- 1School of Animal and Veterinary Sciences, University of Adelaide, Adelaide, SA, Australia
- 2College of Science and Engineering, Flinders University, Adelaide, SA, Australia
Accurate and rapid detection methodologies for monitoring the illicit trafficking of threatened species through highly exploited maritime routes are crucial to support law enforcement and conservation efforts. One of the most prominent trafficked species is the lion (Panthera leo), prized for their pelts, bones, and other derivatives, with the intensity of the trade contributing to their current vulnerable International Union for Conservation of Nature (IUCN) conservation status. The trade in such wildlife products is facilitated by the limited availability of detection technologies at seaports and border crossings to identify and intercept trafficking efforts. Thus, we explored the feasibility of airborne environmental DNA (eDNA) analysis as a novel method to detect lion pelts concealed in shipping containers. Air samples were collected within close proximity of the pelts, as well as from the external air vents of a sealed container using a custom air extraction device. To enhance trace eDNA capture and attempt to overcome the combined challenges of degraded DNA (common in wildlife trade samples) and a confined environment, samples were collected on plasma polymer-coated HEPA F7 filters. All filter samples captured eDNA from the air, with no significant difference in overall yield across filter types (p > 0.05). However, only three surface-modified filters captured amplifiable P. leo mitochondrial DNA using species-specific primers, and only when sampling occurred in close proximity to pelts. Although the adaptation of eDNA-based detection methods shows potential, our findings demonstrate that the current protocol may be unsuitable for law enforcement applications without significant optimisation and validation.
1 Introduction
The exploitation of wildlife for consumables exerts considerable pressure on vulnerable species populations, with resource constraints hindering efforts to prevent the illegal movement of threatened species and their derivatives through complex local, regional and international trade networks. Large carnivores are particularly susceptible to anthropogenic population declines, and trade driven by cultural and economic incentives can intensify these pressures, especially in regions where habitat loss and human-wildlife conflict are prevalent (Arias et al., 2024; Bodasing, 2022). The trade in lion (Panthera leo) parts represents a growing threat to wild populations and conservation efforts across Africa (Almeida et al., 2025; Williams et al., 2025). While bones are the most extensively traded lion derivative, other parts including pelts, teeth and claws are also sought after for zootherapeutic traditional medicines and cultural regalia (Williams et al., 2017; 2025). Despite wildlife protection laws, enforcement challenges and clandestine networks limit effective wildlife trade monitoring (Fukushima et al., 2021), emphasising the need for improved detection measures.
Wildlife products are commonly concealed amongst legitimate goods and trafficked through shipping cargo containers (Duensing et al., 2023; Moloney and Chaber, 2024; Zavagli, 2021). However, customs authorities often lack the capacity to conduct comprehensive inspections. Traditional container inspection methods, including x-ray scanning, are often invasive, resource-intensive and may struggle to detect organic products, highlighting the need for versatile, scalable alternatives (Moloney and Chaber, 2024). Genetic techniques have become invaluable in wildlife forensic investigations, particularly for species identification, trade tracing and population monitoring (Kanthaswamy, 2024; Moore et al., 2021). However, the need for physical samples limits opportunities for trace-based detection in situations where containers cannot be opened or unloaded.
As a non-invasive alternative, environmental DNA (eDNA) analysis has emerged as a powerful tool for detecting trace vertebrate DNA. eDNA refers to genetic material shed by organisms into their surroundings, enabling species identification without the need for direct observation or capture (Thomsen and Willerslev, 2015). Initially gaining traction in aquatic ecosystems, eDNA has revolutionised biodiversity monitoring by offering a highly sensitive, non-invasive method for tracing species populations and monitoring ecosystem changes (Bista et al., 2017; Chucholl et al., 2021; Closek et al., 2019; Fraija-Fernandez et al., 2020). Recent advancements have expanded eDNA applications to monitor terrestrial species through airborne samples (Bohmann and Lynggaard, 2023; Clare et al., 2021; Lynggaard et al., 2022; 2023; Roger et al., 2022; Sohn and Song, 2024; Clare et al., 2022), suitable for species or environments which are challenging to survey using conventional approaches (Garrett et al., 2022; 2023). In biosecurity contexts, airborne eDNA analysis has facilitated the detection of invasive species from debris in shipping containers (Milián-García et al., 2025; Trujillo-González et al., 2022), suggesting the collection of samples for wildlife trafficking monitoring may also be possible.
However, implementing eDNA methodologies remains challenging, particularly in confined spaces where low DNA concentrations and DNA degradation due to poor air circulation and surface exposure limit its effectiveness (Trujillo-González et al., 2022; Milián-García et al., 2025). Additionally, wildlife trade samples are typically aged, processed and/or degraded and provide poor quality genetic material even for traditional analytical methods (Natesh et al., 2019), let alone via airborne sampling. In such cases, innovative strategies may be necessary to improve eDNA binding to air-sampler filters and enhance capture efficiency. For example, surface modification of the material can be considered. One such approach is plasma polymerisation: a versatile technique capable of changing the wetting properties of a substrate from hydrophilic to hydrophobic, and vice versa (Iqbal et al., 2019). This is achieved by fragmentation of volatile monomers into high-energy species in a plasma phase, followed by recombination to form a cross-linked product onto the exposed material: a nanometre thin, conformal coating with surface termination either rich in polar or nonpolar functional groups, depending on the monomer used (Goodman, 1960).
This study investigates the feasibility of capturing airborne eDNA to detect wildlife commodities concealed within shipping containers. Specifically, we aim to provide conceptual proof for the application of airborne eDNA techniques for the detection of P. leo pelts in a containerised environment using a custom air extraction device. Although lion bones are more widely trafficked, pelts were selected to establish this proof-of-concept, given the expectation that they would shed cellular material (i.e. skin, hair) and thus more readily facilitate airborne eDNA detection. We further aim to, i) compare detection success across different sampling configurations, and ii) evaluate the effectiveness of different plasma polymerised filter coatings for eDNA capture. By integrating airborne eDNA analysis with existing screening protocols, this method could provide a scalable, non-invasive approach to improve detection efficiency in settings where traditional container inspections are limited.
2 Materials and methods
2.1 Pelt samples
We acquired three adult African lion (P. leo) pelts from Zoos South Australia (Adelaide, Australia). Additionally, we had access to cheetah (Acinonyx jubatus), leopard (P. pardus), snow leopard (P. uncia) and tiger (P. tigris) pelts. The cheetah pelts were added in 2014 and 2012, the snow leopard in 2019, and one of the female lion pelts in 2019. The records for the remaining pelts were unavailable, but were all likely added before 2012.
2.2 Air extraction device
An air extraction device was developed to collect air samples from shipping containers, inspired by previous eDNA conservation studies (Lynggaard et al., 2022; 2023). The first component of the device consisted of a 3D-printed air vent attachment placed over the container wall vent and held in position by magnets. The attachment was connected via a pipe (43 mm internal diameter, 700 mm long) to a 3D-printed housing directly below. The housing consisted of a sample drawer (108 mm × 108 mm) above two inline 5 V USB-powered battery-operated DC axial fans (120 mm upHere USB fan, model number N12U04, maximum airflow 1.7 m3/min, 1300 rpm per fan). Filters were placed in the sample drawer located 18 mm above the fans. The second component of the device was a 240 V AC powered blower (Ozito, model number BLV-2401, maximum airflow 12.0 m3/min), attached to a 3D-printed nozzle (450 mm long × 7 mm wide) designed to penetrate the gap between the steel doors and the rubber door seal to facilitate the passage of forced air. The 3D-printed components were designed using CAD Inventor 3D modelling software (version 2025.3, Autodesk, USA) and printed with polyethylene terephthalate glycol (PETG) material (Cubic Technology, Australia). The isolated use of the housing component (without pipe and wall vent attachment) for sample collection in this study will be referred to as ‘extraction-only’, while the combined use of both the full extraction and blower components will be called ‘mixed-mode ventilation’.
2.3 Filter development
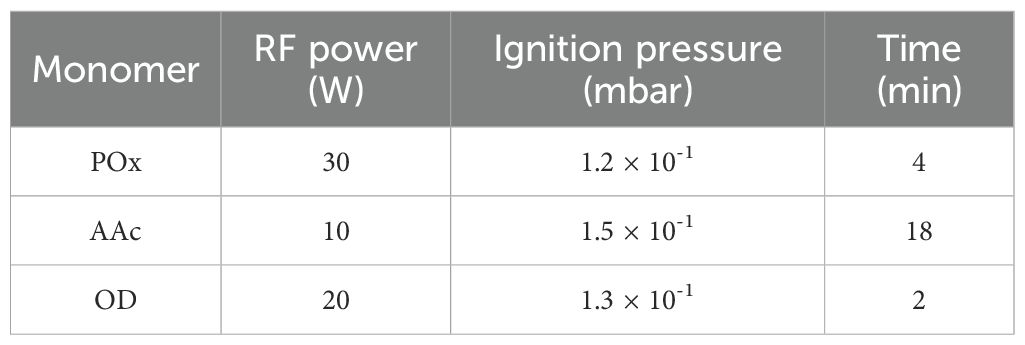
Filters were developed to suit the air extraction device used in this study, with the modifications selected to enhance the capture of eDNA. High Efficiency Particulate Air (HEPA) grade F7 filter material (Merv 13), normally used as a Heating, Ventilation, and Air Conditioning (HVAC) bag filter, was selected based on its previous success in capturing airborne eDNA (Lynggaard et al., 2023). Filters were cut to size to suit the sample drawer. Using plasma polymerisation, filters were coated on both sides with one of three different monomers; 1,7-octadiene (OD, >98% purity), acrylic acid (AAc, 99% purity), or 2-methyl-2-oxazoline (POx, >98% purity) (Merck, Australia).
Plasma polymerisation of the three precursors was carried out in a custom-made radiofrequency plasma reactor operating at 13.56 MHz, as previously described (Chan et al., 2020). The monomers underwent three freeze-pump-thaw cycles prior to use to remove any dissolved gases. The filters were placed in the reactor chamber onto the bottom brass electrode and primed with air plasma at a working pressure of 2.0 x 10–1 mbar, with RF power of 30 W for a duration of three minutes. The reactor was then evacuated to a base pressure of 2.0 x 10–2 mbar with a rotary vane vacuum pump (E2M28, Edwards, UK). Pressure within the reactor was monitored using a Pirani gauge (APG100-XLC, Edwards, UK) and controlled via fine or medium flow needle valves for OD and POx (Chell Instruments, UK), as well as ball valve (ANCORP, USA) for AAc. The plasma polymerisation conditions (Table 1) were optimised based on established procedures (Alvarez de Eulate et al., 2021; Esselbach et al., 2024; Michelmore et al., 2013), and through thorough surface characterisation: spectroscopic ellipsometry for film thickness determination, optical contact angle measurements for wetting properties analysis, as well as x-ray photoelectron spectroscopy (XPS) to confirm their chemical composition. Once the filter surfaces were modified with the plasma polymers, filters were vacuum sealed to prevent exposure to air and stored away from direct sunlight.
2.4 Container configurations
This study evaluated four different sampling configurations (Figure 1). In the first scenario (config. 1), extraction-only samples were collected from within an enclosed space (volume approx. 0.1 - 0.3 m³) where pelts had been stored for one month. Next, extraction-only samples were collected from inside an empty standard ISO 20 ft shipping container (volume 33.2 m³) in the absence of pelts. This negative control is referred to as config. 2. After these samples had been collected, the lion pelts were laid out across the floor on a plastic tarp and sealed inside the container for four days. Extraction-only samples were then collected inside the container within 30 cm of the pelts (air intake facing upwards, not directed towards the pelts), referred to as config. 3. Finally, after the pelts had been sealed in the container for an additional two months, mixed-mode ventilation samples were collected externally from the shipping container vents (config. 4). During container sampling, the doors remained closed and movement was minimized to limit atmospheric disturbance. Average temperature and relative humidity recorded inside the container between 0900h and 1700h on sampling days for each configuration were: a) 14.6 °C, 70.0%, b) 16.8 °C, 60.0%, and c) 29.4 °C, 28.7%, respectively.
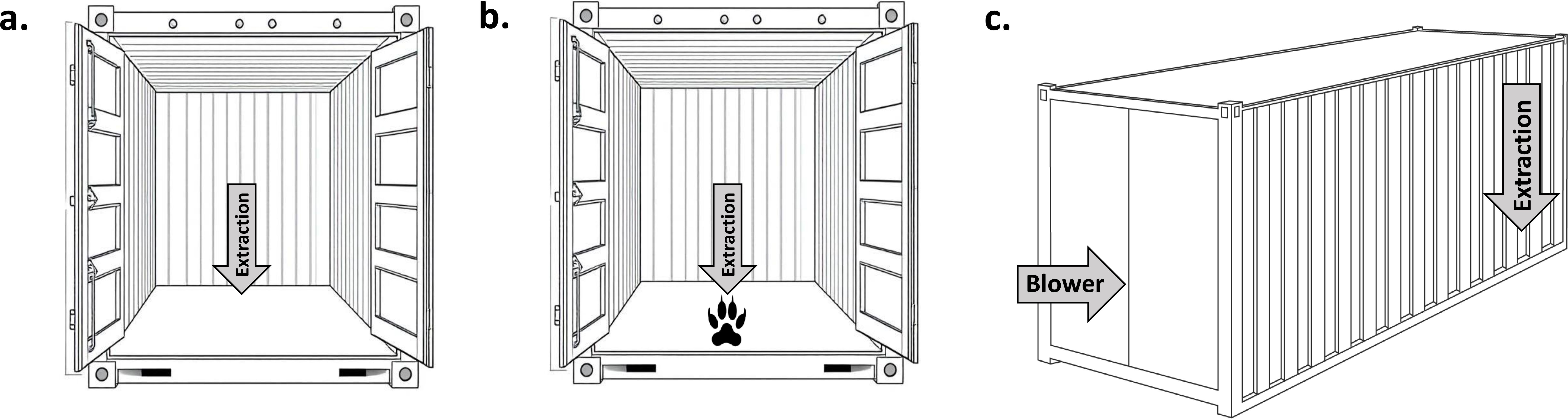
Figure 1. Representation of the 20 ft shipping container configurations tested: (a) extraction-only samples from inside an empty container (config. 2), (b) extraction-only samples from inside a container within 30 cm of the lion pelts (represented by a lion’s pawprint) (config. 3), and (c) mixed-mode ventilation samples collected externally from the container vent with lion pelts sealed inside (config. 4). The arrows represent the direction of airflow generated from either the extraction (airflow towards the device) or blower (airflow away from the device, c only) components. The first sampling configuration (config. 1), in which air samples were extracted from the enclosed storage space, is not shown. The samples in config. 1 were effectively collected in the same manner as shown in config. 3, but from within a smaller volumetric space.
2.5 Filter testing
For each container configuration (see section 2.4), air samples were collected using one non-coated filter and three monomer-coated filters (see section 2.3). Thus, a total of four filters were collected for each configuration, except for config. 4 where two non-coated filters were included - one before and one after all coated filters were collected - to assess whether prolonged airflow altered eDNA conditions within the container, for a total of five filters. All filter samples were collected over a 30-minute period based on the protocol developed by Lynggaard et al. (2022). After collection, samples were immediately stored in sealed plastic sample bags at -20°C until DNA extraction. All sample filters were handled with sterile tweezers, and personnel wore medical gloves and masks to minimise contamination. Between and after sampling events, all equipment was decontaminated with 4-12.5% sodium hypochlorite (bleach) followed by 70% ethanol (Kampmann et al., 2017; Lynggaard et al., 2022; 2023). Containers were not cleaned between treatments.
2.6 DNA extraction
Hair and tissue samples were collected directly from all available pelts (P. leo, A. jubatus, P. pardus, P. uncia, P. tigris). DNA was extracted from all samples in a Physical Containment Level 2 (PC2) laboratory environment using the DNeasy Blood and Tissue Kit (QIAGEN, Germany) following standard laboratory and strict contamination control procedures. Tissues were dissected into 25 mg pieces and placed in microcentrifuge tubes, while for filters a 1.5 cm2 section was selected and cut into very small pieces using scissors. Cells were lysed by adding 400 μL Buffer ATL and 40 μL Proteinase K, then vortexed to mix the solution. The samples were then incubated at 56 °C for five hours (filters) or overnight (tissue and hair) to ensure complete lysis of the cells. After cell lysis, 200 μL Buffer AL was added and vortexed, and the sample was incubated at 70 °C for 10 minutes to further break down cellular components. To facilitate DNA binding, 200 μL of cold 96% ethanol was added, and the samples were mixed and transferred into DNeasy Mini spin columns. These columns were centrifuged at 8000 rpm for one minute to separate the DNA from impurities. The DNA was washed sequentially with 500 μL Buffer AW1 and 500 μL Buffer AW2, with each wash followed by centrifugation at 8000 rpm to ensure removal of contaminants. Finally, DNA was eluted with 100 μL of pre-warmed Buffer AE, incubated at room temperature for five minutes, and then centrifuged at 8000 rpm for one minute to collect the DNA. This elution step was repeated twice to ensure maximal recovery of DNA. DNA concentration was estimated through spectrophotometric measurement (Nanodrop ND-1000, Thermo Fisher Scientific, France) (Supplementary Table S1). DNA extracts were stored at -20 °C prior to PCR.
2.7 Primer selection and validation
Species-specific primers targeting mitochondrial DNA (mtDNA) developed by Henger et al. (2023) were used in this study (Table 2). P. leo primer specificity was assessed in silico using NCBI BLASTn (Altschul et al., 1997). Each primer was queried independently against the NCBI nucleotide database with default parameters. Cross-reactivity was evaluated by examining significant alignments to non-target taxa. To validate assay specificity in vitro, hair and tissue DNA extracts from all available pelts, both target (P. leo) and non-target species, were tested using species-specific primers. Only the P. leo samples showed a positive result at an annealing temperature of 55 °C. The PCR product was extracted from the gel and sent for sequencing at the Australian Genome Research Facility.
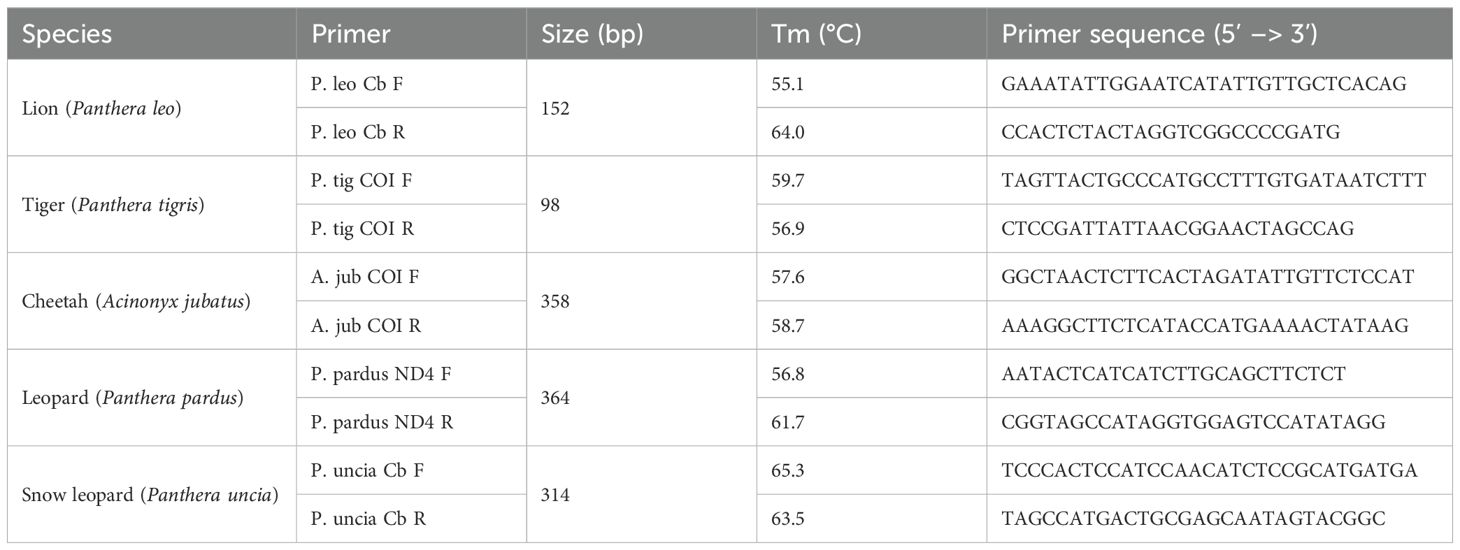
Table 2. Species-specific primers considered in this study (derived from Henger et al., 2023), including length of amplified product, primer melt temperature (Tm) and primer sequences.
2.8 Conventional PCR testing
Conventional PCR was used to analyse both genomic and environmental DNA in this study. Each PCR reaction mixture consisted of 5 µL template DNA, 1 µL forward primer, 1 µL reverse primer, 0.1 µL High Fidelity Platinum™ Taq DNA Polymerase, 0.5 µL 10 mM dNTP Mix, 1 µL 50 mM MgSO4, 2.5 µL 10X High-Fidelity PCR Buffer and Nuclease-Free Water added to a final volume of 25 µL. Each sample was processed in triplicate, and each plate included a positive control P. leo genomic DNA and a non-template control. The PCR conditions included an initial denaturation (94 °C, 2 minutes), followed by 35 cycles of denaturation (94 °C, 30 seconds), annealing (55 °C, 90 seconds) and extension (68 °C, 1 minute). One final extension cycle was performed at 72 °C over 10 minutes. PCR products were visualised on 1% agarose gel under UV light.
3 Results
3.1 Primer evaluation
In silico evaluation of P. leo primers demonstrated high specificity for the target species (no amplification of non-target species at zero nucleotide mismatches for all primers). The forward primer exhibited several low-identity matches, with the highest identity match of these (83% identity) being to an unrelated sea urchin sequence. Meanwhile, the reverse primer exhibited a low-identity match (84% identity) to an extinct Panthera species (P. spelaea). No cross-reactivity of concern against other common DNA found in dust or the general shipping container environment was identified through this search. In vitro validation with hair and tissue samples confirmed successful amplification of P. leo mtDNA, with no cross-amplification observed in non-target species (A. jubatus, P. pardus, P. uncia, P. tigris). Species-specific primers for non-target felids failed to amplify mtDNA from their corresponding samples. Sequencing analysis of lion hair and tissue extracts indicated the presence of target mtDNA, where positive amplicons (152 bp) were confirmed to match against P. leo accessions in NCBI (accession no. XM_042950021.1) with a 99% pairwise similarity with 95% sequence coverage.
3.2 Filter surface characterisation
All plasma polymer films had a thickness of about 30 nm, as determined by spectroscopic ellipsometry. Water contact angle measurements on the plasma polymer coatings revealed that POx and AAc demonstrate hydrophilic wetting behaviour, while OD has hydrophobic wetting properties (Figures 2A–C). This is determined by their chemical composition, confirmed with XPS analysis. OD survey scan demonstrated the coating is almost entirely composed of carbon (C) as expected from its chemical structure (Figure 2F), with low oxygen (O) percentage attributed to adventitious contamination due to exposure to air. Both hydrophilic coatings have O content higher than OD, and POx has an additional heteroatom - nitrogen (N), all in line with their chemical structures (Figures 2D, E). The high-resolution C 1s scans showed that both POx and AAc have a variety of polar functional groups (Figures 2G, H) which govern their hydrophilic wetting behaviour and very similar advancing contact angles just above 60°. The presence of additional amine and imine surface termination on the POx coating determines the slightly lower receding contact angle, and therefore higher contact angle hysteresis (above 48°) in comparison to that of AAc (41°) corresponding to higher adhesion due to the strong interactions between the POx surface and the water droplet. In contrast, OD coating has an insignificant amount of polar functional groups (Figure 2I), and is predominantly terminated with alkyl functional groups, which determines low contact angle hysteresis (12°) and low adhesion between the surface and the water droplet, a consequence of the weak interactions between the two.
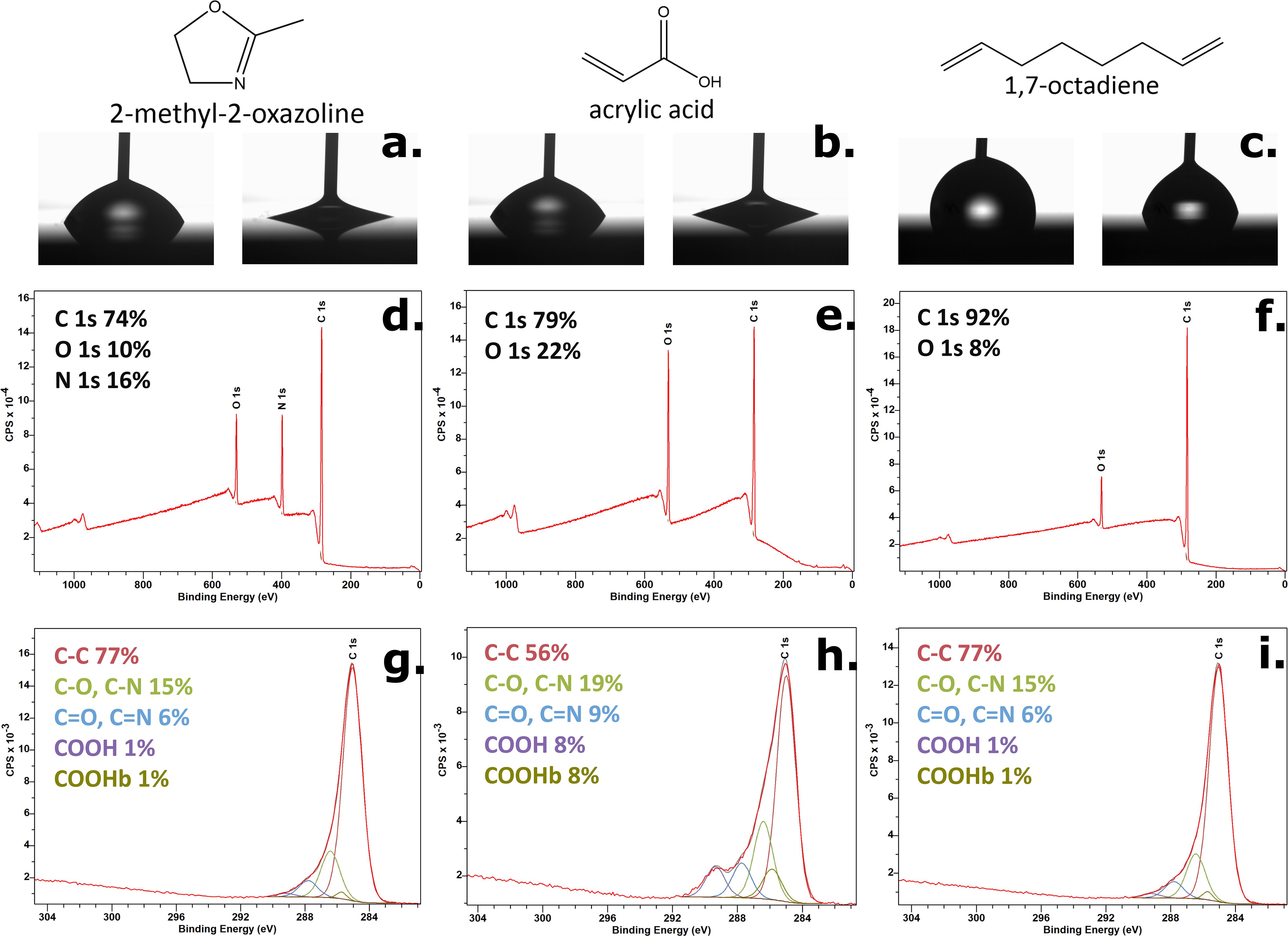
Figure 2. Summary of the surface characterisation of the three plasma polymer coatings used for filter surface modification. Advancing and receding water contact angles on POx (A), AAc (B), and OD (C) with the chemical structures of each monomer above them; XPS survey spectra of POx (D), AAc (E), and OD (F); XPS high-resolution C 1s scans of POx (G), AAc (H), and OD (I).
3.3 Container configuration sampling
Each filter extract was run through gel electrophoresis in triplicate, consistently yielding the same result (Supplementary Table S2). Two coated filters (AAc and OD) collected from the enclosed storage space, config. 1, tested positive for P. leo mtDNA. The non-coated and POx coated filters were negative in this first configuration. Only one filter (AAc) yielded a positive result in the extraction-only internal container configuration, config. 3. In contrast, all filter samples from both the empty extraction-only configuration (config. 2) and the mixed-mode ventilation configuration (config. 4) were negative.
3.4 Filter efficacy
The total mean DNA extract concentration, as measured by spectrophotometry, did not significantly differ between filter coatings (mean 2.8 ng/µL, SD 1.2 ng/µL; Kruskal-Wallis p > 0.05). However, target mtDNA was amplified from three filters in total, comprising two coated filter types: AAc and OD. None of the non-coated or POx coated filters captured detectable levels of P. leo mtDNA.
4 Discussion
This study aimed to explore the feasibility of capturing airborne eDNA to detect lion pelts in a containerised environment. Previous studies have demonstrated successful employment of eDNA techniques to detect trace terrestrial animal DNA in airborne environmental samples (Clare et al., 2021; Garrett et al., 2022; 2023; Lynggaard et al., 2022; 2023; Johnson et al., 2019; Roger et al., 2022; Clare et al., 2022), thus the adaptation of air extraction techniques to overcome the traditional challenges of container searches could provide a unique opportunity to improve detection rates. The availability of advanced detection techniques is essential to disrupt the extensive trade in vulnerable species, such as lions. The primer sets and PCR techniques applied were highly specific for lion eDNA, as validated through in silico testing, in vitro target amplification of genomic DNA, and additional sequencing. In this study, lion mtDNA was successfully extracted from three filter samples, suggesting it was indeed possible to capture eDNA in the air above the pelts. However, techniques suitable for eDNA extraction from containerised environments require further development.
Inconsistencies in eDNA capture may be attributed to the combined challenges of extracting DNA from air sampled from a containerised environment and from degraded samples. Notably, while abiotic (i.e. wind) and biotic (i.e. living organisms) factors significantly influence the dispersion of DNA in the environment (Caza-Allard et al., 2022; Ke et al., 2025), these factors were absent in the current containerised scenario. Air movement in particular is important for the dispersal of eDNA (Métris and Métris, 2023), thus it was hypothesised that the mixed-mode ventilation scenario would increase trace eDNA capture. Additionally, confined spaces are likely less prone to the dilution effect typically observed in environmental sampling and thus should promote better eDNA capture (Clare et al., 2021). However, no positive detections were observed in this configuration, suggesting that either eDNA in the sampled air was below the assay’s limit of detection, or that the number of PCR cycles was insufficient to amplify trace DNA effectively. More sensitive PCR techniques, such as quantitative PCR or digital PCR, could be considered as a means of amplifying trace eDNA suitable for detection, which may overcome some of the limitations associated with DNA availability. Concealment methods used by traffickers would undoubtedly make the detection of trace DNA even more difficult, however this was not assessed in the current study.
The quality of the pelts used in this study further challenged the success of eDNA capture, leading to potential biases in detection accuracy. Previous studies even suggest that, in some contexts, deceased animals or carcasses may not produce detectable eDNA (Curtis and Larson, 2020; Dunker et al., 2017). Whilst there was genetic material captured on all filters, this largely consisted of exogenous DNA. Sample quality likely influenced PCR success, as DNA availability and extraction efficiency depend on the quantity and quality of cellular material present, which is often diminished in wildlife trade-derived samples. Post-mortem processes with regards to cell death, fragmentation and decomposition impact DNA degradation (Alaeddini et al., 2010; Rogers, 2022), while sample collection, storage and handling conditions further influence DNA quality (Rogers, 2022; Zhou et al., 2025). The chemicals used to treat or preserve pelts are also known to degrade host DNA (Casas-Marce et al., 2010; Hebenstreitova et al., 2024), leaving minimal usable DNA for genetic investigations (McDonough et al., 2018; Casas-Marce et al., 2010). While the pelts used were untreated, they were aged and, due to the nature of the dead keratinised cells, DNA yield from direct hair (2.3 ng/µL) and tissue (4.2 ng/µL) analysis was low and of relatively poor quality. Although these samples were notably degraded, this reflects the variable and often poor-quality condition of materials seized across different wildlife trade supply chains, providing a realistic proxy for trace DNA identification. While airborne eDNA detection has its limitations in the context presented, previous studies have demonstrated successful trace DNA deposition detection from packaging materials in direct contact with traded wildlife, hence this may be a more appropriate application (Chan et al., 2024; LeClair et al., 2025).
In response to the expected low quality of DNA in wildlife trade samples, plasma polymerisation was used to modify the surface chemistry of filters to improve airborne eDNA adhesion and retention. While none of the non-coated filters in this study were positive, two coated-filter types – AAc and OD – successfully captured amplifiable target mtDNA. AAc plasma treatment is often used in biosensors and biomedical science as it enables the formation of a chemically active coating which provides surface hydrophilicity and functional carboxylic groups suitable to allow DNA to irreversibly bind (Bitar et al., 2018; Jafari et al., 2009; Ricciardi et al., 2012). AAc was deemed the most successful filter coating, as it was the only coating which produced two positive identifications (configs. 1 and 3). OD is a hydrocarbon monomer which introduces alkyl groups and a hydrophobic surface, and while it is less likely to interact with hydrophilic DNA, it was hypothesised to interact with nonpolar DNA fragments. For example, Cardenas et al. (2003) demonstrated that in aqueous solution single-strand (s-s) DNA adsorbs preferentially and more readily on hydrophobic surfaces than double-stranded (d-s) DNA (Cardenas et al., 2003; Lindman et al., 2021). The s-s DNA molecules are oriented parallel to the substrate and the adsorption occurs via the hydrophobic base fragments. When in air, DNA may undergo partial denaturation which can lead to further exposure of the base fragments, therefore inducing even more hydrophobic properties. Thus, DNA in air could be attracted to the OD coated surface via dispersion forces. Indeed, eDNA was also detected on the OD filter from the enclosed space, but none others. Meanwhile, POx coated filters did not improve sample capture. While the AAc and OD filter coatings overall improved detection efficacy, further scrutinisation is required.
The protocol presented effectively constitutes a presence-absence eDNA survey, and given the high primer specificity any trace mtDNA detection would confirm the presence of the target species. In the context of a suspected trafficking situation, law enforcement merely requires a positive indication of the presence of an illicit commodity to justify a full container search, hence this protocol was intended to provide the first piece of the puzzle in a search operation. In the present study, however, the inability to identify target DNA broadly across filters, especially those obtained from the external container vent, raises questions about the influence of variables affecting eDNA detection and the limitations of our methodology. It is worth noting however that while this protocol did not produce positive results using mixed-mode ventilation, it showed some success through direct sampling - equivalent to opening the container door and directly extracting air or DNA from the commodities, similar to Trujillo-González et al. (2022) and Milián-García et al. (2025). This approach may therefore still be valuable for customs authorities seeking to determine whether illegal goods are mixed with legal ones, especially when visual identification is not possible. While opening the container is more intrusive, it can provide crucial evidence.
This study indicates that eDNA detection for containerised wildlife trafficking requires further investigation. Whilst DNA remains fundamental for identification purposes, it is perhaps not well suited for container screening. Target DNA was successfully recovered from air samples captured using plasma polymer-coated filters, however detection sensitivity was limited by degraded sample quality, methodological shortcomings and constrained airflow, challenges which are also likely to be encountered in operational settings. Our findings highlight the need for further refinement of this method, with future studies focusing on improving sampling efficiency, optimising PCR cycling conditions, and revisiting primer design (i.e. considering primers that target smaller amplicons or conserved regions shared across multiple species). The transition from experimental design to forensic law enforcement practice presents substantial challenges, and at present the method lacks the robust validation required to bridge the gap between experimental feasibility and practical implementation (Frankham et al., 2025; Moore et al., 2021; Ogden et al., 2009). Integrating air sampling technology within existing inspection protocols could in future enhance regulatory efforts, however alternative methods of analysis, such as mass spectrometry (Brown et al., 2021; Coals et al., 2021; Price et al., 2020; Soso and Koziel, 2017; Ueland et al., 2016; 2020), might be more suitable for detecting illegal organic commodities, as the challenges surrounding implementing DNA screening at this stage outweigh the benefits.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because the study used pelt sample and environmental DNA.
Author contributions
GM: Conceptualization, Visualization, Investigation, Methodology, Writing – original draft, Data curation. GB: Writing – review & editing, Investigation. NS: Writing – review & editing, Investigation. AK: Resources, Writing – review & editing, Methodology, Data curation. JM: Investigation, Writing – review & editing, Resources. ID: Writing – review & editing, Methodology, Resources, Data curation, Visualization. MM: Methodology, Writing – review & editing, Resources, Data curation, Funding acquisition, Supervision, Project administration, Visualization. AC: Writing – review & editing, Methodology, Supervision, Conceptualization, Funding acquisition, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by CMA CGM, The University of Adelaide and the Australian Research Council (FT200100301). GM received funding via the CMA CGM SA Supplementary Scholarship and the Australian Government Research Training Program Scholarship.
Acknowledgments
We would like to thank Zoos South Australia for donating the big cat pelts for our trial. The authors would also thank Farhid Hemmatzadeh for their laboratory assistance. The assistance of Mr Christopher Bassell at the XPS facilities at Microscopy Australia enabled by NCRIS at the University of South Australia is also acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcosc.2025.1671488/full#supplementary-material
References
Alaeddini R., Walsh S. J., and Abbas A. (2010). Forensic implications of genetic analyses from degraded DNA—A review. Forensic Sci. International: Genet. 4, 148–157. doi: 10.1016/j.fsigen.2009.09.007
Almeida J., Briers-Louw W. D., Jorge A., Begg C., Roodbol M., Bauer H., et al. (2025). Unsustainable anthropogenic mortality threatens the long-term viability of lion populations in Mozambique. PloS One 20, e0325745. doi: 10.1371/journal.pone.0325745
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alvarez de Eulate E., Gheorghiu A., Amoura C., Whiteley A., Priest C., and MacGregor M. N. (2021). Plasma deposited polyoxazoline thin films for the biofunctionalization of electrochemical sensors. Advanced Materials Technol. 6, 2001292. doi: 10.1002/admt.202001292
Arias M., Coals P., Elves-Powell J., Rizzolo J. B., Ghoddousi A., Boron V., et al. (2024). Reflecting on the role of human-felid conflict and local use in big cat trade. Conserv. Sci. Pract. 6, e13030. doi: 10.1111/csp2.13030
Bista I., Carvalho G. R., Walsh K., Seymour M., Hajibabaei M., Lallias D., et al. (2017). Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat. Commun. 8, 14087. doi: 10.1038/ncomms14087
Bitar R., Cools P., De Geyter N., and Morent R. (2018). Acrylic acid plasma polymerization for biomedical use. Appl. Surface Sci. 448, 168–185. doi: 10.1016/j.apsusc.2018.04.129
Bodasing T. (2022). The decline of large carnivores in Africa and opportunities for change. Biol. Conserv. 274, 109724. doi: 10.1016/j.biocon.2022.109724
Bohmann K. and Lynggaard C. (2023). Transforming terrestrial biodiversity surveys using airborne eDNA. Trends Ecol. Evol. 38, 119–121. doi: 10.1016/j.tree.2022.11.006
Brown A. O., Frankham G. J., Stuart B. H., and Ueland M. (2021). Reptile volatilome profiling optimisation: A pathway towards forensic applications. Forensic Sci. International: Anim. Environments 1, 100024. doi: 10.1016/j.fsiae.2021.100024
Cardenas M., Braem A., Nylander T., and Lindman B. (2003). DNA compaction at hydrophobic surfaces induced by a cationic amphiphile. Langmuir 19, 7712–7718. doi: 10.1021/la026747f
Casas-Marce M., Revilla E., and Godoy J. A. (2010). Searching for DNA in museum specimens: A comparison of sources in a mammal species. Mol. Ecol. Resour. 10, 502–507. doi: 10.1111/j.1755-0998.2009.02784.x
Caza-Allard I., Laporte M., Cote G., April J., and Bernatchez L. (2022). Effect of biotic and abiotic factors on the production and degradation of fish environmental DNA: An experimental evaluation. Environ. DNA 4, 453–468. doi: 10.1002/edn3.266
Chan A. H. J., Gardner M. G., and Linacre A. (2024). Visualisation and detection of latent DNA deposited by pangolin scales onto plastic packaging materials. Forensic Sci. International: Genet. 68, 102975. doi: 10.1016/j.fsigen.2023.102975
Chan K. M., Amoura C., Whiteley A., Rouget J., Shirazi H. S., Cavallaro A., et al. (2020). Functional nanothin films plasma-deposited from 2-isopropenyl-2-oxazoline for biosensor applications. Biointerphases 15, 051005. doi: 10.1116/6.0000499
Chucholl F., Fiolka F., Segelbacher G., and Epp L. S. (2021). eDNA detection of native and invasive crayfish species allows for year-round monitoring and large-scale screening of lotic systems. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.639380
Clare E. L., Economou C. K., Bennett F. J., Dyer C. E., Adams K., McRobie B., et al. (2022). Measuring biodiversity from DNA in the air. Curr. Biol. 32, 693–700.e5. doi: 10.1016/j.cub.2021.11.064
Clare E. L., Economou C. K., Faulkes C. G., Gilbert J. D., Bennett F., Drinkwater R., et al. (2021). eDNAir: proof of concept that animal DNA can be collected from air sampling. PeerJ 9, e11030. doi: 10.7717/peerj.11030
Closek C. J., Santora J. A., Starks H. A., Schroeder I. D., Andruszkiewicz E. A., Sakuma K. M., et al. (2019). Marine vertebrate biodiversity and distribution within the central california current using environmental DNA (eDNA) metabarcoding and ecosystem surveys. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00732
Coals P., Loveridge A., Kurian D., Williams V. L., Macdonald D. W., and Ogden R. (2021). DART mass spectrometry as a potential tool for the differentiation of captive-bred and wild lion bones. Biodiversity Conserv. 30, 1825–1854. doi: 10.1007/s10531-021-02170-2
Curtis A. N. and Larson E. R. (2020). No evidence that crayfish carcasses produce detectable environmental DNA (eDNA) in a stream enclosure experiment. PeerJ 11, e9333. doi: 10.7717/peerj.9333
Duensing S., Schleper M. C., and Busse C. (2023). Wildlife trafficking as a societal supply chain risk: Removing the parasite without damaging the host? J. Supply Chain Manage. 59, 3–32. doi: 10.1111/jscm.12297
Dunker K. J., Sepulveda A. J., Massengill R. L., Olsen J. B., Russ O. L., Wenburg J. K., et al. (2017). Potential of environmental DNA to evaluate northern pike (Esox lucius) eradication efforts: an experimental test and case study. PloS One 12, e0173837. doi: 10.1371/journal.pone.0173837
Esselbach G., Hui K. W., Delcheva I., Jia Z., and MacGregor M. (2024). Plasma coating for hydrophobisation of micro- and nanotextured electrocatalyst materials. Plasma 7, 749–766. doi: 10.3390/plasma7030039
Fraija-Fernandez N., Bouquieaux M.-C., Rey A., Mendibil I., Cotano U., Irigoien X., et al. (2020). Marine water environmental DNA metabarcoding provides a comprehensive fish diversity assessment and reveals spatial patterns in a large oceanic area. Ecol. Evol. 10, 7560–7584. doi: 10.1002/ece3.6482
Frankham G. J., Ogden R., Baker B. W., Ewart K. M., Johnson R. N., Kuiper I., et al. (2025). Standards in wildlife forensic science, with a focus on non-human DNA analysis. Anim. Genet. 56, e70005. doi: 10.1111/age.70005
Fukushima C. S., Tricorache P., Toomes A., Stringham O. C., Rivera-Téllez E., Ripple W. J., et al. (2021). Challenges and perspectives on tackling illegal or unsustainable wildlife trade. Biol. Conserv. 263, 109342. doi: 10.1016/j.biocon.2021.109342
Garrett N. R., Watkins J., Francis C. M., Simmons N. B., Ivanova N., Naaum A., et al. (2022). Out of thin air: surveying tropical bat roosts through air sampling of eDNA. PeerJ 11, e14772. doi: 10.7717/peerj.14772
Garrett N. R., Watkins J., Simmons N. B., Fenton B., Maeda-Obregon A., Sanchez D. E., et al. (2023). Airborne eDNA documents a diverse and ecologically complex tropical bat and other mammal community. Environ. DNA 5, 350–362. doi: 10.1002/edn3.385
Goodman J. (1960). The formation of thin polymer films in the gas discharge. J. Polymer Sci. 44, 551–552. doi: 10.1002/pol.1960.1204414428
Hebenstreitova K., Salaba O., Trubac J., Kufnerova J., and Vanek D. (2024). The influence of tanning chemical agents on DNA degradation: A robust procedure for the analysis of tanned animal hide—A pilot study. Life 14, 147. doi: 10.3390/life14010147
Henger C. S., Straughan D. J., Xu C. C. Y., Nightingale B. R., Kretser H. E., Burnham-Curtis M. K., et al. (2023). A new multiplex qPCR assay to detect and differentiate big cat species in the illegal wildlife trade. Sci. Rep. 13, 9796. doi: 10.1038/s41598-023-36776-z
Iqbal M., Dinh D. K., Abbas Q., Imran M., Sattar H., and Ul Ahmad A. (2019). Controlled surface wettability by plasma polymer surface modification. Surfaces 2, 349–371. doi: 10.3390/surfaces2020026
Jafari R., Arefi-Khonsari F., Tatoulian M., Le Clerre D., Talini L., and Richard F. (2009). Development of oligonucleotide microarray involving plasma polymerized acrylic acid. Thin Solid Films 517, 5763–5768. doi: 10.1016/j.tsf.2009.03.217
Johnson M. D., Cox R. D., and Barnes M. A. (2019). Analysing airborne environmental DNA: A comparison of extraction methods, primer type, and trap type. Environ. DNA 1, 176–185. doi: 10.1002/edn3.19
Kampmann M.-L., Borsting C., and Morling N. (2017). Decrease DNA contamination in laboratories. Forensic Sci. International: Genet. Supplement Ser. 6, e577–e578. doi: 10.1016/j.fsigss.2017.09.223
Kanthaswamy S. (2024). Review: Wildlife forensic genetics—Biological evidence, DNA markers, analytical approaches, and challenges. Anim. Genet. 55, 177–192. doi: 10.1111/age.13390
Ke Y., Liu T., Han C., Yu X., Wang J., Ding L., et al. (2025). A review of eDNA technology in avian monitoring: Current status, challenges and future perspectives. Avian Res. 16, 100235. doi: 10.1016/j.avrs.2025.100235
LeClair G. D., Chatfield M. W. H., and Kinnison M. T. (2025). Environmental DNA as a tool for detecting illegal wildlife trade. Forensic Sci. Int. 370, 112446. doi: 10.1016/j.forsciint.2025.112446
Lindman B., Medronho B., Alves L., Norgren M., and Nordenskiöld L. (2021). Hydrophobic interactions control the self-assembly of DNA and cellulose. Q. Rev. Biophysics 54, e3. doi: 10.1017/S0033583521000019
Lynggaard C., Bertelsen M. F., Jensen C. V., Johnson M. S., Froslev T. G., Olsen M. T., et al. (2022). Airborne environmental DNA for terrestrial vertebrate community monitoring. Curr. Biol. 32, 701–707.e5. doi: 10.1016/j.cub.2021.12.014
Lynggaard C., Froslev T. G., Johnson M. S., Olsen M. T., and Bohmann K. (2023). Airborne environmental DNA captures terrestrial vertebrate diversity in nature. Mol. Ecol. Resour. 24, e13840. doi: 10.1111/1755-0998.13840
McDonough M. M., Parker L. D., McInerney N. R., Campana M. G., and Maldonado J. E. (2018). Performance of destructive museum samples for mammalian genomic studies. J. Mammalogy 99, 789–802. doi: 10.1093/jmammal/gyy080
Métris K. L. and Métris J. (2023). Aircraft surveys for air eDNA: probing biodiversity in the sky. Peer J. 14, e15171. doi: 10.7717/peerj.15171
Michelmore A., Steele D. A., Robinson D. E., Whittle J. D., and Short R. D. (2013). The link between mechanisms of deposition and the physico-chemical properties of plasma polymer films. Soft Matter 9, 6167–6175. doi: 10.1039/C3SM51039E
Milián-García Y., Pyne C., Chen A., Lindsay K., and Hanner R. H. (2025). Uncovering the hidden within shipping containers: Molecular biosurveillance confirms a pathway for introducing multiple regulated and invasive species. Biol. Invasions 27, 91. doi: 10.1007/s10530-025-03549-w
Moloney G. K. and Chaber A.-L. (2024). Where are you hiding the pangolins? Screening tools to detect illicit contraband. PloS One 19, e0299152. doi: 10.1371/journal.pone.0299152
Moore M. K., Baker B. W., Bauman T. L., Burnham-Curtis M. K., Espinoza E. O., Ferrell C. S., et al. (2021). The Society for Wildlife Forensic Science standards and guidelines. Forensic Sci. International: Anim. Environments 1, 100015. doi: 10.1016/j.fsiae.2021.100015
Natesh M., Taylor R. W., Truelove N. K., Hadly E. A., Palumbi S. R., Petrov D. A., et al. (2019). Empowering conservation practice with efficient and economical genotyping from poor quality samples. Methods Ecol. Evol. 10, 853–859. doi: 10.1111/2041-210X.13173
Ogden R., Dawnay N., and McEwing R. (2009). Wildlife DNA forensics – bridging the gap between conservation genetics and law enforcement. Endangered Species Res. 9, 179–195. doi: 10.3354/esr00144
Price E., Larrabure D., Gonzales B., McClure P., and Espinoza E. (2020). Forensic identification of the keratin fibres of South American camelids by ambient ionization mass spectrometry: Vicuña, alpaca and guanaco. Rapid Commun. Mass Spectrometry 34, e8916. doi: 10.1002/rcm.8916
Ricciardi S., Castagna R., Severino S. M., Ferrante I., Frascella F., Celasco E., et al. (2012). Surface functionalization by poly-acrylic acid plasma-polymerized films for DNA diagnostics. Surface Coatings Technol. 207, 389–399. doi: 10.1016/j.surfcoat.2012.07.026
Roger F., Ghanavi H. R., Danielsson N., Wahlberg N., Londahl J., Pettersson L. B., et al. (2022). Airborne environmental DNA metabarcoding for the monitoring of terrestrial insects - A proof of concept from the field. Environ. DNA 4, 790–807. doi: 10.1002/edn3.290
Rogers S. O. (2022). Molecular analyses. 1st ed (Boca Raton, Florida, USA: CRC Press). Available online at: https://www.taylorfrancis.com/books/9781003247432.
Sohn H. and Song Y. (2024). Monitoring of mammal and bird species in an urban ecological park using environmental DNA metabarcoding. Urban Ecosyst. 27, 1891–1904. doi: 10.1007/s11252-024-01557-7
Soso S. B. and Koziel J. A. (2017). Characterizing the scent and chemical composition of Panthera leo marking fluid using solid-phase microextraction and multidimensional gas chromatography–mass spectrometry-olfactometry. Sci. Rep. 7, 5137. doi: 10.1038/s41598-017-04973-2
Thomsen P. F. and Willerslev E. (2015). Environmental DNA: An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Trujillo-González A., Thuo D. N., Divi U., Sparks K., Wallenius T., and Gleeson D. (2022). Detection of Khapra Beetle eDNA using portable technologies in Australian Biosecurity. Front. Insect Sci. 2. doi: 10.3389/finsc.2022.795379
Ueland M., Brown A., Bartos C., Frankham G. J., Johnson R. N., and Forbes S. L. (2020). Profiling volatilomes: A novel forensic method for identification of confiscated illegal wildlife items. Separations 7, 5. doi: 10.3390/separations7010005
Ueland M., Ewart K., Troobnikoff A. N., Frankham G., Johnson R. N., and Forbes S. L. (2016). A rapid chemical odour profiling method for the identification of rhinoceros horns. Forensic Sci. Int. 266, e99–102. doi: 10.1016/j.forsciint.2016.05.011
Williams V. L., Drouilly M., Coals P. G., and Whittington-Jones G. M. (2025). Pan-African review of cultural uses of carnivores. PloS One 20, e0315903. doi: 10.1371/journal.pone.0315903
Williams V. L., Loveridge A. J., Newton D. J., and Macdonald D. W. (2017). Questionnaire survey of the pan-African trade in lion body parts. PloS One 12, e0187060. doi: 10.1371/journal.pone.0187060
Zavagli M. (2021). Red flag indicators for wildlife and timber trafficking in containerized sea cargo: a compendium and guidance for the maritime shipping sector (Hong Kong: TRAFFIC, WWF and United for Wildlife).
Keywords: biosecurity, biodiversity, CITES, conservation, detection, endangered species, illegal wildlife trade, law enforcement
Citation: Moloney GK, Brien GG, Shute NM, Khabiri A, Moloney J, Delcheva I, MacGregor M and Chaber A-L (2025) Assessing the viability of airborne environmental DNA detection for identifying trafficked lion pelts (Panthera leo) in a containerised environment. Front. Conserv. Sci. 6:1671488. doi: 10.3389/fcosc.2025.1671488
Received: 23 July 2025; Accepted: 01 October 2025;
Published: 17 October 2025.
Edited by:
Jessica Bell Rizzolo, Oregon State University, United StatesReviewed by:
Katherine Andrea Solari, Stanford University, United StatesPeter Coals, University of Witwatersrand, South Africa
Copyright © 2025 Moloney, Brien, Shute, Khabiri, Moloney, Delcheva, MacGregor and Chaber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgia Kate Moloney, Z2VvcmdpYS5tb2xvbmV5QGFkZWxhaWRlLmVkdS5hdQ==
 Georgia Kate Moloney
Georgia Kate Moloney Gabrielle Grace Brien1
Gabrielle Grace Brien1 Iliana Delcheva
Iliana Delcheva Anne-Lise Chaber
Anne-Lise Chaber