- 1 Department of Virology and Immunology, Texas Biomedical Research Institute, San Antonio, TX, USA
- 2 Southwest National Primate Research Center, San Antonio, TX, USA
- 3 Microbiology and Immunology Track of the Integrated Multidisciplinary Graduate Program, University of Texas Health Science Center, San Antonio, TX, USA
The herpesviruses are a family of large DNA viruses capable of establishing lifelong infections. Recent reports have shown that herpesviruses package non-coding RNA into virions; this follows earlier observations showing that coding RNAs are detected in virions. Packaging RNAs allows for their function immediately after virus entry and in the absence of de novo transcription. Despite the collective understanding that RNAs are packaged into herpesvirus virions, many questions remain. This review will highlight what is known regarding packaged coding and non-coding RNAs and discuss their potential impact to virus biology.
Herpesviruses (family Herpesviridae) are large double-stranded DNA viruses widely dispersed in nature (Pellet and Roizman, 2007). They are implicated in many human diseases and are divided into three subfamilies: alpha, beta, and gamma-herpesviruses. All three subfamilies share similar characteristics based on the structure of the virion and their ability to maintain a latent state for the life of the host with the potential for period reactivation (Pellet and Roizman, 2007). Herpesviruses have (i) a lipid bilayer envelope decorated with virus-encoded glycoproteins surrounding (ii) a proteinaceous tegument that encases (iii) an icosahedral capsid and (iv) a core with the DNA genome (Pellet and Roizman, 2007). Figure 1 shows a cartoon diagram of a prototypic herpesvirus virion. They differ by their host species, tissue tropism, and pathogenicity. Alphaherpesviruses, for example the prototypic herpes simplex virus (HSV), have short replication cycles and a propensity to establish latency in sensory ganglia. Betaherpesviruses, for example human cytomegalovirus (HCMV), have a slow replication cycle, establish latency in leukocytes and endothelial cells, and have a restricted host range. Finally, gammaherpesviruses, for example Kaposi’s sarcoma associated herpesvirus (KSHV), replicate in cells of the immune system and establish latency in lymphoid tissue (Pellet and Roizman, 2007).
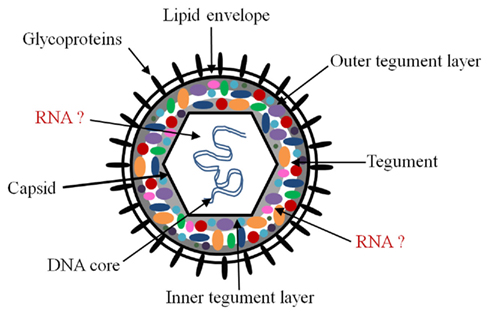
Figure 1. Schematic of herpesvirus virion. The herpesvirus virion is composed of an icosahedral capsid containing the double-stranded DNA genome, a lipid envelope decorated with virally encoded glycoproteins, and a layer filled with proteins termed the tegument which resides between the capsid and the envelope. The likely sites for packaged coding and non-coding RNAs are the tegument and the capsid. There is evidence for RNA–protein interactions in the tegument, but it is unclear whether all packaged RNAs interact with tegument proteins. Although it is possible, we are unaware of evidence for protein–RNA interactions in the nucleus, or RNA–RNA interactions in either the nucleus or tegument.
Non-coding RNAs play important roles in herpesvirus biology. These include longer RNA molecules such as the HSV latency-associated transcript, herpesvirus saimiri U RNAs, murine gamma-herpesvirus 68 (MHV-68) viral tRNA-like molecules (vtRNAs), and virus-encoded microRNAs (miRNAs). Many of these non-coding RNAs are expressed at high levels and have been found in both lytic and latent infections. Despite substantial interest, the functions of many of the herpesvirus non-coding RNAs are not well defined. Following the observation that mRNAs are packaged into herpesvirus virions (Bresnahan and Shenk, 2000; Greijer et al., 2000; Sciortino et al., 2001, 2002; Terhune et al., 2004; Bechtel et al., 2005), recent reports have found evidence that non-coding RNAs are also incorporated into virions (Cliffe et al., 2009; Amen and Griffiths, 2011). These observations suggest a potential role for non-coding RNAs in herpesvirus biology. In this review we will describe what is known about packaged coding and non-coding RNAs and discuss the potential impact of these RNAs to virus biology.
How are the Non-Coding RNAs Packaged into Virus Particles?
In 2000, the discovery of coding RNAs packaged into herpesvirus virions provided convincing evidence that virions of herpesviruses included more than just lipid, genomic DNA, and viral proteins (Bresnahan and Shenk, 2000; Greijer et al., 2000). And by 2009, it was evident that non-coding RNAs were also being packaged (Cliffe et al., 2009; Amen and Griffiths, 2011). While there appears to be agreement in the field that coding and non-coding RNAs are indeed packaged and not an artifact of virion preparation, there is disagreement regarding the selectivity (sequence dependent vs. sequence independent) of the RNA packaging. In the first report describing viral transcripts packaged into HCMV particles, Bresnahan and Shenk (2000) determined that the mRNAs associated with virions were not random; rather, a specific subset of mRNAs appeared to be preferentially associated with virions. Further studies examining the mechanism of RNA incorporation into HCMV virions provided evidence that both cellular and viral transcripts are packaged proportionally to their abundance in the cell (Terhune et al., 2004). It was concluded that RNA packaging is probably mediated through non-specific interactions with proteins found within the HCMV infectious particle (Terhune et al., 2004). Similarly, Greijer et al. (2000) proposed that the viral and cellular mRNA molecules present are incorporated via a non-selective process based on the cellular location of assembly of the HCMV particles. However, there are exceptions. For example, HSV RNAs present in the virions were not uniformly the most abundant RNAs present in infected cells at the time of harvest (Sciortino et al., 2001). The viral RNAs detected in KSHV virions were present in the cell at a range of abundances suggesting a selective process of packaging (Bechtel et al., 2005). It was also demonstrated that non-coding RNAs were selectively packaged within the MHV-68 virion. Here, non-coding small RNAs were not packaged in proportion to their cellular abundance (Cliffe et al., 2009). It was suggested that the viral non-coding RNAs are selectively packaged by binding to RNA-binding proteins, in a similar manner to the way viruses with RNA genomes package their genetic material. However, the non-coding RNAs may also be bound to complementary RNA species that are also packaged. Similarly, in herpes B virus (BV), a close relative of HSV, several miRNAs were present in purified virions (Amen and Griffiths, 2011). While these miRNAs represented some of the more abundant BV-encoded miRNAs, not all of the abundant viral miRNAs were packaged.
Interestingly, the packaged BV-encoded miRNAs were demonstrated to belong to more than one kinetic class. Briefly, the program of herpes viral transcription is as follows: upon infection the virion enters the cell and is transported to the cell nucleus. There, the genome self-circularizes, and the viral genes are transcribed by host RNA polymerases in a cascade-like fashion: immediate-early, early, and late (Figure 2; Pellet and Roizman, 2007). The first are the immediate-early (α) genes that require no prior viral protein synthesis and regulate viral and cell gene expression. The immediate-early genes transactivate early gene expression (β), which are involved with viral DNA synthesis and genome replication. The leaky-late (γ1) genes are transcribed after the onset of viral DNA synthesis, while the true late (γ2) genes are dependent on viral DNA synthesis. These genes are mostly structural and include the envelope glycoproteins and capsid proteins. Packaging of miRNAs from multiple kinetic classes strengthens the hypothesis that the RNAs are selectively packaged in infected cells during the process of viral maturation. Of the nine HSV RNAs reproducibly found in virion preparations, one was an α-RNA, one was an β-RNA, and the rest were γ-RNAs (Sciortino et al., 2001). Therefore, it was noted that the most abundant class of transcripts (true late and leaky-late) present during envelopment do not represent the only kinetic class of RNAs packaged in herpesvirus virions (Sciortino et al., 2001).
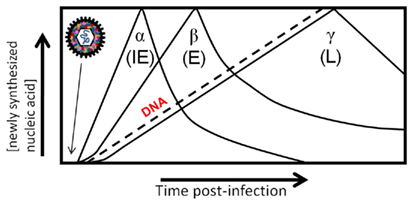
Figure 2. Cascade of herpesvirus gene expression. Herpesvirus genes are expressed in a coordinated temporal cascade: immediate early (α), early (β). There are two subclasses of late gene expression that have been defined, although they are expressed concurrently: leaky-late (γ1) is not dependent on DNA synthesis, and true late (γ2) is dependent on DNA synthesis. Packaging of RNAs in the virion allows immediate expression upon virus entry in the absence of transcription, and thus adds an additional element in the highly regulated cascade of gene expression of herpesviruses.
The observations from studies of HSV, BV, KSHV, and MHV-68 all point to a selective mechanism of packaging based on the various expression levels within infected cells. However, these viruses represent only alpha- and gamma-herpesviruses. Therefore, the selectivity of coding and non-coding RNAs packaged in the virions may be family dependent. Nevertheless, the theory that RNA is selectively packaged is complicated by evidence showing that a large number of cellular RNAs are also packaged (Greijer et al., 2000; Sciortino et al., 2001; Terhune et al., 2004; Amen and Griffiths, 2011; Amen and Griffiths, unpublished data). It is possible that while viruses are selective for their own viral transcripts, the levels of cellular transcripts may “overwhelm” the virus’ selective ability and these transcripts may be non-specifically incorporated. Alternatively, the incorporation of specific cellular transcripts along with important viral transcripts may be functional. Indeed, the virus may regulate the relative levels of RNAs present during the assembly phase of the replication cycle to control the level of packaged RNA. The coding and non-coding RNA species, packaged selectively or not, may still have a biological role in herpesvirus pathogenesis. However, the details concerning selective vs. non-selective incorporation of RNAs into herpesvirus virions remains elusive and must be explored further. In addition next generation sequencing technologies will be required to determine all of the RNAs that are packaged.
Where are the RNAs Located in the Virion?
The location of the non-coding and coding RNAs in the virion has been debated since they were first discovered, and there remains some uncertainty. This may be a consequence of any controversy that remains regarding the different models of herpesvirus virion assembly and egress, or the complexity of the herpesvirus virion, which comprises more than 30 viral proteins, most with unknown function (Mettenleiter et al., 2006; Pellet and Roizman, 2007). Herpesvirus egress has mostly been studied in alphaherpesviruses, but it is becoming apparent that beta- and gamma-herpesviruses use similar mechanisms, although certain distinctions are emerging and different questions remain (Johnson and Baines, 2011). The process of envelopment and egress remains an active area of study (e.g., Campadelli-Fiume et al., 2006; Mettenleiter and Minson, 2006; Mettenleiter et al., 2009; Johnson and Baines, 2011). There are substantial genetic and biochemical data that support a mechanism of primary envelopment in the perinuclear space, followed by de-envelopment in the cytoplasm and subsequent re-envelopment in cytoplasmic membrane compartments (Mettenleiter and Minson, 2006; Pellet and Roizman, 2007; Mettenleiter et al., 2009; Johnson and Baines, 2011). An alternative model involves the dilation of nuclear pores whereby the capsid gains access into the cytoplasm, followed by envelopment in the Golgi compartment (Mettenleiter and Minson, 2006; Mettenleiter et al., 2006). The packaging of coding and non-coding RNAs into virions is compatible with all models; however, the physical location of the RNA in the virion could be different depending on the model. It is thought that RNAs most likely reside inside the capsid or in the tegument (Figure 1); although these locations are not mutually exclusive, association with the tegument is currently favored.
There are several observations that suggest that the tegument is the likely location for packaged RNAs. The tegument fraction of herpesvirus particles is complex, composed of many proteins acquired from the cytoplasm and nucleus of the infected cell and can be divided into inner and outer layers (Figure 1; Johnson and Baines, 2011). The inner tegument proteins of HCMV and HSV are intimately associated with the capsid. Furthermore, these proteins pellet with capsids after extraction with non-ionic detergents (Johnson and Baines, 2011) making it difficult to determine the exact location of the packaged RNAs. Of the known 20–35 tegument proteins in HSV and HCMV respectively (Varnum et al., 2004; Kalejta, 2008; Kelly et al., 2009; Johnson and Baines, 2011), several have been shown to have RNA-binding activity (Sciortino et al., 2002; Terhune et al., 2004). Three HSV tegument proteins have been shown to bind RNA in vitro and in the context of infected cells (Sciortino et al., 2002). Moreover, one of those proteins was shown to mediate the transfer of the RNA between cells (Sciortino et al., 2002). It has been suggested that these tegument protein–RNA interactions may be important for organizing the structure of the tegument domain and will be discussed below. In addition to infectious particles, other particle types including dense bodies and non-infectious particles have been shown to package RNA (Terhune et al., 2004). Dense bodies lack a nucleocapsid, are formed within the cytoplasm of infected cells, and are predominantly composed of tegument proteins (Stinski, 1976). The observation that dense bodies contain packaged RNA supports the notion that transcripts are acquired within the cytoplasm and are located within the viral tegument.
Greijer et al. (2000) identified cellular and viral RNAs in the tegument and viral RNAs in the capsid. It was suggested that in the process of enveloping the tegument-coated nucleocapsid in the cytoplasm, cytoplasmic fluid may be enclosed where a high concentration of RNA from viral and cellular origin is present. However, the purity of the capsid preparations was not tested. They prepared capsids by extraction of virion preparations with Triton-X-100 only, a method which has been shown to not strip tegument proteins or even fully remove glycoproteins (Sciortino et al., 2001; Yu et al., 2005). In sum, while the possibility that coding and non-coding RNAs packaged in capsids cannot be excluded, the majority of the data suggest that they are associated with the tegument.
What are the Functions of Packaged Non-Coding RNAs?
The presence of packaged RNAs in the virion suggests an importance early in infection, as it would permit the RNAs to function prior to the onset of the viral transcription program. During acute replication, the virus modifies the cellular environment to favor infection in several ways, including suppression of the activated host response. Studies of the HCMV UL21.5 mRNA revealed that the packaging of this transcript may be important for modifying the antiviral response before viral transcription occurs (Bresnahan and Shenk, 2000). Following the incorporation into the newly infected cell, the UL21.5 transcript is translated into a protein that is localized to the endoplasmic reticulum and Golgi network. This protein then functions as a viral chemokine decoy receptor specifically interacting with the RANTES chemokine and thereby modulates the host cell response upon infection. Presumably, if the mature protein was incorporated into the virion, that protein would lack the proper signal sequence and not localize to the correct cellular compartment. This CMV-packaged RNA can therefore potentially modify the cellular response before viral transcription occurs. A number of other virion RNAs delivered into the cell are translated (Bresnahan and Shenk, 2000; Sciortino et al., 2001; Bechtel et al., 2005). For KSHV, the protein products of packaged RNAs are antiapoptotic, immunomodulatory, and involved in cellular signal transduction both in the infected cell and the surrounding cells (Bechtel et al., 2005).
Alternatively, the view of packaged RNAs non-selectively incorporated into virions may be more consistent with the idea that they have no role in early viral replication. Rather, they may simply be a result of their local abundance at the time of envelopment. However, it is also possible to imagine that non-selectively incorporated RNAs are serving a purpose. As alluded to earlier, the virus may modulate the relative levels of packaged RNAs by controlling the level of each RNA during envelopment. It is possible that the virus manipulates these levels because the RNAs play a structural role in herpesvirus assembly. Studies in retroviruses suggest that the viral RNA genome and non-specifically incorporated cellular RNAs are critical for virus assembly and particle integrity (Muriaux et al., 2001; Wang and Aldovini, 2002). Perhaps the contribution of the packaged RNAs is to organize the tegument proteins to the tegument domain as suggested with RNA-binding proteins located in the tegument (Sciortino et al., 2002). Furthermore, packaged low-molecular-weight RNAs such as MHV-68 vtRNAs may be important for virion assembly and structure as seen with retroviruses in which tRNAs and other small cellular RNAs are important for particle assembly and integrity (Muriaux et al., 2001).
MiRNAs have recently emerged as regulators of gene expression post-transcriptionally by regulating mRNA translation or stability in the cytoplasm and there is mounting evidence that virus-encoded miRNAs may be important for virus pathogenesis (Boss et al., 2009). While the experimentally determined targets remains modest, herpesvirus miRNAs have been shown to regulate fundamental viral and cellular processes including immune evasion, cell survival and proliferation, and key steps in the herpesvirus life cycle such as latency and the switch from latent to lytic replication (Grey et al., 2007, 2008; Stern-Ginossar et al., 2007; Choy et al., 2008; Murphy et al., 2008; Tang et al., 2008, 2009; Umbach et al., 2008). Deep sequencing revealed 13 BV-encoded miRNAs (Amen and Griffiths, 2011) in addition to three others previously reported (Besecker et al., 2009). To understand the role of BV-encoded miRNAs throughout infection, their expression was analyzed using pharmacological agents (Amen and Griffiths, 2011). Five BV-encoded miRNAs were detected from cells infected in the presence of actinomycin D, an inhibitor of transcription, thereby suggesting that miRNAs were packaged into virions. In the case of BV, the incorporation of viral miRNAs into virions may regulate viral and/or cellular mRNA expression in order for the virus to establish a productive infection in a timely manner due to the short replication cycles of alphaherpesviruses (Amen and Griffiths, 2011). It will be interesting to see if this is a unique property of BV, or whether other herpesviruses incorporate miRNAs into virions. As stated above, herpesviruses regulate their gene expression in a coordinated cascade. The presence of RNAs in the virion adds an additional step to what we recognize as the cascade of gene expression, as they are introduced to infected cells in the absence of viral gene transcription (Figure 2). Further work is underway to determine the importance of these packaged miRNAs to virus infection.
Conclusion
The observation that coding RNAs were packaged into herpesvirus virions over 10 years ago provides an excellent basis for the study of packaged non-coding RNAs. While it is collectively understood that RNAs are packaged into herpesvirus virions, there is debate concerning where they are localized and, more importantly, whether they have a role during viral infection. To determine the function of packaged mRNAs, researchers have employed in vitro translation studies as well as the isolation of their encoded proteins from infected cells (Bresnahan and Shenk, 2000; Sciortino et al., 2001, 2002; Bechtel et al., 2005). However, given that non-coding RNAs do not encode proteins, their functions early during infection may be more difficult to identify. It is likely that non-coding RNAs will need to have a known function before the importance of virion incorporation can be determined. Unfortunately, although this is an area of intense research the functions of most viral non-coding RNAs (miRNAs, etc.) are still unknown. On the positive side, studying the function of packaged non-coding RNAs can be imagined as an easier task especially in knockout studies given that non-coding RNAs commonly have more modest effects that would allow a (at least partially viable) virus to be engineered. It may even be possible that the coding RNA field can gain insight into the function of packaged RNAs from the non-coding RNA field. Lastly, it is possible that packaged non-genomic RNAs play an important role in the biology of viruses other than herpesviruses and we believe that this topic deserves further study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our laboratory has received support from Texas Biomedical Research Institute Virology and Immunology Department startup funds, grants from the Southwest Foundation Forum, Voelcker Foundation, and the National Center for Research Resources (NCRR P51 RR013986 and R21 RR026287). Our research is conducted in facilities constructed with support from Research Facilities Improvement Program grant number C06 RR012087 from NCRR.
References
Amen, M. A., and Griffiths, A. (2011). Identification and expression analysis of herpes B virus-encoded small RNAs. J. Virol. 85, 7296–7311.
Bechtel, J., Grundhoff, A., and Ganem, D. (2005). RNAs in the virion of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 79, 10138–10146.
Besecker, M. I., Harden, M. E., Guanglin, L., Wang, X. J., and Griffiths, A. (2009). Discovery of herpes B virus-encoded microRNAs. J. Virol. 83, 3413–3416.
Boss, I. W., Plaisance, K. B., and Renne, R. (2009). Role of virus-encoded microRNAs in herpes virus biology. Trends Microbiol. 17, 544–553.
Bresnahan, W. A., and Shenk, T. (2000). A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288, 2327–2328.
Campadelli-Fiume, G., Roizman, B., Wild, P., Mettenleiter, T. C., and Minson, T. (2006). The egress of herpes viruses from cells: the unanswered questions. J. Virol. 80, 6716–6719.
Choy, E. Y., Siu, K. L., Kok, K. H., Lung, R. W., Tsang, C. M., To, K. F., Kwong, D. L., Tsao, S. W., and Jin, D. Y. (2008). An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205, 2551–2560.
Cliffe, A. R., Nash, A. A., and Dutia, B. M. (2009). Selective uptake of small RNA molecules in the virion of murine gamma herpesvirus 68. J. Virol. 83, 2321–2326.
Greijer, A. E., Dekkers, C. A. J., and Middeldorp, J. M. (2000). Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 14, 640–651.
Grey, F., Hook, L., and Nelson, J. (2008). The functions of herpesvirus-encoded microRNAs. Med. Microbiol. Immunol. 197, 261–267.
Grey, F., Meyers, H., White, E. A., Spector, D. H., and Nelson, J. (2007). A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3, e163. doi:10.1371/journal.ppat.0030163
Johnson, D. C., and Baines, J. D. (2011). Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394.
Kalejta, R. F. (2008). Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72, 249–65.
Kelly, B. J., Fraefel, C., Cunningham, A. L., and Diefenbach, R. J. (2009). Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145, 173–186.
Mettenleiter, T. C., Klupp, B. G., and Granzow, H. (2006). Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9, 423–429.
Mettenleiter, T. C., Klupp, B. G., and Granzow, H. (2009). Herpesvirus assembly: an update. Virus Res. 143, 222–234.
Mettenleiter, T. C., and Minson, T. (2006). Egress of alphaherpesviruses. J. Virol. 80, 1610–1601; author reply 1611–1612.
Muriaux, D., Mirro, J., Harvin, D., and Rein, A. (2001). RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U.S.A. 98, 5246–5251.
Murphy, E., Vanicek, J., Robins, H., Shenk, T., and Levine, A. J. (2008). Suppression of immediate-early viral gene expression by herpes virus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U.S.A. 105, 5453–5458.
Pellet, P. E., and Roizman, B. (2007). “The family Herpesviridae: a brief introduction,” in Field’ Virology, 5th Edn, eds D. M. P. H. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (New York, NY: Lippincott-Williams and Wilkins). 2479–2499.
Sciortino, M. T., Suzuki, M., Taddeo, B., and Roizman, B. (2001). RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75, 8105–8116.
Sciortino, M. T., Taddeo, B., Poon, A. P., Mastino, A., and Roizman, B. (2002). Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. U.S.A. 99, 8318–8323.
Stern-Ginossar, N., Elefant, N., Zimmermann, A., Wolf, D. G., Saleh, N., Biton, M., Horwitz, E., Prokocimer, Z., Prichard, M., Hahn, G., Goldman-Wohl, D., Greenfield, C., Yagel, S., Hengel, H., Altuvia, Y., Margalit, H., and Mandelboim, O. (2007). Host immune system gene targeting by a viral miRNA. Science 317, 376–381.
Stinski, M. F. (1976). Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J. Virol. 19, 594–609.
Tang, S., Bertke, A. S., Patel, A., Wang, K., Cohen, J. I., and Krause, P. R. (2008). An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U.S.A. 105, 10931–10936.
Tang, S., Patel, A., and Krause, P. R. (2009). Novel less-abundant viral miRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83, 1433–1442.
Terhune, S. S., Schroer, J., and Shenk, T. (2004). RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 78, 10390–10398.
Umbach, J. L., Kramer, M. F., Jurak, I., Karnowski, H. W., Coen, D. M., and Cullen, B. R. (2008). MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783.
Varnum, S. M., Streblow, D. N., Monroe, M. E., Smith, P., Auberry, K. J., Pasa-Tolic, L., Wang, D., Camp, D. G. II., Rodland, K., Wiley, S., Britt, W., Shenk, T., Smith, R. D., and Nelson, J. A. (2004). Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78, 10960–10966.
Wang, S. W., and Aldovini, A. (2002). RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76, 11853–11865.
Keywords: virion, herpesvirus
Citation: Amen MA and Griffiths A (2011) Packaging of non-coding RNAs into herpesvirus virions: comparisons to coding RNAs. Front. Gene. 2:81. doi: 10.3389/fgene.2011.00081
Received: 06 October 2011;
Accepted: 29 October 2011;
Published online: 17 November 2011.
Edited by:
Muller Fabbri, The Ohio State University, USAReviewed by:
Patrick Provost, Université Laval, CanadaBernadette Dutia, University of Edinburgh, UK
Copyright: © 2011 Amen and Griffiths. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Anthony Griffiths, Department of Virology and Immunology, Texas Biomedical Research Institute, P.O. Box 760549, San Antonio, TX 78227, USA. e-mail:YWdyaWZmaXRoc0B0eGJpb21lZC5vcmc=
 Melanie A. Amen1,3
Melanie A. Amen1,3