- 1 Siomi Lab, Department of Molecular Biology, Keio University School of Medicine, Tokyo, Japan
- 2 Japan Science and Technology Agency, Core Research for Evolutional Science and Technology, Saitama, Japan
PIWI-interacting RNAs (piRNAs) are germline-specific small non-coding RNAs that form piRNA-induced silencing complexes (piRISCs) by associating with PIWI proteins, a subclade of the Argonaute proteins predominantly expressed in the germline. piRISCs protect the integrity of the germline genome from invasive transposable DNA elements by silencing them. Multiple piRNA biogenesis factors have been identified in Drosophila. The majority of piRNA factors are localized in the nuage, electron-dense non-membranous cytoplasmic structures located in the perinuclear regions of germ cells. Thus, piRNA biogenesis is thought to occur in the nuage in germ cells. Immunofluorescence analyses of ovaries from piRNA factor mutants have revealed a localization hierarchy of piRNA factors in female nuage. However, whether this hierarchy is female-specific or can also be applied in male gonads remains undetermined. Here, we show by immunostaining of both ovaries and testes from piRNA factor mutants that the molecular hierarchy of piRNA factors shows gender-specificity, especially for Krimper (Krimp), a Tudor-domain-containing protein of unknown function(s): Krimp is dispensable for PIWI protein Aubergine (Aub) nuage localization in ovaries but Krimp and Aub require each other for their proper nuage localization in testes. This suggests that the functional requirement of Krimp in piRNA biogenesis may be different in male and female gonads.
Introduction
PIWI-interacting RNAs (piRNAs) are germline-specific small RNAs that function in RNA silencing. In Drosophila, piRNAs arise mostly from transposable elements and associate with germline-specific Argonaute proteins (PIWI proteins), Aubergine (Aub), Piwi, and Ago3, to guide PIWI proteins to their targets (Saito et al., 2006, 2009; Vagin et al., 2006; Aravin et al., 2007; Brennecke et al., 2007; Gunawardane et al., 2007; Nishida et al., 2007). Mutations in the piwi genes lead to de-repression of transposable elements (Cox et al., 1998; Schmidt et al., 1999; Vagin et al., 2004; Kalmykova et al., 2005; Klattenhoff et al., 2007; Li et al., 2009). Mutations in the piRNA-producing genomic loci, the piRNA clusters, show a similar phenotype (Brennecke et al., 2007). Thus, both PIWI and piRNAs are necessary for transposon silencing in the germline. Through this mechanism, the PIWI–piRNA complex, also called the piRNA-induced silencing complex (piRISC), protects the integrity of the germline genome from invasive transposable DNA elements and ensures oogenesis and spermatogenesis.
PIWI-interacting RNAs are produced through two mechanisms, the primary processing pathway and the amplification loop (or the Ping-Pong cycle; Aravin et al., 2007; Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009; Malone et al., 2009; Siomi et al., 2011). Primary piRNAs are first produced through the primary processing pathway; then, the majority of primary piRNAs are amplified by the Ping-Pong cycle to maintain a high expression level of piRNAs in the germline. Maternally deposited piRNAs also serve as primary piRNAs to induce the Ping-Pong cycle in embryos.
Primary piRNAs are mostly antisense to transposon mRNAs, show a strong bias for uracil at the 5′ end (1U bias) and are favorably loaded onto Aub and Piwi (Li et al., 2009; Malone et al., 2009). Primary piRNAs in ovarian somatic cells require the female sterile (1) Yb (Yb) gene for their accumulation (Olivieri et al., 2010; Saito et al., 2010; Qi et al., 2011). Yb is necessary for self-renewal of germline and somatic stem cells in ovaries (Szakmary et al., 2009). Germ cells do not express Yb and, therefore, Yb is unnecessary for primary piRNA production in germ cells. Zucchini (zuc) and armitage (armi) are necessary for primary piRNA production in both ovarian somas (Haase et al., 2010; Olivieri et al., 2010; Saito et al., 2010) and germ cells (Malone et al., 2009). Vasa, maelstrom (mael), krimper (krimp), armi, and spindle-E (spn-E) are necessary for primary piRNA accumulation in testes (Nagao et al., 2010). Other genes necessary for primary piRNA production remain elusive.
The Ping-Pong cycle performs piRNA amplification and transposon silencing, simultaneously; this is because transposon mRNAs serve as substrates for the Aub–piRNA complexes. The Aub-Slicer activity, the endonuclease activity required for cleaving the target mRNAs (Gunawardane et al., 2007; Nishida et al., 2007), determines the 5′ end of mature piRNAs in the sense orientation. Mature piRNAs are then loaded onto Ago3. Thus, Ago3-associating piRNAs are mostly in the sense orientation. The Ago3–piRNA complex, in turn, cleaves antisense transcripts of transposons and the resultant piRNAs are loaded onto Aub. Other factors required for the amplification loop include Vasa, Tudor (Tud), Krimp, Spn-E, and Mael (Findley et al., 2003; Lim and Kai, 2007; Malone et al., 2009).
Tud is one of the Tud domain-containing proteins expressed in fly gonads (Boswell and Mahowald, 1985; Thomson and Lasko, 2004; Arkov et al., 2006). The Tud domains in Tud specifically interact with the symmetrical dimethylarginines (sDMAs) in Aub and Ago3 (Kirino et al., 2009; Nishida et al., 2009; Liu et al., 2010a,b; Siomi et al., 2010). The association between Tud and Aub or Ago3 occurs simultaneously and the Tud–Aub–Ago3 complex contains piRNA intermediate-like RNA molecules but, in the complex, Aub and Ago3 are free from mature piRNAs. Thus, Tud might serve as a platform for the amplification loop (Nishida et al., 2009). Spn-E has a Tud domain (Gillespie and Berg, 1995; Ponting, 1997); however, it does not seem to associate with Aub and Ago3 (Nishida et al., 2009). Krimp also contains a Tud domain (Lim and Kai, 2007). Whether Krimp has the ability to associate with PIWI members through their sDMAs remains undetermined.
Until now, the piRNA biogenesis factors that have been identified in Drosophila are localized in the cytoplasm and are preferentially accumulated in specific cytoplasmic perinuclear structures, the nuage, and Yb bodies in germ and somatic cells, respectively. Hence, both nuage and Yb bodies are considered to be the locations for piRNA biogenesis (Findley et al., 2003; Lim and Kai, 2007; Olivieri et al., 2010; Saito et al., 2010; Qi et al., 2011; for review see Siomi et al., 2011). Zuc is localized to mitochondria (Olivieri et al., 2010; Saito et al., 2010). How Zuc localization to mitochondria is mechanistically involved in piRNA biogenesis remains unknown.
Accumulated evidence has shown that in ovaries, nuage localization of the piRNA factors occurs in a molecular hierarchical fashion. The current understanding is that Vasa, a DEAD-box RNA helicase that is detected specifically in the germ cells, is the first to be located to the nuage (Findley et al., 2003): Vasa localization is followed by Spn-E, Aub, Ago3, and Mael. Whether this hierarchy is female-specific or occurs also in males remains unexamined. To address this question, we performed immunofluorescence analyses using monoclonal antibodies against individual piRNA factors and found that the nuage localization hierarchy was gender-specific; an obvious difference was seen for Krimp. Krimp is placed between Aub and Ago3 in ovaries, but is placed at the same level with Aub, but upstream of Ago3 in testes. These results suggest that the functional requirement of Krimp in piRNA biogenesis may be different between male and female gonads.
Materials and Methods
Drosophila Strains
The yellow white (y w) and Oregon-R strains were used as wild-type (wt) strains. The tud allele used was tud1 bw sp/CyO l(2)DTS5131 (Drosophila Genetic Resource Center stock number: 106505). The aub alleles used were aubHN2 cn bw/CyO and aubQC42 cn bw/CyO. aubHN2 cn bw/CyO and aubQC42cn bw/CyO were crossed to yield aub heterozygous mutant flies; aubQC42/aubHN2 (Nishida et al., 2007). The ago3 alleles used were ago3t2/TM6B Tb and ago3t3/TM6B Tb. ago3t2/TM6B Tb and ago3t3/TM6B Tb were crossed to yield ago3 heterozygous mutant flies; ago3t2/ago3t3 (Li et al., 2009). The krimp allele used was w1118; PBac{WH}krimpf06583/CyO (Bloomington Drosophila Stock Center; 18990). w1118; PBac{WH}krimpf06583/CyO and w1118; Df(2R)BSC309/CyO (Bloomington Drosophila Stock Center; 23692) were crossed to yield krimp transheterozygote mutant flies; krimpf06583/Df. Transheterozygotes krimpf06583/Df exhibited male and female sterility and an extent of loss in krimp perinuclear staining similar to that exhibited by homozygous krimpf06583 (Lim and Kai, 2007, and data not shown). Hence, krimpf06583 was used as a loss-of-function allele to characterize krimp phenotypes in this study. All stocks were maintained at 25°C.
Production of Antibodies and Western Blotting
An anti-Tudor monoclonal antibody was raised specifically against the C-terminus of the protein. A 327 amino acid fragment at the C-terminal end of Tudor (amino acid 2189 to the end) was fused with glutathione S-transferase (GST) and used to immunize mice. An anti-Krimp monoclonal antibody was raised specifically against the N-terminus of the protein. A 200 amino acid fragment at the N-terminal end of Krimp was fused with GST and used to immunize mice. Anti-Tudor and anti-Krimp monoclonal antibodies were produced essentially as described previously (Ishizuka et al., 2002) and purified from culture supernatant of hybridoma cells under standard procedures using Thiophilic-Superflow resin (BD Biosciences). Western blot analysis was performed as described previously (Miyoshi et al., 2005). Anti-Aub and anti-Ago3 antibodies were used as described previously (Nishida et al., 2007, 2009).
Immunohistochemistry
Testes and ovaries were stained with the various antibodies as previously described (Nishida et al., 2007; Nagao et al., 2010). An anti-Aub antibody was directly labeled using the HiLyte Fluor 555 Labeling Kit-NH2 (Dojindo Molecular Technologies). Anti-Ago3 (1:5,000 dilution; Nagao et al., 2010), anti-Tud (1:250 dilution; Nishida et al., 2009), and anti-Krimp (1:250 dilution) antibodies were used as primary antibodies. Alexa 488-conjugated or Alexa 546-conjugated anti-mouse IgG (Molecular Probes; 1:1,000 dilution) were used as secondary antibodies. DNA was stained with DAPI. All images were collected using a confocal microscope (Zeiss LSM5 EXCITER). All immunohistochemical analyses were performed at least three times and representative images are shown in the figures.
Results and Discussion
To ensure that we had a constant supply of anti-Tud and anti-Krimp antibodies for this study, we produced Tud and Krimp monoclonal antibodies. We used recombinant Tud and Krimp peptide fragments expressed in and purified from E. coli to immunize mice. Western blotting of total ovary and cultured Schneider 2 (S2) lysates showed that the antibodies reacted with proteins of the expected sizes of Tud and Krimp (Figures 1A,B, respectively). The Tud band in S2 lysate migrated slightly faster than that in ovary lysate (Figure 1A). This may reflect post-translational modification of Tud protein, although clear evidence for this is still required. We also performed western blotting on tud and krimp mutant ovaries (Figures 1A,B). The bands corresponding to Tud and Krimp were not present in the mutants, indicating a high specificity of the antibodies for the individual antigens.
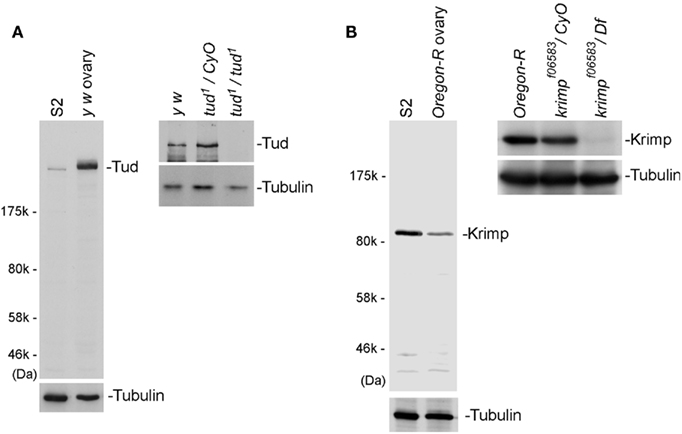
Figure 1. Specificity of anti-Tud and anti-Krimp antibodies. (A) Western blotting was performed on Schneider 2 (S2) and total ovary lysates using an anti-Tud monoclonal antibody raised in our laboratory. A band of the size predicted for Tud was detected in both lysates. In tud mutant (tud1/tud1) ovaries, the bands corresponding to Tud disappeared, indicating that the antibody was specific to the antigen. Tubulin was used as a loading control. (B) Western blotting was performed on S2 and total ovary lysates using an anti-Krimp monoclonal antibody. A band of the size predicted for Krimp was detected. In krimp mutant (krimpf06583/Df) ovaries, the bands corresponding to Krimp disappeared, indicating that the antibody was specific to Krimp.
We previously examined the subcellular localization of Tud in aub mutant ovaries using an anti-Tud antibody that was a kind gift from A. Nakamura (CDB-RIKEN, Japan; Nishida et al., 2009). Here, we used the newly produced anti-Tud antibody to repeat this immunofluorescence study on both control and aub mutant ovaries. Very little Tud was localized to the nuage when Aub was not expressed (Figure 2A), confirming the previous observation that Aub is necessary for Tud nuage localization (Nishida et al., 2009). Tud localization has not been previously examined in ago3 mutant ovaries. We immunostained for Tud in ago3 mutant ovaries and detected Tud in the nuage (Figure 2A), suggesting that Ago3 is dispensable for Tud nuage localization. Thus, the nuage localization of Aub, but not of Ago3, is necessary for Tud nuage localization. Aub localization to nuage is not affected by loss of ago3 expression (Figure 2A), although Ago3 localization to nuage is affected by loss of aub expression (Figure 2A). Hence, for nuage localization, Ago3 requires Aub but Aub does not require Ago3.
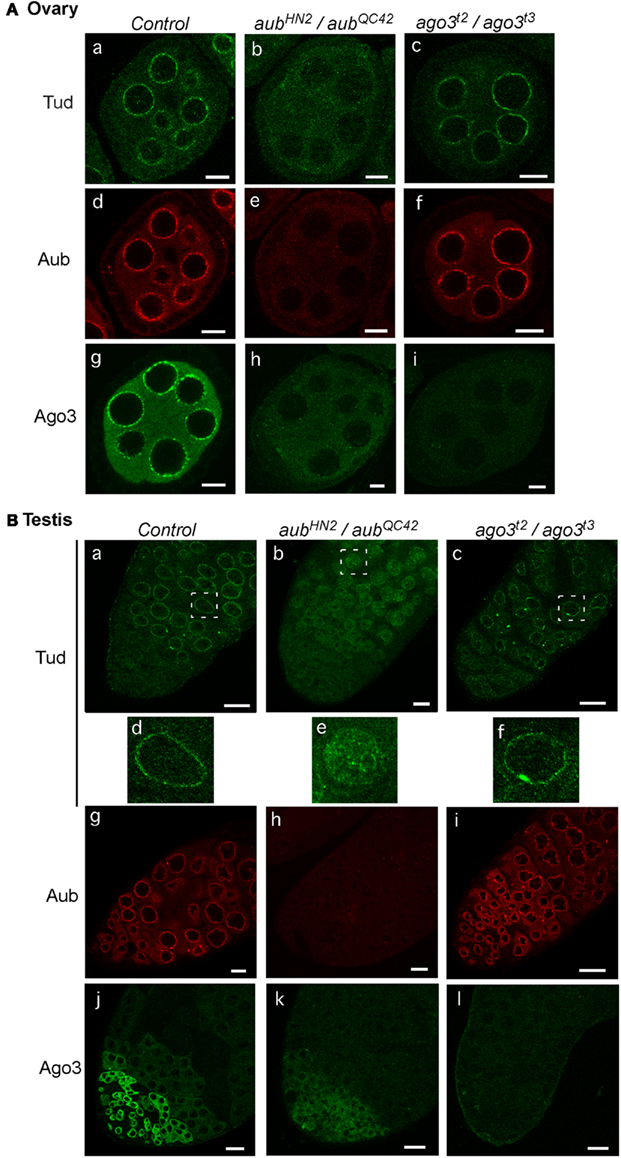
Figure 2. Tud subcellular localization in aub and ago3 mutant ovaries and testes. (A) Ovaries: the control (a), and aub (aubHN2/aubQC42) (b), and ago3 (ago3t2/ago3t3) (c) mutant ovaries were stained with anti-Tud antibody; the control (d), and aub (aubHN2/aubQC42) (e) ago3 (ago3t2/ago3t3) (f) mutant ovaries were stained with anti-Aub antibody; and the control (g), and aub (aubHN2/aubQC42) (h), and ago3 (ago3t2/ago3t3) (i) mutant ovaries were stained with anti-Ago3 antibody. Scale bars in all ovary images indicate 10 μm. (B) Testes: the control (a), and aub (aubHN2/aubQC42) (b), and ago3 (ago3t2/ago3t3) (c) mutant testes were stained with anti-Tud antibody. Enlarged images of the regions that are boxed with white dotted lines in (B)(a–c) are shown in (B) (d–f), respectively. The control (g), and aub (aubHN2/aubQC42) (h), and ago3 (ago3t2/ago3t3) (i) mutant testes were stained with anti-Aub antibody; and the control (j); and aub (aubHN2/aubQC42) (k); and ago3 (ago3t2/ago3t3) (l) mutant testes were stained with anti-Ago3 antibody. Scale bars in all testis images indicate 20 μm. All immunohistochemical analyses were performed at least three times and representative images are shown.
We then examined the subcellular localization of Tud in aub and ago3 mutant testes. The Tud signal was detected in the nuage in ago3 mutant testes (Figure 2B) the same as in control. In aub mutant testes, however, the Tud signal was hardly detected in the nuage; instead, it appeared to be in the nucleus in particular in spermatogonia cells (Figure 2B). These results suggest that Aub controls the Tud nuage localization even in testes. Aub was localized to the nuage in ago3 testes (Figure 2B), confirming that Aub nuage localization does not require Ago3 in both female and male gonads. Ago3 is not localized to nuage in aub mutant testes (Figure 2B). Thus, Ago3 nuage localization requires Aub in both males and females.
We then investigated how the subcellular localization of Aub and Ago3 was affected by loss of tud function. In ovaries, Aub was detected only slightly in the nuage in tud mutant flies (Figure 3A), as has been reported previously (Nishida et al., 2009). In contrast, Ago3 was localized to the nuage, the same as in the control. These results suggest that under conditions where tud function is absent, Ago3 finds an alternative way to be localized to the nuage in a Tud-independent manner. Another possible interpretation is that a small amount of Aub in the nuage is sufficient for Ago3 to be localized to the nuage. Also in testes, Ago3 localization was barely affected by loss of tud function, whereas Aub localization was drastically changed (Figure 3B); Aub was sparsely localized in the nuage and instead was dispersed in the cytosol. These results indicate that the requirement and dispensability of Tud for nuage localization of Aub and Ago3, respectively, are similar in male and females.
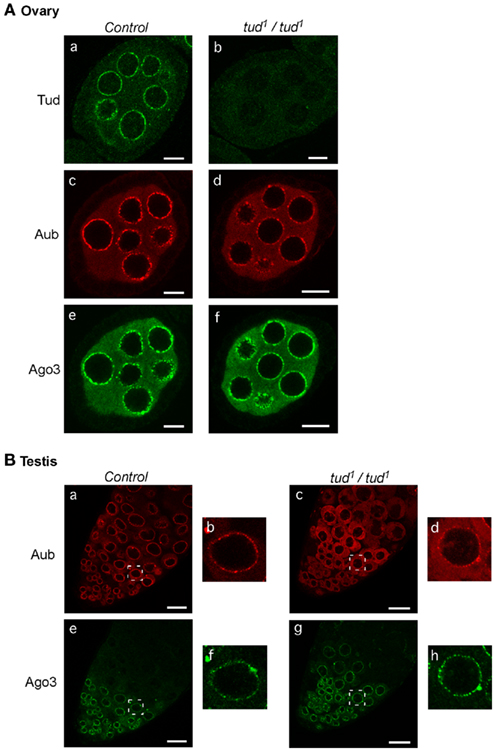
Figure 3. Loss of tud function affects the subcellular localization of Aub and Ago3 in ovaries and testes similarly. (A) Ovaries: tud heterozygous and homozygous mutant ovaries were immunostained with anti-Tud, anti-Aub, and anti-Ago3 antibodies [(a): tud hetero/anti-Tud; (b): tud homo/anti-Tud; (c): tud hetero/anti-Aub; (d): tud homo/anti-Aub; (e): tud hetero/anti-Ago3; (f): tud homo/anti-Ago3]. Scale bars in all ovary images indicate 10 μm. (B) Testes: tud mutant and control testes were immunostained with anti-Aub and anti-Ago3 antibodies [(a): tud hetero/anti-Aub; (b): enlarged image of the region boxed with a white line in (a); (c): tud homo/anti-Aub; (d): enlarged image of indicated region in (c); (e): tud hetero/anti-Ago3; (f): enlarged image of indicated region in (e); (g): tud homo/anti-Ago3; (h): enlarged image of indicated region in (g)]. Scale bars in all testis images indicate 20 μm. All immunohistochemical analyses were performed at least three times and representative images are shown.
We also examined how loss of krimp affects the localization of Aub and Ago3 in the germline. In krimp mutant ovaries, Ago3 lost its ability to localize to the nuage, although Aub was accumulated in the nuage the same as in the control (Figure 4A). Thus, Krimp is necessary for nuage localization of Ago3, but not of Aub. Tud nuage localization was maintained even after krimp expression was lost (Figure 4A). In contrast, in krimp mutant testes, Aub, Ago3, and Tud nuage localization was abolished (Figure 4B). The requirement of Krimp for Aub and Tud nuage localization is different in ovaries and testes.
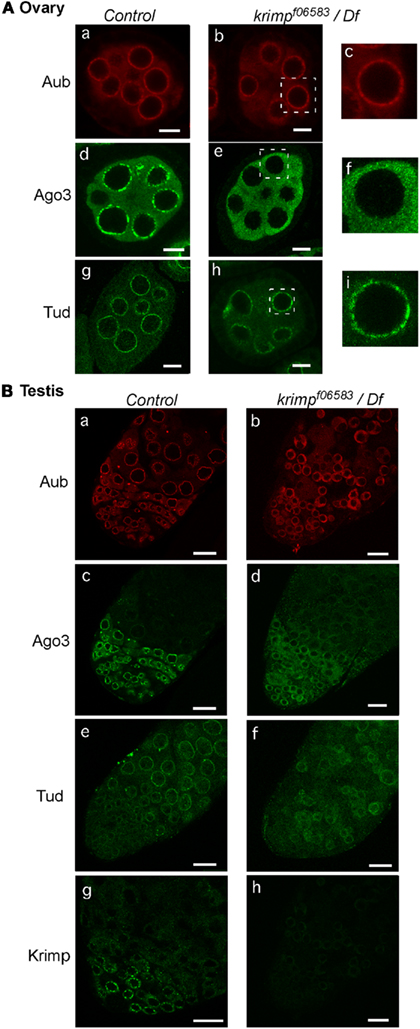
Figure 4. Loss of krimp function affects the subcellular localization of Aub and Ago3 in ovaries and testes differently. (A) Ovaries: the control and krimp mutant ovaries were immunostained with anti-Aub, anti-Ago3, and anti-Tud antibodies [(a): control/anti-Aub; (b): krimp mutant/anti-Aub; (c): enlarged image of the part boxed with a white line in (b); (d): control/anti-Ago3; (e): krimp mutant/anti-Ago3; (f): enlarged image of indicated region in (e); (g): control/anti-Tud; (h): krimp mutant/anti-Tud; (i): enlarged image of indicated region in (h)]. Scale bars in all ovary images indicate 10 μm. (B) Testes: the control and krimp mutant testes were immunostained with anti-Aub, anti-Ago3, anti-Tud, and anti-Krimp antibodies [(a): control/anti-Aub; (b): krimp mutant/anti-Aub; (c): control/anti-Ago3; (d): krimp mutant/anti-Ago3; (e): control/anti-Tud; (f): krimp mutant/anti-Tud; (g): control/anti-Krimp; (h): krimp mutant/anti-Krimp]. Scale bars in all testis images indicate 20 μm. All immunohistochemical analyses were performed at least three times and representative images are shown.
An earlier study showed that Krimp is localized to the nuage in ovaries (Lim and Kai, 2007). In the present study, we confirmed this localization (Figure 5A) using our newly produced anti-Krimp antibody (Figure 1B). We then examined by immunostaining of ovaries and testes how Krimp nuage localization is affected by loss of other piRNA factors. We found that loss of ago3 and tud expression in ovaries did not affect Krimp localization (Figure 5A). However, in aub mutant ovaries, Krimp signals were hardly detected in the nuage; instead, Krimp accumulated to unknown cytoplasmic structures (Figure 5A). Thus, Aub, but not Ago3 or Tud, is required for Krimp nuage localization. Why Krimp accumulates in cytoplasmic structures remains unknown. The Krimp signal of immunostaining was lower in aub and tud mutant testes than that in the control, while Krimp expression was unchanged by loss of ago3 expression (Figure 5B). Western blotting confirmed a severe reduction in the expression level of Krimp in aub and tud mutant testes (Figure 5C). Thus, the requirement of Tud in Krimp nuage localization is different between males and females. Why loss of aub and tud expression causes a severe reduction in the Krimp expression requires further investigation. Krimp was hardly detected in the tud mutant testes. However, Ago3 was able to accumulate in the nuage, possibly because Aub was, at least partially, localized to the nuage in tud mutant testes. It could be postulated that Ago3 nuage localization necessitates either Aub or Krimp in the nuage.
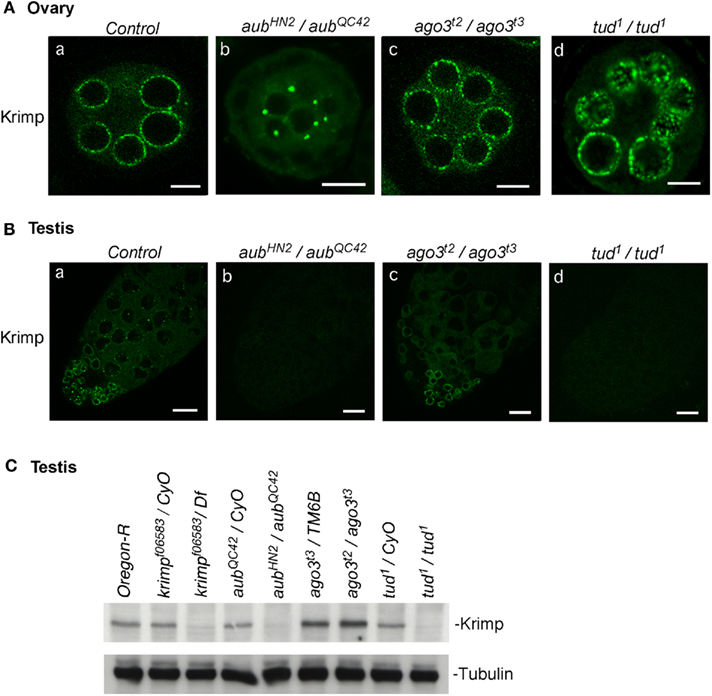
Figure 5. Krimp nuage localization in the piRNA mutant ovaries and testes. (A) Ovaries: the control (a) and aub (b), ago3 (c), and tud (d) mutant ovaries were immunostained with an anti-Krimp antibody. Scale bars in all ovary images indicate 10 μm. (B) Testes: the control (a) and aub (b), ago3 (c), and tud (d) mutant testes were immunostained with an anti-Krimp antibody. (C) Western blotting shows that the expression of Krimp is hardly detected in aub and tud mutant testes. Scale bars in all testis images indicate 20 μm. All immunohistochemical analyses were performed at least three times and representative images are shown.
The nuage localization of piRNA factors, Aub, Tud, Krimp, and Ago3, in ovaries and testes from wt and the piRNA factor mutant flies was summarized (Figure 6A) and their molecular hierarchy was compared (Figure 6B). Their expression patterns in wt ovary and testis were also summarized (Figure 6C). An obvious difference in the hierarchy is the location of Krimp between Aub and Ago3 in ovaries but at the same level as Aub and upstream of Ago3 in testes. Krimp expression in testis is limited to developmentally early stage cells, such as germline stem cells, gonialblasts, and spermatogonia (Figure 6C) and this may be the cause of the gender difference observed in the location of Krimp in the molecular hierarchy.
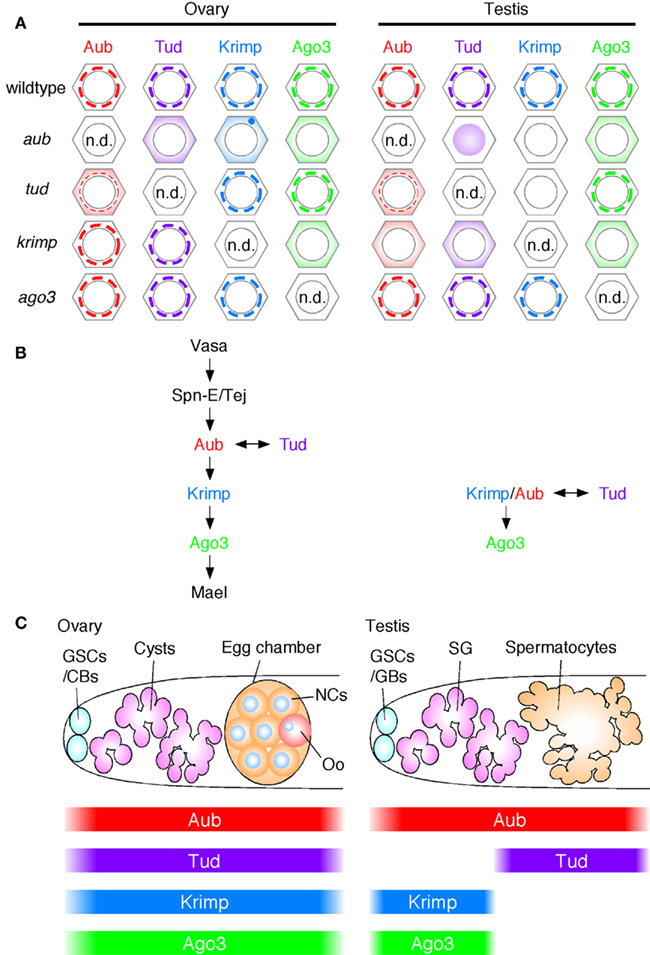
Figure 6. Molecular hierarchy of the piRNA factors in their localization to nuage in ovaries and testes. (A) The nuage localization of piRNA factors, Aub, Tud, Krimp, and Ago3 in wild-type (wt) and their mutant ovaries and testes: n.d.: none detected. Since none of the piRNA factors in this figure were detected in the nucleus of wt ovaries and testes, therefore they were considered to be cytoplasmic proteins. In tud mutant ovaries and testes, Aub is only weakly accumulated in the nuage; thus, the nuage is indicated with a thinner line. The Krimp signals are hardly detected in aub and tud mutant testes. (B) The nuage localization hierarchy: Aub is placed upstream of Ago3 in both ovaries and testes. However, Krimp shows gender-specificity; in ovaries Krimp lies between Aub and Ago3, in testes Krimp is placed at the same level as Aub in testes. Tud is required for Aub localization in both male and female nuage. However, Ago3 can be localized to the nuage without tud function in both males and females. It is likely that Ago3 nuage localization occurs in both Aub-dependent and Tud-independent manners. (C) The expression patterns of Aub, Tud, Krimp, and Ago3 in ovaries and testes. GSCs, germline stem cells; CBs, cystoblasts; NCs, nurse cells; Oo, oocyte; GBs, goniablasts; SG, spematogonia.
We reported previously that, in testes, Ago3 functions in the amplification loop with Aub for producing transposon-derived piRNAs (Nagao et al., 2010). However, most Ago3-associated testis piRNAs corresponding to Suppressor of Stellate [Su(Ste)] genes that function in silencing the Stellate gene, are antisense-oriented and are also detected in piRNAs associated with Aub (Nagao et al., 2010). In addition, the vast majority of Ago3-associated piRNAs derived from the AT-chX locus on chromosome X, some of which may function in silencing vasa, are also antisense-oriented and are found among Aub-associated piRNAs (Nagao et al., 2010). The piRNAs that show characteristics similar to those of AT-chX and Su(Ste) piRNAs have so far not been found in ovaries (Nagao et al., 2010). Our current finding, that Krimp is located in a different position in the nuage localization hierarchy in ovaries and testes, may further support the idea that some mechanisms of piRNA biogenesis differ in ovaries and testes. Our interpretation is that Krimp may be one of the important factors for producing AT-chX and Su(Ste) primary piRNAs and may enabling their loading into not only Aub, but also Ago3, in testes. In ovaries, Krimp is placed between Aub and Ago3 in the hierarchy and this may be why primary piRNAs are specifically loaded into Aub, but not into Ago3. Krimp has a Tud domain and, thus, it is reasonable to postulate that Krimp associates with Aub and/or Ago3 through their sDMA modifications in both ovaries and testes. However, the mode of their associations could be different in the two tissues: the extent of sDMA modification of Aub and Ago3 may be different and/or the sDMA requirements in the association between Krimp and Aub/Ago3 may be different. These possible differences could be the cause of the gender difference for Krimp in the piRNA factor hierarchy. We are currently investigating the biochemical function of Krimp in piRNA biogenesis and its relationship with Aub and Ago3, as well as the modification status of Aub and Ago3 in both ovaries and testes, which will help further our understanding of piRNA biogenesis in general and of the gender differences in the molecular mechanisms in particular.
Acknowledgments
We thank T. Okada, A. Shibuya, M. Komai, and H. Kotani for technical assistance and other members of the Siomi laboratory for discussions and comments on the manuscript. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) to Kazumichi M. Nishida, Kaoru Sato, Haruhiko Siomi, and Mikiko C. Siomi. Mikiko C. Siomi is supported by Core Research for Evolutional Science and Technology (CREST) from Japan Science and Technology Agency (JST).
References
Aravin, A. A., Hannon, G. J., and Brennecke, J. (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764.
Arkov, A. L., Wang, J. Y., Ramos, A., and Lehmann, R. (2006). The role of Tudor domains in germline development and polar granule architecture. Development 133, 4053–4062.
Boswell, R. E., and Mahowald, A. P. (1985). Tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97–104.
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., and Hannon, G. J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103.
Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727.
Findley, S. D., Tamanaha, M., Clegg, N. J., and Ruohola-Baker, H. (2003). Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130, 859–871.
Gillespie, D. E., and Berg, C. A. (1995). Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 9, 2495–2508.
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., Siomi, H., and Siomi, M. C. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590.
Haase, A. D., Fenoglio, S., Muerdter, F., Guzzardo, P. M., Czech, B., Pappin, D. J., Chen, C., Gordon, A., and Hannon, G. J. (2010). Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24, 2499–2504.
Ishizuka, A., Siomi, M. C., and Siomi, H. (2002). A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16, 2497–2508.
Kalmykova, A. I., Klenov, M. S., and Gvozdev, V. A. (2005). Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 33, 2052–2059.
Kirino, Y., Kim, N., de Planell-Saguer, M., Khandros, E., Chiorean, S., Klein, P. S., Rigoutsos, I., Jongens, T. A., and Mourelatos, Z. (2009). Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 11, 652–658.
Klattenhoff, C., Bratu, D. P., McGinnis-Schultz, N., Koppetsch, B. S., Cook, H. A., and Theurkauf, W. E. (2007). Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55.
Li, C., Vagin, V. V., Lee, S., Xu, J., Ma, S., Xi, H., Seitz, H., Horwich, M. D., Syrzycka, M., Honda, B. M., Kittler, E. L., Zapp, M. L., Klattenhoff, C., Schulz, N., Theurkauf, W. E., Weng, Z., and Zamore, P. D. (2009). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521.
Lim, A. K., and Kai, T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 6714–6719.
Liu, H., Wang, J. Y., Huang, Y., Li, Z., Gong, W., Lehmann, R., and Xu, R. M. (2010a). Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 24, 1876–1881.
Liu, K., Chen, C., Guo, Y., Lam, R., Bian, C., Xu, C., Zhao, D. Y., Jin, J., MacKenzie, F., Pawson, T., and Min, J. (2010b). Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. U.S.A. 107, 18398–18403.
Malone, C. D., Brennecke, J., Dus, M., Stark, A., McCombie, W. R., Sachidanandam, R., and Hannon, G. J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535.
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H., and Siomi, M. C. (2005). Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848.
Nagao, A., Mituyama, T., Huang, H., Chen, D., Siomi, M. C., and Siomi, H. (2010). Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16, 2503–2515.
Nishida, K. M., Okada, T. N., Kawamura, T., Mituyama, T., Kawamura, Y., Inagaki, S., Huang, H., Chen, D., Kodama, T., Siomi, H., and Siomi, M. C. (2009). Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28, 3820–3831.
Nishida, K. M., Saito, K., Mori, T., Kawamura, Y., Nagami-Okada, T., Inagaki, S., Siomi, H., and Siomi, M. C. (2007). Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922.
Olivieri, D., Sykora, M. M., Sachidanandam, R., Mechtler, K., and Brennecke, J. (2010). An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29, 3301–3317.
Ponting, C. P. (1997). Tudor domains in proteins that interact with RNA. Trends Biochem. Sci. 22, 51–52.
Qi, H., Watanabe, T., Ku, H. Y., Liu, N., Zhong, M., and Lin, H. (2011). The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 286, 3789–3797.
Saito, K., Inagaki, S., Mituyama, T., Kawamura, Y., Ono, Y., Sakota, E., Kotani, H., Asai, K., Siomi, H., and Siomi, M. C. (2009). A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299.
Saito, K., Ishizu, H., Komai, M., Kotani, H., Kawamura, Y., Nishida, K. M., Siomi, H., and Siomi, M. C. (2010). Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24, 2493–2498.
Saito, K., Nishida, K. M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., and Siomi, M. C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222.
Schmidt, A., Palumbo, G., Bozzetti, M. P., Tritto, P., Pimpinelli, S., and Schäfer, U. (1999). Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 151, 749–760.
Siomi, M. C., Mannen, T., and Siomi, H. (2010). How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24, 636–646.
Siomi, M. C., Sato, K., Pezic, D., and Aravin, A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258.
Szakmary, A., Reedy, M., Qi, H., and Lin, H. (2009). The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J. Cell Biol. 185, 613–627.
Thomson, T., and Lasko, P. (2004). Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40, 164–170.
Vagin, V. V., Klenov, M. S., Kalmykova, A. I., Stolyarenko, A. D., Kotelnikov, R. N., and Gvozdev, V. A. (2004). The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 1, 54–58.
Keywords: nuage, piRNA, PIWI, Drosophila, germline
Citation: Nagao A, Sato K, Nishida KM, Siomi H and Siomi MC (2011) Gender-specific hierarchy in nuage localization of PIWI-interacting RNA factors in Drosophila. Front. Gene. 2:55. doi: 10.3389/fgene.2011.00055
Received: 08 July 2011;
Accepted: 12 August 2011;
Published online: 30 August 2011.
Edited by:
Muller Fabbri, The Ohio State University, USAReviewed by:
Patrick Provost, Université Laval, CanadaFrancesca Fanini, Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori, Italy
Michael Frohman, Stony Brook University, USA
Copyright: © 2011 Nagao, Sato, Nishida, Siomi and Siomi. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Mikiko C. Siomi, Keio University School of Medicine, 3 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan. e-mail:c2lvbWltQHo2LmtlaW8uanA=
 Akihiro Nagao1
Akihiro Nagao1