- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
Cathepsin L genes, which belonged to cysteine proteases, were a series of multifunctional protease and played important roles in a lot of pathological and physiological processes. In this study, we analyzed the characteristics a cathepsin L (named Mn-CL2) in the female oriental river prawn, Macrobrachium nipponense which was involved in ovary maturation. The Mn-CL2 was1,582 bp in length, including a 978 bp open reading frame that encoded 326 amino acids. The Mn-CL2 was classified into the cathepsin L group by phylogenetic analysis. Real-time PCR (qPCR) analysis indicated that Mn-CL2 was highly expressed in the hepatopancreas and ovaries of female prawns. During the different ovarian stages, Mn-CL2 expression in the hepatopancreas and ovaries peaked before ovarian maturation. In situ hybridization studies revealed that Mn-CL2 was localized in the oocyte of the ovary. Injection of Mn-CL2 dsRNA significantly reduced the expression of vitellogenin. Changes in the gonad somatic index also confirmed the inhibitory effects of Mn-CL2 dsRNA on ovary maturation. These results suggest that Mn-CL2 has a key role in promoting ovary maturation.
Introduction
Macrobrachium nipponense, also called oriental river prawn, is an important freshwater commercial prawn in China. Female prawns have the characteristics of “Short period of sexual maturity” (1). After entering into the breeding season, females can reach to sexual maturity after being raised for about 45 days, and can reproduce multiple generations in the same year. During the breeding season, especially the water temperature reached above 28°C, it takes only 15 days for females to complete an ovarian maturation cycle (from ovarian emptying to the next spawning). Although it makes an ideal materials for studying crustacean reproduction, however, it has been an important problem for the prawn industry for many years. Frequent reproduction leads to multiple generations living together, causing high breeding density, feed consumption and risk of hypoxia. Therefore, it is very important to find out the key genes of ovary maturation.
The generation and accumulation of yolk is not only a necessary prerequisite for the maturation of oocytes, but also a decisive factor in the female reproductive cycle during the periodic maturation of the ovary of crustaceans (2). Subsequently, we constructed transcriptomes of I–V stages of ovary to detect the key genes that play important roles in the maturation (3). According to the comparison analysis of differentially expressed genes and KEGG enrichment in five stages of ovarian transcriptomes, several genes closely related to rapid ovary maturation were screened. The most significant enriched pathway in vitellogenesis stages was lysosomal pathway and we got several cathepsin L genes which might involve in ovary maturation from this pathway (4).
Lysosomal pathway, which contained a variety of hydrolyases, namely, cathepsin, nuclease, phosphatase, glycosidase, lipase, etc., played a main role in decomposing various exogenous and endogenous macromolecules (5). Some lysosomal cathepsin, such as aspartic protease and cysteine proteases identified during ovarian development, were found to be involved in the Vn hydrolysis process of tetrapods and chicken oocytes (6–8). Cathepsin L, which belonged to cysteine proteases, was stored in lysosomes as a proenzyme. They were a series of multifunctional protease and played key roles in many pathological and physiological processes (9–11). In mammals, cathepsin L participated in not only in proteolysis, but also in antigen presentation, tissue regeneration, metastasis, bone apoptosis, and other important life activities (12–15). So far, Cathepsin L genes have been reported in many crustaceans, such as L. vannamei, Metapenaeus ensis, Eriocheir sinensis, Macrobrachium rosenbergii, and M. nipponense (16–20). These Cathepsin L genes all played key roles in immune system of aquatic animal. In insects and fishes, some Cathepsin Ls were proved to be related to the hydrolysis of vitellogenin (21, 22). The role in reproductive system of Cathepsin L were rarely reported in crustaceans.
In this paper, a cathepsin L2 was characterized from the M. nipponense ovary transcriptomes. We analyzed its cDNA sequence and described evolutionary relationship. The expression profiles and tissue location were also processed. An RNAi was used to illustrate the function of cathepsin L2 in regulating the ovary maturation. The aim of this study is to provide a new idea to solve the “rapid sexual maturation” problem in M. nipponense.
Materials and Methods
Ethics Approval and Consent to Participate
In this study, no endangered or protected species were involved. The experimental protocols, methods of this study was approved by the Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
Experimental Animals
Adult healthy female M. nipponense (weighted 0.73 ± 0.16 g) were collected from the Dapu scientific experimental base of Freshwater Fisheries Research Center (Wuxi, China). All prawns were kept in a recirculating freshwater system under the same environment.
Sequence Validation and Bioinformatics Analysis
The full-length cathepsin L2 cDNA sequence of M. nipponense was obtained from the transcriptomes of ovary. All sequences of the ovary transcriptome data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession SAMN11603268-SAMN11603282) under Bioproject PRJNA541783. A pair of primers (listed in Table 1) were designed to verify the open reading frame sequence accuracy. Sequence analysis were carried out according to previous research and the phylogenetic tree was performed by MEGA 7.0 using the neighbor-joining method (23).
Spatio-Temporal Expression of Mn-CL
qPCR was used to quantify mRNA expression levels of Mn-CL with EIF gene as the internal reference and the primers were listed in Table 1 (24). The cerebral ganglion (Cg), eyestalks (E), heart (H), muscles (M), ovaries (O), hepatopancreas (He) and gills (G) were collected from adult female prawns (n = 5). Ovary stages were divided into 5 stages according to previous study (1). Hepatopancreas and ovaries of different ovary stages were collected (n = 5).
In Situ Hybridization (ISH)
The 5 stages ovaries samples of female prawns were collected and fixed in formalin. In situ hybridization (ISH) was carried out as reported previously (25). The probes sequences were listed in Table 1.
RNA Interference
The RNAi primers of Mn-CL were designed containing T7 promoter (listed in Table 1). A GFP gene (green fluorescent protein) was selected as a control and its RNAi primers were listed in Table 1 (26). Ds-RNA of Mn-CL and GFP were produced by Transcript AidTMT7 High Yield Transcription kit (Fermentas, Inc., USA).
The prawn ovaries of the RNA interference for ovarian development are better to be in the same development period at the beginning of experiment, so that the variation of gonadal development index (GSI) can be counted more easily and visually in the end. At the beginning of this experiment, most female prawns were in ovary stage IV. Therefore, in order to select a large number of females whose ovaries were developing at the same time, stage IV prawns were selected. One hundred healthy female prawns (0.73 ± 0.16 g) in stage IV were randomly divided into two groups and a three-week RNAi experiment was carried out (water temperature was 25 °C). The experiment group (N = 50) was injected with 4 μg/g.b.w of ds-Mn-CL each and the injection site is pericardial cavity membrane (25). The same volumes of ds-GFP were applied in control group (N = 50). Ds-RNA of Mn-CL and GFP were injected every five days and five prawns from each group were randomly collected at 1st, 9th, and 17th days after the injection (N =.5). The interference efficiency of was detected in both hepatopancreas and ovary by qPCR. The Vitellogenin (VG) mRNA level was also tested to evaluate the regulation of Mn-CL on vitellogenin gene. Gonad Somatic Index (GSI = gonadal weight/body weight × 100%) was also calculated to illustrate the role of Mn-CL gene in ovarian maturation (1). Ten prawns of the experiment and control groups were randomly sampled on days 1st, 9th, and 17th and the weight of body and ovary were record.
Expression Detection and Statistical Analysis
The relative expression levels of Mn-CL, Vg mRNA and GFP were calculated using the 2−ΔΔCT method with EIF (eukaryotic translation initiation factor 5A) as the reference gene (24, 27). SPSS 23.0 software was used to do statistical analyses and One-way ANOVA and two-tailed t-test were used to analyze statistical differences. All quantitative data described as mean ± standard deviation, and a significant difference was indicated by P <0.05.
Results
Characterization of Cdna Encoding Mn-CL2
The cathepsin L2 of M. nipponense were obtained and named Mn-CL2 (Genbank: OL422141).The Mn-CL2 gene was1,582 bp long, including a 282 bp and 322 bp of 5′and 3′untranslated region, respectively, and a 978 bp open reading frame (ORF) which encoded a 326 amino acid. The estimated molecular mass was 36.68 kDa. The theoretical pI was 4.65. The Mn-CL2 pro-peptide contained a 16 amino acid signal peptide and a 310 amino acid mature peptide. Structural prediction analysis indicated three conserved cysteine protease functional sites, a cysteine at 126–137aa, histidine at 270–279aa, and asparagine at 287–306aa (Figure 1). Mn-CL2 belonged to the cysteine protease family of typical cathepsin, containing conserved sequences GCXGG and E-X3-I-X2-I-F-X3-N-X3-I-X3-N.

Figure 1 Full cDNA sequence and predicted amino acid sequence of Mn-CL2 in M. nipponense. The shaded area indicates the signal peptide. The black underlined boxes represent the active sites of cysteine, histidine, and asparagine. The cathepsin L signature sequences E-X3-R-X2-I-F-X3-N-X3-I-X3-N were in bold and bigger fonts with double line and GCXGG were shown in shadow underlined. The conserved catalytic triad residues (Cys, His, Asn) are boxed, and its amino sequences are underlined. The asterisk (*) represents the termination codon. The polyadenylation tail signal is underlined.
Phylogenetic Analysis and Sequence Alignment of Mn-CL2
Several amino acid sequences of cathepsin L, F, O, C, and B in insects and crustaceans were deduced and a phylogenetic tree was built using the NJ method to elucidate the phylogenetic relationships (Figure 2). The tree contained two distinct branches, which suggested that cathepsin L, F, and O shared common ancestor, as well as cathepsin B and C shared common ancestor. Cathepsin Ls were divided into four branches, which do not fully support traditional taxonomic relationships. Cathepsin L of M. nipponense (AEC22811.1) was clustered with most Cathepsin Ls of crustaceans and insects. However, the Cathepsin L1 (MW684082) and Mn-CL2 of M. nipponense, which obtained from ovary transcriptomes, were clustered in distinct branches, indicating significant differences in structure.
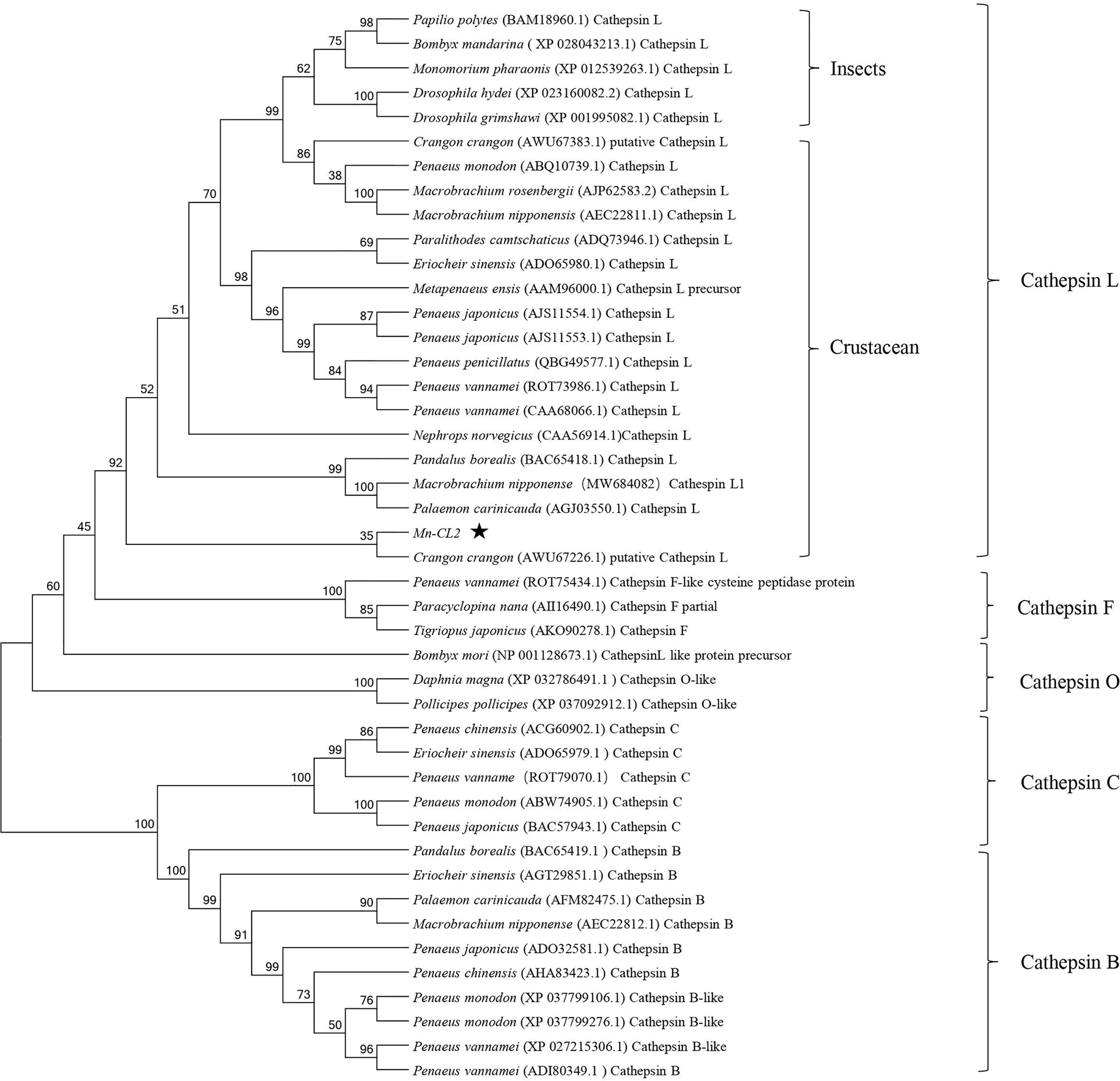
Figure 2 Multiple amino acid alignment and structure prediction of Mn-CL2 amino acid sequence with cathepsin L from other crustaceans. Cathepsin L1 of M. nipponense, MW684082; Cathepsin Ls (M. nipponense, AEC22811.1; M. rosenbergii, AHW49157.1; Litopenaeus vannamei, CAA68066; P. camtschaticus, AGJ03550.1; Penaeus japonicus, AJS11554.1; Penaeus monodon, ABQ10739.1; Eriocheir sinensis, ADO65980.1). ERFNIN, GCNGG, and GNFD motifs are represented by black boxes. The active sites are marked with red arrows. The symbol star indicated Mn-CL2.
Multiple alignments of Cathepsin L of crustaceans were built with DNAMAN 6.0 (Figure 3).Homology analysis showed that Mn-CL2 had the highest homology with Penaeus japonicus (53.19%). Multiple alignments of cathepsin Ls from other crustaceans were performed. The Mn-CL2 shared amino acid identity with other crustaceans (M. nipponense, M. rosenbergii, L. vannamei, Paralithodes camtschaticus, P. japonicus, P. monodon, and E. sinensis) were 45.32, 50.77, 51.37, 50.3, 46.04, and 51.98%, respectively. Comparison of amino acid sequences with other crustaceans showed that the three catalytic active sites of cysteine protease were highly conserved.
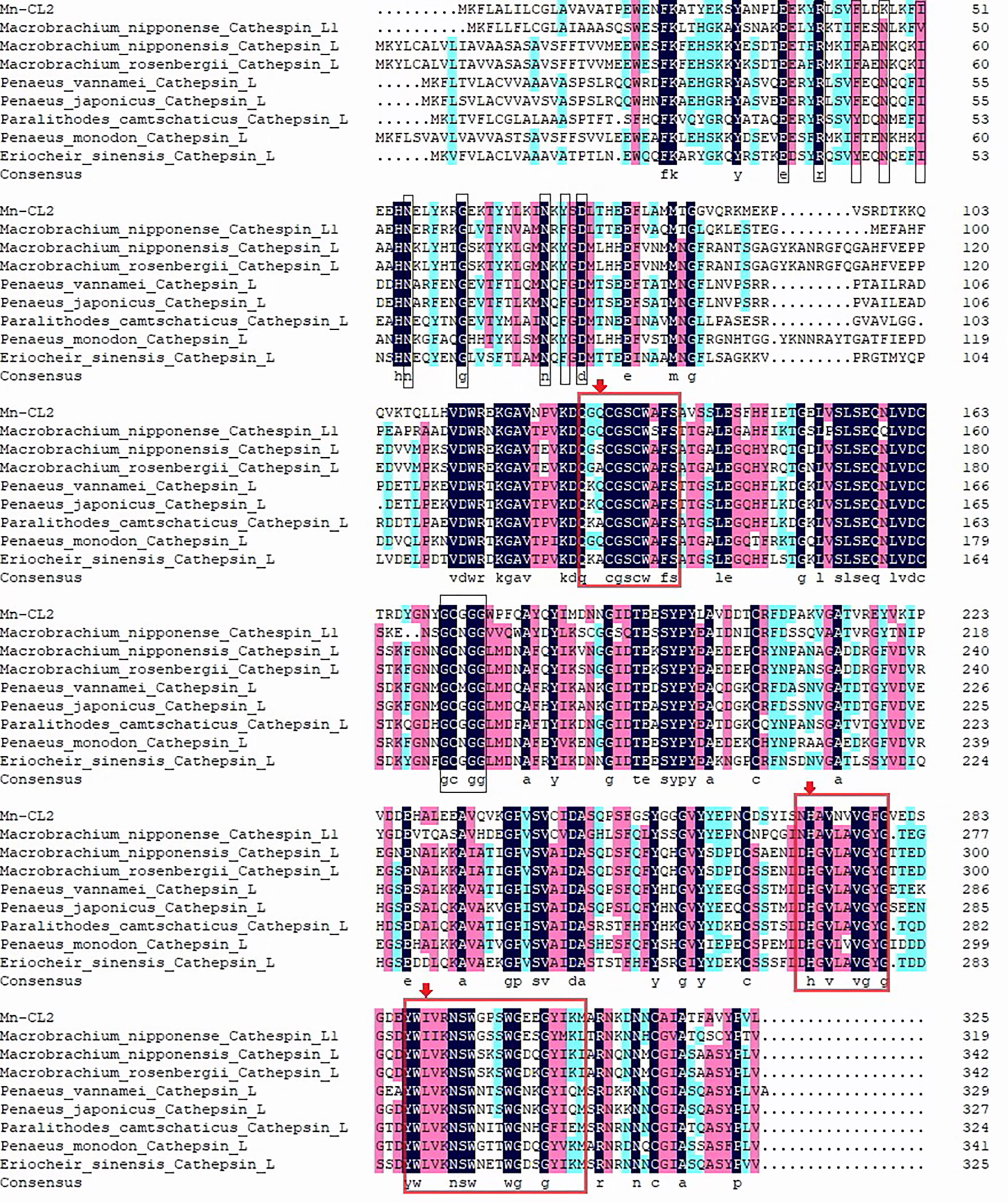
Figure 3 Phylogenetic tree of Mn-CL2 amino acid sequence in different groups. The numbers below the node indicate the bootstrap value.
Tissue Distribution and Expression Patterns of Mn-CL2 in Different Ovary Stages
The spatio-temporal expression was analyzed in female prawns. Tissue distribution expression was showed in Figure 4. The Mn-CL2 were detected in eyestalks (E), cerebral ganglion (Cg), heart (H), hepatopancreas (He), gills (G), muscles (M), and ovaries (O). The results showed that Mn-CL2 hardly expressed in cerebral ganglion, heart, eyestalks, and gill. It expressed in muscle weakly. The extremely high expression were detected in hepatopancreas and followed by ovary (P <0.01).
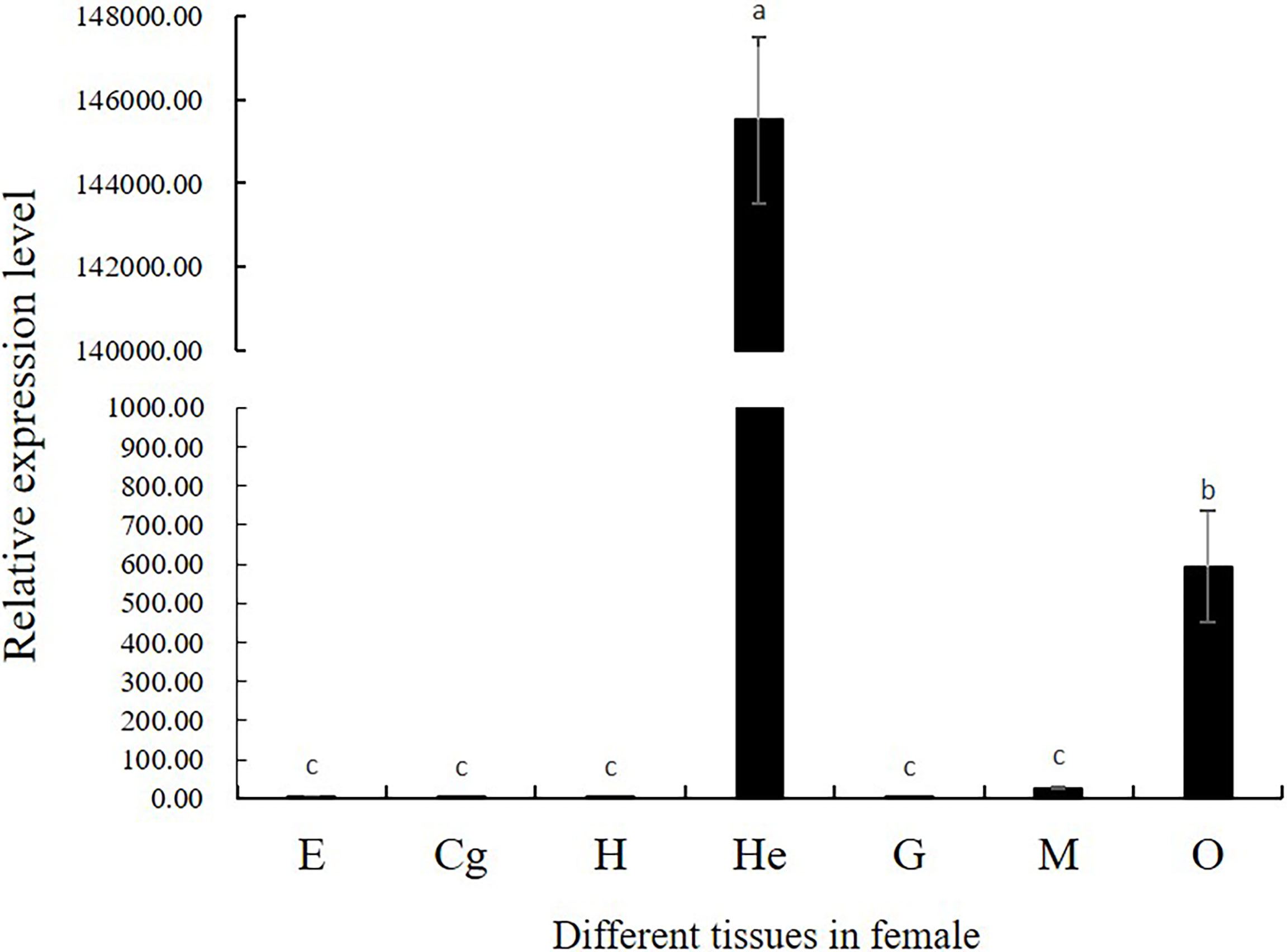
Figure 4 Tissue distribution of Mn-CL2. E, eyestalk; Cg, cerebral ganglion; H, heart; He, hepatopancreas; G, gill; M, muscle; O, ovary. Bars with different small letters indicated significant differences (P < 0.05).
According to the results of tissue distribution expression patterns of Mn-CL2, we further analyzed Mn-CL2 expression profiles of hepatopancreas and ovary in five ovarian stages. In ovaries (Figure 5), the expression of Mn-CL2 was relatively stable without significant change from stage I to stage II (P >0.05). It increased from stage II to stage III significantly, and the expression level in stage III was 10 times higher than that in stage II (P <0.05). Subsequently, its expression dropped dramatically during stage IV, which was 1/15 of what it was at stage III (P <0.05). Mn-CL2 had the lowest expression in ovarian stage V (P <0.05). The expression profiles of Mn-CL2 in hepatopancreas was very different from that in ovary (Figure 5). Mn-CL2 of hepatopancreas maintained a high level expression in each stages of ovary. We found that Mn-CL2 was relative lower in stages III and V (P <0.05) and its expression peaked was detected in stages I and IV (P >0.05).
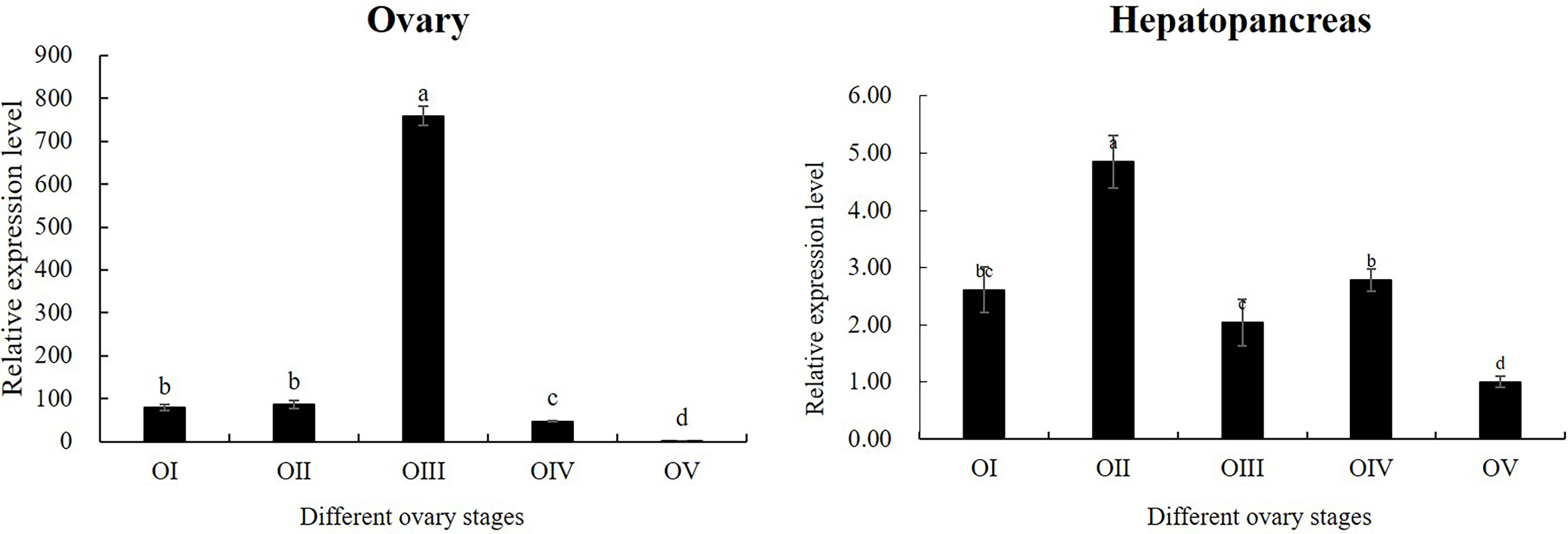
Figure 5 Expression patterns of Mn-CL2 in the hepatopancreas and ovary at different ovarian stages. Different ovarian stages expressions, OI undeveloped stage, OII developing stage, OIII nearly-ripe stage, OIV ripe stage, OV spent stage. Statistical analyses were performed with one-way ANOVA analysis. Data are shown as mean ± SD (n = 5). Different letters denote significant differences (P < 0.05). Bars with different small letters indicated significant differences (P < 0.05).
Mn-CL2 Localization in the Ovary by ISH
Mn-CL2 was detected by ISH in five stages of ovaries to show the cellular localizations (Figure 6). The ISH results detected Mn-CL2 signal was in all ovary stages. The Mn-CL2 was visualized in oocyte including yolk granule, follicle cell, cytoplasmic membrane, nucleus, and follicle membrane. It mainly expressed in nucleus and follicle membrane. It showed that the signal of Mn-CL2 in the ovary was significantly enhanced during stage II (primary vitellogenesis) to stage III (secondary vitellogenesis), and gradually weakened after stage IV. The signal of Mn-CL2 was weakest in stage I (oogonium proliferation stage).
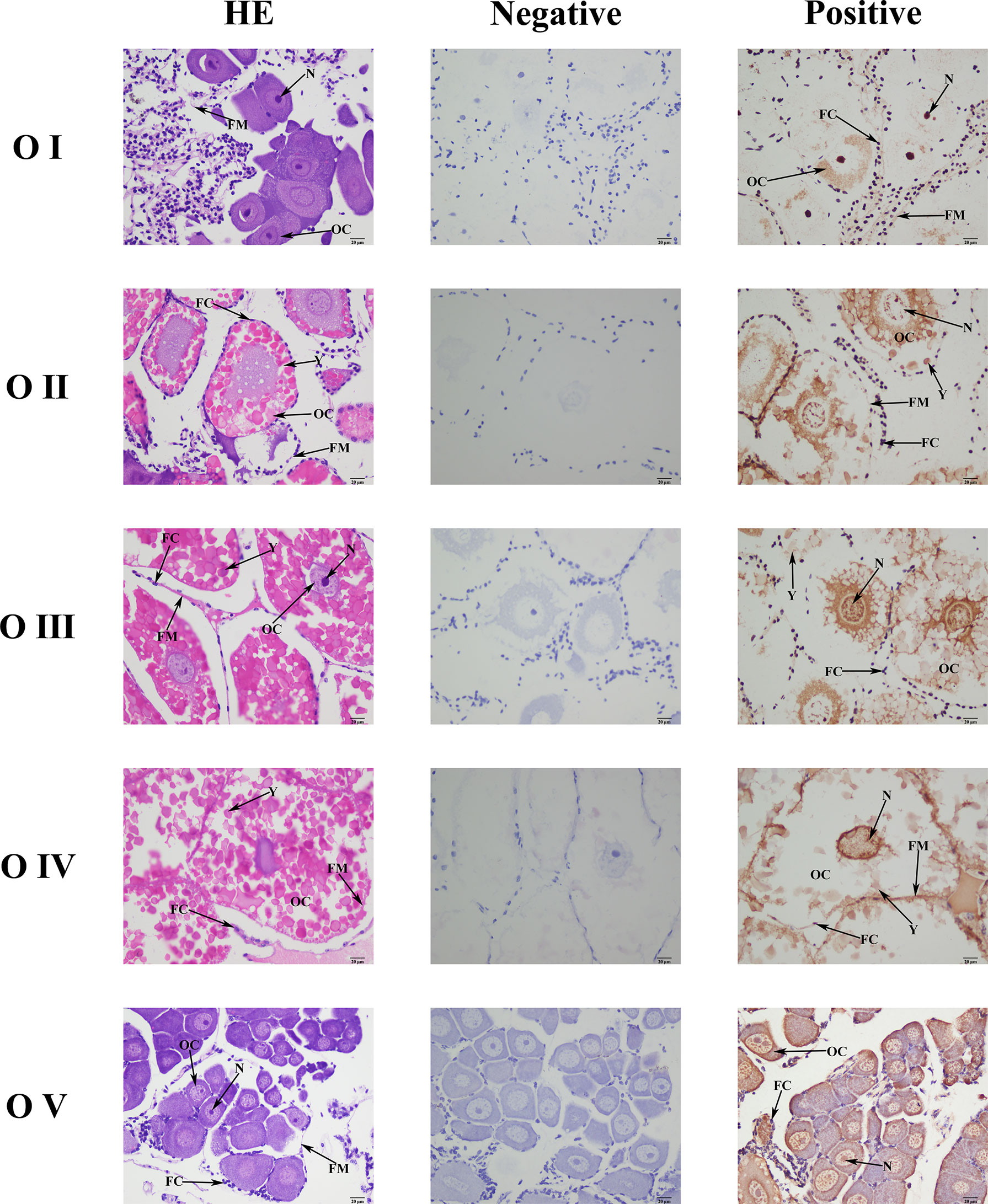
Figure 6 Location of Mn-CL2 detected in the ovary by in situ hybridization. A Photograph of M. nipponense ovary in ovarian cycle. OC, oocyte; N, nucleus; CM, cytoplasmic membrane; Y, yolk granule; FC, follicle cell; FM, follicle membrane; Scale bars: ×400.
The Role of Mn-CL2 in Ovary Maturation
The function of Mn-CL2 in ovary maturation was revealed by a three-week RNA interference. The results showed that the dsRNA of Mn-CL2 had a remarkable effect on the ninth day after injection, the expression level of Mn-CL2 was downregulated remarkably by 99.67% on the 9th day after injection (P <0.01) (Figure 7A). The Mn-CL2 expression remained at a very low level which was less than 8% of the control group, till the end of the experiment (on the 17th day). We also tested Vitellogenin (Vg) transcript after dsRNA of Mn-CL2 injection. The Vg expression decreased by 98.74% after RNA interference on the 9th day (P <0.01). It was almost 200 times higher in the control group than in the interference group (P <0.01) at the end of the experiment (Figure 7B).
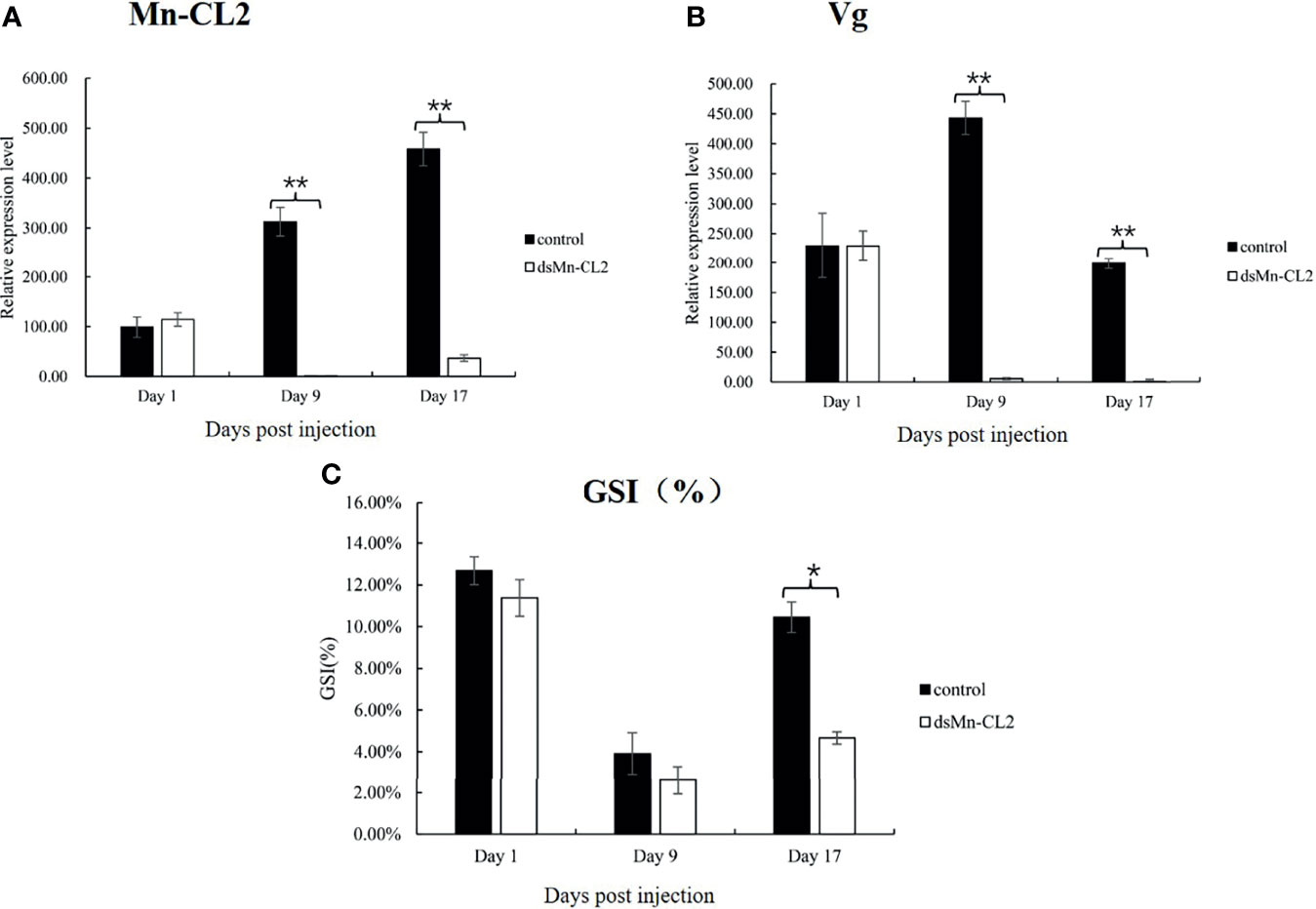
Figure 7 Function analysis of Mn-CL2 by RNAi. (A) Efficiency of RNAi-Mn-CL2 knockdown in ovary. (B) Vg transcript after Mn-CL2 dsRNA injection. (C) Changes in GSI (%) of female M. nipponense after injection with Mn-CL2 dsRNA. Data are presented as mean ± SD (n = 6); *denotes statistical significance of P < 0.05. **denotes statistical significance of P < 0.01.
During the interference experiment, GSI (Gonad Somatic Index) was also tracked (Figure 7C). Before injection, the average GSI was between 11 and 12% approximately in both control and interference groups while the ovaries of the prawns were almost in stage IV. Later, the ovaries developed from full to empty and enters the next ovarian maturation cycle. The GSI results indicated that on the 1st and 9th days after injection, the ovaries development had no difference between control and interference group (P >0.05). At the end of the experiment, the GSI in interference group was only half that of control group (P <0.05).
Discussion
In this study, we identified a novel cathepsin L gene from M. nipponense named Mn-CL2, and put insight into the Mn-CL2 changes during ovary development, which are the most crucial physiological and morphological processes. Mn-CL2 was suggested to be involved in a secretary mechanism by containing a typical amino acid signal peptide (28). It belonged to the cysteine family of typical cathepsin, and its precursor domain contains typical cathepsin L conserved sequences GCXGG. The 4 amino acids in the GCXGG motif are conserved, while the X represents different amino acids. The X in cathepsin L and L1 of M. nipponense, M. rosenbergii, and P. monodon were aspartic acid N, in M. ensis is cysteine C, in L. vannamei is methionine M and in E. sinensis is glycine G. In Mn-CL2, The X was glycine G, which was completely different from cathepsin L and L1 of M. nipponense. It is speculated that the cysteine residues in this motif are located at the corner position and are involved in disulfide bonds formation, which are related to the stability of the enzyme activity region structure and play an important role in the formation of protein structure (29). Therefore Mn-CL2 may have similar functions and effects of other species cathepsin L. Crustacean cathepsin Ls contained 4 distinct branches. The high percent identity in residues across taxa indicated that cathepsin L, F and O evolved from gene duplication events (18). Three cathepsin L of M. nipponense belonged to different branches separately, suggesting the functional variation of Cathepsin L in the same species group.
Tissue distribution results proved that the most important tissue for Mn-CL2 expression is hepatopancreas. Hepatopancreas is the most important site of proteolysis and apoptosis in crustaceans and cathepsin L not only participates in proteolysis, but also plays key roles in life activities such as apoptosis, tissue regeneration and antigen presentation (30). This is similar to the results of cathepsin L studies in other species (18, 22, 31). In these studies, cathepsin L was highly expressed not only in hepatopancreas, but also in brain, heart, gill, hemolymph, and other tissues, which suggested it was closely related to the growth and immune functions. The big difference was that Mn-CL2 found in this study, as well as Mn-CTS L1 (another cathepsin L detected in the ovary transcriptomes of M. nipponense form our lab) expressed highly in ovary. Especially, Mn-CL2 had rarely expression in other tissues except for hepatopancreas and ovary (4). These results gave evidence that Mn-CL2 play a special role in the ovaries.
In order to further prove the function of Mn-CL2 in ovarian development of M. nipponense, qPCR was performed in hepatopancreas and ovaries at different ovarian stages. Previous studies (2) suggested that the changes from stages I to II of ovarian development of M. nipponense were due to the development of oocytes and cytoplasmic synthesis. It belonged to the early stage of ovarian development. Stages II–III were the rapid ovarian development stage, oocytes continue to grow significantly and yolk granules in the cytoplasm accumulate rapidly. Stage IV was the ovary mature stage in preparing for spawning and Stage V was empty stage after spawning. Mn-CL2 expression of a marked increase in the amount and reach the maximum, in stage III presumably because Stage III was rapid vitellogenesis stage. It was also a period of rapid growth and proliferation of oocytes, mature egg form requires a lot of yolk proteins provide nutrients and energy. In hepatopancreas, Mn-CL2 maintained high expression level continuously, suggesting that hepatopancreas is the main production site. Previous studies in freshwater prawn and crab species gave proves that Vg was synthesized in hepatopancreas, and then transported to the oocyte through the hemolymph (32, 33). The cellular localizations was detected by ISH and Mn-CL2 mRNA were examined in all in five ovary stages. We found Mn-CL2 mRNA signals were stronger in stages II and III which is well distributed in the nucleus and around yolk. cathepsin L was reported to be distributed around the cytoplasm and yolk granules in Xenopus (34) which was consistent with the results of this study. All the results indicated that as the lysosome protein, Mn-CL2 was produced in hepatopancreas and played a key role in ovarian maturation.
The cathepsin L, and also B and D, have been proved to play crucial roles in yolk formation in teleosts. In teleosts, vitellogenin (Vg) was processed into smaller yolk proteins after it was entered into oocyte by incorporated receptor (22, 35). In the oocyte of marine and freshwater teleosts, the cathepsin L and B were involved in yolk proteolysis (36, 37). These cathepsins were supposed to be essential enzymes in yolk compartments mediated by H+-ATPase-acidification (38, 39). It was also reported that cathepsin L participated in fish and amphibian embryos and larvae yolk protein degradation (40–43). Cathepsin L have also been reported in many crustacean species, such as E. sinensis, M. rosenbergii, and L. vannamei, mainly functioned for food digestion and protein degradation to provide energy (17, 18, 44). Researches in other aquatic organisms focuses on physiological processes such as growth, immunity, embryonic development and energy storage (15, 16, 31). In crustaceans, the role of cathepsin L involved in ovary maturation were rarely reported.
From the transcriptomes of Stages I–V stages of ovary in M. nipponense, we characterized several cathepsin L genes which showed evidences that involve in ovary maturation (3, 4) and more details about their roles in crustaceans needs to be clarified and demonstrated. In this paper, RNAi was used to illustrate the special roles of Mn-CL2 in ovary maturation of M. nipponense. RNAi results showed that Mn-CL2 could downregulated Vg expression, which was different from Mn-CTS L1. Vg is usually expressed in large quantities during specific developmental periods of individuals, and its production, processing and even transportation depend on the regulatory effects of corresponding hormones. We hypothesized that Mn-CL2 is an important upstream response element of genes involved in regulating ovarian development and Vg production. Mn-CL2 expression in ovary was significantly inhibited from the 9th day after the dsRNA injection and the inhibitory effects lasted until the end of the experiment (17th day). The results proved that ds-Mn-CL2 was effective and specific for Mn-CL2 function analysis. The ds-Mn-CL2 also showed extremely inhibitory effects on Vg gene expression suggesting Mn-CL2 may play an important role in inhibiting production of yolk protein. GSI result directly showed the inhibitory effect of ds-Mn-CL2 on ovary maturation. At the end of the experiment, the GSI in control group increased almost to the peak, while the GSI in interference group was only half that of control group. All these results suggested that Mn-CL2 played an important role in yolk generation, and inhibiting the expression of Mn-CL2 gene can effectively inhibit ovarian maturation.
Conclusion
In this study, we reported new basic knowledge about a novel cathepsin L in crustaceans. The function analysis of Mn-CL2 showed that inhibiting Mn-CL2 can effectively inhibit ovarian maturation. Our data provided new information about the regulation of ovarian maturation in M. nipponense and may be helpful for solving the problem of rapid M. nipponense development in the aquaculture industry. However, the relationship between other regulatory genes and ovarian maturation requires further study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
Author Contributions
Designed the study, HQ and HF. Carried out the experiments and writing original draft, SJ and WX. Provided technical assistance, WZ and JZ. Methodology and data curation, SJ and WX. Resources, DC and YG. Software, YW. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the Jiangsu Agricultural Industry Technology System (JATS [2021] 515); the National Key R&D Program of China (2018YFD0901303); the Central Public-interest Scientific Institution Basal Research Fund CAFS (2020TD36); the China Agriculture Research System-48 (CARS-48); and the New cultivar breeding Major Project of Jiangsu province (PZCZ201745).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thank you for the Jiangsu Province Platform for the Conservation and Utilization of Agricultural Germplasm.
References
1. Qiao H, Xiong YW, Zhang WY, Fu HT, Jiang SF, Sun SM, et al. Characterization, Expression, and Function Analysis of Gonad-Inhibiting Hormone In Oriental River Prawn, Macrobrachium Nipponense and Its Induced Expression by Temperature. Comp Biochem Physiol Molecular Integr Physiol (2015) 185:1–8. doi: 10.1016/j.cbpa.2015.03.005
2. Bai HK, Qiao H, Li FJ, Fu HT, Sun SM, Zhang WY, et al. Molecular Characterization and Developmental Expression of Vitellogenin in the Oriental River Prawn Macrobrachium Nipponense and the Effects of RNA Interference and Eyestalk Ablation on Ovarian Maturation. Gene (2015) 562:22–31. doi: 10.1016/j.gene.2014.12.008
3. Zhang YN, Fu Y, Jiang SF, Qiao H, Xiong YW, Fu HT, et al. Comparative Metabolomics Analysis of Ovarian Developmental Stages in Macrobrachium Nipponense. Comp Biochem Physiol Part D Genomics Proteomics (2019) 34:5. doi: 10.1016/j.cbd.2019.100648
4. Zhu JP, Fu HT, Qiao H, Jin SB, Zhang WY, Jiang SF, et al. Expression and Functional Analysis of Cathepsin L1 in Ovarian Development of the Oriental River Prawn, Macrobrachium Nipponense. Aquacult Rep (2021) 20:10. doi: 10.1016/j.aqrep.2021.100724
5. Wang MJ, Hu YH, Li MX, Xu Q, Zhang XL, Wang XS, et al. A Proteomics Analysis of the Ovarian Development in Females of Haemaphysalis Longicornis. Exp Appl Acarol (2020) 80(2):289–309. doi: 10.1007/j.cbpa10493-020-00469-3
6. Opresko LK, Karpf RA. Specific Proteolysis Regulates Fusion Between Endocytic Compartments in Xenopus Oocytes. Cell (1987) 51:557–68. doi: 10.1016/j.cell0092-8674(87)90125-5
7. Raikhel AS, Dhadialla TS. Accumulation of Yolk Proteins in Insect Oocytes. Annu Rev Entomol (1992) 37:217–51. doi: 10.1146/annurev.en.37.010192.001245
8. Abreu LA, Valle D, Manso PP, Façanha AR, Pelajo-Machado M, Masuda H, et al. Proteolytic Activity of Boophilus Microplus Yolk Pro-Cathepsin D (BYC) Iscoincident With Cortical Acidification During Embryogenesis. Insect Biochem Mol Biol (2004) 34(5):443–9. doi: 10.1016/j.ibmb.2004.01.006
9. Terra WR, Dias RO, Ferreira C. Recruited Lysosomal Enzymes as Major Digestive Enzymes in Insects. Biochem Soc Trans (2019) 47:615–23. doi: 10.1042/bst20180344
10. Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Feng WU, et al. Cathepsin L Upregulation-Induced EMT Phenotype Is Associated With the Acquisition of Cisplatin or Paclitaxel Resistance in A549 Cells. Acta Pharmacol Sin (2016) 37:1606–22. doi: 10.1038/aps.2016.93
11. Karrer KM, Peiffer SL. Ditomas ME. Two Distinct Gene Subfamilies Within the Family of Cysteine Protease Genes. Proc Natl Acad Sci (1993) 90:3063–7. doi: 10.1073/pnas.90.7.3063
12. Dohchin A, Suzuki J, Seki H, Masutani M, Shiroto H, Kawakami Y. Immunostained Cathepsins B and L Correlate With Depth of Invasion and Different Metastatic Pathways in Early Stage Gastric Carcinoma. Cancer (2000) 89:482–7. doi: 10.1002/1097-0142(20000801)89:3<482::Aid-cncr2>3.0.Co;2-5
13. Furuyama N, Fujisawa Y. Distinct Roles of Cathepsin K and Cathepsin L in Osteoclastic Bone Resorption. Endocr Res (2000) 26:189–204. doi: 10.3109/07435800009066161
14. Lindeman JH, Hanemaaijer R, Mulder A, Dijkstra S, Szuhai K, Bromme D, et al. Cathepsin K Is the Principal Protease in Giant Cell Tumor of Bone. Am J Pathol (2004) 165:593–600. doi: 10.1016/s0002-9440(10)63323-8
15. Chen J, Zhang L, Yang N, Cao M, Tian M, Fu Q, et al. Characterization of the Immune Roles of Cathepsin L in Turbot (Scophthalmus Maximus L.) Mucosal Immunity. Fish Shellfish Immunol (2020) 97:322–35. doi: 10.1016/j.fsi.2019.12.005
16. Hu KJ, Leung PC. Shrimp Cathepsin L Encoded by An Intronless Gene Has Predominant Expression in Hepatopancreas, and Occurs in the Nucleus of Oocyte. Comp Biochem Physiol B-Biochem Mol Biol (2004) 137:21–33. doi: 10.1016/j.cbpc.2003.09.010
17. Zhao ZY, Yin ZX, Weng SP, Guan HJ, Li SD, Xing K, et al. Profiling of Differentially Expressed Genes in Hepatopancreas of White Spot Syndrome Virus-Resistant Shrimp (Litopenaeus Vannamei) by Suppression Subtractive Hybridisation. Fish Shellfish Immunol (2007) 22:520–34. doi: 10.1016/j.fsi.2006.07.003
18. Li WW, Jin XK, He L, Jiang H, Gong YN, Xie YN, et al. Molecular Cloning, Characterization, Expression and Activity Analysis of Cathepsin L in Chinese Mitten Crab Eriocheir sinensis. Fish Shellfish Immunol (2010) 29:1010–8. doi: 10.1016/j.fsi.2010.08.007
19. Zhao W, Chen L, Zhang F, Wu P, Li E, Qin J. Molecular Characterization of Cathepsin L Cdna and Its Expression During Oogenesis and Embryogenesis in the Oriental River Prawn Macrobrachium Nipponense (Palaemonidae). Genet Mol Res (2013) 12:5215–25. doi: 10.4238/2013.October.30.6
20. Shen JD, Cai QF, Yan LJ, Du CH, Liu GM, Su WJ, et al. Cathepsin L Is An Immune-Related Protein in Pacific Abalone (Haliotis Discus Hannai) - Purification and Characterization. Fish Shellfish Immunol (2015) 47:986–95. doi: 10.1016/j.fsi.2015.11.004
21. Zhao XF, An XM, Wang JX, Dong DJ, Du XJ, Sueda S, et al. Expression of the Helicoverpa Cathepsin B-Like Proteinase During Embryonic Development. Arch Insect Biochem Physiol (2005) 58:39–46. doi: 10.1002/arch.20030
22. Tingaud-Sequeira A, Cerdà J. Phylogenetic Relationships and Gene Expression Pattern of Three Different Cathepsin L (Ctsl) Isoforms in Zebrafish: Ctsla Is the Putative Yolk Processing Enzyme. Gene (2007) 386:98–106. doi: 10.1016/j.gene.2006.08.018
23. Wang YB, Jin SB, Fu HT, Qiao H, Sun SM, Zhang WY, et al. Identification and Characterization of the DMRT11E Gene in the Oriental River Prawn Macrobrachium Nipponense. Int J Mol Sci (2019) 20(7):1734. doi: 10.3390/ijms20071734
24. Hu YN, Fu HT, Qiao H, Sun SM, Zhang WY, Jin SB, et al. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium Nipponense. Int J Mol Sci (2018) 19:2258. doi: 10.3390/ijms19082258
25. Li F, Qiao H, Fu HT, Sun SM, Zhang WY, Jin SB, et al. Identification and Characterization of Opsin Gene and Its Role in Ovarian Maturation in the Oriental River Prawn Macrobrachium Nipponense. Comp Biochem Physiol B-Biochem Mol Biol (2018) 218:1–12. doi: 10.1016/j.cbpb.2017.12.016
26. Zhang SB, Jiang P, Wang ZQ, Long SR, Liu RD, Zhang X, et al. Dsrna-Mediated Silencing of Nudix Hydrolase in Trichinella Spiralis Inhibits the Larval Invasion and Survival in Mice. Exp Parasitol (2016) 162:35–42. doi: 10.1016/j.exppara.2016.01.005
27. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(T)(-Delta Delta C) Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
28. Le Boulay C, Sellos D. Van Wormhoudt A. Cathepsin L Gene Organization in Crustaceans. Gene (1998) 218:77–84. doi: 10.1016/s0378-1119(98)00385-0
29. Wang Y, Zhao B, Ding F. Jiang, X. Gut-Specific Expression of Cathepsin L and B in Amphioxus Branchiostoma Belcheri Tsingtauense Larvae. Eur J Cell Biol (2008) 87:185–93. doi: 10.1016/j.ejcb.2007.10.002
30. Caceci T, Neck KF, Lewis DH, Sis RF. Ultrastructure of the Hepatopancreas of the Pacific White Shrimp, Penaeus-Vannamei (Crustacea, Decapoda). J Mar Biol Assoc UK (1988) 68:323–37. doi: 10.1017/s002531540005222x
31. Hu KJ, Leung PC. Food Digestion by Cathepsin L and Digestion-Related Rapid Cell Differentiation in Shrimp Hepatopancreas. Comp Biochem Physiol B-Biochem Mol Biol (2007) 146:69–80. doi: 10.1016/j.cbpb.2006.09.010
32. Ara F, Damrongphol P. Vitellogenin Gene Expression at Different Ovarian Stages in the Giant Freshwater Prawn, Macrobrachium Rosenbergii, and Stimulation by 4-Nonylphenol. Aquacult Res (2014) 45:320–6. doi: 10.1111/j.1365-2109.2012.03229.x
33. Urtgam S, Treerattrakool S, Roytrakul S, Wongtripop S, Prommoon J, Panyim S, et al. Correlation Between Gonad-Inhibiting Hormone and Vitellogenin During Ovarian Maturation in the Domesticated. Penaeus Monodon Aquacult (2015) 437:1–9. doi: 10.1016/j.aquaculture.2014.11.014
34. Miyata S, Nishibe Y, Kihara HK. Effects on Properties of A Thiol Protease From Xenopus Embryos of Changes in Substrate and Assay Conditions. Cell Biol Int (1995) 19:333–8. doi: 10.1006/cbir.1995.1076
35. Fabra M, Raldua D, Bozzo MG, Deen PMT, Lubzens E, Cerda J. Yolk Proteolysis and Aquaporin-1o Play Essential Roles to Regulate Fish Oocyte Hydration During Meiosis Resumption. Dev Biol (2006) 295:250–62. doi: 10.1016/j.ydbio.2006.03.034
36. Carnevali O, Cionna C, Tosti L, Lubzens E, Maradonna F. Role of Cathepsins in Ovarian Follicle Growth and Maturation. Gen Comp Endocrinol (2006) 146:195–203. doi: 10.1016/j.ygcen.2005.12.007
37. LaFleur GJ, Raldua D, Fabra M, Carnevali O, Denslow N, Wallace RA, et al. Derivation of Major Yolk Proteins From Parental Vitellogenins and Alternative Processing During Oocyte Maturation in Fundulus Heteroclitus. Biol Reprod (2005) 73:815–24. doi: 10.1095/biolreprod.105.041335
38. Matsubara T, Nagae M, Ohkubo N, Andoh T, Sawaguchi S, Hiramatsu N, et al. Multiple Vitellogenins and Their Unique Roles in Marine Teleosts. Fish Physiol Biochem (2003) 28:295–9. doi: 10.1023/b:Fish.0000030559.71954.37
39. Raldua D, Fabra M, Bozzo MG, Weber E, Cerda J. Cathepsin B-Mediated Yolk Protein Degradation During Killifish Oocyte Maturation Is Blocked by An H+-Atpase Inhibitor: Effects on the Hydration Mechanism. Am J Physiol-Regul Integr Comp Physiol (2006) 290:R456–66. doi: 10.1152/ajpregu.00528.2005
40. Sire MF, Babin PJ. Vernier JM. Involvement of the Lysosomal System in Yolk Protein Deposit and Degradation During Vitellogenesis and Embryonic-Development in Trout. J Exp Zool (1994) 269:69–83. doi: 10.1002/jez.1402690109
41. Yoshizaki N, Yonezawa S. Cysteine Proteinase Plays A Key Role for the Initiation of Yolk Digestion During Development of Xenopus Laevis. Dev Growth Differ (1998) 40:659–67. doi: 10.1046/j.1440-169X.1998.t01-4-00010.x
42. Kestemont P, Cooremans J, Abi-Ayad A, Melard C. Cathepsin L in Eggs and Larvae of Perch Perca Fluviatilis: Variations With Developmental Stage and Spawning Period. Fish Physiol Biochem (1999) 21:59–64. doi: 10.1023/a:1007714314072
43. Kwon JY, Prat F, Randall C, Tyler CR. Molecular Characterization of Putative Yolk Processing Enzymes and Their Expression During Oogenesis and Embryogenesis in Rainbow Trout (Oncorhynchus Mykiss). Biol Reprod (2001) 65:1701–9. doi: 10.1095/biolreprod65.6.1701
Keywords: Macrobrachium nipponense, cathepsin L, ovary maturation, mRNA expression, RNA interference
Citation: Jiang S, Xiong Y, Zhang W, Zhu J, Cheng D, Gong Y, Wu Y, Qiao H and Fu H (2022) Molecular Characterization of a Novel Cathepsin L in Macrobrachium nipponense and Its Function in Ovary Maturation. Front. Endocrinol. 12:816813. doi: 10.3389/fendo.2021.816813
Received: 17 November 2021; Accepted: 13 December 2021;
Published: 10 January 2022.
Edited by:
Jie Ma, University of Idaho, United StatesCopyright © 2022 Jiang, Xiong, Zhang, Zhu, Cheng, Gong, Wu, Qiao and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtuo Fu, fuht@ffrc.cn; Hui Qiao, qiaoh@ffrc.cn
†These authors have contributed equally to this work and share first authorship
 Sufei Jiang
Sufei Jiang Yiwei Xiong1†
Yiwei Xiong1† Hui Qiao
Hui Qiao Hongtuo Fu
Hongtuo Fu