- 1Department of Agricultural, Food and Forest Sciences, University of Palermo, Palermo, Italy
- 2Research Centre for Plant Protection and Certification, Council for Agricultural Research and Economics, Palermo, Italy
- 3Department of Architecture (DARCH), University of Palermo, Palermo, Italy
- 4Research Centre for Olive, Fruit and Citrus Crops, Council for Agricultural Research and Economics, Rende, Italy
The long-lived and evergreen olive tree dominates the Mediterranean landscape, representing an agroecological and cultural symbol and a genetic heritage of inestimable value. Sicily, for historical, geographical, and cultural reasons, has a very rich and distinctive olive germplasm. In this work, a large survey was conducted to discover, collect, and characterize the genetic diversity of centennial monumental olive trees from historical sites, such as the Greek Temple Valley (Agrigento), ancient gardens, or farmland present in the western part of the island. Trees were chosen based on their height, trunk, stump size, and presumed age; particularly, only olive trees with an age estimated at more than 400 years old were taken into consideration. For the morphological characterization, the leaf, fruit, and endocarp traits were analyzed. For the molecular characterization, 11 polymorphic microsatellite markers largely used for fingerprinting analysis were used. Reference cultivars were included in the analysis for comparison. Nuclear DNA was extracted from different parts of the plant (young leaves of shoots taken from the canopy and young leaves taken from suckers, which arose from the basal part of the tree) to check if the trees were grafted and to explore their diversity. Most of the monumental trees have been grafted at least one time during their long life, and some genotypes showed unique genetic profiles combined with peculiar phenotypic traits. Suckers (rootstock of the trees) showed a strict genetic relationship with an ancient monumental oleaster tree, also included in the study. “Patriarch” (original mother plants) trees of local cultivars were also identified. This research revealed a high level of the still unexplored genetic diversity of the Sicilian olive germplasm and highlighted its importance as a gene reservoir, which could support new breeding programs for the evaluation and possible selection of traits linked to putative resilience to abiotic and biotic stresses (particularly Xylella fastidiosa subsp. pauca ST53 or soil- borne diseases or insects). The results will be useful for improving the conservation process, enriching existing collections of olive genetic resources, and supporting on-farm conservation projects.
1 Introduction
The olive (Olea europaea L.), originating from the geographical area extending from the Southern Caucasus (Iran) to Mesopotamia and Palestine, is a tree well adapted to warm temperate regions, characterized by long and dry summer seasons (Lavee, 1992), as occurs in the Mediterranean area. Domesticated at least six thousand years ago from a Near Eastern and Aegean gene pool and spread by the Phoenicians and Greeks (Newton et al., 2014), the olive tree can be considered the symbol of civilization and an integral part of the Mediterranean landscape, strongly linked to human activities, traditions, and commercial trades. Exploited since the Mesolithic and cultivated for oil production at least since the Bronze Age, it has a wide distribution in all the countries bordering the Mediterranean Sea, where a population of approximately 800 million individuals live (Schicchi et al., 2021).
The olive tree has great genetic variability originating from the fact that it is a predominantly allogamous species, leading to high levels of heterozygosity and DNA polymorphism among individuals (Angiolillo et al., 1999; Rallo et al., 2000; Petruccelli et al., 2021). The longevity of the olive tree and the strong genetic stability have allowed to preserve its variability, leading to the constitution of a dynamic pool of cultivated plant (landraces) and cultivars that are locally adapted and suitable for traditional farming systems. As olive is easily vegetatively propagated by self-rooting organs (mainly ovules and cuttings) and by grafting onto seedlings, many landraces and cultivars were spread (Fiorino and Lombardo, 2002). The occurrence of gamic crosses with the wild olive population (Olea europaea subsp. oleaster, Hoffmanns & Link) and among genotypes has increased its variability (Marra et al., 2013; Caruso et al., 2014). Therefore, the adaptation to different pedoclimatic contexts has favored the emergence of new genotypes or heterogeneous regional populations that are highly different at the morphological, molecular, and agronomic levels.
Among fruit trees, the olive has one of the richest varietal platforms (Marra, 2009); according to FAO data (2010), 2629 accessions are known in the ex situ collections around the world. Bartolini et al. (2014) estimate that the world variety heritage consists of 1626 cultivars grown in 59 countries and that 5300 names are known (Bartolini et al., 1998). The richness of the olive germplasm finds a clear representation in the extraordinary and heterogeneous Italian varietal platform. The number of native Italian cultivars amounts to 631, with 1849 synonyms and 827 names not yet identified (Bartolini et al., 2014); however, this number is underestimated (Perri et al., 2015). The olive sector in Italy is spread over an area of 1,141,019 hectares with an annual production of 29,910,074 quintals of olives (ISTAT 2022), placing it at second place in the Mediterranean basin after Spain (FAOSTAT 2023). The main Italian region for oil and table olive production is Puglia, followed by Calabria, Sicily, and Campania (ISTAT 2022). Among the Italian regions, Sicily is characterized by a vast germplasm of olive trees, including many monumental trees. In addition, the island has the highest number of Protected Denomination of Origin (PDO) extra- virgin olive oil designations.
Due to its geographical position, Sicily has always been a crossroads of the Mediterranean. The historical and cultural diversity of Sicily finds analogies in the large variability of the pedoclimatic environments that have inevitably contributed to creating biodiversity in many crop species. The natural and anthropical selective pressure, the frequent occurrence of spontaneous crossing between cultivated and wild forms, and the exchange of seeds and material for vegetative propagation have led to the creation of a vast olive heritage, subject to continuous evolutionary changes.
The vast Sicilian varietal platform is based on 25 cultivars, well characterized at the morphological, molecular, and agronomical levels, thanks to the intense studies conducted in the last 30 years, which also established ex situ collections (La Mantia et al., 2005; Caruso et al., 2007; Marra et al., 2013; Barone et al., 2014; Caruso et al., 2014; Marra et al., 2014; Marino et al., 2017; Sala et al., 2021). However, so far, no work has been carried out to characterize the monumental trees of which the island is rich. Therefore, there is the need to characterize them since they represent a genetic source potentially of high value.
The value of centuries-old olive trees lies in their incredible longevity and resistance, which has allowed them to survive climatic variations, natural disasters such as fires, floods, biotic, and abiotic adversities, and that they still can be productive and healthy. These trees are often characterized by large sizes, shaped as real art works by atmospheric phenomena and biotic agents over the time. Thus, they evoke a sense of eternity, which earned them the definition of “Monumental olives” or “Patriarch” trees as they could be the ancestor mother plants of some well-known cultivars.
Molecular markers allow to detect the relationships between cultivars and wild genotypes and between Olea species and subspecies. Among the molecular markers, microsatellite or SSR (simple sequence repeats) are still considered the marker of choice for genotyping, and they are believed to be a standard tool for germplasm management in many species, as significant efforts have been made to align a range of SSR data to allow comparison among standardized databases (e.g., apple — Denancé et al., 2020; sweet cherry— Ordidge et al., 2021; peach— Marchese et al., 2005; Trifonova et al., 2021; almond— Dangl et al., 2009; Cimò et al., 2017; hazelnut— Fiore et al., 2022; palm— Giovino et al., 2020; Giovino et al., 2023). SSRs have been proved to be very effective in identifying and discriminating olive cultivars, including Sicilian ones, thanks to their transferability, high variability, and co-dominance, as widely documented in the literature (Baldoni et al., 2000; Rallo et al., 2000; Carriero et al., 2002; Cipriani et al., 2002; Caruso et al., 2005; La Mantia et al., 2005; Caruso et al., 2007; Doveri et al., 2008; Marra et al., 2013; Caruso et al., 2015; Atrouz et al., 2021). Haouane et al. (2011) and El Bakkali et al. (2019) unequivocally identified more than 500 accessions from the world collection of Cordoba and Marrakech, while Trujillo et al. (2014) discriminated 322 olive varieties out of 499 analyzed accessions from 21 Mediterranean countries. In addition to the aforementioned fingerprinting analyses for which microsatellites represent one of the best solutions, they are also widely used in paternity test analyses (De la Rosa et al., 2004; Marchese et al., 2016a; Lombardo et al., 2019), traceability studies (Martins-Lopes et al., 2008; Alba et al., 2009), mapping (De la Rosa et al., 2003; Wu et al., 2004; Marchese et al., 2016b), population genetics, and phylogenetic studies (e. g. Belaj et al., 2007; Belaj et al., 2010; Erre et al., 2010).
Recently, many fingerprinting studies have been focusing on the analysis of the genetic diversity of monumental olive trees believed to represent peculiar gene pools probably related to resilience to biotic and abiotic stress. Anestiadou et al. (2017) explored the molecular diversity of 43 monumental olive trees on the island of Cyprus, discovering a large genetic diversity never described before. Other studies conducted on monumental olive trees have revealed historical–cultural links between genotypes from different geographical areas but low genetic diversity. Aksehirli-Pakyurek et al. (2017) investigated genetic relationships between 112- centuries-old genotypes from Crete and Turkey, discovering that many of them correspond to the well- known cultivars Koroneiki, Throubolia, and Mastoidis from Greece and Samanli and Gemlik from Turkey. Valeri et al. (2022) analyzed 190 ancient trees from Malta and discovered 46 unique Maltese genotypes, finding that almost 30% of trees were grafted and, in some cases, rootstocks presented unique profiles. In Spain, over 4,500 monumental olive trees were genotyped with SSR markers, revealing limited genetic variability as most of them corresponded to the cultivar Farga, except for 10 genotypes that presented unknown profiles (Ninot et al., 2018). Similarly in Italy, Rotondi et al. (2018) analyzed 206 secular olive trees from Emilia- Romagna using SSR markers, reporting that most of the olive trees belonged to 10 cultivars that were already characterized.
Given the importance of secular olive trees, the activities of identification, characterization, in situ conservation, and census are essential for their protection.
The main aim of this work was to identify and characterize monumental olive tree accessions from the western part of Sicily found in two archaeological areas—the Valley of the Temples Archaeological Park and Kolymbethra Garden located in Agrigento— and farms from three of the six EVO oil PDO areas of the island (Valli Trapanesi, Valle del Belice, and Val di Mazara) to assess whether the analyzed genotypes are still unknown or represent a possible “Patriarch” (“mother plants”) of traditional (indigenous) cultivars already characterized and, for some of them, extensively grown on the island. To achieve this goal, we also performed the alignment of SSR data from many accessions of olive germplasm collections from the Mediterranean Basin (MB) present in the literature and in our own databases by using a standard set of nuclear SSR markers and standard reference cultivars.
2 Materials and methods
2.1 Selection of trees and plant material
The identification and choice of the olive trees was carried out reflecting the aspect of “monumentality”, which considers tree size and longevity (Schicchi et al., 2021). The trunk diameter of the trees, measured one meter above the ground, was longer than 1.5 m. This size is considered an important index to obtain an idea of the presumed age of the tree. Unlike the diameter, since longevity is not a directly measurable parameter unless sophisticated instruments are used, estimating the age of olive trees is carried out using indirect methods, such as extrapolating the age based on growth rate estimations.
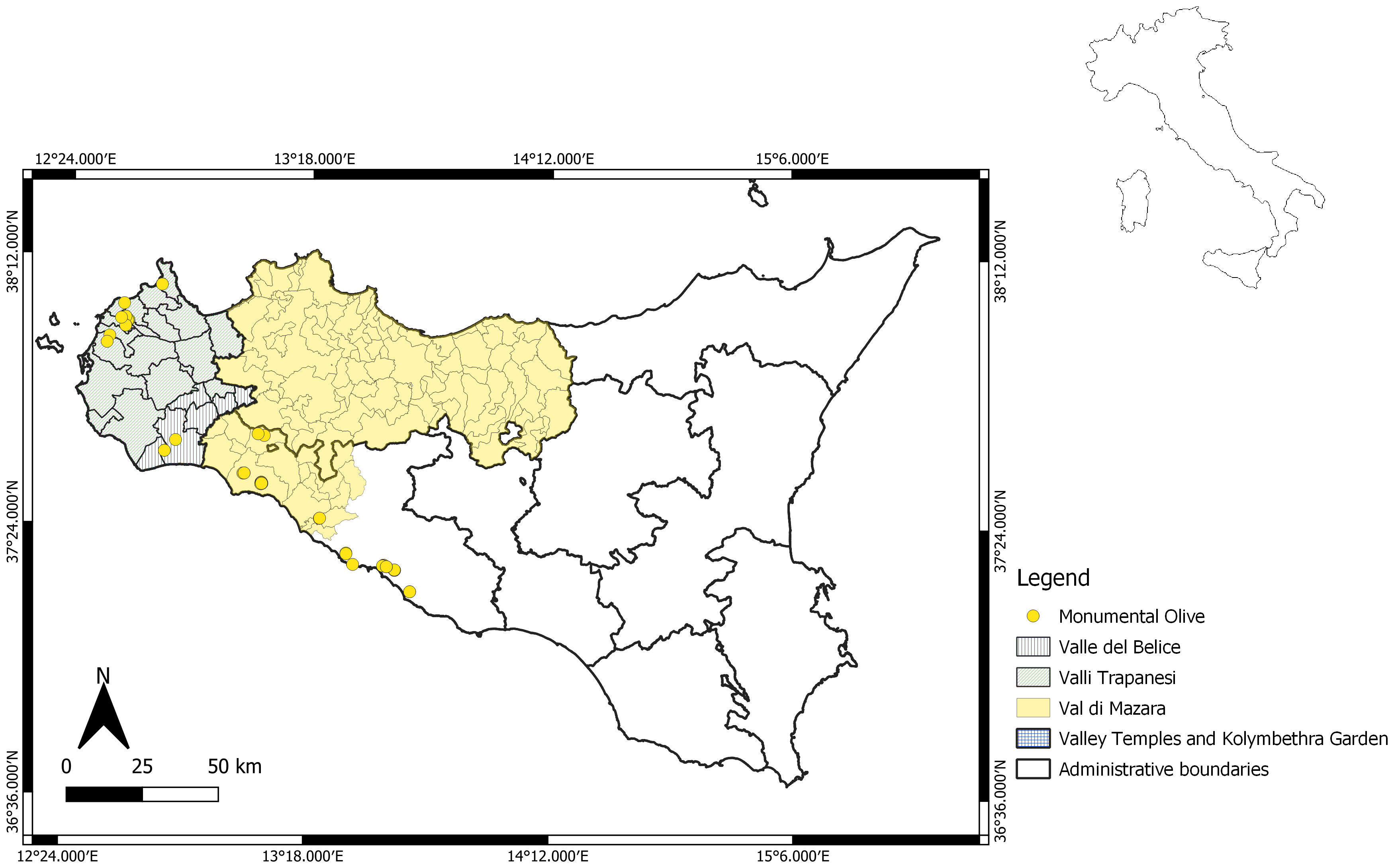
A total of 73 trees were included in the analysis, and many of them could produce fruits (Table S1). Some, although still productive, were abandoned, while 10 trees were collected from two archaeological areas: the Valley of the Temples Archaeological Park and Kolymbethra Garden, both located in the Agrigento province. For molecular characterization, one monumental tree of Olea europaea var. sylvestris, known as Oleastro Inveges, was also included. The georeferentiation of trees was performed (Figure 1).
2.2 Molecular characterization
For the molecular characterization 73 monumental trees, including 1 tree of Olea europaea var. sylvestris (Oleastro Inveges — indicated as Mon_33_OI) and 8 Sicilian standard cultivars (Table S1) were screened with 11 SSR markers (Table 1).
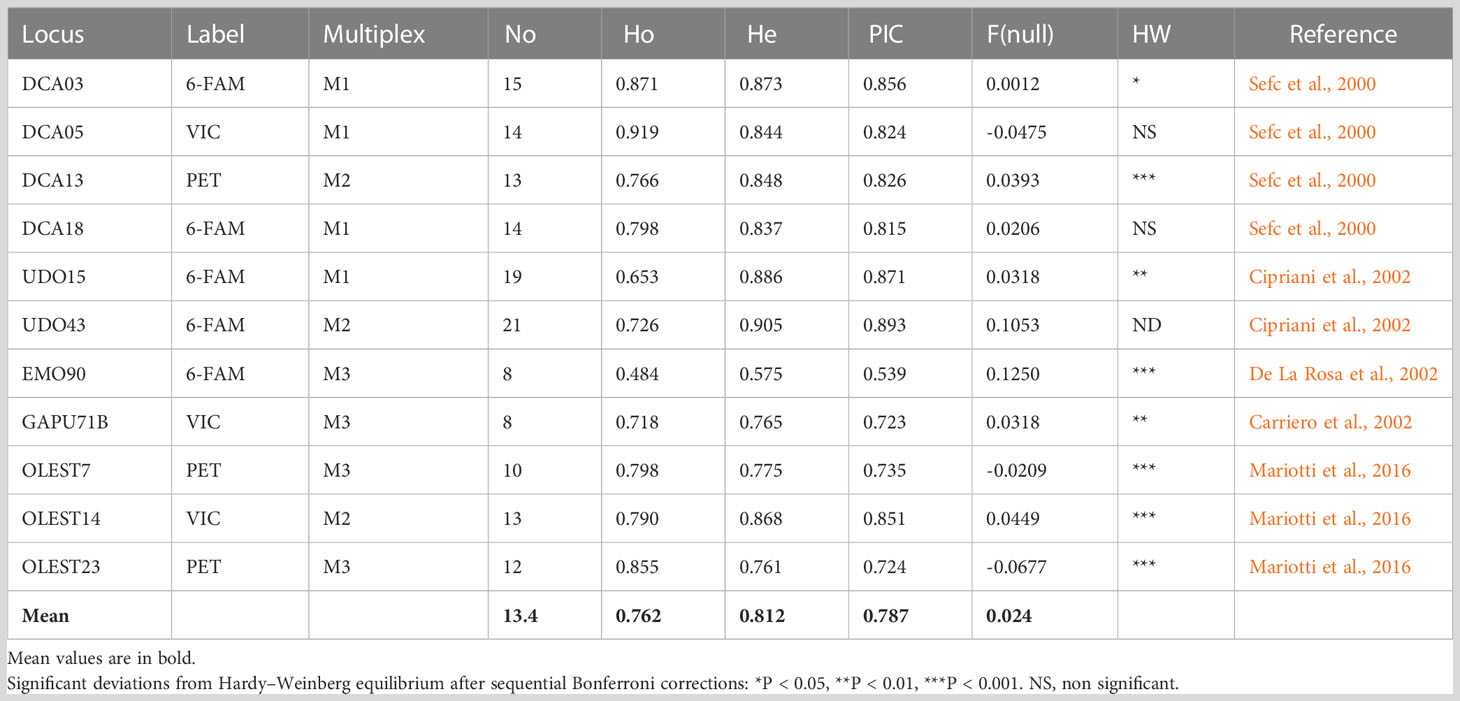
Table 1 SSR statistical parameters: names of the locus, fluorescent dye label, PCR multiplex mix, number of alleles (No), observed heterozygosity (Ho), expected heterozygosity (He), polymorphic information content (PIC), null allele value F(null), deviation from Hardy–Weinberg equilibrium, and reference.
For three monumental trees (Mon_32C, Mon_35C, and Mon_37C) showing morphological differences at the branch levels, more samples were collected from the branches (Mon_32B1, Mon_32B2, Mon_32B3_ Mon_32B4, Mon_35B1, Mon_35B2, Mon_37B1, Mon_37B2).
For the 53 selected monumental trees, including the “Oleastro Inveges”, samples from the suckers were also collected, indicated in the analysis with the letter S (sucker), while samples from the upper part of the trees were indicated with the letter C (canopy).
2.3 DNA extraction
Genomic DNA was extracted from approximately 20 mg of finely ground powder of young leaves collected from the upper part of the crown and leaves taken from the suckers using the CTAB method by Doyle and Doyle (1987), without beta-mercaptoethanol addition and with other minor modifications. DNA quality and concentration were checked with a NanoDrop spectrophotometer 2000 (Thermo Scientific), diluted to 50 ng/ul and stored at -20°C until used.
2.4 SSR genotyping
SSR amplifications were performed in three multiplex PCRs (M1, M2, M3), and forward primers were labeled at their 5′ end with three dyes, FAM, VIC, and PET, as follows: DCA3 (-6FAM), DCA5 (VIC), DCA13 (PET), DCA18 (-6FAM), EMO90 (-6FAM), GAPU71b (VIC), UDO15(-6FAM), UDO43 (-6FAM), OLEST7 (PET), OLEST14 (VIC), and OLEST23 (PET) (Table 1).
Most of these markers are in common with other olive genotyping studies (Marra et al., 2013; Trujillo et al., 2014; El Bakkali et al., 2019; Atrouz et al., 2021).
Each multiplex PCR reaction was carried out in a total volume of 8 μL, including 1.5 μL DNA (50 ng/μL), 2x Qiagen Multiplex PCR master mix buffer, and 0.2 μM primer mix (Caruso et al., 2014; Fiore et al., 2022), using the following thermal cycles: the first 10 cycles consisted of a denaturation step at 94°C for 15 min, followed by denaturation at 94°C for 30 s, annealing at 60°C for 1 min and 30 s, and a one-minute extension at 72°C, which was lowered by one-degree centigrade for each cycle; the second thermal regime, repeated for 25 cycles, consisted of a denaturation step at 94°C for 30 s, an annealing cycle at 50°C for 1 min and 30 s, and finally a one- minute extension at 72°C. Allele detection was performed using a 3500 XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) loaded with POP-7 polymer (Applied Biosystem). The GeneScan 500LIZ (Life Technologies, Carlsbad, CA, USA) was used as an internal standard for allele size detection using the software GeneMapper 4.0.
2.5 SSR statistic parameters
The software CERVUS program version 3.0.7 (Kalinowski et al., 2007) was employed to compute the genetic analysis of the microsatellite profiles. The number of alleles (No), the observed and expected heterozygosity (Ho, He) values, the polymorphic information content (PIC), the occurrence of null alleles (Fnull), and deviation from the Hardy–Weinberg equilibrium, corrected using the Bonferroni method, were calculated (Table 1).
2.6 Cluster analysis
The genetic distance and relationships among the genotypes, based on 11 microsatellite markers, were computed with SISTAT 10 software (SPSS, Inc., Chicago, IL, USA), using distance percent and the linkage average method, and a dendrogram was built with the UPGMA algorithm.
2.7 Structure analysis
In order to get a more complete framework of the olive relationships among Mediterranean countries, SSR data of these Sicilian monumental trees were compared with those of 64 local Sicilian accessions and 270 accessions from the MB and beyond (Tables S3, S4), using common SSR markers (in a range of 6 to 8 loci) found in the literature (Trujillo et al., 2014; El Bakkali et al., 2019; Atrouz et al., 2021) and included in the internal database of olive genetic resources of the Department SAAF, University of Palermo, Italy, and the Council for Agricultural Research and Economics, Research Centre for Olive, Fruit and Citrus Crops, (CREA-OFA), Italy. Before comparison, all SSR markers were subjected to a “binning” following the methodology described by Ben Mohamed et al. (2017). The analysis of the genetic structure populations was computed using a Bayesian-based clustering model implemented in STRUCTURE v.2.3.4 (Pritchard et al., 2000) using 8 SSR markers. A burn-in period of 100,000 generations and 100,000 Markov chain Monte Carlo replications was used under an admixture model and correlated allele frequencies. Three replicate simulations were run for each K value, ranging from 1 to 12. To select the optimal K value, the STRUCTURE HARVESTER v.0.9.94 was used while employing the Evanno et al. (2005) transformation method. To assign samples to a particular population, a membership coefficient q > 0.8 limits was used, while coefficients ≤ 0.8 were considered genetically admixed.
Although we did not examine the plastid (pt) DNA haplotypes of monumental trees in this study, we used our knowledge of ptDNA haplotypes of common authenticated varieties (data not shown) from the olive germplasm banks (Department SAAF, University of Palermo, Italy; CREA-OFA, Rende, Italy, Marrakech, Morocco, and Cordoba, Spain) to better understand the genetic relationships between these trees and the olive accessions from abroad. Information on published ptDNA haplotypes (Besnard et al., 2013b) is reported in Table S4.
2.8 Morphological traits
The morphological characterization was carried out in October and November of 2021 and 2022 following the indications contained in the “Manual for the primary characterization of the Olive Cultivars of the Sicilian Germplasm” (Caruso et al., 2007).
For each tree, 30 leaves, taken from the middle portion of the branches of the year inserted on the fruiting branches, and 30 drupes, collected from the intermediate sector of the fruiting branches, which were randomly selected in the median portion of the crown, were collected. A total of 13 traits — four for the leaves (shape, length, width, and area), five for the fruits (shape, length, width, weight, and flesh/pit ratio), and four for the endocarps (shape, length, width, and weight) — were measured.
For the evaluation of fruit and endocarp, the position used was that of the maximum asymmetry, in which the carpellar suture is visible to the observer.
As some of the trees presented branches with different leaves and fruits traits and thus resulted in being grafted with different genotypes, morphological characterization was carried out for the putative different genotypes grafted in the same tree.
All data were used for the realization of well-illustrated elaiographic cards with the indication of their position that could be included in naturalistic itinerary guides, of which some examples are provided in Supplementary Figures S1, S2.
2.9 Cluster analysis based on morphological traits.
Overall, 13 morphological traits were used for the cluster analysis, using the SYSTAT10 software (SPSS, Inc., Chicago, IL, USA) and employing the Euclidian distance and “single linkage” method. This software allowed the comparison between the monumental trees being analyzed and eight reference cultivars characterized in previous work and present in the reference database established and updated by the Department of Agriculture, Food and Forestry Science (SAAF), University of Palermo (Caruso et al., 2014) and sited at the collection field (P. Messina - Sciacca) partner of the SPREMO project.
3 Results
3.1 SSR polymorphism
A total of 147 alleles were found, with allelic richness ranging from 8 (EMO90 and GAPU71B) to 21 (UDO47) and with a mean of 13,4 alleles, revealing a high level of genetic diversity. The mean expected (He) heterozygosity was 0.81, and the polymorphic information content (PIC) value was 0.79, both indicating the high level of polymorphism of the chosen SSR markers. Seven loci (DCA03, DCA13, EMO90, GAPU71b, OLEST7, OLEST14, and OLEST23) significantly deviated from HW, with a probability of null alleles F (FNull) of 0.0012, 0.125, 0.0318, -0.0209, 0.0449, -0.068, and 0.0393, respectively (Table 1). The SSR profiles are depicted in Table S2.
3.2 Cluster analysis, cases of synonyms, and relationship
In the dendrogram, two main clusters were present, which were divided into additional sub-clusters (Figure 2). The first cluster included trees identical to the cv. Santagatese or somatic mutants.
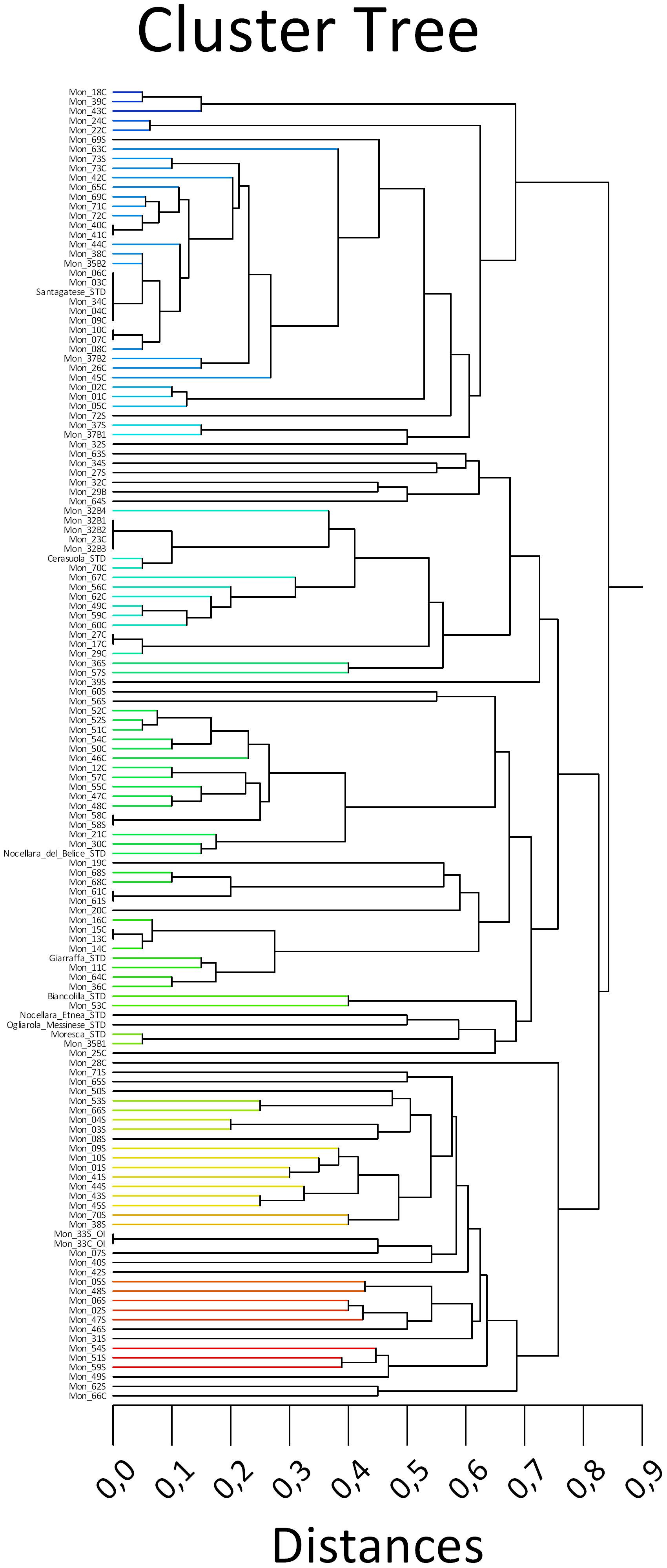
Figure 2 UPGMA dendrogram of 73 Sicilian monumental trees, including one Olea europaea var. sylvestris and eight reference cultivars, based on 11 SSR markers.
The second cluster presented two main sub-clusters. One of each sub-cluster was divided into six small sub-groups containing the eight reference cultivars and many centennial trees falling within them showing minimal mutations; therefore, they could represent clones of polyclonal cultivars present in certain territorial areas. The second sub-cluster grouped almost all the suckers and the wild monumental tree Oleaster Inveges (Mon_33) and two trees (Mon_28C and Mon_66C) with unique profiles.
The set of 11 SSR primers successfully discriminated most of the analyzed genotypes, except for the following cases of identity groups, as depicted in Figure 2: Mon_03C/Mon_04C/Mon_06C/MOn_09C/Mon_40C/Mon_41C, which showed an identical SSR profile with the Sicilian cultivar Santagatese. Mon_34C/Mon_35B2 and Mon_44C were undistinguishable and they grouped not far from the ‘Santagatese’ group as well as Mon_07C/Mon_10C, and Mon_08C; Mon_69C, Mon_71C and Mon72C. Mon_36C and Mon_11C can be considered somatic mutants of the Sicilian cultivar Giarraffa.
The Mon_70C tree had an identical result to the cultivar Cerasuola and also the two branches of the tree Mon32 (Mon_32B2, Mon32b3). Two other branches of Mon _32B1 and Mon_32b4, as well as Mon_23C, presented close genetic relationships with cv. Cerasuola, constituting probable somatic mutants.
The trees Mon17C, Mon _27C, and Mon_29C showed identical profiles, while Mon _49C and Mon _59C were undistinguishable, as well as Mon 10C and Mon 07C.
The molecular identity between the crown and sucker for the trees Mon_37, Mon_58, Mon_61, Mon_68, Mon_73, and Oleastro Inveges (Mon_33_OI) was evident.
Apart from the above-reported exceptions, it was clear that most of the monumental trees were grafted at least once as there was no correspondence between the molecular profiles of the two parts of the same tree: the crown and the sucker. Due to the large size of the trunk and the branches, the grafting may likely have been carried out in situ a long time ago.
When the comparison was performed among the accessions used in this study and the olive fingerprinting profiles present in the molecular databases of the CREA- OFA, Rende, Italy, and of the Department SAAF, University of Palermo, Italy, additional cases of identity and/or probable somatic mutants were discovered.
The monumental tree Mon_19C had an identical result to the Sicilian accession Demoniata (CREA-OFA, Rende collection); Mon_21C was also undistinguishable from the Tunisian accession Fakhari (Ben Mohamed et al., 2017), showing only two little somatic mutations; Mon_24C = Sic_052; Mon_28C was identical to the accession number Sic_048; the tree Mon_29C and the accession Zimbimbo (CREA-OFa) may be somatic mutants; Mon_11C and Mon_36C were identical to the Sicilian cultivar Giarraffa and to the two Algerian accessions Grosse du Hamma and Balbale Azzaba (Atrouz et al., 2021); Moresca from the CREA-OFA collection was identical to Mon_35B1; Mon_18C; Mon_39C and Mon_43C could not be discriminated from the Sicilian accession Aitana of the CREA-OFA collection, which, in turn, differed from the accession Aitana existing in the database of the Department SAAF, University of Palermo.
3.3 Morphological analysis
The morphological cluster tree (Figure 3) was not exhaustive and just in a few cases supported the molecular analysis, confirming the similarity among the trees genetically close to the cv. Santagatese and some trees genetically close to ‘Cerasuola’. At the morphological level, trees genetically identical to the cv. Santagatese and the possible somatic mutants shared a remarkable similarity regarding the silvery coloring of the lower leaf page and the high vegetative vigor; however, more data and observations are needed to confirm possible cases of identity and mutations to affirm that it could be a polyclonal cultivar. As concerns the inconsistencies between the morphological and molecular analyses, this was not surprising since the morphological traits rarely associate with the molecular ones if measured in different environmental conditions.
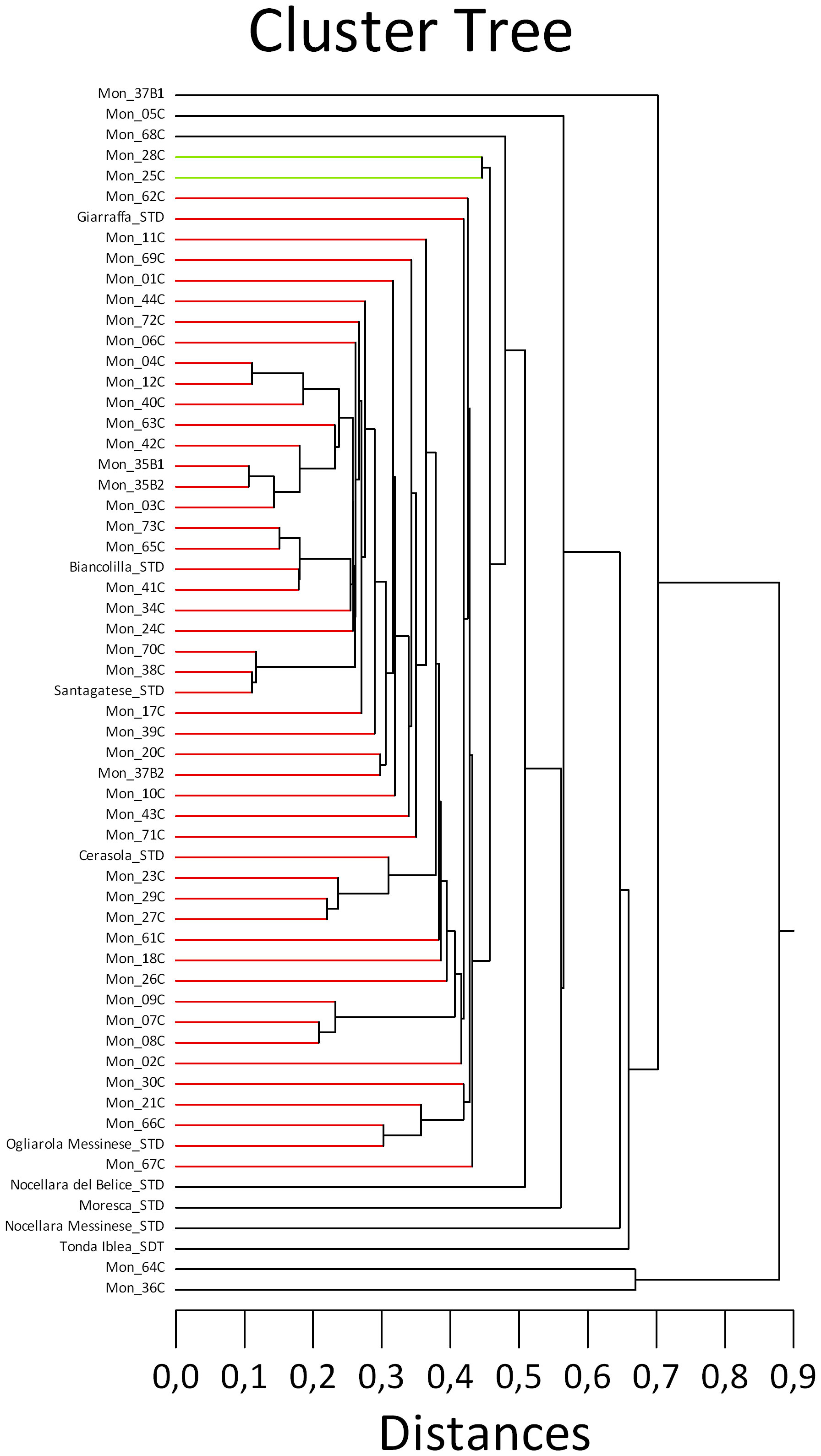
Figure 3 Cluster analysis using 13 morphological traits of the studied Sicilian monumental trees, including reference cultivars, based on the Euclidian distance and “single linkage” method. .
3.4 Genetic structure and relationship analysis
One of the main aims of this study was to assess the genetic structure of the Sicilian monumental trees by comparing their molecular profiles with those of reference Sicilian cultivars and local unknown accessions and 270 varieties from the Mediterranean Basin.
Three populations were identified using the ΔK method (Figure 4).
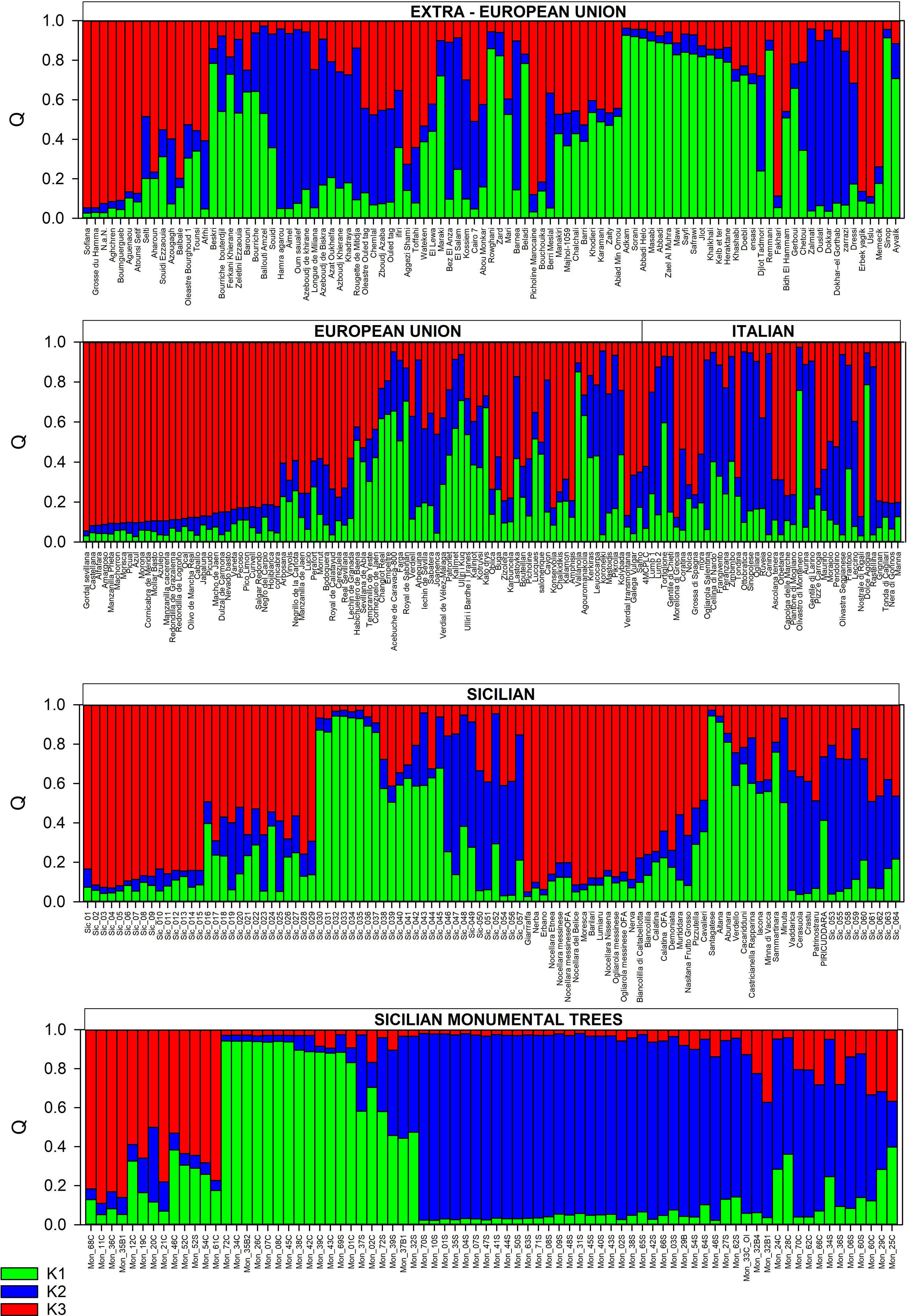
Figure 4 Structure analysis (K = 3) of the centennial Sicilian olive trees, 64 local Sicilian accessions, and 270 accessions from the Mediterranean Basin and beyond.
All the monumental accessions were distributed in all the differentiated populations and admixed groupings. The threshold value ≥ 0.80 was assigned for 12 monumental trees (Mon_01C, Mon_07C, Mon_08C, Mon_26C, Mon_34C, Mon_35C, Mon_38C, Mon_39C, Mon_42C, Mon_43C, Mon_45C, Mon_72C) and one rootstock (Mon_69S) to the K1 population (green). Other cultivars (Santagatese, Aitana, Abunara) and 8 local accessions were included in this group. Most of the varieties from eastern part of MB (14 from Syria, 1 from Turkey, 2 from Iran, 1 from Lebanon) were included in the K1 population, suggesting a clear ancestry of the monumental accessions from the Middle East, particularly from Syria.
With very few exceptions, all the samples (34) collected from the suckers and the 2 samples collected from the canopy of the Oleastro d’Inveges (Mon_33C_OI/Mon_33S_OI) were included in the K2 population (blue), which also included the Greek self-compatible cv. Koroneiki (Marchese et al., 2016a; Marchese et al., 2016b) and 6 Italian (Canino, Ogliarola Salentina, Aurina, Rastellina, Ottobratica, and Olivastra Seggianese), 4 Algerian (Hamra agarou, Aimel, Oum saualef, and Azheboudi de Khirane), and 4 Tunisian varieties (Zalmati, Dokhar el Ghortab, Ouslati, and Dokhar), all sharing the small drupe trait (Taamalli et al., 2006; Perri et al., 2015; Atrouz et al., 2021; Kafkaletou et al., 2021).
Four monumental trees (Mon_11C, Mon_36C, Mon_68C, and Mon_35B1) belonged to the K3 population (red), including most of the main known/economically important Sicilian cultivars of Giarraffa, Nocellara Etnea, Nocellara Messinese, Nocellara del Belice, Ogliarola Messinese, and Moresca, as well as minor and neglected cultivars (Lumiaru, Erbano, Nerba, and Barilari) and many local genotypes that are often present in farms as unique specimens (15). In this group were also included 5 Italian cultivars, most of the Spanish cultivars (32), 1 from Portugal, 1 from Greece, 7 from Algeria, 1 from Tunisia, and only 2 from Turkey. Three Sardinian cultivars (Tonda di Cagliari, Manna, and Nera di Gonnos) were found in this group, but these varieties seem to show a high genetic similarity to the Sicilian cv. Nocellara Messinese (Belaj et al., 2022).
As expected, a very large set of accessions showed an admixed genotype (Figure 3), and most of these genotypes (82) were included in a grouping sharing more alleles with the K3 population (Figure 3; Tables S3, S4). Many of Sicilian genotypes (13 monumental accessions: 12 samples from canopy and 1 from rootstocks; 7 known cultivars and 14 local accessions) were included in this group, as were a few varieties from the central–eastern MB (1 from Cyprus, 2 from Greece, 5 from the Balkan Peninsula, and 1 from Turkey) and several cultivars from the central–western MB (8 from Algeria, 2 from Egypt, 2 from France, 12 from Spain, and 1 from Portugal).
Another large set of monumental accessions (11), 7 samples from the canopy and 4 from the rootstock, and 3 known Sicilian cultivars (Cerasuola, Vaddarica, and Crastu) showing a membership value near 0,8 for the K2 population fell in the admixture grouping together with 15 other local accessions. Admixed clones of known cultivars also fell in this grouping, such as 11 and 2 Italian varieties and local accessions (from Lombardy), respectively, 1 from Syria, 1 from Israel, 2 from Greece, 4 from Algeria, 3 from Egypt, 4 from Tunisia, 1 from Morocco, 1 from France, and 2 from Spain. Admixed genotypes sharing most of alleles with the K1 population included 4 monumental accessions (3 rootstocks and 1 from canopy), 6 minor Sicilian accessions (Verdello, Cacaridduni, Castricianella rapparina, Iacona, Minna di Vacca, and Sammartinara), 8 local genotypes, 3 cultivars from Syria, 1 from Lebanon, 1 from Turkey, 2 from Greece, 3 from the Balkan Peninsula, 3 from Italy, 6 from Algeria, 1 from Tunisia, 1 from Egypt, and 5 from Spain. These results confirmed that an extended diversification process occurred around the entire MB for the olive species (Khadari et al., 2019).
4 Discussion
The importance of monumental olive trees as valuable genetic resources of resilience and hardiness traits has only been appreciated in the last decade, and an extensive survey of centenarian olive trees has been initiated throughout the Mediterranean Basin in Italy (Salimonti et al., 2013), Spain (Díez et al., 2011; Ninot et al., 2018), Cyprus (Anestiadou et al., 2017), Israel/Palestine (Barazani et al., 2014; Petruccelli et al., 2014), and Lebanon (Chalak et al., 2015).
Sicily, as a crossroads of Mediterranean populations, has a very significant olive heritage (Caruso et al., 2007; Marra et al., 2013; Caruso et al., 2014) and is rich in monumental olive trees, which are worth being deeply studied since they have resisted through the centuries, presumably overcoming biotic and abiotic adversities. The area under study falls within the specification of three of the six EVO oil PDO areas of the island (Valli Trapanesi, Valle del Belice, and Val di Mazara) and two famous archaeological areas (the Valley of the Temples Archaeological Park and Kolymbethra Garden), located in Agrigento, and shows a great wealth of monumental trees, which were inventoried and characterized for the first time in this current work.
The SSR fingerprints revealed a wide genetic variability of the monumental trees. Overall, the average number of alleles (Na =13.4), PIC value (0.79), observed heterozygosity (Ho =0.76), and expected heterozygosity (He = 0.81) were higher compared to the values reported in similar studies. For instance, a survey of 190 monumental trees from Malta investigated with 10 SSRs revealed 11.6 alleles per locus He = 0.72, Ho = 0.76, and PIC = 0.69 (Valeri et al., 2022). The study of 52 centennial olives from Cyprus showed 7.79 alleles per locus and a PIC value of 0.499. The survey of 4500 ancient Spanish trees showed 9.1 alleles per locus, He 0.545, and PIC = 0.46 (Ninot et al., 2018). Meanwhile, the analysis of 112 cultivated and wild Greek and Turkish olive accessions showed higher values of 19 alleles per locus, He = 0.81, Ho = 0.99, and PIC = 0.88 (Aksehirli-Pakyurek et al., 2017). Differences in the SSR variability parameters of Sicilian trees further highlight their diversity and the potential importance of this gene pool.
The SSR profiles showed that almost all the trees were grafted at least once, as evidenced by the different molecular profiles between the crown and suckers originating from the rootstock. Our findings are in agreement with similar studies conducted to explore the genetic diversity of ancient olive trees in other Italian regions (Rotondi et al., 2018) and other countries, such as Cyprus (Anestiadou et al., 2017), Malta (Valeri et al., 2022), Spain (Díez et al., 2011; Ninot et al., 2018), Israel/Palestine (Barazani et al., 2014), and Turkey (Yoruk and Taskin, 2014; Aksehirli-Pakyurek et al., 2017). Only in a few cases did the two parts of the tree (crown and rootstock) share identical profiles (Mon_37C/S, Mon_61C/S, Mon_68C/S, Mon_73C/S, Oleastro Inveges— Mon_33_OIC/S).
In some cases, the monumental trees corresponded to well-known cultivars: Mon_03C/Mon_04C/Mon_06C, found in the two archaeological areas of Agrigento (Italy), and Mon_40C/Mon_41C/Mon_69C/Mon_72C/Mon_73C = ‘Santagatese’, Mon_36C/Mon_11C = ‘Giarraffa’, and Mon_70C/Mon_32B2/Mon_32B3 = ‘Cerasuola’.
The comparison with the SSR profiles of olive from Mediterranean countries enabled the identification of additional cases of identities. The tree Mon_19C could not be distinguished from ‘Demoniata’ (CREA-OFA- Rende), and Mon_21C can be considered a somatic mutant of Tunisian accession ‘Fakhari’ (Ben Mohamed et al., 2017). Moreover, Mon_24C = Sic_052, Mon_28C is identical to Sic_048, and the tree Mon_29C and the accession ‘Zimbimbo’ from the CREA-OFA collection may be somatic mutants, while Mon_36C/Mon_11C were identical to the Sicilian cultivar Giarraffa and to the two Algerian accessions ‘Grosse du Hamma’ and ‘Balbale Azzaba’ (Atrouz et al., 2021).
‘Moresca’ from the CREA-OFA collection was identical to Mon_35B1, and Mon_39C and Mon_43C could not be discriminated from the Sicilian accession ‘Aitana’ of the CREA-OFA collection. However, this last one differed from the accession ‘Aitana’, present in the database of the Department SAAF, University of Palermo.
At the morphological level, the degree of similarity was very high, but more data and observations are needed to confirm possible cases of identity and somatic variants. The visual comparison of the morphological characters, with reference to the catalogue of Sicilian olive cultivars (Caruso et al., 2005), confirmed the remarkable similarity of some genotypes attributable to the cultivar Santagatese, which would therefore be a polyclonal cultivar. The presence of monumental trees grafted with the cv. Santagatese in this part of the island (Agrigento and Trapani) is surprising since this cultivar is very widespread in the north-eastern part of Sicily, especially in the province of Messina in the Nebrodi mountains, areas where the cultivar likely originated. It is well known that the cv. Santagatese is a high- vigor tree, and because of that it is often used as a windbreak or as an ornamental plant due to the silvery coloring of its lower leaf page. Not far from the archaeological areas of Agrigento and also in the Trapani areas, there are settlements and monasteries of Benedictine monks, famous as olive growers and for producing olive oil for religious ceremonies (Caracuta, 2020). Thus, it is likely that from the 12th century they planted very extensive olive groves by introducing grafting material from the northern part of Sicily.
Ten monumental trees (Mon_22C, Mon_24C, Mon_25C, Mon_28C, Mon_46C, Mon_55C, Mon_59C, Mon_63C, Mon66_C, and Mon_68C)showed a quite unique genetic profile, which can be further studied and exploited as a resource of genes.
The cultivated olive diversification process and the elucidation of their origins represent a priority to ensure the efficient management of local olive genetic resources (Khadari et al., 2019). In this work, to get a more complete outline of olive genetic relationships and the diversification process in the Mediterranean Basin (MB), molecular data results from Sicilian monumental samples were compared with 334 molecular profiles, including local Sicilian accessions and cultivars from across the MB (Table S3; S4). Diez et al. (2015) evaluated the ancestry of a large set of cultivated and wild olives from across the MB by analyzing their population genetic structure, substantially highlighting three populations (Q1, Q2, and Q3) and a wide grouping of mosaic genotypes. These three main genetic pools pair the geographical areas of west (namely Q1), center (Q2), and east (Q3) of the Mediterranean Basin (Diez et al., 2015), where olive cultivation developed around 5,000 years ago (e. g. Besnard et al. 2013a).
A similar approach was used by Atrouz et al. (2021), who analyzed a large set of cultivated olive trees from the central–western MB. Similar results were outlined, showing a wide grouping of cultivars from this region assigned to the Q2 population and most of the Spanish varieties assigned to the Q1 population. These results reflect the previously described framework of the genetic pattern of diversification of cultivated olive in the Mediterranean Basin and the Middle East (Baldoni et al., 2006; Díez et al., 2011; Besnard et al., 2013a; Besnard et al., 2013b; di Rienzo et al., 2018; Belaj et al., 2022).
Our structure analysis more closely reflected the pattern of differentiation found by Diez et al. (2015), as monumental accessions and varieties from the Mediterranean countries and beyond were assigned to three populations (K1, K2, and K3), and a large set of cultivars showed an admixed grouping. A high number of well-known and economically important Sicilian varieties (Giarraffa, Moresca, Nocellara Etnea, Nocellara Messinese, and Nocellara del Belice, as well as Barilari and Erbano) were included in the K3 population (cluster red). Most of the Spanish varieties fell in the K3 population (Figure 4; Table S4), previously included in the Q1 group by Atrouz et al. (2021) and Diez et al. (2015). Diez et al. (2015), exploring the plastid DNA diversity, found that there is a strict genetic relationship between the Q1 population and oleasters from the eastern Mediterranean Basin, presenting the ptDNA haplotype E1.1 (bbb), largely spread around all the Mediterranean countries. We did not determine the plastid DNA lineages of monumental accessions in this study, but several common authenticated varieties with known ptDNA haplotypes from the olive germplasm banks (Department SAAF, University of Palermo, Italy; CREA-OFA, Rende, Italy; Marrakech, Morocco; and Cordoba, Spain) were included in the dataset (Table S4). Almost all the cultivars with a known ptDNA haplotype belonging to K3 population showed the E1.1 lineage, except the Sicilian varieties Giarraffa and Moresca, showing the variant E1.2. This single variant (E1.2) is a minor variant of E1.1, separated by one or two mutation events (Diez et al., 2015). In this study, two monumental trees were found to genetically correspond to the cv. Giarraffa. The cvs. Giarraffa and Moresca were reported as putative parentals of many local Sicilian varieties accessions in a few offspring combinations (Atrouz et al., 2021). Besnard et al. (2013b) observed the co-occurrence of plastid DNA haplotypes E1.1, E1.2, and E1.3 in populations close to the Syrian–Turkish border. All these observations suggest the same origin from the Middle East and eastern Mediterranean of the monumental trees and cultivars belonging to the same K3 population. It is known that olive cultivation was significantly developed in the eastern Mediterranean at least 6000 years ago, spreading toward Anatolia and the island of Cyprus and from the island of Crete toward Egypt (Langgut et al., 2019).
The common accessions analyzed here led us to speculate a possible correspondence of the K2 population to the Q2 group, as reported by Diez et al. (2015) to substantially include cultivars from the western and central MB, representing a product of historical olive–oleaster admixture that seemed to have diversified earlier than the Q1 population and originated probably from Q3 and wild west oleasters (WW). Most of rootstocks were included in the K2 population clustering with the wild olive form ‘Oleastro d’Inveges’ but surprisingly not with the Sicilian varieties and/or accessions. However, a few known Sicilian varieties (Vaddarica, Crastu, and Cerasuola) and some monumental accessions such as Mon_28C, which genetically corresponds to the accession Sic_048, were found in the group of admixed genotypes, with the highest allelic frequency attributable to the K2 population. It was evident that the rootstocks showed considerable diversity. Grafting is a very ancient practice dating back more than 2000 years ago, as reported by Theophrastus (Bombarely et al., 2021), and considering the plants grafted are very ancient and the overlapping of geographic sites of the diffusion of wild and cultivated olives, it could be speculated that the rootstocks analyzed here belong to the wild var. sylvestris. Similar results were observed for the cv. Cerasuola: some monumental accessions were identical or very similar to this variety, once again indicating its genetic authenticity and probable ancient origin. However, it is difficult to understand whether the origin of this variety is attributable to a local domestication process or a migration process via vegetative propagation, considering the genetic correspondence between the cv. Cerasuola and the Tunisian cv. Zarrazi Zarzis (Belaj et al., 2022).
Diez et al. (2015) found in the Q2 group substantial cultivars from the western and central MB. For instance, other cultivars from abroad included in the K2 population were the Italian Canino, Olivastra seggianese, Ottobratica, Ogliarola Salentina, Rastellina, and Aurina; the Greek Koroneiki; the Tunisian Zalmati, Oueslati, Dokhar, and Dokhar–el Gorthab; and the Algerian Hamra agarou, Aimel, Oum saualef, and Azeboudj de khirane. All these cultivars show a small fruit size, commonly considered a “wild” morphological trait. Our results seem to corroborate and further highlight the possibility of introgression between cultivated and wild olive trees as well. It has been proposed that allele introgressions from wild olive genotypes, followed by human selection, may have improved the adaptation traits of olive cultivars to local environments (Julca et al., 2020).
Almost all the common varieties from the east (Syria), analyzed by Diez et al. (2015), belonging to the Q3 group fell in our population K1 and in an admixed group (Table S4). All the known chlorotypes of varieties from the K1 population belonged to E1.1 type, indicating a clear ancestry from the east (Syria), presumably one of the most ancient. Recent sequencing of cultivated and wild olive nuclear genomes confirmed the hypothesis of a close relationship between cultivars of distant locations (southern Spain) and cultivars from the Levant (Syria, Iran, Lebanon, and Turkey) originating from a common primary domestication process (Julca et al., 2020). The Sicilian cv. Santagatese, in addition to many monumental trees with high genetic and morphological similarity to it, fell in the K1 population. Atrouz et al. (2021) found that the Spanish cv. Cirujal, belonging to the Q3 group of Diez et al. (2015), shows a high level of similarity with the Sicilian cultivar Santagatese.
As expected, the widest group discovered here included mosaic genotypes (242), most of them mainly showing the allele pattern of the K3 (82) and K2 (61) populations. The Sicilian germplasm was found equally distributed among these groupings.
The results of this research underline Sicily as a Mediterranean hot spot for biodiversity conservation, considering its rich olive germplasm that shows different ancestries. A large part of the Sicilian germplasm probably originated from human migration processes or commercial trades; however, wild forms of olive “oleaster” are native to the island and may have originated from seeds spread by animals. These wild forms almost disappeared because they were used as a rootstock of native and better performing cultivars selected by growers. The large size of the monumental trees indicates the very ancient origins of olive growing in Sicily. Many of the cultivars currently grown on the island descend from monumental trees, and some of them found in this study, called “Patriarch”, are the original mother plants of the Sicilian cultivars. This means that olive oil produced in Sicily from these cultivars is exclusive to the island and can be surely recognized as a Protected Origin Olive (PDO).
5 Conclusion
The valorization of historic agricultural landscapes and the protection of germplasm resources from genetic erosion is considered a priority by the international community. The Mediterranean Basin is very rich in monumental olive trees. In some cases they are present within important archaeological sites, while others are scattered along rural areas, placing them at risk of erosion and extinction and under threat by human activities. These long-lived trees may represent a source of genes of resistance to biotic agents or even to adverse climatic conditions that have occurred during their long life. This gene pool has a very important value and can be exploited by genetic improvement programs to produce new genotypes suitable for new conditions and emergent diseases such as Xylella fastidiosa subsp. pauca (ST53) and again to obtain increasingly sustainable productions. Given that the value of centuries-old trees is intrinsic to the territory in which they are located, the protection of centuries-old trees is fundamental. The Italian state delegates to the individual regions the protection and selection of protected areas, including natural monuments. There is still no national law on the safeguarding, enhancement, and protection of territories that are characterized by the presence of monumental olive trees. Although at the regional level there is a project supporting the “Guardian Farmers”, who care for and protect olive genetic resources in situ, there is a lack of more incisive actions such as the drawing up of a map of the sites where these ancient trees are located and the organization of itineraries aimed at visiting these old “Patriarchs”. The Department SAAF, University of Palermo, has recently started the vegetative propagation by the cutting, rooting, and grafting of these genotypes for future agronomical characterization in addition to the realization of well-illustrated elaiographic cards with the indication of their position that could be included in naturalistic itinerary guides (prototype examples are available for consultation in Figures S1, S2). Moreover, some genotypes have been chosen and included in genome resequencing projects to discover traits of resilience to biotic and abiotic stresses.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
CT, MA, and MP conceived the study and were in charge of overall planning; BF, IV, MA, RS, TA, and ZS performed the molecular experiment and analyzed the data; IA, SG, GA, TA, and GI conducted the morphological analysis; MA, MP, and ZS performed the statistical analyses; MA took the lead in writing the manuscript in consultation with CT and ZS; CT, MP, GA and MA oversaw the funding acquisition; all authors have reviewed, read, and agreed to the published version of the manuscript.
Funding
The “Fondo Finalizzato alla Ricerca di Ateneo” (FFR2021), University of Palermo, Italy, was employed by CT, MA, and MF to conduct this research. ‘PSR SICILIA 2014-2020—Programma di Sviluppo Rurale— Misura 16—Cooperazione—Sottomisura 16.1—“Sostegno per la costituzione e la gestione dei gruppi operativi del PEI in materia di produttività e sostenibilità dell’agricoltura”—Gruppo Operativo: ATS ProOlivo—Project title: “Applicazione di tecnologie “smart” per il monitoraggio, prevenzione e diagnosi precoce delle malattie di interesse economico dell’olivo’ - SPREMO - “Assessorato Agricoltura - Regione Sicilia”.
Acknowledgments
The authors are grateful to the students Adamo Giacomo, Cacioppo Francesco, Catanzaro Giuseppe, Festosi Salvatore, Sorrentino Vincenzo, and Vassallo Marco for helping with the sample collection and GPS localization of trees. Authors thank ‘PSR SICILIA 2014-2020 - Project SPREMO.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2023.1206832/full#supplementary-material
References
Aksehirli-Pakyurek M., Koubouris G. C., Petrakis P. V., Hepaksoy S., Metzidakis I. T., Yalcinkaya E., et al. (2017). Cultivated and wild olives in Crete, Greece-genetic diversity and relationships with major Turkish cultivars revealed by SSR markers. Plant Mol. Biol. Rep. 35, 575–585. doi: 10.1007/s11105-017-1046-y
Alba V., Sabetta W., Blanco A., Pasqualone A., Montemurro C. (2009). Microsatellite markers to identify specific alleles in DNA extracted from monovarietal virgin olive oils. Eur. Food Res. Technol. 229, 375–382. doi: 10.1007/s00217-009-1062-8
Anestiadou K., Nikoloudakis N., Hagidimitriou M., Katsiotis A. (2017). Monumental olive trees of Cyprus contributed to the establishment of the contemporary olive germplasm. PloS One 12 (11), e0187697. doi: 10.1371/journal.pone.0187697
Angiolillo A., Mencuccini M., Baldoni L. (1999). Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor. Appl. Genet. 98, 411–421. doi: 10.1007/s001220051087
Atrouz K., Bousba R., Marra F. P., Marchese A., Conforti F. L., Perrone B., et al. (2021). Algerian Olive germplasm and its relationships with the central-Western Mediterranean varieties contributes to clarify cultivated olive diversification. Plants 10 (4), 678. doi: 10.3390/plants10040678
Baldoni L., Pellegrini M., Mencuccini M., Angiolillo A., Mulas M. (2000). Genetic relationships among cultivated and wild olives revealed by AFLP markers”. Acta Hortic. 521, 275–284. doi: 10.17660/ActaHortic.2000.521.30
Baldoni L., Tosti N., Ricciolini C., Belaj A., Arcioni S., Pannelli G., et al. (2006). Genetic structure of wild and cultivated olives in the central Mediterranean basin. Ann. Bot. 98, 935–942. doi: 10.1093/aob/mcl178
Barazani O., Westberg E., Hanin N., Dag A., Kerem Z., Tugendhaft Y., et al. (2014). A comparative analysis of genetic variation in rootstocks and scions of old olive trees–a window into the history of olive cultivation practices and past genetic variation. BMC Plant Biol. 14, 1–8. doi: 10.1186/1471-2229-14-146
Barone E., La Mantia M., Marchese A., Marra F. P. (2014). Improvement in yield and fruit size and quality of the main Italian table olive cultivar 'Nocellara del belice'. Sci. Agric. 71, 2–57. doi: 10.1590/S0103-90162014000100007
Bartolini G., Cerreti S., Bati C. B., Stefani F., Zelasco S., et al. (2014). "Oleadb: Worldwide database for the management of genetic resources of olive (Olea europaea L.)". In Proceedings of the X National Congress on Biodiversity, Rome,Italy, pp. 18-23.
Bartolini G., Prevost G., Messeri C., Carignani G. (1998). Olive germplasm: cultivars and world-wide collections (Rome, Italy: FAO).
Belaj A., Muñoz-Diez C., Baldoni L., Porceddu A., Barranco D., Satovic Z. (2007). Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Ann. Bot-London 100 (3), 449–458. doi: 10.1093/aob/mcm132
Belaj A., Muñoz-Diez C., Baldoni L., Satovic Z., Barranco D. (2010). Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Sci. Hortic. 124, 323–330. doi: 10.1016/j.scienta.2010.01.010
Belaj A., Ninot A., Gómez-Gálvez F. J., El Riachy M., Gurbuz-Veral M., Torres M., et al. (2022). Utility of EST-SNP markers for improving management and use of olive genetic resources: a case study at the worldwide olive germplasm bank of córdoba. Plants 921, 1–22. doi: 10.3390/plants11070921
Ben Mohamed B., Zelasco S., Ben Ali S., Guasmi F., Triki T., Conforti F. L., et al. (2017). Exploring olive trees genetic variability in the southeast of Tunisia. Genet. Mol. Res. 16, 4. doi: 10.4238/gmr16039850
Besnard G., El Bakkali A., Haouane H., Baali-Cherif D., Moukhli A., Khadari B. (2013b). Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generations of admixture. Ann. Bot. 112, 1293–1302. doi: 10.1093/aob/mct196
Besnard G., Khadari B., Navascueí s M., Fernaíndez-Mazuecos M., El Bakkali A., Arrigo N., et al. (2013a). The complex history of the olive tree: from late quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proc. R. Soc. London. Ser. B. Biol. Sci. 1756, 20122833. doi: 10.1098/rspb.2012.2833
Bombarely A., Doulis A. G., Lambrou K. K., Zioutis C., Margaritis E., Koubouris G. (2021). Elucidation of the origin of the monumental olive tree of vouves in Crete, Greece. Plants 10, 2374. doi: 10.3390/plants10112374
Caracuta V. (2020). Olive growing in puglia (South-eastern italy): a review of the evidence from the Mesolithic to the middle ages. Veg. Hist. Archaeobot. 29 (5), 595–620. doi: 10.1007/s00334-019-00765-y
Carriero F., Fontanazza G., Cellini F., Giorio G. (2002). Identification of simple sequence repeats (SSRs) in olive (Olea europaea l.). Theor. Appl. Genet. 104, 301–307. doi: 10.1007/s001220100691
Caruso T., Campisi G., Motisi A., La Mantia M., Occorso G., Testolin R. (2005). “Morphological, phenological and molecular characterization of the Sicilian indigenous olive (Olea europaea sativa l.) germplasm,” in 5th International Symposium on Olive Growing, Izmir, Turkey. 406.
Caruso T., Cartabellotta D., Motisi A., Campisi G., Occorso G., Bivona G., et al. (2007). Cultivar di olivo siciliane. identificazione validazione, caratterizzazione morfologica e molecolare e qualità degli oli. contiene manuale per la caratterizzazione primaria di cultivar di olivo siciliane. 1st ed (Palermo, Italy: Regione Siciliana, Assessorato Agricolturae Foreste), 1–202.
Caruso T., D’anna F., Campisi G., Romano C., Macaluso L., Di Benedetto G., et al. (2015). Catalogo accessioni di olivo, pesco, fragolina di bosco (Caltanissetta, Italy: Fondo Europeo agricolo per lo sviluppo rurale: l’Europa investe nelle zone rurali).
Caruso T., Marra F. P., Costa F., Campisi G., Macaluso L., Marchese A. (2014). Genetic diversity and clonal variation within the main Sicilian olive cultivars based on morphological traits and microsatellite markers. Sci. Hortic. 180, 130–138. doi: 10.1016/j.scienta.2014.10.019
Chalak L., Haouane H., Essalouh L., Santoni S., Besnard G., Khadari B. (2015). Extent of the genetic diversity in Lebanese olive (Olea europaea l.) trees: a mixture of an ancient germplasm with recently introduced varieties. Genet. Resour. Crop Ev. 62, 621–633. doi: 10.1007/s10722-014-0187-1
Cimò G., Marchese A., Germanà M. A. (2017). Microspore embryogenesis induced through in vitro anther culture of almond (Prunus dulcis mill.). Plant Cell Tiss. Org. 128, 85–95. doi: 10.1007/s11240-016-1086-2
Cipriani G., Marrazzo M. T., Marconi R., Cimato A., Testolin R. (2002). Microsatellite markers isolated in olive (Olea europaea l.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor. Appl. Genet. 104, 223–228. doi: 10.1007/s001220100685
Dangl G. S., Yang J., Golino D. A., Gradziel T. (2009). A practical method for almond cultivar identification and parental analysis using simple sequence repeat markers. Euphytica 168, 41–48. doi: 10.1007/s10681-008-9877-0
De la Rosa R., Angiolillo A., Guerrero C., Pellegrini M., Rallo L., Besnard G., et al. (2003). A first linkage map of olive (Olea europaea l.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theor. Appl. Genet. 106 (7), 1273–1282. doi: 10.1007/s00122-002-1189-5
De La Rosa R., James C. M., Tobutt K. R. (2002). Isolation and characterization of polymorphic microsatellites in olive (Olea europaea l.) and their transferability to other genera in the oleaceae. Mol. Ecol. Notes 2, 265–267. doi: 10.1046/j.1471-8286.2002.00217.x
De la Rosa R., James C. M., Tobutt K. R. (2004). Using microsatellites for paternity testing in olive progenies. HortScience 39 (2), 351–354. doi: 10.21273/HORTSCI.39.2.351
Denancé C., Muranty H., Durel C. E. (2020). “MUNQ - malus UNiQue genotype code for grouping apple accessions corresponding to a unique genotypic profile,” (France: Portail Data INRAE). doi: 10.15454/HKGMAS
Díez C. M., Trujillo I., Barrio E., Belaj A., Barranco D., Rallo L. (2011). Centennial olive trees as a reservoir of genetic diversity. Ann. Bot-London 108 (5), 797–807. doi: 10.1093/aob/mcr194
Diez C. M., Trujillo I., Martinez-Urdiroz N., Barranco D., Rallo L., Marfil P., et al. (2015). Olive domestication and diversification in the Mediterranean basin. New Phytol. 206, 436–447. doi: 10.1111/nph.13181
di Rienzo V., Sion S., Taranto F., D’Agostino N., Montemurro C., Fanelli V., et al. (2018). Genetic flow among olive populations within the Mediterranean basin. PeerJ 6, e5260. doi: 10.7717/peerj.5260
Doveri S., Gil F. S., Díaz A., Reale S., Busconi M., da Câmara Machado A., et al. (2008). Standardization of a set of microsatellite markers for use in cultivar identification studies in olive (Olea europaea l.). Sci. Hortic. 116 (4), 367–373. doi: 10.1016/j.scienta.2008.02.005
Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
El Bakkali A., Essalouh L., Tollon C., Rivallan R., Mournet P., Moukhli A., et al. (2019). Characterization of worldwide olive germplasm banks of marrakech (Morocco) and Cordoba (Spain): towards management and use of olive germplasm in breeding programs. PloS One 14, 10. doi: 10.1371/journal.pone.0223716
Erre P., Chessa I., Muñoz-Diez C., Belaj A., Rallo L., Trujillo I. (2010). Genetic diversity and relationships between wild and cultivated olives (Olea europaea l.) in Sardinia as assessed by SSR markers. Genet. Resour. Crop Ev. 57, 41–54. doi: 10.1007/s10722-009-9449-8
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 (8), 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
FAOSTAT (2023). Crops and Livestock Products Rome. Available at: https://www.fao.org/faostat/en/#data/QCL (accessed March, 2023).
Fiore M. C., Marchese A., Mauceri A., Digangi I., Scialabba A. (2022). Diversity assessment and DNA-based fingerprinting of Sicilian hazelnut (Corylus avellana l.) germplasm. Plants 11 (5), 631. doi: 10.3390/plants11050631
Fiorino P., Lombardo N. (2002). “Germoplasma, materiale vivaistico e certificazione,” in Atti Convegno Internazionale Olivicoltura, Spoleto, Italy. 82–90
Giovino A., Guarino C., Marchese A., Sciarillo R., Domina G., Tolone M., et al. (2023). Genetic variability of Chamaerops humilis (Arecaceae) throughout its native range highlights two species movement pathways from its area of origin. Bot. J. Linn. Soc 201, 361–376. doi: 10.1093/botlinnean/boac053
Giovino A., Marchese A., Domina G. (2020). Morphological and genetic variation of Chamaerops humilis (Arecaceae) in relation to the altitude. Caryologia 73 (4), 85–98. doi: 10.13128/caryologia-1011
Haouane H., El Bakkali A., Moukhli A., Tollon C., Santoni S., Oukabli A., et al. (2011). Genetic structure and core collection of the world olive germplasm bank of marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica 139, 1083–1094. doi: 10.1007/s10709-011-9608-7
ISTAT (2022). Italian National Institute of Statistics Cultivations. Available at: http://dati.istat.it/Index.aspx?DataSetCode=DCSP_COLTIVAZIONI# (accessed March, 2023).
Julca I., Marcet-Houben M., Cruz F., Gómez-Garrido J., Gaut B. S., Díez C. M., et al. (2020). Genomic evidence for recurrent genetic admixture during the domestication of Mediterranean olive trees (Olea europaea l.). BMC Biol. 18 (1), 1–25. doi: 10.1186/s12915-020-00881-6
Kafkaletou M., Ouzounidou G., Tsantili E. (2021). Fruit ripening, antioxidants and oil composition in koroneiki olives (Olea europaea l.) at different maturity indices. Agronomy 11 (1), 122. doi: 10.3390/agronomy11010122
Kalinowski S. T., Taper M. L., Marshall T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16 (5), 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Khadari B., El Bakkali A., Essalouh L., Tollon C., Pinatel C., Besnard G. (2019). Cultivated olive diversification at local and regional scales: evidence from the genetic characterization of French genetic resources. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01593
La Mantia M., Lain T., Caruso T., Testolin R. (2005). SSR-based DNA fingerprints reveal the genetic diversity of Sicilian olive (Olea europaea l.) germplasm. J. Hortic. Sci. Biotech. 80 (5), 628–632. doi: 10.1080/14620316.2005.11511989
Langgut D., Cheddadi R., Carrión J. S., Cavanagh M., Colombaroli D., Eastwood W. J., et al. (2019). The origin and spread of olive cultivation in the Mediterranean basin: the fossil pollen evidence. Holocene 29 (5), 902–922. doi: 10.1177/0959683619826654
Lavee S. (1992). “Evolution of cultivation techniques in olive growing,” in Olive oil quality, accademia di georgofili (Firenze: Editrice Polistampa), 37–44.
Lombardo L., Fila G., Lombardo N., Epifani C., Duffy D. H. III, Godino G., et al. (2019). Uncovering olive biodiversity through analysis of floral and fruiting biology and assessment of genetic diversity of 120 Italian cultivars with minor or marginal diffusion. Biology 62, 1–26. doi: 10.3390/biology8030062
Marchese A., Marra F. P., Caruso T., Mhelembe K., Costa F., Fretto S., et al. (2016b). The first high-density sequence characterized SNP-based linkage map of olive (Olea europaea l. subsp. europaea) developed using genotyping by sequencing. Aust. J. Crop Sci. 10 (6), 857–863. doi: 10.3316/INFORMIT.323243229612850
Marchese A., Marra F. P., Costa F., Quartararo A., Fretto S., Caruso T. (2016a). An investigation of the self-and inter-incompatibility of the olive cultivars' arbequina' and 'Koroneiki' in the Mediterranean climate of Sicily. Aust. J. Crop Sci. 10 (1):88-93. doi: 10.3316/informit.936851291841108
Marchese A., Tobutt K. R., Caruso T. (2005). Molecular characterisation of Sicilian Prunus persica cultivars using microsatellites. J. Hortic. Sci. Biotech. 80 (1), 121–129. doi: 10.1080/14620316.2005.11511902
Marino G., Macaluso L., Marra F. P., Ferguson L., Marchese A., Campisi G., et al. (2017). Horticultural performance of 23 Sicilian olive genotypes in hedgerow systems: vegetative growth, productive potential and oil quality. Sci. Hortic. 217, 217–225. doi: 10.1016/j.scienta.2017.01.046
Mariotti R., Cultrera N. G. M., Mousavi S., Baglivo F., Rossi M., Albertini E., et al. (2016). Development, evaluation, and validation of new EST-SSR markers in olive (Olea europaea l.). Tree Genet. Genomes 12, 120. doi: 10.1007/s11295-016-1077-9
Marra F. P. (2009). “Ambiente di coltivazione”. l’ulivo e l’olio (Angelini R. Bayer Crop Science. Script. Coltura e cultura). (Italy: Città di Castello (PG))
Marra F. P., Caruso T., Costa F., Di Vaio C., Mafrica R., Marchese A. (2013). Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea l. subsp. europaea) cultivated in southern Italy revealed by SSR markers. Tree Genet. Genomes 9 (4), 961–973. doi: 10.1007/s11295-013-0609-9
Marra F. P., Marchese A., Campisi G., Guzzetta G., Caruso T., Mafrica R., et al. (2014). Intra-cultivar diversity in southern Italy olive cultivars depicted by morphological traits and SSR markers. Acta Hortic. 1057, 571–576. doi: 10.17660/ActaHortic.2014.1057.73
Martins-Lopes P., Gomes S., Santos E., Guedes-Pinto H. (2008). DNA Markers for Portuguese olive oil fingerprinting. J. Agr. Food Chem. 56 (24), 11786–11791. doi: 10.1021/jf801146z
Newton C., Lorre C., Sauvage C., Ivorra S., Terral J. F. (2014). On the origins and spread of Olea europaea l. (olive) domestication: evidence for shape variation of olive stones at ugarit, late bronze age, Syria–a window on the Mediterranean basin and on the westward diffusion of olive varieties. Veg. Hist. Archaeobot. 23, 567–575. doi: 10.1007/s00334-013-0412-4
Ninot A., Howad W., Aranzana M. J., Senar R., Romero A., Mariotti R., et al. (2018). Survey of over 4,500 monumental olive trees preserved on-farm in the northeast Iberian peninsula, their genotyping and characterization. Sci. Hortic. 231, 253–264. doi: 10.1016/j.scienta.2017.11.025
Ordidge M., Litthauer S., Venison E., Blouin-Delmas M., Fernandez-Fernandez F., Höfer M., et al. (2021). Towards a joint international database: alignment of SSR marker data for European collections of cherry germplasm. Plants 10 (6), 1243. doi: 10.3390/plants10061243
Perri E., Zelasco S., Vizzarri V., Benincasa C., Carbone F., Lo Feudo G., et al. (2015). Il germoplasma olivicolo del campo collezione del centro di ricerca per l’olivicoltura e l’industria olearia (CREA-OLI) di rende. (Italy: Rende). pp. 404.
Petruccelli R., Giordano C., Salvatici M. C., Beghè D., Rodolfi M., Fabbri A., et al. (2021). Characterization and conservation of “Olivo della strega”: an ancient olive tree, precious resource for natural and cultural heritage. Rend. Fis. Acc. Lincei 32, 311–324. doi: 10.1007/s12210-021-00989-z
Petruccelli R., Giordano C., Salvatici M. C., Capozzoli L., Ciaccheri L., Pazzini M., et al. (2014). Observation of eight ancient olive trees (Olea europaea l.) growing in the garden of Gethsemane. Comptes Rendus Biol. 337 (5), 311–317. doi: 10.1016/j.crvi.2014.03.002
Pritchard J. K., Stephens M., Donnelly P. J. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Rallo P., Dorado G., Martin A. (2000). Development of simple sequence repeats (SSRs) in olive tree (Olea europaea l.). Theor. Appl. Genet. 101, 984–989. doi: 10.1007/s001220051571
Rotondi A., Ganino T., Beghè D., Di Virgilio N., Morrone L., Fabbri A., et al. (2018). Genetic and landscape characterization of ancient autochthonous olive trees in northern Italy. Plant Biosyst. 152 (5), 1067–1074. doi: 10.1080/11263504.2017.1415993
Sala G., Caruso T., Marra F. P., Zafonte F., Amico Roxas A., Schiavo B., et al. (2021). Study of energetic properties of different tree organs in six Olea europaea l. cultivars. Sci. Rep. 11 (1), 17047. doi: 10.1038/s41598-021-96436-y
Salimonti A., Simeone V., Cesari G., Lamaj F., Cattivelli L., Perri E., et al. (2013). A first molecular investigation of monumental olive trees in apulia region. Sci. Hortic. 162, 204–212. doi: 10.1016/j.scienta.2013.08.005
Schicchi R., Speciale C., Amato F., Bazan G., Di Noto G., Marino P., et al. (2021). The monumental olive trees as biocultural heritage of Mediterranean landscapes: the case study of Sicily. Sustainability 13 (12), 6767. doi: 10.3390/su13126767
Sefc K. M., Lopes M. S., Mendonça D., Rodrigues Dos Santos M., Da Câmara Machado M., Da Câmara Machado A. (2000). Identification of microsatellites loci in olive (Olea europaea l.) and their characterisation in Italian and Iberian olive trees. Mol. Ecol. 9, 1171–1193. doi: 10.1046/j.1365-294x.2000.00954.x
Taamalli W., Geuna F., Banfi R., Bassi D., Daoud D., Zarrouk M. (2006). Agronomic and molecular analyses for the characterisation of accessions in Tunisian olive germplasm collections. Elect. J. Biotechn. 9 (5). doi: 10.2225/vol9-issue5-fulltext-12
Trifonova A. A., Boris K. V., Mesyats N. V., Tsiupka V. A., Smykov A. V., Mitrofanova I. V. (2021). Genetic diversity of peach cultivars from the collection of the Nikita botanical garden based on SSR markers. Plants 10 (12), 2609. doi: 10.3390/plants10122609
Trujillo I., Ojeda M. A., Urdiroz N. M., Potter D., Barranco D., Rallo L., et al. (2014). Identification of the worldwide olive germplasm bank of córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 10, 141–155. doi: 10.1007/s11295-013-0671-3
Valeri M. C., Mifsud D., Sammut C., Pandolfi S., Lilli E., Bufacchi M., et al. (2022). Exploring olive genetic diversity in the Maltese islands. Sustainability 14, 10684. doi: 10.3390/su141710684
Wu S. B., Collins G., Sedgley M. (2004). A molecular linkage map of olive (Olea europaea l.) based on RAPD, microsatellite, and SCAR markers. Genome 47 (1), 26–35. doi: 10.1139/g03-091
Keywords: Olea europaea, olive genetic resources, microsatellite markers, on-farm and in-situ conservation strategy, putative biotic and abiotic stress resilience
Citation: Marchese A, Bonanno F, Marra FP, Trippa DA, Zelasco S, Rizzo S, Giovino A, Imperiale V, Ioppolo A, Sala G, Granata I and Caruso T (2023) Recovery and genotyping ancient Sicilian monumental olive trees. Front. Conserv. Sci. 4:1206832. doi: 10.3389/fcosc.2023.1206832
Received: 16 April 2023; Accepted: 16 May 2023;
Published: 30 May 2023.
Edited by:
Jesus V Jorrin Novo, University of Cordoba, SpainReviewed by:
Claudio Di Vaio, University of Naples Federico II, ItalyValentina Fanelli, University of Bari Aldo Moro, Italy
Copyright © 2023 Marchese, Bonanno, Marra, Trippa, Zelasco, Rizzo, Giovino, Imperiale, Ioppolo, Sala, Granata and Caruso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Marchese, YW5uYWxpc2EubWFyY2hlc2VAdW5pcGEuaXQ=
 A. Marchese
A. Marchese F. Bonanno2
F. Bonanno2 F. P. Marra
F. P. Marra D. A. Trippa
D. A. Trippa S. Zelasco
S. Zelasco A. Giovino
A. Giovino V. Imperiale
V. Imperiale G. Sala
G. Sala T. Caruso
T. Caruso