- 1Department of Obstetrics and Gynecology, Uniformed Services University of the Health Sciences, Bethesda, MD, United States
- 2Division of Pharmacology and Toxicology, Department of Psychology, Institute for Neuroscience, The University of Texas at Austin, Austin, TX, United States
Gonadotropin-releasing hormone (GnRH) is a key regulatory molecule of the hypothalamus–pituitary (PIT)–gonadal (HPG) axis that ultimately leads to the downstream release of estradiol (E2) and progesterone (P). These gonadal steroids feed back to the hypothalamus and PIT to regulate reproductive function and behavior. While GnRH is thought to be the master regulator of reproduction, its metabolic product GnRH-(1–5) is also biologically active. Thimet oligopeptidase 1 (also known as EP24.15) cleaves GnRH to form GnRH-(1–5). GnRH-(1–5) is involved in regulation of the HPG axis, exerting its actions through a pair of orphan G protein-coupled receptors, GPR101 and GPR173. The physiological importance of GnRH-(1–5) signaling has been studied in several contexts, but its potential role during reproductive senescence is poorly understood. We used an ovariectomized (OVX) rat model of reproductive senescence to assess whether and how GnRH-(1–5) signaling genes in hypothalamic subnuclei change in response to aging and/or different estradiol replacement regimens designed to model clinical hormone replacement in women. We found that Gpr101 and Gpr173 mRNA expression was increased with age in the arcuate nucleus, while expression of Gpr173 and EP24.15 increased with age in the medial preoptic area. Treatment with E2 in younger OVX animals increased expression of Gpr101, Gpr173, and EP24.15. However, older animals treated with E2 showed decreased expression of these GnRH-(1–5) signaling genes, displaying an age-related decline in responsiveness to E2. To our knowledge, this is the first study to systematically assess the effects of age and different clinically relevant regimens of E2 replacement on GnRH-(1–5) signaling genes.
Introduction
Gonadotropin-releasing hormone (GnRH) is a key regulatory molecule of the hypothalamus–pituitary (PIT)–gonadal (HPG) axis. Neurons in the hypothalamus release GnRH which acts downstream on the PIT to stimulate transcription and secretion of luteinizing hormone (LH) and follicle stimulating hormone (FSH) (1). In turn, LH and FSH stimulate follicular maturation and release of the steroids estradiol (E2) and progesterone (P). These steroids feed back to the HPG axis to maintain homeostatic regulation of reproductive function and behavior.
While GnRH is thought to be the master regulator of reproduction, its metabolic product GnRH-(1–5) is also shown to be biologically active. GnRH-(1–5) is produced after thimet oligopeptidase 1 (also known as EP24.15) cleaves the covalent bond linking the fifth and sixth amino acids of GnRH (2, 3). GnRH-(1–5), like its parent peptide, is involved in the regulation of the HPG axis. Both GnRH gene expression (4) and secretion (5) are stimulated by GnRH-(1–5). Additionally, the facilitation of lordosis by GnRH is mediated by its metabolism to GnRH-(1–5) (6). However, GnRH-(1–5) binds to alternative receptors than its parent GnRH peptide (6, 7). The actions of GnRH-(1–5) are mediated through a pair of orphan G protein-coupled receptors, GPR101 and GPR173 (8–10). The downstream signaling actions of GnRH-(1–5) occur through traditional G-protein signaling pathways (GPR101) and non-canonical pathways in which β-arrestin 2 is rapidly recruited (GPR173) (8, 11).
It is thought that during aging, changes in the hypothalamic GnRH system, as well as PIT and ovarian processes, are key components that contribute to reproductive senescence (12). The hypothalamic changes include decreased GnRH release and neural activation, as well as diminution of the preovulatory GnRH/LH surge. The subsequent PIT and ovarian changes result in diminished E2 and P secretion and changes in the positive feedback system on GnRH-induced LH surge [reviewed in Ref. (13)]. Additionally, GnRH cleavage enzyme (EP24.15) immunoreactivity within the median eminence where GnRH axons terminate is sensitive to hormonal changes, as its expression decreases from the early proestrous period (high circulating E2 and low LH) to the late proestrous period (low circulating E2 and high LH) (3). It is possible that GnRH-(1–5) may have additional peripheral effects (14). However, whether and how the GnRH-(1–5) signaling pathway changes during reproductive aging within the brain is unknown.
Utilizing a reproductive aging female rat model (15, 16), we sought to determine the impact of age and hormone treatment duration and timing on genes crucial for GnRH-(1–5) signaling, particularly in the medial preoptic area (mPOA) and arcuate nucleus (ARC) of the hypothalamus. The mPOA is of interest as GnRH cell bodies are mainly found here, and it is a major site for the regulation of reproductive function (17–19). The ARC is responsible for regulating the negative feedback response to E2, as well as assisting in generation of the pulsatile release of GnRH (20, 21). Importantly, Gpr101, Gpr173, and EP24.15 are all expressed within these regions (22–24). In addition, gene expression within motor cortex (MC) and PIT were used as comparisons to the hypothalamus. Our goal is to provide mechanistic insights into GnRH and GnRH-(1–5) signaling pathway regulation by E2 deficiency and treatment during reproductive aging.
Materials and Methods
Tissue samples assayed in this study were previously generated and utilized for separate publications (15, 16). This study utilized the cDNA generated by these previous publications.
Animals
Female Sprague-Dawley rats (Harlan, Indianapolis, IN, USA) were purchased at 3–4 months [reproductively mature (MAT); virgin] and 10–11 months old [reproductively aging (AG); retired breeder]. On arrival, rats were pair housed at random with same-age partners in a controlled room temperature (22°C) and light cycle (12-h light, 12-h dark, lights on at 7 a.m.). Food and water were available ad libitum. All animal experiments were conducted following protocols approved by the Institutional Animal Care and Use Committee at the University of Texas at Austin and in accordance with The Guide for the Care and Use of Experimental Animals.
The animal procedures were previously described (15, 16). Briefly, animals were acclimated to the new housing environment for 1 week prior to 2 weeks of estrous cycle monitoring by vaginal lavage with sterile saline. Females age 3–4 months with regular 4–5 days cycles were used for the MAT group. Females age 10–11 months with regular cycles (50%), irregular estrous cycles (30%), or persistent estrus (20%) were randomly assigned to different treatments for the AG groups (Figure 1). Upon determination of estrous cyclicity, all animals underwent bilateral ovariectomy (OVX) under isoflurane inhalation anesthesia, and each animal was administered a non-steroidal anti-inflammatory drug (Rimadyl; 5 mg/kg) at the beginning of surgery for analgesia. Animals were randomly assigned to one of eight treatment groups (Figure 1, groups 1–8), allowing for the examination of different temporal regimens of hormone treatment (16). Animals within the same cage always received the same treatment.
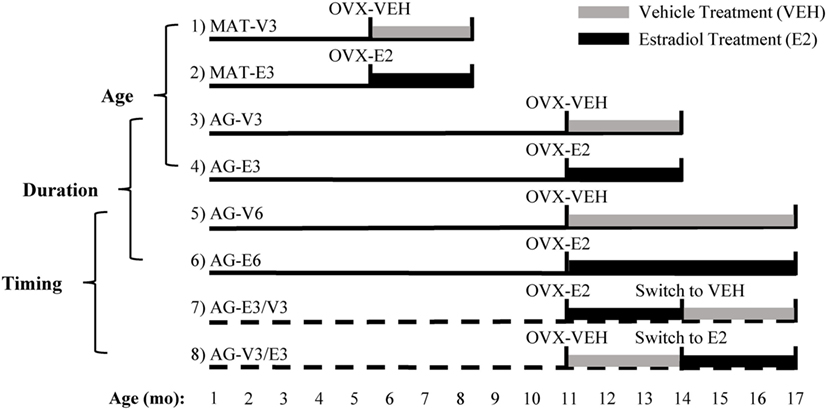
Figure 1. Experimental design used to study the effects of age, timing, and duration of hormone treatment. Ovariectomy (OVX) surgery, followed by vehicle (VEH) or estradiol (E2) capsule implantation, was performed at age 4–5 [mature (MAT)] and 11–12 months [aging (AG)]. Animals in groups 1–6 were MAT or AG rats that were given VEH or E2 for 3 or 6 months. Animals in groups 7 and 8 were AG rats that were given VEH or E2 post-OVX and then switched after 3 months to the opposite treatment for an additional 3 months. Comparisons between groups 1–4, 3–6, and 5–8 were made as described in Section “Materials and Methods.” Figure adapted from Ref. (16).
At the time of surgery, Silastic capsules containing either 100% cholesterol (VEH) or 5% 17β-estradiol/95% cholesterol (E2) were implanted subcutaneously between the shoulder blades. Delivery of E2 via Silastic capsule was previously shown to last for at least 6 months without loss of integrity (25). After OVX, all animals received new identifiers to allow for blinded data collection. Groups 1 and 2 consisted of MAT animals treated with VEH or E2 for 3 months, respectively (MAT-V3 and MAT-E3). Groups 3 and 4 consisted of AG animals treated with VEH or E2 for 3 months, respectively (AG-V3 and AG-E3). Groups 5 and 6 consisted of AG animals treated with VEH or E2 for 6 months, respectively (AG-V6 and AG-E6). Groups 7 and 8 consisted of AG animals treated with E2 followed by VEH for 3 months each (group 7, AG-E3/V3) or VEH followed by E2 for 3 months each (group 8, AG-V3/E3).
Tissue Collection
After 3 or 6 months of hormone treatment, animals were euthanized by rapid decapitation between 1 and 3 p.m. (4–6 h before lights off at 7 p.m.). Brains were quickly extracted and briefly cooled on ice. Coronal brain sections (eight total) were taken at 1 mm intervals throughout the entire hypothalamus using an ice-cold brain matrix (Ted Pella, Inc., Redding, CA, USA). Sections were quickly immersed in 1.5 mL of RNAlater (cat. no. AM7021M, Invitrogen, Waltham, MA, USA) and stored overnight at 4°C. After overnight storage, each section was mounted on plain glass slides and stored at −20°C before micropunching. Additionally, at time of euthanasia, the PIT was removed, immersed in RNAlater overnight at 4°C, and stored at −20°C prior to RNA extraction.
RNA Extraction and Real-time PCR
Brains treated with RNAlater were thawed once for micropunching. Hypothalamic regions containing the mPOA (bregma −0.26 to −1.80) and ARC (bregma −2.12 to −4.52) (26) were micropunched using a 1.2-mm diameter punch (cat. no. 57399, Stoelting, Wood Dale, IL, USA). As a control (non-hypothalamic) region, MC was punched, using a 1.2-mm diameter punch, from regions related to motor control of the forelimbs and forepaws. The entire PIT was used for extraction of RNA. Total RNA, from all regions and the PIT, was extracted using the RNeasy Mini Kit (cat. no. 74104, Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. DNase digestion was performed on-column using the RNase-Free DNase Set (cat. no. 79254, Qiagen). RNA was eluted in 30 µL RNase-free water, and RNA quality and concentration were determined with the Agilent RNA 6000 Nano kit (cat. no. 5067-1511, Agilent Technologies, Santa Clara, CA, USA) on the bioanalyzer. For each region, 200 ng of total RNA was reverse-transcribed to single-stranded cDNA using a high capacity cDNA reverse transcription kit (cat. no. 4374966, Applied Biosystems, Foster City, CA, USA).
The mRNA expression of Gpr101, Gpr173, EP24.15, and the control gene Gapdh was assessed in each region by real-time PCR using the iQ SYBR Green Supermix (cat. no. 1708884, Bio-Rad, Hercules, CA, USA). Additionally, mRNA expression of Gnrh1 and Gnrhr were assessed in the mPOA and PIT, respectively. Each sample was assayed in duplicate using 400 nM of the appropriate primer pair on the CFX Connect Real-time System (Bio-Rad). Mature adult male rat hypothalamus was included in each assay as an intra- (0.281 ± 0.044%) and interassay (0.601 ± 0.084%) control. The following cycling parameters were used: initial denaturation and enzyme activation at 95°C for 3 min followed by 40 cycles of denaturation (95°C, 15 s), annealing (60°C, 30 s), extension (72°C, 30 s), and reading. Melt curve analysis was conducted after each real-time reaction to demonstrate the presence of a single amplicon. Amplified products were purified using the QIAquick PCR Purification Kit (cat. no. 28104, Qiagen) and verified post-purification by agarose gel analysis and sequenced with the ABI 3500xL Genetic Analyzer (Applied Biosystems). Sequences were verified using NCBI BLAST and comparing sequences to the Reference RNA sequences (refseq_rna) database. Primers specific for Gapdh, Gnrh1, Gnrhr, Gpr101, Gpr173, and EP24.15 are shown in Table 1. Relative expression of each gene was determined using the delta delta Ct (ΔΔCt) method (27–29), normalizing each sample to Gapdh. In a previous publication (15), additional brain regions from these animals were analyzed and Gapdh was used as the housekeeping gene. In both that study and the current study, there were no effects of age or E2 treatment on the expression of Gapdh. Therefore, Gapdh was determined to be a valid normalizing gene. All data were expressed relative to the mature, vehicle-treated group (MAT-V3; Figure 1). For each group, cDNA for up to seven animals was analyzed, and deviation from this number in each figure is the result of lack of expression, exhaustion of samples, or the removal of outliers after analysis via Grubb’s test.
Statistical Analyses
Statistical analyses were conducted using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Differential tissue expression of Gpr101, Gpr173, and EP24.15 was compared using a one-way ANOVA followed by a Bonferroni post hoc test. Based on the study design (Figure 1), three different sets of comparisons were performed: (1) The effects of age (MAT vs. AG) and hormone (VEH vs. E2) were analyzed by two-way ANOVA (groups 1–4; Figure 1). (2) The effects of treatment duration (3 vs. 6 months) and hormone (VEH vs. E2) were analyzed by two-way ANOVA (groups 3–6; Figure 1). (3) The effect of the timing of hormone treatment was analyzed by a one-way ANOVA with a Bonferroni post hoc test (groups 5–8; Figure 1). Interactions among variables were also analyzed for the two-way ANOVA analyses. For each analysis, significant main or interaction effects were followed by a Fisher least significant difference post hoc test. A value of p < 0.05 was considered significant. Gnrh1 expression data in the mPOA were transformed utilizing the ratio transform in GraphPad. Prior to statistical analysis, these transformed data were log-transformed for analysis. Assistance with statistical analyses was provided by the USUHS Biostatistics Consulting Center.
Results
Relative Abundance of GnRH-(1–5) Signaling Genes
The relative abundance of Gpr101, Gpr173, and EP24.15 within the mPOA, ARC, PIT, and MC was first compared in mature (MAT-V3) and aging (AG-V3) rats (Figure 2). Expression of each gene is shown relative to levels in the mPOA. In both mature and aging animals, Gpr101 expression was significantly greater in the mPOA and ARC versus the PIT and MC (Figures 2A,B, p < 0.05). In mature animals, the expression of Gpr173 in the ARC was greater than in the PIT and MC (p < 0.05). However, in mature animals, the expression of Gpr173 in the mPOA was only significantly increased compared with the MC (Figure 2C, p < 0.05). Among aging animals, the expression of Gpr173 was greater in the ARC than in the mPOA (p < 0.05), and both regions had higher expression than the PIT and MC (Figure 2D, p < 0.05). There were no differences in EP24.15 expression levels among tissues analyzed (Figures 2E,F). The average Ct values from the MAT-V3 group were graphed to compare the relative expression of each gene within the different tissues analyzed (Figures S1A–D in Supplementary Material).
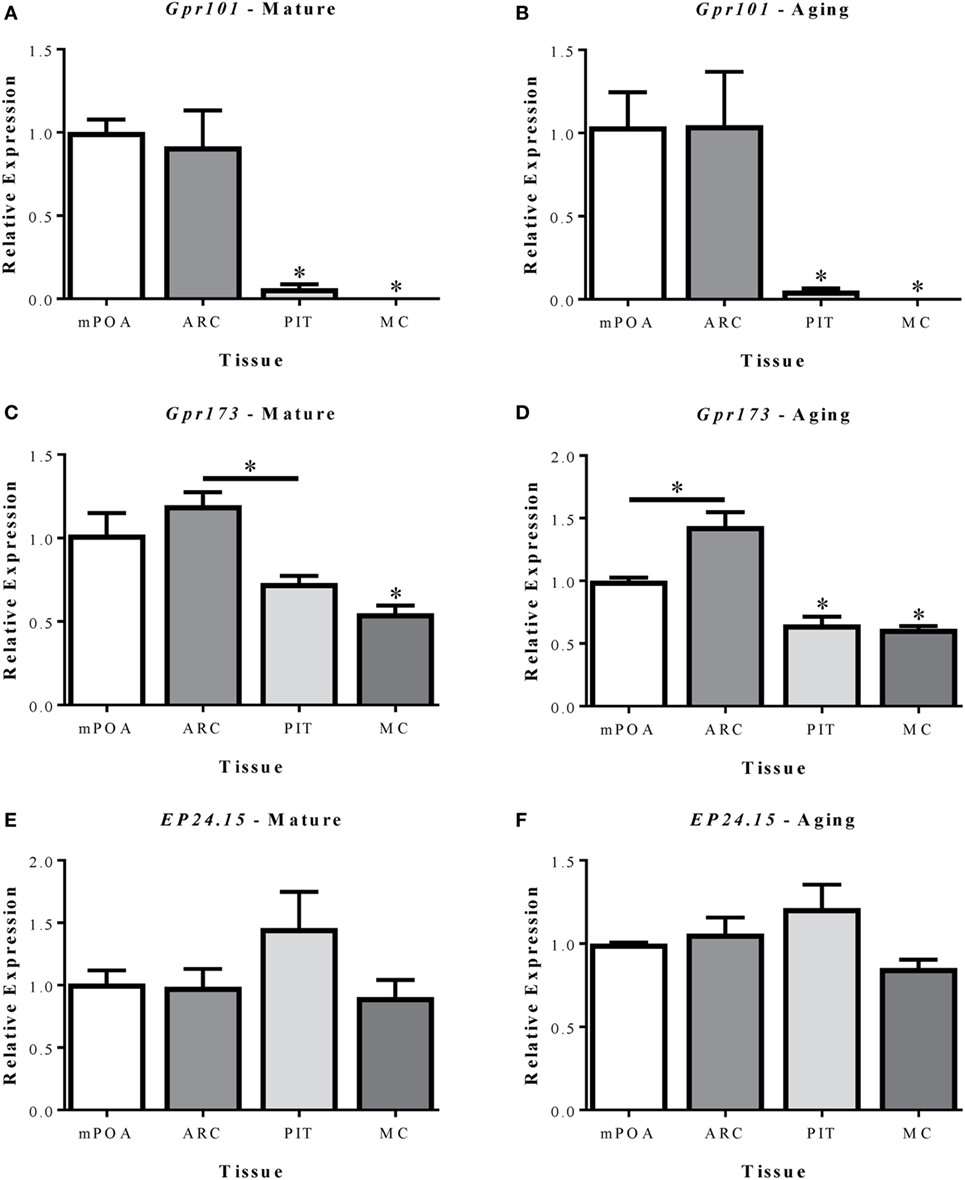
Figure 2. Relative mRNA expression of Gpr101 (A,B), Gpr173 (C,D), and EP24.15 (E,F) in three brain regions and the pituitary of mature (7–8 months; left column) and aging (14–17 months; right column) female Sprague-Dawley rats. (A–F) The tissues analyzed included the medial preoptic area (mPOA), arcuate nucleus (ARC), pituitary (PIT), and motor cortex (MC). (A,B) Expression of Gpr101 in mature and aging female Sprague-Dawley rats. (C,D) Expression of Gpr173 in mature and aging female Sprague-Dawley rats. (E,F) Expression of EP24.15 in mature and aging female Sprague-Dawley rats. Data shown are mean ± SEM (n = 3). *p < 0.05 versus mPOA and ARC unless otherwise specified.
Effects of Age and Estradiol on GnRH-(1–5) Receptors mRNA Expression
To determine the mechanisms by which GnRH-(1–5) signaling was altered after aging, ovarian hormone loss, and E2 treatment, we quantified mRNA expression of its receptors, Gpr101 and Gpr173, within the mPOA, ARC, and PIT (Figures 3 and 4). We found no significant changes in Gpr101 mRNA expression within the mPOA (Figure 3A). However, within the ARC, there were significant effects of age [F(1,22) = 15.24, p < 0.05], hormone [F(1,22) = 8.325, p < 0.05], and a significant interaction between treatment duration and hormone [F(1,23) = 5.179, p < 0.05]. There was a significant increase in Gpr101 expression after 3 months of E2 treatment in the MAT-E3 rats (Figure 3B, p < 0.05). Expression of Gpr101 consistently increased with age, as AG-V3 rats showed greater expression than MAT-V3 rats, and AG-V6 rats had greater expression than AG-V3 rats (Figure 3B, p < 0.05). Additionally, AG-E3 rats showed greater Gpr101 mRNA expression than MAT-E3 rats, demonstrating the age-related increases in expression (Figure 3B, p < 0.05). Interestingly, treatment with E2 for 6 months in the aged rats (AG-E6) decreased the expression of Gpr101 within the ARC, relative to the AG-V6 group (Figure 3B, p < 0.05). There were no significant changes in Gpr101 expression within the PIT (Figure 3C), and the MC control also showed no significant changes (Figure 3D; Figure S2 in Supplementary Material).
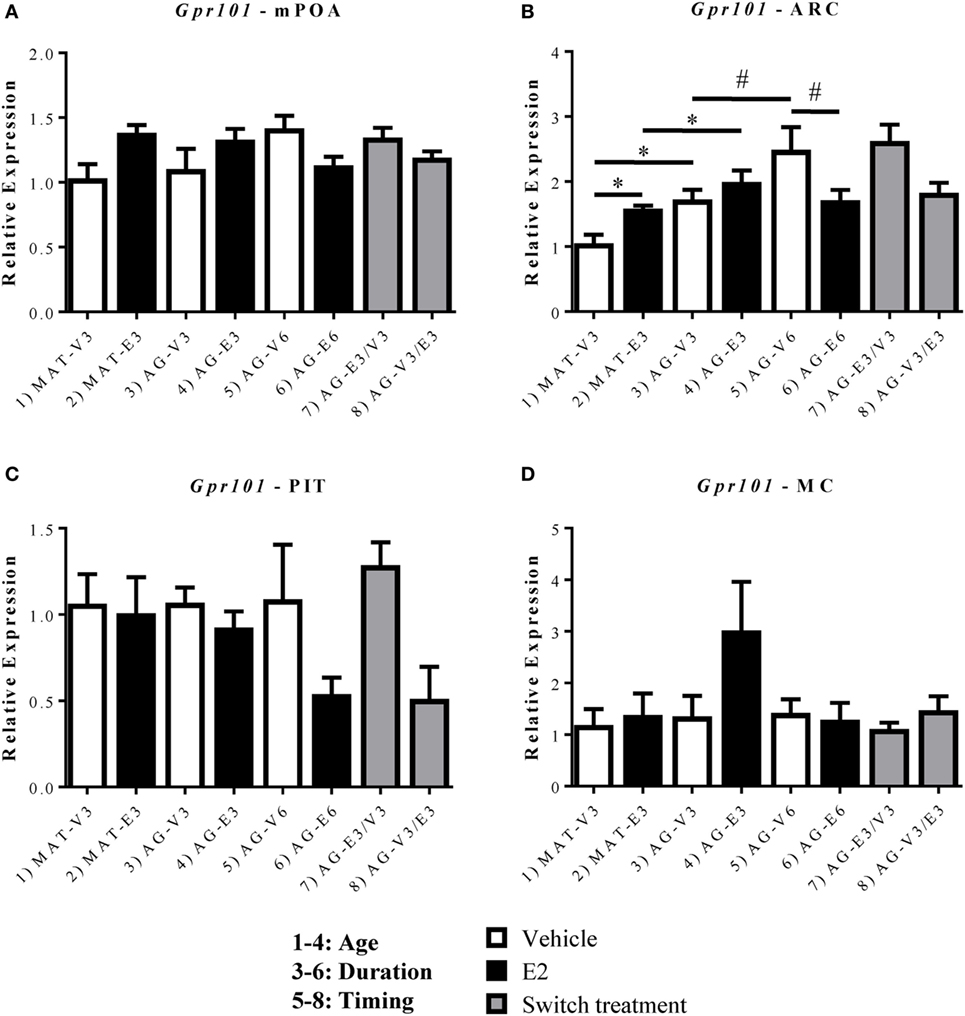
Figure 3. Effects of age and estradiol on mRNA expression of Gpr101. (A–D) The expression of Gpr101 mRNA was analyzed within the (A) medial preoptic area (mPOA), (B) arcuate nucleus (ARC), (C) pituitary (PIT), and (D) motor cortex (MC). Comparisons of age were made between groups 1–4 (analysis 1), duration of hormone treatment between groups 3–6 (analysis 2), and timing of hormone treatment between groups 5–8 (analysis 3). All comparisons were made as described in Section “Materials and Methods.” Data shown are mean ± SEM (n = 4–7). *p < 0.05 (analysis 1); #p < 0.05 (analysis 2).
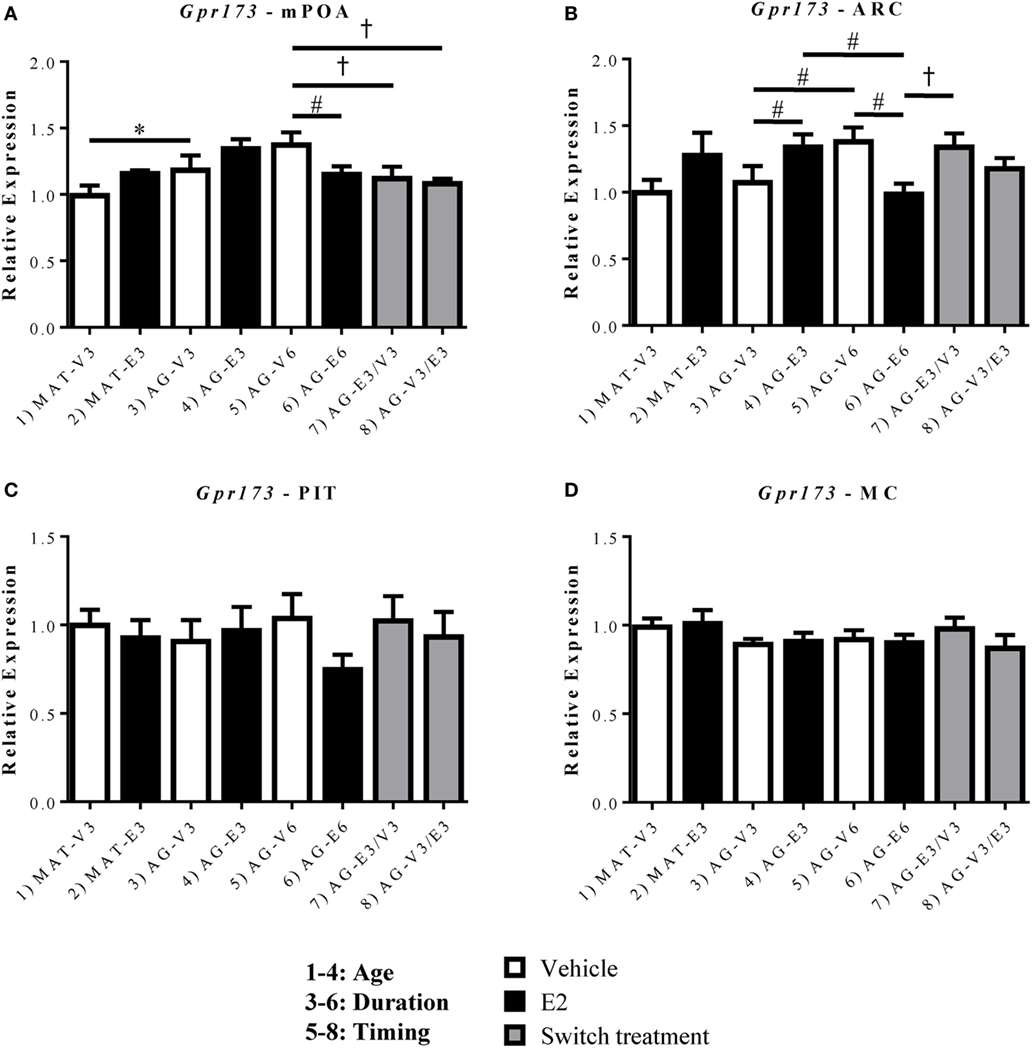
Figure 4. Effects of age and estradiol on mRNA expression of Gpr173. (A–D) The expression of Gpr173 mRNA was analyzed within the (A) medial preoptic area (mPOA), (B) arcuate nucleus (ARC), (C) pituitary (PIT), and (D) motor cortex (MC). Comparisons of age were made between groups 1–4 (analysis 1), duration of hormone treatment between groups 3–6 (analysis 2), and timing of hormone treatment between groups 5–8 (Analysis 3). All comparisons were made as described in Section “Materials and Methods.” Data shown are mean ± SEM (n = 5–7). *p < 0.05 (analysis 1); #p < 0.05 (analysis 2); †p < 0.05 (analysis 3).
Unlike Gpr101, Gpr173 displayed significant effects of age and/or hormone replacement within both the mPOA and ARC (Figures 4A,B). Within the mPOA, there was a significant effect of age [F(1,22) = 8.485, p < 0.05] and a significant interaction between treatment duration and hormone [F(1,24) = 8.12, p < 0.05]. The mRNA expression of Gpr173 increased with age between the AG-V3 and MAT-V3 rats (Figure 4A, p < 0.05). There were no significant effects of E2 in younger rats, however, those treated with E2 for 6 months (AG-E6) displayed decreased Gpr173 expression relative to the AG-V6 rats (Figure 4A, p < 0.05). Additionally, there was no effect of the order of E2 treatment in the aged rats, as both the AG-E3/V3 and AG-V3/E3 rats displayed decreased Gpr173 expression relative to the AG-V6 rats (Figure 4A, p < 0.05). Within the ARC, Gpr173 expression displayed a significant interaction between treatment duration and hormone [F(1,24) = 15.57, p < 0.05; Figure 4B]. Rats treated with VEH for 6 months (AG-V6) had greater Gpr173 expression than the AG-V3 rats (Figure 4B, p < 0.05). Additionally, the aged rats treated with E2 for 3 months (AG-E3) showed increased Gpr173 expression compared to the AG-V3 group (Figure 4B, p < 0.05). As seen in the mPOA, rats treated with E2 for 6 months (AG-E6) actually showed a decrease in Gpr173 relative to the AG-V6 group (Figure 4B, p < 0.05). Interestingly, compared to the mPOA, the timing and duration of E2 treatment appears to be important in the ARC, as the AG-E3/V3 rats had higher Gpr173 expression than the AG-E6 rats, while there was no difference from the expression in the AG-V6 group (Figure 4B, p < 0.05). There were no significant changes in Gpr173 expression within the PIT (Figure 4C), and the MC control also showed no significant changes (Figure 4D).
Effects of Age and Estradiol on GnRH-(1–5) Converting Enzyme, EP24.15, mRNA Expression
EP24.15 expression was assessed within the mPOA, ARC, and PIT (Figure 5). Within the mPOA, there was a significant interaction between treatment duration and hormone [F(1,24) = 6.867, p < 0.05]. EP24.15 expression increased with age in the AG-V6 versus AG-V3 rats and decreased with E2 treatment in the AG-E6 versus AG-V6 rats (Figure 5A, p < 0.05). Within the ARC, there was a significant effect of hormone [F(1,24) = 10.51, p < 0.05], as E2 replacement increased EP24.15 expression in the MAT-E3 versus MAT-V3 rats (Figure 5B, p < 0.05). As seen in the mPOA, there was a significant interaction between treatment duration and hormone [F(1,24) = 6.998, p < 0.05], and treatment with E2 for 6 months decreased EP24.15 expression in AG-E6 versus AG-V6 rats (Figure 5B, p < 0.05). The timing of E2 treatment was also found to be important, as the AG-E3/V3 rats had higher EP24.15 expression than the AG-E6 rats, yet there was no difference from the AG-V6 rats (Figure 5B, p < 0.05). Unlike Gpr101 and Gpr173, EP24.15 expression in the PIT showed a significant interaction between treatment duration and hormone [F(1,20) = 4.463, p < 0.05]. Treatment with E2 for 6 months decreased EP24.15 expression in the AG-E6 versus the AG-V6 rats (Figure 5C, p < 0.05). There were no significant changes seen in the MC control (Figure 5D).
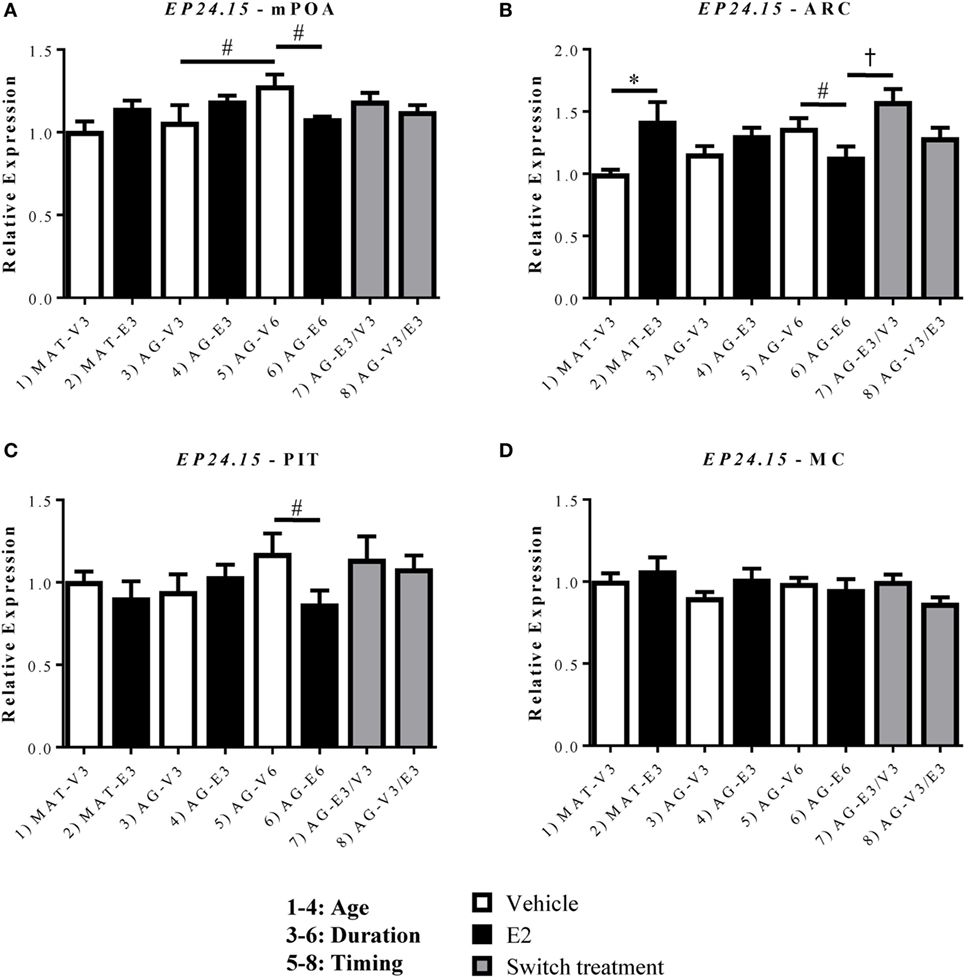
Figure 5. Effects of age and estradiol on mRNA expression of EP24.15. (A–D) The expression of EP24.15 mRNA was analyzed within the (A) medial preoptic area (mPOA), (B) arcuate nucleus (ARC), (C) pituitary (PIT), and (D) motor cortex (MC). Comparisons of age were made between groups 1–4 (analysis 1), duration of hormone treatment between groups 3–6 (analysis 2), and timing of hormone treatment between groups 5–8 (analysis 3). All comparisons were made as described in Section “Materials and Methods.” Data shown are mean ± SEM (n = 5–7). *p < 0.05 (analysis 1); #p < 0.05 (analysis 2); †p < 0.05 (analysis 3).
Effects of Age and Estradiol on Gnrh1 and Gnrhr Expression
In order to better understand the changes in key GnRH-(1–5) signaling genes, it was important to also assess whether there were changes in the expression of Gnrh1 and its receptor, Gnrhr, within this paradigm. Due to these molecules’ more limited expression, this study focused on the mPOA for Gnrh1 (where GnRH cell bodies are found) and the PIT for Gnrhr (where GnRH exerts its effects on the HPG axis). Within the mPOA, there were no significant changes in the expression of Gnrh1 mRNA with age or E2 treatment (Figure 6A). However, there were significant effects of age [F(1,19) = 4.978, p < 0.05], hormone [F(1,19) = 89.43, p < 0.05], and treatment duration [F(1,20) = 39.47, p < 0.05] on the expression of Gnrhr mRNA. Gnrhr was consistently downregulated by E2 treatment at all ages and durations (Figure 6B, p < 0.05). As the rats aged, Gnrhr expression also decreased, as the AG-V3 rats showed lower expression than the MAT-V3 rats (Figure 6B, p < 0.05). The timing and duration of E2 replacement became important in the aging rats as treatment for 3 months followed by VEH for 3 months (AG-E3/V3) did not show any difference from 6 months of VEH treatment (AG-V6); however, Gnrhr expression in this group was significantly higher than in rats treated with E2 for 6 months (AG-E6) and those treated with VEH followed by E2 (AG-V3/E3; Figure 6B, p < 0.05).
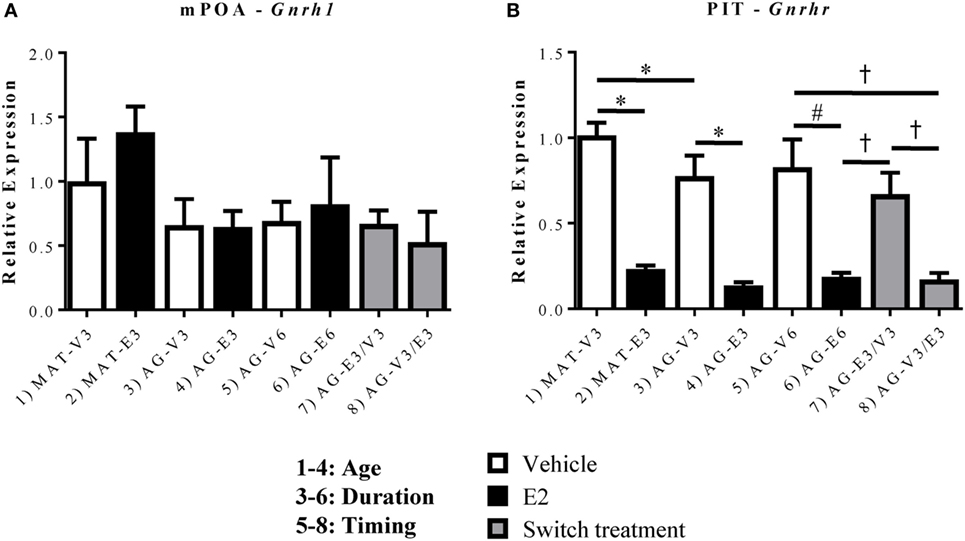
Figure 6. Effects of age and estradiol on mRNA expression of Gnrh1 and Gnrhr. (A) The expression of Gnrh1 mRNA was analyzed within the medial preoptic area (mPOA). (B) The expression of Gnrhr mRNA was analyzed within the pituitary (PIT). Comparisons of age were made between groups 1–4 (analysis 1), duration of hormone treatment between groups 3–6 (analysis 2), and timing of hormone treatment between groups 5–8 (analysis 3). All comparisons were made as described in Section “Materials and Methods.” Data shown are mean ± SEM (n = 6–7). *p < 0.05 (analysis 1); #p < 0.05 (analysis 2); †p < 0.05 (analysis 3).
Discussion
This study analyzed the effects of aging and the timing and duration of E2 treatment on the expression of key GnRH-(1–5) signaling genes in the OVX rat. Gene expression of receptors that GnRH-(1–5) binds to, Gpr101 and Gpr173, was affected by both age and E2 in the mPOA (Gpr173) and ARC (Gpr101 and Gpr173). Additionally, gene expression of the enzyme that generates GnRH-(1–5), EP24.15, was affected by both age and E2 in the mPOA and by E2 alone in the ARC and PIT. To our knowledge, this is the first study to systematically assess the effects of age and different clinically relevant regimens of E2 replacement on GnRH-(1–5) signaling genes.
Aging Affects Relative Distribution of GnRH-(1–5) Signaling Genes
The present study showed tissue-specific expression of Gpr101, Gpr173, and EP24.15 mRNA. Presently, there is a paucity in quantitation of Gpr101 and Gpr173 expression in specific neural nuclei. Gpr101 mRNA is most highly expressed in the hypothalamus, specifically in the ARC (22, 30, 31) and mPOA (22, 30), across multiple species (22, 32–35). These mRNA findings have been supported by protein studies of GPR101 in the ARC and whole hypothalamus (35). Trivellin et al. also showed much higher expression of Gpr101 mRNA in whole mouse hypothalamus versus PIT by RT-qPCR (35), a finding consistent with our study, as our results show that the expression of Gpr101 is highest in the mPOA and ARC versus the PIT and MC. We also found that there was no apparent effect of aging on the relative distribution of Gpr101 between the mPOA, ARC, PIT, and MC.
Similar to Gpr101, Gpr173 mRNA has been detected in multiple species (23, 31, 33, 34, 36), with the highest expression in the hypothalamus compared with other central and peripheral regions (34). Gpr173 mRNA in particular is highly expressed in areas related to the control of reproductive function, including areas with dense expression of estrogen receptor-α and kisspeptin, both important regulators of reproduction (23). The present study is consistent with these studies, showing that expression of Gpr173 is highest in the hypothalamic regions we studied, including the ARC and mPOA, compared to the MC and the PIT. Detection of GPR173 protein has proven to be difficult, as effective antibodies have yet to be produced (23). Our results do show that there is an effect of age on the relative expression of Gpr173 mRNA between tissues. The mature animals (MAT-V3) show similar expression between the mPOA and ARC, however, the aging animals (AG-V3) have significantly higher expression in the ARC compared to the mPOA. This finding should be expanded on in future studies to determine whether there is a functional role for this shift in gene expression. EP24.15 has also been detected throughout the brain and across species (3, 24, 37–40), and consistent with our results, there are no tissue-dependent differences in expression reported. Future studies are needed to examine the connection between mRNA and protein expression in this model, to determine whether the effects of age translate to the protein level.
Aging and E2 Alter Expression of GnRH-(1–5) Signaling Genes
Previous studies demonstrated that GnRH-(1–5) is biologically active and has important roles in the facilitation of lordosis (6) and regulating the amplitude of GnRH pulsatile release (5). Intracerebroventricular injection of GnRH-(1–5) into the third ventricle induced lordosis, and the effects were not blocked by EP24.15 antiserum or Antide, a potent GnRH receptor antagonist (6). Additionally, treatment of hypothalamic explants, dispersed primary cells from the hypothalamus, and GT1–7 cells using GnRH-(1–5) all increased the amplitude of GnRH pulsatile release (5). Based on these results, it was important to understand the control of GnRH-(1–5) signaling gene expression after ovarian hormone loss within the mPOA, ARC, and PIT, regions crucial for HPG axis signaling.
Unlike Gpr101, the expression of Gpr173 displayed significant changes in the mPOA. Within the mPOA, Gpr173 expression increased with age after ovarian hormone loss. However, there was no significant effect of E2 replacement until after 6 months of treatment, at which time E2 decreased the expression of Gpr173. Gpr173 expression 6 months post-ovarian hormone loss was also repressed by E2 treatment independent of the timing of treatment, as both the AG-E3/V3 and AG-V3/E3 groups had significantly lower expression than the AG-V6 animals. Similar to Gpr173, the expression of EP24.15 increased with age in the mPOA. Again, there was no significant effect of E2 replacement until after 6 months of treatment, at which time E2 decreased EP24.15 expression. Unlike the expression of Gpr173, there were no effects of the duration of hormone treatment on EP24.15.
It is within the ARC that our study identified the most significant changes in Gpr101 mRNA expression. Our results demonstrate that as the rat ages after ovarian hormone loss, the expression of Gpr101 mRNA increases within the ARC, and E2 replacement increases expression in younger animals. The expression of Gpr173 also increased within the ARC with age after ovarian hormone loss. A similar effect was seen in the ARC for Gpr173 and EP24.15, as in both cases, treatment with E2 in younger animals led to an increase in mRNA expression. Importantly, after 6 months of E2 replacement, this effect is reversed, and E2 actually decreases expression of Gpr101, Gpr173, and EP24.15 within the ARC. These results are also similar to recently reported data that demonstrated a reversal in E2-induced gene expression in castrated young and aging male rats (41). Interestingly, the expression of Gpr173 and EP24.15 within the ARC was found to be dependent on the duration of E2 treatment after long-term ovarian hormone loss. Animals treated with E2 then switched to VEH (AG-E3/V3) displayed a significant increase in mRNA expression versus those treated with E2 for 6 months (AG-E6). This is an interesting result, as it differs from the effects in the mPOA for both genes. It appears that there are some similarities between Gpr101, Gpr173, and EP24.15 in the ARC, however, after prolonged ovarian hormone loss, the effects of E2 replacement on Gpr173 and EP24.15 are tissue dependent. Identifying the functional significance of this tissue-dependence will be crucial to future studies.
Serum hormone levels in the rats used in the present study were measured previously. Serum LH levels were lowered by E2 treatment, independent of age, consistent with expected estrogen negative feedback effects (16). Similarly, Gnrhr expression in the PIT was significantly decreased with E2 treatment, independent of age, as shown in previous studies (42, 43). By contrast, our current data on the expression of Gnrh mRNA in the mPOA demonstrated no significant changes in expression with E2 treatment or age; however, previous work (in intact female rats) has shown that Gnrh mRNA levels may change independently of transcription and secretion (44). Previous work in OVX animals undergoing E2 treatment has also shown either no change or a small, significant decrease in Gnrh mRNA expression between young and middle-aged OVX animals (45, 46). The finding of little change in Gnrh expression, in the context of significant decreases in Gnrhr mRNA levels with E2 treatment, suggests the possibility of a reduced role for GnRH and an increased role for its metabolite GnRH-(1–5), as well as the potential for altered GnRH release at the median eminence.
This study assessed the effects of age, hormone, treatment duration, and the timing of treatment on the expression of key GnRH-(1–5) signaling genes. The serum E2 levels for the rats studied (see Figure 1 for group numbering), as published previously (15, 16), are as follows (mean ± SEM, in pg/mL): (1) 24 ± 3, (2) 105 ± 11, (3) 20 ± 1, (4) 63 ± 7, (5) 17 ± 2, (6) 63 ± 5, (7) 20 ± 2, and (8) 64 ± 6. The potential mechanisms by which E2 regulates the expression of Gpr101 and Gpr173, the receptors for GnRH-(1–5), remain to be discovered. Evaluation of the known promoter sequences suggests no known classical estrogen response elements (EREs). However, this does not rule out the possibility of putative weak estrogen responsive motifs, or ERE-independent signaling through association with other DNA-binding transcription factors. The evaluation of the promoters for Gpr101 and Gpr173 is the subject of ongoing projects. Despite this lack of apparent EREs, these genes were shown to be responsive to E2 within the current study. The most intriguing E2 effects were seen in Gpr173 expression in the ARC. It was here that AG-E3 animals displayed an increase in expression with E2 treatment, while AG-E6 animals displayed a decrease (serum E2 levels of 63 ± 7 and 63 ± 5 pg/mL, respectively), versus their respective vehicle controls. Since the AG-V6 animals already displayed increased Gpr173 expression compared to AG-V3 animals, these effects may be a result of aging and a gain or loss of specific feedback mechanisms. Effects like these, as well as the mechanism of E2-responsiveness of these receptors, are yet to be elucidated.
Conclusion
The results reported here are the first to systematically assess the effects of E2 replacement regimens on the mRNA expression of Gpr101, Gpr173, and EP24.15 within tissues crucial to HPG regulation. Previous work has shown that OVX followed by immediate implantation with E2 in young rats mimics the changes in GnRH neurons and LH surges that occur at middle age in intact rats (47, 48). As such, our results suggest that rats entering middle age have increased expression of Gpr173 and EP24.15 in the mPOA and increased expression of Gpr101 and Gpr173 in the ARC. Future studies should examine the protein expression of these genes within a similar paradigm and begin to delineate the functional significance of these changes. Recent work has demonstrated that Gpr173 mRNA is expressed in both immortalized hypothalamic GnRH and kisspeptin (Kiss1) neurons (49), a finding that should be confirmed by protein colocalization studies in vivo. The immunoreactivity of EP24.15 was also recently examined in relation to GnRH and Kiss1. Woitowich et al. found that EP24.15 was colocalized with GnRH neurons in the mPOA and both the internal and external zones of the median eminence in adult male rats (50). The authors also found that EP24.15 immunoreactivity was colocalized with that of Kiss1 within the ARC in metestrous female rats, however, no regions of colocalization were seen in the anteroventral periventricular nucleus (AVPV) (50), despite the identification of Kiss1 mRNA in this region (51). The localization of GPR101 in relation to GnRH or Kiss1 neurons is still to be determined.
The importance of the colocalization of EP24.15 and GPR173 with GnRH and Kiss1 neurons is slowly emerging as recent research suggests a more complex regulation of reproduction and GnRH release by a multitude of factors. Aside from cleaving GnRH to GnRH-(1–5), EP24.15 is responsible for the cleavage of Kiss1 (50) and Phoenixin (PNX) (52), both of which are peptides capable of stimulating LH secretion in vivo (23, 53). Immunohistochemical analysis has found PNX expressed in multiple regions of the hypothalamus, including the ARC and AVPV, both of which contain Kiss1 neurons (54). Further, these regions are particularly important for Kiss1 signaling in the female rat, as Kiss1 neurons in both the ARC and AVPV project to and modulate the negative and positive feedback effects of estrogen on GnRH neurons, respectively. Understanding the interplay between Kiss1, PNX, and GnRH-(1–5) within intact models of aging will be crucial to determining the role of each in the regulation of GnRH expression and secretion, and ultimately, their combined changes that lead to reproductive senescence.
In summary, we have systematically assessed the change in expression of genes crucial to GnRH-(1–5) signaling in response to aging and different estradiol replacement regimens designed to model clinical hormone replacement in women. Examining expression of GnRH-(1–5) signaling genes in the mPOA, ARC, and PIT is crucial as these regions are associated with regulation of GnRH (as well as LH and FSH) via the kisspeptin pathway [reviewed in Ref. (55).]. As the female rat ages, the E2-induced increases in GnRH-(1–5) signaling genes disappear, and E2 treatment eventually decreases the expression of Gpr101, Gpr173, and EP24.15 in both the mPOA and ARC. Use of this rat model may be clinically relevant, as the primary outstanding question of the Women’s Health Initiative is determination of the optimal temporal E2 regimen to benefit health and well-being in women. Further studies are needed to determine the potential overlap between GnRH-(1–5) signaling, and other components of the reproductome, including Kiss1 and PNX.
Ethics Statement
This study was carried out in accordance with the recommendations of The Guide for the Care and Use of Experimental Animals. The protocol was approved by the Institutional Animal Care and Use Committee at the University of Texas at Austin.
Author Contributions
Conception or design of the study: WY, AG, and TW. Data collection: BB, WY, and AG. Data analysis and interpretation: BB, WY, AG, and TW. Drafting and critical revision of the article: BB, WY, AG, and TW. Final approval of the version to be published: WY, AG, and TW.
Disclaimer
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Brian Pham for technical expertise on this project and Dr. Cara Olsen of the USUHS Biostatistics Consulting Center for her support in statistical analyses. The authors also thank Dr. Zhao Zhang Li of the USUHS Biomedical Instrumentation Center for her support in sequencing qPCR amplicons for primer verification.
Funding
This research was supported by the National Institutes of Health R03HD078645 (to TW) and P01AG016765 (to AG) and the Uniformed Services University Intramural grant G1852488 (to TW).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/article/10.3389/fendo.2017.00282/full#supplementary-material.
References
1. Clarke IJ, Cummins JT. The temporal relationship between gonadotropin releasing hormone (GnRH) and lutenizing hormone (LH) secretion in ovariectomized ewes. Endocrinology (1982) 111(5):1737–9. doi:10.1210/endo-111-5-1737
2. Molineaux CJ, Lasdun A, Michaud C, Orlowski M. Endopeptidase-24.15 is the primary enzyme that degrades luteinizing hormone releasing hormone both in vitro and in vivo. J Neurochem (1988) 51(2):624–33. doi:10.1111/j.1471-4159.1988.tb01084.x
3. Wu TJ, Pierotti AR, Jakubowski M, Sheward WJ, Glucksman MJ, Smith AI, et al. Endopeptidase EC 3.4.24.15 presence in the rat median eminence and hypophysial portal blood and its modulation of the luteinizing hormone surge. J Neuroendocrinol (1997) 9(11):813–22. doi:10.1046/j.1365-2826.1997.00637.x
4. Wu TJ, Mani SK, Glucksman MJ, Roberts JL. Stimulation of luteinizing hormone-releasing hormone (LHRH) gene expression in GT1–7 cells by its metabolite, LHRH-(1–5). Endocrinology (2005) 146(1):280–6. doi:10.1210/en.2004-0560
5. Larco DO, Williams M, Schmidt L, Sabel N, Lange J, Woller MJ, et al. Autoshortloop feedback regulation of pulsatile gonadotropin-releasing hormone (GnRH) secretion by its metabolite, GnRH-(1–5). Endocrine (2015) 49(2):470–8. doi:10.1007/s12020-014-0492-7
6. Wu TJ, Glucksman MJ, Roberts JL, Mani SK. Facilitation of Lordosis in rats by a metabolite of luteinizing hormone releasing hormone. Endocrinology (2006) 147(5):2544–9. doi:10.1210/en.2005-1646
7. Roberts JL, Mani SK, Woller MJ, Glucksman MJ, Wu TJ. LHRH-(1–5): a bioactive peptide regulating reproduction. Trends Endocrinol Metabol (2007) 18(10):386–92. doi:10.1016/j.tem.2007.09.005
8. Cho-Clark M, Larco DO, Semsarzadeh NN, Vasta F, Mani SK, Wu TJ. GnRH-(1–5) transactivates EGFR in Ishikawa human endometrial cells via an orphan G protein-coupled receptor. Mol Endocrinol (2014) 28(1):80–98. doi:10.1210/me.2013-1203
9. Cho-Clark M, Larco DO, Zahn BR, Mani SK, Wu TJ. GnRH-(1–5) activates matrix metallopeptidase-9 to release epidermal growth factor and promote cellular invasion. Mol Cell Endocrinol (2015) 415:114–25. doi:10.1016/j.mce.2015.08.010
10. Larco DO, Semsarzadeh NN, Cho-Clark M, Mani S, Wu TJ. The novel actions of the metabolite GnRH-(1-5) are mediated by a G protein-coupled receptor (GPCR). Front Endocrinol (2013) 4:83. doi:10.3389/fendo.2013.00083
11. Larco DO, Semsarzadeh NN, Cho-Clark M, Mani SK, Wu TJ. β-arrestin 2 is a mediator of GnRH-(1–5) signaling in immortalized GnRH neurons. Endocrinology (2013) 154(12):4726–36. doi:10.1210/en.2013-1286
12. Kermath BA, Gore AC. Neuroendocrine control of the transition to reproductive senescence: lessons learned from the female rodent model. Neuroendocrinology (2012) 96(1):1–12. doi:10.1159/000335994
13. Rudolph LM, Bentley GE, Calandra RS, Paredes AH, Tesone M, Wu TJ, et al. Peripheral and central mechanisms involved in hormonal control of male and female reproduction. J Neuroendocrinol (2016) 28(7):1–12. doi:10.1111/jne.12405
14. Baldwin EL, Wegorzewska IN, Flora M, Wu TJ. Regulation of type II luteinizing hormone-releasing hormone (LHRH-II) gene expression by the processed peptide of LHRH-I, LHRH-(1–5) in endometrial cells. Exp Biol Med (2007) 232(1):146–55. doi:10.3181/00379727-207-2320146
15. Garcia AN, Depena CK, Yin W, Gore AC. Testing the critical window of estradiol replacement on gene expression of vasopressin, oxytocin, and their receptors, in the hypothalamus of aging female rats. Mol Cell Endocrinol (2016) 419:102–12. doi:10.1016/j.mce.2015.10.004
16. Yin W, Maguire SM, Pham B, Garcia AN, Dang N-V, Liang J, et al. Testing the critical window hypothesis of timing and duration of estradiol treatment on hypothalamic gene networks in reproductively mature and aging female rats. Endocrinology (2015) 156(8):2918–33. doi:10.1210/en.2015-1032
17. Sternberger LA, Hoffman GE. Immunocytology of luteinizing hormone-releasing hormone. Neuroendocrinology (1978) 25(2):111–28. doi:10.1159/000122734
18. Witkin JW, Paden CM, Silverman AJ. The luteinizing hormone-releasing hormone (LHRH) systems in the rat brain. Neuroendocrinology (1982) 35(6):429–38. doi:10.1159/000123419
19. Powers B, Valenstein ES. Sexual receptivity: facilitation by medial preoptic lesions in female rats. Science (1972) 175(4025):1003–5. doi:10.1126/science.175.4025.1003
20. Maeda KI, Adachi S, Inoue K, Ohkura S, Tsukamura H. Metastin/Kisspeptin and control of estrous cycle in rats. Rev Endocr Metab Disord (2007) 8(1):21–9. doi:10.1007/s11154-007-9032-6
21. Goodman RL, Hileman SM, Nestor CC, Porter KL, Connors JM, Hardy SL, et al. Kisspeptin, neurokinin B, and dynorphin act in the arcuate nucleus to control activity of the GnRH pulse generator in ewes. Endocrinology (2013) 154(11):4259–69. doi:10.1210/en.2013-1331
22. Bates B, Zhang L, Nawoschik S, Kodangattil S, Tseng E, Kopsco D, et al. Characterization of Gpr101 expression and G-protein coupling selectivity. Brain Res (2006) 1087(1):1–14. doi:10.1016/j.brainres.2006.02.123
23. Stein LM, Tullock CW, Mathews SK, Garcia-Galiano D, Elias CF, Samson WK, et al. Hypothalamic action of phoenixin to control reproductive hormone secretion in females: importance of the orphan G protein-coupled receptor Gpr173. Am J Physiol Regul Integr Comp Physiol (2016) 311(3):R489–96. doi:10.1152/ajpregu.00191.2016
24. Pierotti AR, Lasdun A, Ayala JM, Roberts JL, Molineaux CJ. Endopeptidase-24.15 in rat hypothalamic/pituitary/gonadal axis. Mol Cell Endocrinol (1991) 76(1–3):95–103. doi:10.1016/0303-7207(91)90264-S
25. Daniel JM, Hulst JL, Berbling JL. Estradiol replacement enhances working memory in middle-aged rats when initiated immediately after ovariectomy but not after a long-term period of ovarian hormone deprivation. Endocrinology (2006) 147(1):607–14. doi:10.1210/en.2005-0998
26. Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates (2nd). New York: Academic Press (1986).
27. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods (2001) 25(4):402–8. doi:10.1006/meth.2001.1262
28. Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res (2001) 29(9):e45. doi:10.1093/nar/29.9.e45
29. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc (2008) 3(6):1101–8. doi:10.1038/nprot.2008.73
30. Nilaweera KN, Wilson D, Bell L, Mercer JG, Morgan PJ, Barrett P. G protein-coupled receptor 101 mRNA expression in supraoptic and paraventricular nuclei in rat hypothalamus is altered by pregnancy and lactation. Brain Res (2008) 1193:76–83. doi:10.1016/j.brainres.2007.11.048
31. Rønnekleiv OK, Fang Y, Zhang C, Nestor CC, Mao P, Kelly MJ. Research resource: gene profiling of G protein-coupled receptors in the arcuate nucleus of the female. Mol Endocrinol (2014) 28(8):1362–80. doi:10.1210/me.2014-1103
32. Lee DK, Nguyen T, Lynch KR, Cheng R, Vanti WB, Arkhitko O, et al. Discovery and mapping of ten novel G protein-coupled receptor genes. Gene (2001) 275(1):83–91. doi:10.1016/S0378-1119(01)00651-5
33. Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A (2003) 100(8):4903–8. doi:10.1073/pnas.0230374100
34. Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell (2008) 135(3):561–71. doi:10.1016/j.cell.2008.08.040
35. Trivellin G, Daly AF, Faucz FR, Yuan B, Rostomyan L, Larco DO, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med (2014) 371(25):2363–74. doi:10.1056/NEJMoa1408028
36. Matsumoto M, Saito T, Takasaki J, Kamohara M, Sugimoto T, Kobayashi M, et al. An evolutionarily conserved G-protein coupled receptor family, SREB, expressed in the central nervous system. Biochem Biophys Res Commun (2000) 272(2):576–82. doi:10.1006/bbrc.2000.2829
37. Smith AI, Tetaz T, Roberts JL, Glucksman M, Clarke IJ, Lew RA. The role of EC 3.4.24.15 in the post-secretory regulation of peptide signals. Biochimie (1994) 76(3–4):288–94. doi:10.1016/0300-9084(94)90160-0
38. Molineaux CJ, Yu B, Ayala JM. Distribution of endopeptidase-24.15 in rat brain nuclei using a novel fluorogenic substrate: comparison with endopeptidase-24.11. Neuropeptides (1991) 18(1):49–54. doi:10.1016/0143-4179(91)90163-D
39. Healy DP, Orlowski M. Immunocytochemical localization of endopeptidase 24.15 in rat brain. Brain Res (1992) 571(1):121–8. doi:10.1016/0006-8993(92)90517-D
40. Massarelli EE, Casatti CA, Kato A, Camargo ACM, Bauer JA, Glucksman MJ, et al. Differential subcellular distribution of neurolysin (EC 3.4.24.16) and thimet oligopeptidase (EC 3.4.24.15) in the rat brain. Brain Res (1999) 851(1–2):261–5. doi:10.1016/S0006-8993(99)02135-6
41. Nutsch VL, Bell MR, Will RG, Yin W, Wolfe A, Gillette R, et al. Aging and estradiol effects on gene expression in the medial preoptic area, bed nucleus of the stria terminalis, and posterodorsal medial amygdala of male rats. Mol Cell Endocrinol (2017) 442:153–64. doi:10.1016/j.mce.2016.12.023
42. Kaiser UB, Jakubowiak A, Steinberger A, Chin WW. Regulation of rat pituitary gonadotropin-releasing hormone receptor mRNA levels in vivo and in vitro. Endocrinology (1993) 133(2):931–4. doi:10.1210/endo.133.2.8393779
43. Seong JY, Kang SS, Kam K, Han Y-G, Kwon HB, Ryu K, et al. Differential regulation of gonadotropin-releasing hormone (GnRH) receptor expression in the posterior mediobasal hypothalamus by steroid hormones: implication of GnRH neuronal activity. Mol Brain Res (1998) 53(1–2):226–35. doi:10.1016/S0169-328X(97)00297-0
44. Gore AC, Oung T, Yung S, Flagg RA, Woller MJ. Neuroendocrine mechanisms for reproductive senescence in the female rat. Endocrine (2000) 13(3):315–23. doi:10.1385/endo:13:3:315
45. Gore AC, Oung T, Woller MJ. Age-related changes in hypothalamic gonadotropin-releasing hormone and N-methyl-d-aspartate receptor gene expression, and their regulation by oestrogen, in the female rat. J Neuroendocrinol (2002) 14(4):300–9. doi:10.1046/j.1365-2826.2002.00777.x
46. Miller BH, Gore AC. Alterations in hypothalamic insulin-like growth factor-I and its associations with gonadotropin releasing hormone neurones during reproductive development and ageing. J Neuroendocrinol (2001) 13(8):728–36. doi:10.1046/j.1365-2826.2001.00686.x
47. Tsai HW, Legan SJ. Loss of luteinizing hormone surges induced by chronic estradiol is associated with decreased activation of gonadotropin-releasing hormone neurons. Biol Reprod (2002) 66(4):1104–10. doi:10.1095/biolreprod66.4.1104
48. Tsai HW, Legan SJ. Chronic elevation of estradiol in young ovariectomized rats causes aging-like loss of steroid-induced luteinizing hormone surges. Biol Reprod (2001) 64(2):684–8. doi:10.1095/biolreprod64.2.684
49. Treen AK, Luo V, Belsham DD. Phoenixin activates immortalized GnRH and kisspeptin neurons through the novel receptor GPR173. Mol Endocrinol (2016) 30(8):872–88. doi:10.1210/me.2016-1039
50. Woitowich NC, Philibert KD, Leitermann RJ, Wungjiranirun M, Urban JH, Glucksman MJ. EP24.15 as a potential regulator of kisspeptin within the neuroendocrine hypothalamus. Endocrinology (2016) 157(2):820–30. doi:10.1210/en.2015-1580
51. Kauffman AS, Gottsch ML, Roa J, Byquist AC, Crown A, Clifton DK, et al. Sexual differentiation of kiss1 gene expression in the brain of the rat. Endocrinology (2007) 148(4):1774–83. doi:10.1210/en.2006-1540
52. Glucksman M, Philibert K, Woitowich N, Urban J, DeJoseph G. Elucidating the reproductome: system-wide regulation of reproductive neuropeptides. FASEB J (2017) 31(1 Suppl):936.15.
53. Navarro VM, Castellano JM, Fernández-Fernández R, Barreiro ML, Roa J, Sanchez-Criado JE, et al. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology (2004) 145(10):4565–74. doi:10.1210/en.2004-0413
54. Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, et al. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology (2004) 145(9):4073–7. doi:10.1210/en.2004-0431
Keywords: gonadotropin-releasing hormone, gonadotropin-releasing hormone (1–5), GPR101, GPR173, EP24.15, aging, estradiol
Citation: Bauman BM, Yin W, Gore AC and Wu TJ (2017) Regulation of Gonadotropin-Releasing Hormone-(1–5) Signaling Genes by Estradiol Is Age Dependent. Front. Endocrinol. 8:282. doi: 10.3389/fendo.2017.00282
Received: 13 July 2017; Accepted: 09 October 2017;
Published: 27 October 2017
Edited by:
Ishwar Parhar, Monash University Malaysia, MalaysiaReviewed by:
Toni R. Pak, Loyola University Chicago, United StatesGustavo M. Somoza, Instituto de Investigaciones Biotecnologicas-Instituto Tecnologico de Chascomus (IIB-INTECH), Argentina
Copyright: © 2017 Bauman, Yin, Gore and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: T. John Wu, twu@usuhs.edu
 Bradly M. Bauman
Bradly M. Bauman Weiling Yin2
Weiling Yin2 T. John Wu
T. John Wu