014; Macleod et al., 2014; Salman et al., 2014; Begley and Ioannidis, 2015). For our discussion, we define reproducibility of a study as the property of obtaining the same results from conducting an independent study of a published experiment, as long as the methods are closely matched to the original study (Chan et al., 2014; Glasziou et al., 2014; Ioannidis et al., 2014; Macleod et al., 2014). The burden of reproducibility increases with new computational tools as commercial development essentially limits how much of the internal operations can be divulged in an open-source format (Macleod et al., 2014). In the era of digital medicine, the usual barriers of peer-review and statistical analysis are not enough – this holds true especially for machine learning-type classifiers where the end-product is built on private libraries (Blockeel et al., 2013; Lemley, 2019; Lu, 2019; Stupple et al., 2019). The blockchain provides a unique approach to this problem by serving as an immutable recording device whereby a sandboxed container can interface with a new classifier, apply new testing data to verify this classifier, and publish the results along with the methods to a shared data layer (Stupple et al., 2019). A group of established validators can reference these published results, quantify them into numerical reputation points, and make their final recommendations available to a publicly accessible data layer. The reputation points can stratify reliability within the network (Stupple et al., 2019). Of note, the technical components necessary for this new form of peer-review have already been deployed in practice among different blockchain implementations. For instance – Quorum and the Golem Network are two distributed ledger protocols that have the operational infrastructure to support payload privacy and sandboxed off-chain computations, respectively. In this article, the salient features necessary for a blockchain based peer-review are reviewed from three protocols: a reputation system, off-chain computations, and private transaction states. The operational principles and design considerations behind these implementations are reviewed in order to highlight how peer-review can be a pertinent use-case. The article is organized as follows: I begin with an introduction to reputation systems by discussing Augur, a prediction-markets powered reputation system built on the Ethereum blockchain. This is followed by a review of sandboxing and support for off-chain computations available in the Golem Network. Finally, the article ends with a brief overview of the dual public/private transaction states in Quorum, with a focus on how the dual states can be applied to a peer-review of private classifiers. The goal throughout this article is to highlight how existing components can be repurposed for peer-review and scientific validation of digital health applications.
Augur Network: Reputation Tokens
To understand how reputation parameters embed and propagate through a blockchain network, let us begin with a brief overview of Augur. A decentralized predictions system requires four fundamental components: a platform for participating members to submit predictions for an outcome, a mechanism to reward points for accurate predictions, a consensus framework to consolidate reward points into a longer-standing parameter such as reputation, and a group of validators that maintain the integrity of the network. Augur is an ERC-20 token built on top of the Ethereum blockchain as a platform for prediction markets (Peterson et al., 2015). Augur is powered by a token called Reputation (REP), which is used by members for event reporting. By owning REP, and participating in the accurate reporting on the outcomes of events, token holding members of the network receive a portion of the settlement fees from the platform. The incentive structure is such that accurate reporting of outcomes has the highest return on investment for REP token holders (Peterson et al., 2015). The more REP a reporter owns, and reports correctly with, the more fees they will earn for their work in maintaining the integrity of the platform (Peterson et al., 2015). There is no monetary value associated with REP, these tokens only propagate reputation across the network, and act as a multiplier for return on network rewards. The market outcome settled by a majority of the reporters is validated against the canonical outcome from the external world, a consensus is reached and REP tokens are rewarded for accurate predictions (Peterson et al., 2015). Augur has an oracle that can migrate information on external events to a blockchain without having to rely on a trusted intermediary. The validators can use this oracle to adjudicate any reputation conflicts that arise on the network (Peterson et al., 2015). Over time, a record of continuous change in REP of token holders and internal validation lead to a market of reliable set of users with a high reputation and long-standing history of accurate predictions network-wide (Peterson et al., 2015). In this setting, reputation becomes the backbone of long-term reliability and users with high REP can be attached to editorial and reviewer positions.
Golem Network: Decentralized Computation Farm
Golem is a peer-to-peer network that connects providers with computing power to requestors who submit computationally heavy tasks for processing. The network functions as a Platform-as-a-Service and provides computational resources in a decentralized fashion in return for payment in tokens. Golem has three main players: the suppliers of computational resources (providers), requestors who submit tasks to be computed by the network, and finally software developers who add new features to the network (Wood, 2014; The Golem Project, 2016).
In Golem, one of the most crucial components of the decentralized ecosystem is the Application Registry. It provides developers with tools to deploy software running on the Ethereum blockchain, and toolkits to monetize use cases (Wood, 2014). The main functions of the registry include giving developers a platform to publish applications, give requestors a directory to search and request specific tools for their tasks, and to give providers a mechanism to control the code they execute for security concerns. Golem allows requesters to execute the code of an application on a host computer, but this code is sandboxed and executed with the minimal required privileges (Wood, 2014; The Golem Project, 2016). The process of off-chain computing involves running an application on a host machine linked to the blockchain, once the computational task is finished, the host machine submits the results to the blockchain along with the processing time. In order to protect the host from executing malicious code, the application runs in a sandboxed environment within a container designed to limit any external access. At present, the only application configured for off-chain computation with sandboxing is decentralized animation rendering developed by Golem, but more microservices are planned (Wood, 2014; The Golem Project, 2016).
Due to the halting problem, it remains technically infeasible to evaluate a segment of code and heuristically determine whether any malicious components are present. To that end, the Application Registry has three classes of access control: authors, validators, and providers. Authors publish applications to the registry, validators review and certify applications as trustworthy by adding them to a whitelist (The Golem Project, 2016). Now these applications are considered sanctioned, and available to a provider for execution. Providers requisition the hardware to execute a requestor’s task and are given the right to choose the applications they want to run, based on the validators they select (The Golem Project, 2016). A provider can take advantage of this mechanism, managing her own whitelist/blacklists, or simply using whitelists of validators she trusts. Payments between requesters and providers are facilitated by a transaction framework in Golem (The Golem Project, 2016). This framework along with the Application Registry make it possible to seamlessly request hardware from a computational farm, submit a task, and obtain the final results. In the future, scientific applications on Application Registry would have the potential to operate on a device-agnostic computational farm, and report on the final outcome of computations along with the methods (The Golem Project, 2016). This can be bound under a smart contract, so that no operator intervention is required from the start of computation till the end with the results submitted (The Golem Project, 2016).
Quorum: Dual Transaction States
Quorum has native support for transaction privacy, this manifests in the form of transaction states with public and private payloads on the network. Broadly speaking, the lifecycle of a private transaction on Quorum involves two components: a Quorum node and Tessera (Chase, 2016). A review of the Quorum node is provided elsewhere (Chase, 2016), the remainder of this section will focus on the design and components of Tessera, the privacy engine of Quorum. Tessera enables the encryption/decryption of payloads and propagation of private transactions on the network. It in turn has two main components: a transaction manager and an Enclave (Chase, 2016). Both are presented in the upcoming sections.
Nodes can determine if a transaction is private or public, one quick method of doing this involves looking at the header for a parameter called v-value, for instance, a higher v-value points to a private transaction (Chase, 2016). If the transaction is deemed private, a node can only process the associated payload if it has the private keys needed to decrypt and unpack the payload (Chase, 2016). Nodes who are not party to the transaction payload will simply skip the transaction, and therefore not execute the private payload. In order to support this bifurcation of transaction states, Quorum stores the public payloads in a common public state that is synchronized globally, and the private payloads are synchronized locally with the involved parties (Chase, 2016). Additionally, this model allows for granular control over modifying the state during the cross-communication between the public and private contracts. There are well-specified software locks on the state of a virtual machine to prevent sync conflicts (Chase, 2016). Here, is an example: when a private payload refers being executed makes a reference to external or public information, the virtual machine has software locks that forcibly enforce a read-only mode. In this manner, if a call derived from the public-private interaction requests a change to the internal state of virtual machine, it throws an exception. In this manner, Quorum limits the number of actors that can update the internal state, ultimately reducing friction between subsequent contracts running on the virtual machine (Chase, 2016). Next, a discussion of private payloads and the instrument used by Tessera for performing encryption and crypto-related operations is presented.
Tessera Enclave
In network security terms, an Enclave is defined as a secure computing asset that has no interactions with the remainder of the network, or any other systems. The main application of an Enclave in Quorum is to limit network access and protect information that exists inside of an Enclave from external malicious attacks (Chase, 2016). Most distributed ledger protocols rely on cryptographic techniques to maintain transaction validity, member verification, and the network state through a chain of cryptographically hashed headers. In order to achieve real-time performance enhancements for crypto-specific operations and technical isolation to create a crypto-space, much of the cryptographic work including public/private key generation and data encryption/decryption is designated to the Enclave. It holds the private keys and functions essentially as a virtual hardware security module on the Quorum network. The argument for a crypto-specific space becomes relevant in the context of memory leaks: Enclave works asynchronously with the transaction manager to help strengthen privacy by managing the encryption/decryption of operations in an isolated way (Chase, 2016). This enables sensitive operations to be handled in a single container with layers of memory protection, without any demonstrable potential for leakage into areas of execution memory that may be visible to the remainder of the network. The Enclave is designed to manage all types of key management for Quorum, as well as supporting any operations required by private payloads given that a particular node is participating in the executing the payload on a virtual machine. In the most simplistic operational model, each Enclave interacts only with the transaction manager local to that node, but there is support for more complex interactions between transaction managers from different nodes involving multiple Enclaves (Chase, 2016). Let’s briefly talk about transaction managers next.
Tessera Transaction Manager
In the lifecycle of a private transaction and payloads, the transaction manager is the central point of communication and distribution of private payloads (Chase, 2016). It interfaces with most other components of the network and manages the directional movement of private data. Transaction manager provides access to transaction data, exchanges payloads on Quorum blockchain with transaction managers of other nodes involving new participants, but does not have access to decode any sensitive data because it lacks private keys. Even though the transaction manager has database access, it utilizes the Enclave for operations involving private key cryptography. This design principle has been constituted to limit access to an attacker that may have gained access to a node or a transaction manager from reaching private keys (Chase, 2016). Additionally, two important network features of the transaction manager include forming a peer-to-peer network of transaction managers such that peer/key information can be broadcasted, and interfacing with the Enclave for cryptographic tasks even between nodes (Chase, 2016).
A network powered by Tessera that enables private payloads and dual transaction states has the potential for creating a special off-chain testing environment specifically for proprietary software acting as payloads. Only the involved parties will have access to the payload being tested, and the entire verification process can be automated by a smart contract. Once the verification begins, an isolated container can test new data, publish the results to a blockchain, and then self-destruct. This ensures no unauthorized access to payloads or any proprietary software. A panel of validators interested in proving the reproducibility and generalizability of a classifier can use the published results from an off-chain test in making final recommendations. Such a system can pave the way to development of a standardized peer-review pathway for verification of digital health applications and proprietary software leading to an overall reduction of the burden of replicability on the original developers (Chase, 2016). Figure 1 graphically summarizes this concept. A new blockchain protocol encompassing these features can lead to a new era of blockchain-enabled peer-review of scientific studies, particularly in machine learning for evidence based medicine.
FIGURE 1
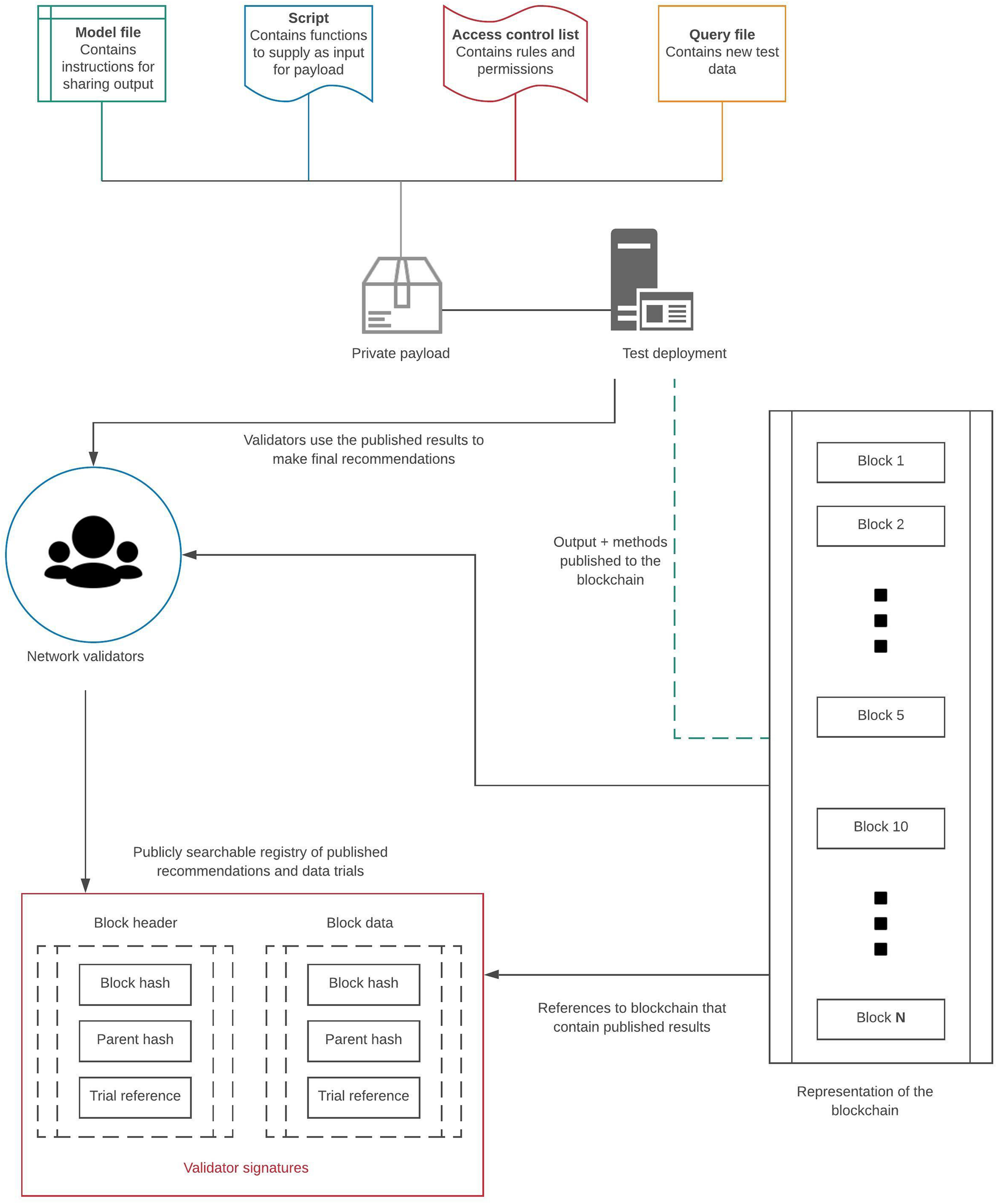
Use-case for blockchain as a peer-review network. The four components dictate how verification of a private payload should be carried out. The model file would have instructions for how to publish the results along with the methods used. The script will dictate how new data is provided to the classifier. The access control list dictates user permissions, and finally, the query file contains new data that can be used to test a private machine learning classifier. The results are published to the blockchain along with a reference to the network validators. The results are reviewed along with the methods by the validators, and final recommendations are published to a publicly searchable registry. This registry serves as a public interface for searchable results that reference specific blockchain instances when the trials were carried out.
Discussion
The “reproducibility crisis” has come to define a period of error-correction standards put forth by consortiums, journals, and grant-awarding institutions (Ioannidis et al., 2014). Designing a universal verification scheme to enhance experimental methods is an incredibly challenging task, from a technical standpoint. In the limited scope of digital medicine and health, blockchain provides a unique opportunity for a peer-review model that can be automated and is powered for public testing and reporting. Moreover, governance structure in the context of Decentralized Autonomous Organization (DAOs) enabled by blockchain protocols is another interesting area to explore (Norta, 2016). A Technical Steering Committee (TSC) elected from the network can oversee the verification protocols, experimental design and data reporting. They can provide a collaborative link with researchers and study authors to ensure the verification experiments are carried out appropriately. This committee can further provide recommendations once the testing has been completed, and award reputation points to research groups that have been consistently publishing highly reproducible results.
Here, I propose three recommendations that aim to improve data-storage, communication, and the robustness of research findings by focusing on specific barriers to reproducible science. These measures are examples of policies that are already in place by different projects and organizations. They are not intended to be exhaustive, but instead, provide examples of practical approaches to actions that can be implemented by researchers, journals and funding institutions:
Data storage: Research-data repositories such as Figshare have RESTFUL APIs that make it very easy to reference data stored on the platform (Thelwall and Kousha, 2016). These references can be attached as metadata to a decentralized data structures such a block on a blockchain. The blockchain allows for the creation of an immutable metadata trail that contains references to off-chain locations of data (Thelwall and Kousha, 2016). Although research data and supplemental information would be stored as tags attached to the blockchain, the information is hosted externally to prevent bloating of the blockchain (Thelwall and Kousha, 2016). Journals can require the wallet addresses containing this metadata and make it publicly available for anyone to verify. Moreover, new protocols such as IPFS allow for more permanent storage of data in a decentralized fashion and allow for easy sharing of files over a peer-to-peer network (Benet, 2014).
Decentralized endorsement: The Academic Endorsement System (AES) is built on the blockchain as a reputation system that uses Academic Endorsement Points (AEP) instead of REP (in case of Augur) to reward scientific work that is worthy of endorsement (b8d5ad9d974a44e7e2882f986467f4d3,2016). This established framework provides a more comprehensive alternative to existing reputation systems such as Augur, when applied to scientific research. The amount of AEP credited to a scientist is based on AEP received for prior work. In that sense, researchers who have produced significant endorsed output in the past will have a greater influence on the community at large (b8d5ad9d974a44e7e2882f986467f4d3,2016). Additionally, any kind of research can be endorsed, for instance blog posts, data sets, and even code-segments (b8d5ad9d974a44e7e2882f986467f4d3,2016). The adoption of such a system provides a new set of metrics that can be calculated by a journal, and presented alongside traditional metrics such as number of citations and H-index (Jacsó, 2008).
Fluid communication: Journals are rather inflexible and limited vehicles for post-publication communication with authors. A blockchain based social media platform called Steem is creating a space where content creators to be rewarded (in cryptocurrency) directly by the readers for posting new materials (Larimer et al., 2016). Built on top of Steem, PEvO (Publish and Evaluate Onchain) recently emerged as a commenting platform for scientific papers where readers can communicate with the authors (Wolf et al., 2016). At the same time, authors can share updates, reply to comments and integrate constant reviews into new information extending the paper as a living document – dynamic features that are lacking in the established peer-review process (Wolf et al., 2016). The interactions on PEvO such as likes and comments result in a pay out in tokens to the authors which incentivizes quality conversations and result sharing (Wolf et al., 2016). As opposed to traditional commenting platform, PEvO offers a greater incentive for researchers to engage in meaningful conversations with readers and answer questions or provide clarification which may highlight the strengths or weaknesses of a study.
In this article, we highlighted three key features necessary for blockchain peer-review: reputation systems necessary for development of a network-wide index of reproducibility, off-chain computations for the actual verification protocols, and private payloads to ensure the safety of intellectual property. Such a blockchain network is not in practice yet, but this article serves to highlight a new use-case for existing implementations such as Quorum or Golem. Ultimately, new forms of peer-review powered by blockchain are largely design problems: all the infrastructure needed is already in practice. Can we incentivize and sustain a network that takes advantage of automation and reliability provided by blockchain and smart contracts? The very near future will only present us with more challenges rather than solutions, but blockchain remains a very promising direction for building the next generation of peer-review systems.
Statements
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
b8d5ad9d974a44e7e2882f986467f4d3 (2016). Towards Open Science: The Case for a Decentralized Autonomous Academic Endorsement System.10.5281/zenodo.60054(accessed December 29, 2019).
2
BeamA. L.KohaneI. S. (2018). Big data and machine learning in health care.JAMA3191317–1318.
3
BegleyC. G.EllisL. M. (2012). Drug development: raise standards for preclinical cancer research.Nature483531–533. 10.1038/483531a
4
BegleyC. G.IoannidisJ. P. (2015). Reproducibility in science: improving the standard for basic and preclinical research.Circ. Res.116116–126. 10.1161/circresaha.114.303819
5
BenetJ. (2014). Ipfs-content addressed, versioned, p2p file system.arXiv Preprint.
6
BlockeelH.KerstingK.NijssenS.ŽeleznýF. (2013). Machine Learning and Knowledge Discovery in Databases.Berlin: Springer.
7
ChalmersI.BrackenM. B.DjulbegovicB.GarattiniS.GrantJ.GülmezogluA. M.et al (2014). How to increase value and reduce waste when research priorities are set.Lancet383156–165. 10.1016/S0140-6736(13)62229-1
8
ChanA. W.SongF.VickersA.JeffersonT.DickersinK.GøtzscheP. C.et al (2014). Increasing value and reducing waste: addressing inaccessible research.Lancet383257–266. 10.1016/S0140-6736(13)62296-5
9
ChaseJ. P. M. (2016). Quorum White Paper. Available at: https://github.com/jpmorganchase/quorum/blob/master/docs/Quorum%20Whitepaper%20v0.2.pdf(accessed November, 18, 2019).
10
DarcyA. M.LouieA. K.RobertsL. W. (2016). Machine learning and the profession of medicine.JAMA315551–552.
11
GlasziouP.AltmanD. G.BossuytP.BoutronI.ClarkeM.JuliousS.et al (2014). Reducing waste from incomplete or unusable reports of biomedical research.Lancet383267–276. 10.1016/S0140-6736(13)62228-X
12
GoodmanS. N.FanelliD.IoannidisJ. P. (2016). What does research reproducibility mean?Sci. Transl. Med.8341s1210.1126/scitranslmed.aaf5027
13
HeadM. L.HolmanL.LanfearR.KahnA. T.JennionsM. D. (2015). The extent and consequences of p-hacking in science.PLoS Biol.13:e1002106. 10.1371/journal.pbio.1002106
14
IoannidisJ. P. (2005). Why most published research findings are false.PLoS Med.2:e124. 10.1371/journal.pmed.0020124
15
IoannidisJ. P.GreenlandS.HlatkyM. A.KhouryM. J.MacleodM. R.MoherD.et al (2014). Increasing value and reducing waste in research design, conduct, and analysis.Lancet383166–175. 10.1016/s0140-6736(13)62227-8
16
JacsóP. (2008). The pros and cons of computing the h-index using Scopus.Online Inform. Rev.32524–535. 10.1108/14684520810897403
17
KononenkoI. (2001). Machine learning for medical diagnosis: history, state of the art and perspective.Artif. Intell. Med.389–109. 10.1016/s0933-3657(01)00077-x
18
LarimerD.ScottN.ZavgorodnevV.JohnsonB.CalfeeJ.VandebergM. (2016). Steem: An Incentivized, Blockchain-Based Social Media Platform. Available at: https://steem.io/SteemWhitePaper.pdf
19
LemleyK. V. (2019). Machine learning comes to nephrology.J. Am. Soc. Nephrol.301780–1781. 10.1681/asn.2019070664
20
LuD. (2019). DeepMind’s medical AI.New Sci.243:15.
21
MacleodM. R.MichieS.RobertsI.DirnaglU.ChalmersI.IoannidisJ. P.et al (2014). Biomedical research: increasing value, reducing waste.Lancet383101–104.
22
NortaA. (2016). “Designing a smart-contract application layer for transacting decentralized autonomous organizations.” In Proceedings of the International Conference on Advances in Computing and Data Sciences (Singapore: Springer), 595–604. 10.1007/978-981-10-5427-3_61
23
PetersonJ.JosephK.MicahZ.WilliamsA. K.StephanieA. (2015). Augur: a decentralized oracle and prediction market platform.arXiv Preprint
24
RahalR. M.Open Science Collaboration (2015). Estimating the reproducibility of psychological science.Science349:aac4716. 10.1126/science.aac4716
25
SalmanR. A. S.BellerE.KaganJ.HemminkiE.PhillipsR. S.SavulescuJ.et al (2014). Increasing value and reducing waste in biomedical research regulation and management.Lancet383176–185. 10.1016/s0140-6736(13)62297-7
26
StoddenV.LeischF.PengR. D. (eds) (2014). Implementing Reproducible Research.Boca Raton, FL: CRC Press.
27
StuppleA.SingermanD.CeliL. A. (2019). The reproducibility crisis in the age of digital medicine.NPJ Digit. Med.21–3.
28
The Golem Project (2016). The Golem Project Crowdfunding Whitepaper. Available at: https://golem.network/doc/Golemwhitepaper.pdf(accessed November, 18, 2019).
29
ThelwallM.KoushaK. (2016). Figshare: a universal repository for academic resource sharing?Online Inform. Rev.40333–346. 10.1108/oir-06-2015-0190
30
TomaševN.GlorotX.RaeJ. W.ZielinskiM.AskhamH.SaraivaA.et al (2019). A clinically applicable approach to continuous prediction of future acute kidney injury.Nature572116–119. 10.1038/s41586-019-1390-1
31
WolfM.WiegandM.DrichelA. (2016). PEvO: Decentralized Open Access and Evaluation. Available at: https://pevo.science/files/pevo_whitepaper.pdf(accessed December 29, 2019).
32
WoodG. (2014). Ethereum: a secure decentralised generalised transaction ledger.Ethereum Proj. Yellow Pap.1511–32.
33
ZhangZ. (2019). Machine learning method for the management of acute kidney injury: more than just treating biomarkers individually.Biomark. Med.131251–1253. 10.2217/bmm-2019-0363
Summary
Keywords
blockchain, social good, smart contract, peer-review, digital medicine
Citation
Dhillon V (2020) Blockchain Based Peer-Review Interfaces for Digital Medicine. Front. Blockchain 3:8. doi: 10.3389/fbloc.2020.00008
Received
20 November 2019
Accepted
04 February 2020
Published
05 March 2020
Volume
3 - 2020
Edited by
Dean Korošak, University of Maribor, Slovenia
Reviewed by
Jonathan M. Fuchs, FACHE, The Kidder Street Consulting Group, United States; Erika Beerbower, Independent Researcher, Denver, United States
Updates
Copyright
© 2020 Dhillon.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vikram Dhillon, dhillonv10@gmail.com
This article was submitted to Blockchain for Science, a section of the journal Frontiers in Blockchain
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.