- 1 Computer Science, University of Birmingham, Birmingham, UK
- 2 Department of Biology, Sonoma State University, Rohnert Park, CA, USA
- 3 Princess Alexandra Hospital, The University of Queensland Diamantina Institute, Woolloongabba, QLD, Australia
Evolution is often characterized as a process involving incremental genetic changes that are slowly discovered and fixed in a population through genetic drift and selection. However, a growing body of evidence is finding that changes in the environment frequently induce adaptations that are much too rapid to occur by an incremental genetic search process. Rapid evolution is hypothesized to be facilitated by mutations present within the population that are silent or “cryptic” within the first environment but are co-opted or “exapted” to the new environment, providing a selective advantage once revealed. Although cryptic mutations have recently been shown to facilitate evolution in RNA enzymes, their role in the evolution of complex phenotypes has not been proven. In support of this wider role, this paper describes an unambiguous relationship between cryptic genetic variation and complex phenotypic responses within the immune system. By reviewing the biology of the adaptive immune system through the lens of evolution, we show that T cell adaptive immunity constitutes an exemplary model system where cryptic alleles drive rapid adaptation of complex traits. In naive T cells, normally cryptic differences in T cell receptor reveal diversity in activation responses when the cellular population is presented with a novel environment during infection. We summarize how the adaptive immune response presents a well studied and appropriate experimental system that can be used to confirm and expand upon theoretical evolutionary models describing how seemingly small and innocuous mutations can drive rapid cellular evolution.
Introduction: The Role of Cryptic Genetic Variation in Evolution
Cryptic genetic variation (CGV) describes genetic diversity that is hidden within populations that occupy their native habitats but that is revealed as trait diversity within new environments (Gibson and Dworkin, 2004; Le Rouzic and Carlborg, 2008; Schlichting, 2008; McGuigan and Sgrò, 2009). CGV has been revealed in populations through changes in pH, temperature, nutrient concentrations, moisture, salinity, oxygen, altitude, habitat, and hormone levels. In Drosophila melanogaster, the presence of cross veins in wings, wing shape, scutellar bristle number, and the patterning of photoreceptors in the eye are each highly polygenic traits with little variation in native environments but considerable variation under modified environmental conditions.
Cryptic genetic variation is of considerable scientific interest due to its potential contribution to evolution (see Cryptic Genetic Variation and Evolution in Appendix) and particularly for rapid evolutionary responses to changing environments. Rapid evolution – evolutionary change that is orders of magnitude faster than expected based on inferences from the fossil record – has been reported in dozens of species where local populations are presented with novel environments (Hendry and Kinnison, 1999; Kinnison and Hendry, 2001; Reznick and Ghalambor, 2001). Rapid evolution is unlikely to be driven by de novo mutations due to the relatively slow pace that new genotypes can be discovered and fixed in a population. On the other hand, standing genetic variation is more likely to support rapid evolution in novel environments because it is available immediately at the time that selection changes and because multiple copies of a beneficial allele may already exist thereby increasing both the speed and likelihood of fixation (Barrett and Schluter, 2008). Because cryptic genetic variation is selectively neutral in the original environment, it can accumulate and be maintained, while its release of heritable trait diversity could provide fuel for rapid evolutionary responses to environmental change.
In line with this paradigm, a recent study by Wagner and colleagues, using a ribozyme RNA enzyme as their experimental model, confirmed that ribozyme populations containing cryptic variation more rapidly adapted to new substrates compared to ribozyme populations that did not contain cryptic mutations (Hayden et al., 2011). This study provides the first direct experimental confirmation that cryptic genetic changes can promote rapid evolution. While additional studies strongly implicate CGV in the rapid evolution of complex phenotypes (for example see Palmer, 2004; McGuigan et al., 2011), a direct connection between cryptic alleles and novel traits has not been confirmed in other studies. Directly demonstrating the contribution of CGV in rapid evolution of a complex organism remains a challenging milestone (McGuigan and Sgrò, 2009), in part because genetic diversity can be distributed over a large genome, traits can have a large but unknown genetic footprint, and several factors such as mutations, stochastic effects, and environmental heterogeneity can influence allele frequencies and confound links between CGV and adaptation.
To formally confirm the role of cryptic variation in the evolution of complex phenotypes it is essential to identify model systems where specific and well-characterized cryptic alleles can be shown to drive trait differences with adaptive significance in new environments. In this review we explain how the adaptive immune response of T cells provides an ideal model system that clearly demonstrates the potentially important relationship between CGV and rapid evolution.
The Adaptive Immune System: A Primer
The adaptive immune system provides vertebrates with the ability to recognize and respond to a variety of pathogens including parasites, bacteria, fungi, and viruses. The primary effector cells of the adaptive immune system are B cells and T cells, which are derived from the same multi-potent hematopoietic stem cells. B cells mature in the bone marrow and are involved primarily in the creation of antibodies. B cells express the B cell receptor (BCR), which is a membrane-bound antibody molecule. Different BCRs recognize and bind to their respective foreign molecules (antigens). Once a B cell binds its cognate antigen and receives an additional signal from a helper T cell, it activates proximal signaling events triggering its differentiation into a short lived effector plasma cell that functions to secrete antibodies (soluble forms of the BCR) that bind to its cognate antigen. Antibodies bound to foreign molecules make them easy targets for phagocytosis and other clearance mechanisms. Sub-populations of activated B cells differentiate into long-lived antigen-specific memory B cells, which remain in circulation and can respond quickly if the host is re-infected by the same pathogen.
T cells mature in the thymus and are involved primarily in cell-mediated immune responses. They differ from B cells in that their antigen binding receptor, known as the T cell receptor (TCR) can only recognize peptide antigens that have been proteolytically cleaved and presented in the context of a host cell major histocompatibility complex (MHC) protein (Figure A2 in Appendix). All nucleated cells are capable of expressing antigen in an MHC class I complex and activating a T cell response. However B cells, dendritic cells, and macrophages additionally express MHC class II molecules, as well as other immune-stimulatory molecules, that allow enhanced activation of T cells, and are classified as professional antigen-presenting cells (APCs).
There are two primary types of T cells, cytotoxic T cells (CTLs), and helper T cells. Naive CTLs are activated when they recognize their cognate antigenic peptide presented in the context of an infected host cell’s MHC class I molecule. Once activated, the CTL undergoes clonal expansion where it simultaneously differentiates into a mature CTL and divides rapidly, generating a large number of activated effector cells that travel through the body and scan for infected cells expressing the same antigenic peptide. Active CTLs induce lysis and apoptosis in infected cells. Most effector CTLs undergo apoptosis and are cleared away by phagocytic cells once the infection is resolved, however a subset of CTLs differentiate into memory cells that can rapidly respond to subsequent infections by the same pathogen. Helper T cells have no cytotoxic activity, and instead function to regulate the adaptive immune response. The TCR of T helper cells recognize antigen peptide bound to class II MHC molecules on the host cell surface. The activation of naive T helper cells results in differentiation and clonal expansion similar to CTLs. Unlike CTLs, activation of helper T cells triggers them to release cytokines that regulate the activity of many types of cells including APCs, B cells, and CTLs.
Together, the components of the adaptive immune system respond rapidly and specifically to infection from previously un-exposed pathogens (Raff, 1973). Importantly, in most cases, during subsequent exposure to the same pathogen, the memory response is so protective that no outward signs of re-exposure may be seen (Sallusto et al., 2004). Although many of the characteristics of the adaptive immune response were demonstrated decades ago (North, 1969; Paul and Benacerraf, 1977), the elegance and efficiency of antigen recognition was not fully appreciated until the genetic and molecular basis of this response was determined (Rajewsky, 1996).
The Immunological Role of the T Cell Receptor
Under conditions where foreign proteins are present, such as during a viral infection or phagocytosis of a pathogen, foreign peptides become available to be processed and loaded onto MHC molecules. It is the replacement of a proportion of endogenous self-peptides by foreign peptides that activates the T cells in the periphery. T cell activation involves an elaborate series of events between the T cell and the MHC-expressing APC (Summary of T Cell Selection in the Thymus in Appendix). Several receptor–ligand interactions between T cells and APCs are important for activation, however activation differences between T cells are determined by whether TCR recognizes the foreign peptide loaded onto the MHC molecule. Interestingly, T cell activation has been shown to be digital in nature: a T cell can only exist in an “Off” (resting) state or an “On” (activated) state (Altan-Bonnet and Germain, 2005; Daniels et al., 2006; Chakraborty et al., 2009; Das et al., 2009; Lin et al., 2009; Prasad et al., 2009). This means that as long as the threshold of activation is achieved, relatively weak stimuli (low affinity) will induce the same response as strong stimuli (high affinity) at the single cell level. In other words, selectively relevant trait differences in T cells can be captured by a single binary variable (Altan-Bonnet and Germain, 2005; Chakraborty et al., 2009). Although there are some differences in how activation takes place for different classes of T cells and the different types of MHC molecules, TCR recognition of the peptide loaded MHC remains the defining step in all cases of T cell activation.
The Genetic Basis of the TCR Repertoire
The development of a diverse T cell repertoire is a critical step for maintaining the overall health of an organism as it allows the adaptive immune system to respond to many different pathogenic antigens. Genetic variation of the TCR across the T cell population provides the requisite variety of cell response options that is necessary for the adaptive immune response. These genetic differences among T cells arise through the stochastic recombination of a diverse set of gene segments that make up the TCR. The TCR itself is composed of several subunits ultimately expressed on the surface of the cell. The α and β chains of the TCR are directly responsible for ligand recognition and therefore represent the variable and unique portion of each TCR complex that interacts with peptide–MHC complexes. The other subunits that comprise the TCR (collectively called CD3 chains) are non-variable and are responsible for propagating signals in the cytoplasm of the cell. These signaling subunits contain highly conserved motifs that are critical for initiating the intracellular events leading to T cell activation.
T cell development begins with the entry of T cell progenitors into the thymus, which initiates a program of differentiation that includes TCR gene re-arrangement and expression (Figure 1 and Summary of T Cell Selection in the Thymus in Appendix). The variability of each α and β chain is generated by the random selection and imprecise joining of different germline encoded gene segments (Schatz, 2004). The recombination of these gene segments also involves the addition of random nucleotides at each gene segment junction further increasing the variability seen in TCR repertoires. The α and β chains, if recombined successfully, are then co-expressed and assembled with the CD3 chains to form a TCR with specificity that is unique to that cell. Studies have suggested that this mechanism can potentially generate 1012–1015 different possible TCRs (Davis and Bjorkman, 1988). The stochastic generation of the TCR by recombination thus constitutes a large, pre-established genotype space for TCR and along with its associated antigenic specificity provides a relatively simple and well-defined fitness landscape of T cell activation.
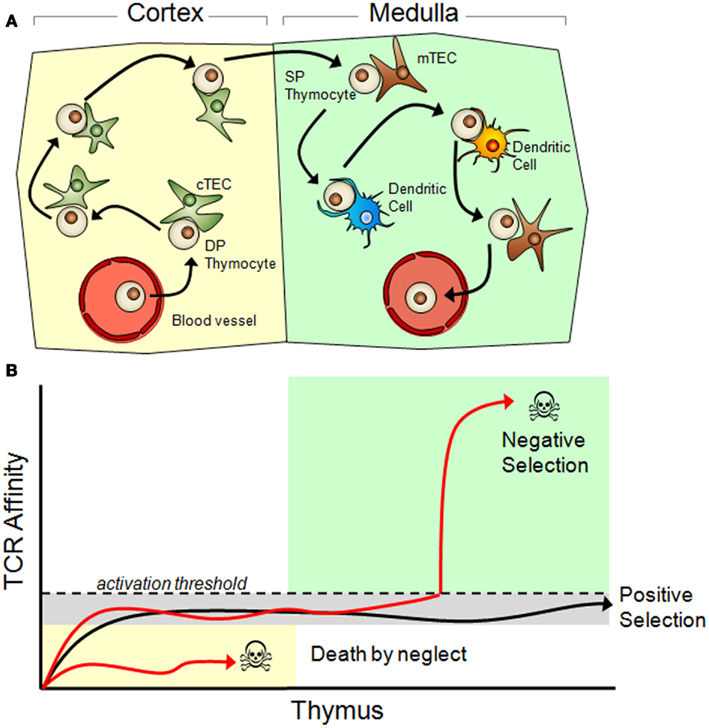
Figure 1. Thymic selection of T cells. (A) Immature T cells (called thymocytes) enter the thymic cortex and, supported by factors secreted by cortical thymic epithelial cells (cTECs, green), undergo differentiation. During this time TCR re-arrangement occurs until an active TCR is expressed along with both the CD4 and CD8 co-receptors (double positive/DP stage). Those DP thymocytes that have a functional TCR and survive commit to a single positive (SP) state (SP thymocytes express either CD4 or CD8, but not both) and migrate to the thymic medulla. Here, SP thymocytes systematically scan medullary thymic epithelial cells (mTECs, shown in brown) and dendritic cells presenting self-peptides in their MHC molecules. During the SP thymocyte stage, any thymocyte that expresses a TCR that has an above threshold affinity to self-peptides undergoes apoptosis and dies. Surviving thymocytes eventually exit the thymus and enter the periphery as mature, naïve T cells. (B) Positive and negative selection is determined by TCR affinity to their peptide/MHC ligand. Once a TCR is expressed on the cell surface it immediately begins interacting with MHC molecules presenting self-peptides. Functional TCR’s generate a basal signal output that, although insufficient to cross the activation threshold, is still necessary for thymocyte survival. TCR’s that cannot recognize peptide/MHC complexes ultimately die by neglect. Surviving thymocytes then migrate to the thymic medulla where they are exposed to self antigen through promiscuous gene expression in mTECs and DCs. Any TCR that reacts with a self-peptide with too high of an affinity will induce T cell activation and apoptosis. Only thymocytes with a functional TCR, which is competent to trigger low-level signaling but does not cross the activation threshold to self-peptides, is positively selected and permitted to exit the thymus.
TCR/MHC Interactions within the Thymus Dictate Selection of the Naïve T Cell Population
Mature T cells present in the periphery must be able to carefully distinguish between cells presenting harmless endogenous self-peptides and cells presenting foreign peptides derived from potentially dangerous pathogens. Once the TCR has been assembled through stochastic DNA recombination, it is rigorously tested within the thymus to ensure that it is functional but will not respond to self-peptides (Figure 1 and Summary of T Cell Selection in the Thymus in Appendix). Defects in this selection process can lead to either profound immunodeficiencies or debilitating autoimmune diseases. At the heart of this selection process is the determination of how the newly formed TCR interacts with a peptide loaded MHC molecule. First, TCRs are screened for their ability to weakly recognize endogenous “self” peptides displayed on an MHC molecule within the cortex of the thymus (Figure 1 and reviewed in Starr et al., 2003).
Those mature T cells that survive thymic selection represent fewer than 5% of the starting pre-selected thymocyte population and are exported from the thymus, entering the circulatory system to ultimately reside in the peripheral lymphoid organs. It is believed that following selection in the thymus, the range of TCR variation is between 106 and 107 (Arstila et al., 1999).
T Cells Provide an Unambiguous Example of Evolution Driven by Environment-Exposed CGV
With evidence of cryptic genetic variants being co-opted into beneficial functions (Waddington, 1942, 1957a; for example through exaptation; Badyaev, 2005), there could potentially be many circumstances where novel environments reveal CGV and drive rapid evolution in natural populations (Palmer, 2004; Hayden et al., 2011; McGuigan et al., 2011). To clearly demonstrate this evolutionary pathway, several conditions must be met. Populations typically must occupy habitats that are stable over many generations (CGV condition 1) and accumulate considerable genetic diversity while exhibiting only small trait differences (CGV condition 2). When presented with novel environments, the same population needs to display new quantitative traits (CGV condition 3) that have a heritable basis (CGV condition 4) and thus may contribute to evolution within the novel environment (Gibson and Dworkin, 2004; Whitacre, 2012).
Each of these conditions corresponds with well-characterized features in the adaptive immune system. T cells develop in a stable thymic environment where a sub-population is selected to mature and leave the thymus based on positive/negative selection. To satisfy the selection conditions, TCRs must have an affinity to host cell protein fragments that is strong enough to result in non-trivial adhesion between the T cell and host cell but is not so strong as to elicit T cell activation. The result is a thymic environment that efficiently selects for mature T cells with similar behavior under “normal” conditions that are devoid of foreign antigens. These selection conditions within the thymus are stable and remain the same throughout the lifetime of the organism (CGV condition 1).
The genome:proteome landscape of TCR contains many distinct proteins that are functionally equivalent for the selection conditions of the thymus. Because thymic selection cannot distinguish between these neutral variants, this allows for the accumulation of a genetically diverse repertoire of receptors across the naïve T cell population (CGV condition 2). However, these TCRs retain functional differences that, while hidden in the initial selection context of the thymus, can lead to the expression of highly distinct cell responses (activation) when T cells are removed from their initial developmental environment and presented with antigens. Each novel (antigen) environment leads to the activation of a small unique subset of naïve T cells, which at the population level appears as a widening of the T cell population’s affinity distribution (Figure 1). The antigen-specific exposure of T cell affinity differences is an example of trait differences released under novel environmental conditions and is a hallmark of CGV (CGV condition 3). Antigen-specific T cell response differences have a clear genetic basis that is found in the variation of alpha and beta chains (CGV condition 4). Moreover, in a properly functioning immune system, the exposure of response differences leads to activation and subsequent clonal expansion for a small selected set of T cells, demonstrating the evolutionary relevance of this CGV. Defining characteristics of the innate and adaptive immune response in T cells thus correspond with defining characteristics of evolutionarily relevant CGV phenomena (summarized in Figure 2 and A Summary of CGV’s Role in T-cell Activation in Appendix). Together, these features formally establish a role for cryptic genetic variation in a well-defined evolving cell population.
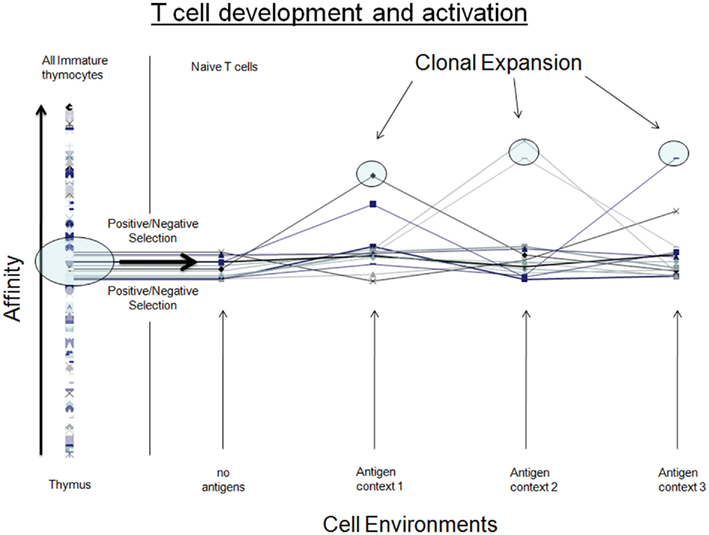
Figure 2. Comparing attributes of T cells (cell development and activation) with CGV in natural populations. In both cases, genetic diversity arises in a stable environment that selects cells/organisms that fall within a narrow band of trait values. When exposed to a novel environment, trait differences are revealed in the population. In adaptive immunity, we consistently observe a subset of cells that are selected to undergo clonal expansion. In natural populations, a change in the environment does not always immediately reveal trait differences that are selectively relevant; however selection of sub-populations with selective advantage resulting from CGV is expected to take place over sufficiently long timescales.
An Ideal Experimental System
T cell adaptive immunity provides ideal genotypic, phenotypic, and environmental conditions that allow links between CGV and environment-induced adaptation to be clearly established. For one, the adaptive immune response preserves selected phenotypes (memory T cells) without continued stimulation from the environment and without the need for additional genetic accommodation. CGV can thus be isolated from future environmental changes and genetic changes that would continue to take place when CGV is revealed in natural populations such as genetic accommodation and drift.
Adaptive immunity also has a well-characterized genetic basis with a small genetic footprint, despite T cell activation being a highly polygenic trait. Relevant genetic differences between T cells are constrained to a single TCR receptor with a genetic sequence that is highly constrained to a predefined space of recombined alpha/beta chains. TCR is generated through a stochastic recombination of gene segments. As a result of this recombination, each gene segment can contribute to activation differences in the T cell repertoire yet remains cryptic within a number of genetic backgrounds. This provides an important similarity to CGV in sexually reproducing populations without the presence of other features of sexual reproduction that would otherwise arise, such as linkage disequilibrium and assortative mating. Furthermore, with the immune system there is no need to isolate CGV from other causes of genetic diversity such as environmental heterogeneity or density dependent selection.
The environmental changes that induce T cell responses are also well-characterized by changes in antigens attached to the surfaces of APCs. Because the time between infections is typically much longer than the immune response time (T cell activation and transition to long-lived memory cells), environmental volatility is not a confounding factor. In addition, environment-induced trait differences are nicely characterized by a digital on/off activation response.
In contrast, evolution in natural populations proceeds through a concurrent stochastic population process where both standing genetic variation and de novo mutations contribute to adaptation, and where non-selective factors such as genetic drift and genetic hitchhiking also influence allele frequencies. Studying the contribution of CGV in evolution is also difficult because its role is typically revealed in a heterogeneous and dynamic environment where genetic assimilation (Schmalhausen and Dobzhansky, 1949; Waddington, 1953, 1957b) and genetic accommodation (West-Eberhard, 2003, 2005) are sometimes needed to preserve the inheritance of phenotypic adaptations in the face of ongoing environmental change (West-Eberhard, 2003; Pigliucci et al., 2006). By isolating CGV from these factors, T cell adaptive immunity provides an experimental system where the contribution of CGV toward rapid evolution of cellular phenotypes can be clearly established and confirmed.
It is worth clarifying why we focus on adaptation in T cells and not B cells. The adaptive response in naive T cells arises from CGV exposure within a novel environment and does not involve a classic genetic search process where the onset of adaptation would take place by the discovery of a novel allele. The story is more complicated in B cells because adaptations can arise from both CGV and a genetic search process. In particular, after the environment-dependent activation of B cells, a genetic search occurs that involves clonal expansion, hyper-mutation, and selection, with the result being a more finely tuned memory B cell. Clearly both gene-induced and environment-induced adaptations are therefore relevant to the immune system. By focusing on T cells we remove any ambiguity as to the origins of adaptation and also in the subsequent proof of principles that were discussed.
Concluding Remarks
Evolution is traditionally characterized as a process involving incremental genetic changes that are discovered and fixed in a population through genetic drift and selection. While the genetic basis of heredity is not disputed, it is increasingly recognized that the introduction of novel alleles is not the only way that heritable phenotypes originate in species. A growing body of evidence suggests that changes in the environment induce heritable and evolutionarily relevant trait differences; revealing previously cryptic genetic diversity that drives rapid evolution (Schmalhausen and Dobzhansky, 1949; Waddington, 1953, 1957b; West-Eberhard, 2003, 2005; Gibson and Dworkin, 2004; Palmer, 2004; Barrett and Schluter, 2008; Schlichting, 2008).
However, multiple confounding factors normally make it difficult to demonstrate the evolutionary significance of cryptic genetic variation (Barrett and Schluter, 2008; McGuigan and Sgrò, 2009). Evidence of environment-exposed CGV driving heritable changes in a deme is sporadic at best (McGuigan and Sgrò, 2009); it remains difficult to systematically uncover, and its plausibility is thus far only indirectly supported by the controlled manipulation of artificial environments (Barrett and Schluter, 2008; Hayden et al., 2011; McGuigan et al., 2011) or phylogenetic analysis (Palmer, 2004; Barrett and Schluter, 2008).
In this article we have recounted how T cell maturation and antigen-specific activation provide a clear and unambiguous example of environmental exposure of cryptic genetic variation that drives complex phenotypic changes in a cell population. CGV within the TCR repertoire facilitates rapid evolutionary responses in the adaptive immune system and is fundamental to the survival of vertebrates in the face of infections from diverse pathogenic sources.
Cryptic genetic variation-driven evolution may have broad repercussions to scientific research investigating the preservation of species and the eradication of evolvable pathogens. In conservation biology for instance, intraspecific genetic diversity is a well known factor in extinction risk. However, only genetic variants that produce trait differences under future environmental stresses can mitigate extinction risks. Quantifying G × E interactions for stresses that a species may encounter in the future, for instance by selecting stresses based on predictive models of regional climate change, should reveal biodiversity with the greatest significance for species conservation efforts. Moreover, with limited conservation budgets, strategies for preserving this much smaller CGV footprint may provide a more realistic and realizable conservation goal.
On the other hand, in highly evolvable diseases such as aggressive cancers, the accumulation of CGV may contribute to the rapid evolution of therapy resistance that is seen in the latest generation of targeted therapies (Tian et al., 2011). Tumors with high genetic diversity have already been shown to more rapidly evolve resistance to therapies (Gerlinger and Swanton, 2010; Tian et al., 2011). Interestingly, the therapeutic strategies used today may inadvertently play to the strengths of evolvable cancers (Read et al., 2011). For instance, therapies are often applied long after therapy resistance is first established in the cancer cell population, thereby allowing a founder population to grow and acquire the genetic diversity needed for further adaptation to the next therapy that is eventually applied. If the therapy were instead changed at the point of lowest CGV, the cell population would be highly vulnerable to new environmental stresses in a manner analogous to the extinction risks posed by low genetic diversity in natural populations. CGV will not readily accumulate in a rapidly changing environment with strong directional selection and we propose therefore that dynamic therapeutic strategies could provide a new approach to reducing the evolutionary potential of cancers containing high levels of genetic diversity.
Using an ideal model system, this paper formally establishes links between cryptic alleles and environment-induced adaptation in a complex cellular phenotype. As confirmation of these principles is further documented in other biological contexts and becomes more widely accepted, we anticipate that CGV will act as a focal point in efforts to preserve or eradicate evolving populations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Angus Harding is supported by an NHMRC project grant, an ARC project grant, and The Brain Foundation Australia. James M. Whitacre is supported by a DSTO grant and an EPSRC grant (No. EP/E058884/1).
References
Altan-Bonnet, G., and Germain, R. N. (2005). Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 3, e356. doi:10.1371/journal.pbio.0030356
Arstila, T. P., Casrouge, A., Baron, V., Even, J., Kanellopoulos, J., and Kourilsky, P. (1999). A direct estimate of the human alphabeta T cell receptor diversity. Science 286, 958.
Badyaev, A. V. (2005). Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proc. Biol. Sci. 272, 877.
Barrett, R. D. H., and Schluter, D. (2008). Adaptation from standing genetic variation. Trends Ecol. Evol. (Amst.) 23, 38–44.
Chakraborty, A. K., Das, J., Zikherman, J., Yang, M., Govern, C. C., Ho, M., Weiss, A., and Roose, J. (2009). Molecular origin and functional consequences of digital signaling and hysteresis during Ras activation in lymphocytes. Sci. Signal. 2, pt2.
Daniels, M. A., Teixeiro, E., Gill, J., Hausmann, B., Roubaty, D., Holmberg, K., Werlen, G., Hollander, G. A., Gascoigne, N. R., and Palmer, E. (2006). Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729.
Das, J., Ho, M., Zikherman, J., Govern, C., Yang, M., Weiss, A., Chakraborty, A. K., and Roose, J. P. (2009). Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136, 337–351.
Davis, M. M., and Bjorkman, P. J. (1988). T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402.
Gerlinger, M., and Swanton, C. (2010). How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br. J. Cancer 103, 1139–1143.
Gibson, G., and Dworkin, I. (2004). Uncovering cryptic genetic variation. Nat. Rev. Genet. 5, 681–690.
Hayden, E. J., Ferrada, E., and Wagner, A. (2011). Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474, 92–97.
Hendry, A. P., and Kinnison, M. T. (1999). Perspective: the pace of modern life: measuring rates of contemporary microevolution. Evolution 53, 1637–1653.
Kinnison, M. T., and Hendry, A. P. (2001). The pace of modern life II: from rates of contemporary microevolution to pattern and process. Genetica 112, 145–164.
Le Rouzic, A., and Carlborg, Ö. (2008). Evolutionary potential of hidden genetic variation. Trends Ecol. Evol. (Amst.) 23, 33–37.
Lin, J., Harding, A., Giurisato, E., and Shaw, A. S. (2009). KSR1 modulates the sensitivity of mitogen-activated protein kinase pathway activation in T cells without altering fundamental system outputs. Mol. Cell. Biol. 29, 2082–2091.
McGuigan, K., Nishimura, N., Currey, M., Hurwit, D., and Cresko, W. A. (2011). Cryptic genetic variation and body size evolution in threespine stickleback. Evolution 65, 1203–1211.
McGuigan, K., and Sgrò, C. M. (2009). Evolutionary consequences of cryptic genetic variation. Trends Ecol. Evol. (Amst.) 24, 305–311.
North, R. J. (1969). Cellular kinetics associated with the development of acquired cellular resistance. J. Exp. Med. 130, 299.
Paul, W. E., and Benacerraf, B. (1977). Functional specificity of thymus-dependent lymphocytes. Science 195, 1293.
Pigliucci, M., Murren, C., and Schlichting, C. (2006). Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 209, 2362.
Prasad, A., Zikherman, J., Das, J., Roose, J. P., Weiss, A., and Chakraborty, A. K. (2009). Origin of the sharp boundary that discriminates positive and negative selection of thymocytes. Proc. Natl. Acad. Sci. U.S.A. 106, 528–533.
Read, A. F., Day, T., and Huijben, S. (2011). The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 2), :10871.
Reznick, D. N., and Ghalambor, C. K. (2001). The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica 112, 183–198.
Sallusto, F., Geginat, J., and Lanzavecchia, A. (2004). Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763.
Schlichting, C. D. (2008). Hidden reaction norms, cryptic genetic variation, and evolvability. Ann. N. Y. Acad. Sci. 1133, 187–203.
Schmalhausen, I., and Dobzhansky, T. (1949). Factors of Evolution: The Theory of Stabilizing Selection. Philadelphia: Blakiston Co.
Starr, T., Jameson, S., and Hogquist, K. (2003). Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139.
Tian, T., Olson, S., Whitacre, J. M., and Harding, A. (2011). The origins of cancer robustness and evolvability. Integr. Biol. (Camb.) 3, 17–30.
Waddington, C. H. (1942). Canalization of development and the inheritance of acquired characters. Nature 150, 563.
Waddington, C. H. (1957b). The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. London: Allen & Unwin.
West-Eberhard, M. (2003). Developmental Plasticity and Evolution. New York: Oxford University Press.
West-Eberhard, M. (2005). Developmental plasticity and the origin of species differences. Proc. Natl. Acad. Sci. U.S.A. 102(Suppl. 1), :6543.
Whitacre, J. M. (2012). Genetic and environment-induced pathways to innovation: on the possibility of a universal relationship between robustness and adaptation in complex biological systems. Evol. Ecol. 25, 965–975.
Appendix
Cryptic Genetic Variation and Evolution
Trait robustness toward mutations and environmental variations is conditional upon both the genetic and environmental background. Within a particular environment, trait robustness toward mutations enables considerable genetic diversity to accumulate under mutation-selection balance. While many genetic variants may appear silent or cryptic with this environment, the conditionality of trait robustness on the environmental background means that such populations will reveal heritable trait differences within novel environmental conditions. This phenomenon is referred to as cryptic genetic variation (CGV) and has been hypothesized to be a source of rapid evolutionary adaptation in new environments.
Confusion about the role of CGV in evolution can be attributed to long-standing difficulties in understanding relationships between robustness and adaptation and also due to historical descriptions of evolution as a genetic search process. For example, Mutational Robustness – the stability of the phenotype to mutations – was originally predicted to impede evolvability (Frank, 2007; Wagner, 2008) because it lowers the mutational accessibility of distinct heritable phenotypes from a single genotype and it reduces selective differences within a genetically diverse population (Wagner, 2008). Only in the last decade has a resolution to this paradox been established for mutational (Ciliberti et al., 2007; Wagner, 2008; Whitacre and Bender, 2010) and environmental (Whitacre, 2012) forms of robustness. For instance, mutational robustness is now believed to facilitate evolvability in two ways: first by establishing genotypic neutral networks from which large numbers of distinct genotypes can be sampled (Wagner, 2008) and second by allowing genetic diversity to arise in a population with subsequent trait differences revealed in an environment-dependent manner (Whitacre, 2012). Both of these pathways to adaptation occur due to the presence of CGV in populations (see Figure A1). However, what is not yet well appreciated is the important role that CGV can play when evolution occurs in a dynamic environment. In particular, CGV may provide the necessary fuel for adaptation under new stressful environments while bypassing the negative selection that limits phenotypic variability under stable conditions.
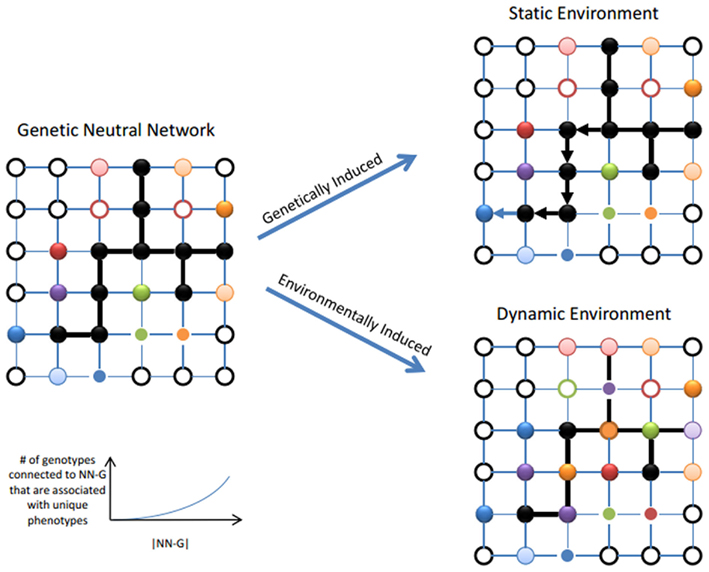
Figure A1. The relationship between cryptic genetic variation and a population’s propensity to evolve. Left Panel: When many genotypes with similar fitness are present in a fitness landscape, these can connect to form a neutral network (NN, see black nodes). Distinct heritable phenotypes are still accessible through mutations that lead to genotypes that are not members of NN (see colored nodes). Larger NN can extend through larger regions of the fitness landscape and have access to a greater range of distinct heritable phenotypes (Ciliberti et al., 2007; Wagner, 2008; Whitacre and Bender, 2010; Whitacre, 2012). Right Panel, Genetically induced adaptation: If the environment is stable, a population will diffuse and drift over the NN allowing it to sample many heritable phenotypes over time and thereby supporting evolution. Right Panel, Environmentally induced adaptation: Both the properties of individual phenotypes and entire fitness landscapes depend on the environment. In a stable environment, a population is able spread over a region of the NN because these genetic differences are selective cryptic. However a change in the environment can influence each genotype differently so that genetic distinctions in the population are revealed as phenotypic distinctions as well. At the fitness landscape level, this appears as a partial collapse of the NN.
Summary of T Cell Selection in the Thymus
Thymic development of T cells begins with the entry of T cell progenitors into the thymus through blood vessels near the cortico-medullary junction (Le Douarin and Jotereau, 1975). Thymic entry into the cortex initiates a program of differentiation that includes TCR gene rearrangement and expression, in combination with changes in expression of cell surface proteins (Nitta et al., 2008). The α and β chains of the TCR are directly responsible for ligand recognition and therefore represent the variable and unique portion of each TCR complex that interacts with peptide–MHC complexes. The variability of each α and β chain is generated by the random selection and imprecise joining of different germline encoded gene segments. During development, thymocytes first begin forming the TCR β chain by the assembling one variable (V), one diversity (D), and one joining (J) gene segment to an invariant constant region (C) from a “library” of V (n = 52), D (n = 2), and J (n = 13) segments that are all initially encoded within each β gene locus (Schatz, 2004). The recombination of these gene segments also involves the addition of random nucleotides at each gene segment junction further increasing the variability seen in TCR repertoires. The newly formed β chain pairs up with a surrogate, pre-formed α chain called the pre-T cell α (pTα), as well as with the CD3 chains, forming a pre-TCR. The pre-TCR has all the signaling components of a mature TCR, leading to cellular proliferation, the arrest of β chain gene re-arrangements, and the expression of CD4 and CD8. These CD4/CD8 double positive (DP) thymocytes comprise the majority of thymocytes within the thymus.
Once the pre-TCR DP thymocytes cease proliferating they become smaller and begin the genetic re-arrangement of the TCR α chain. The α chain rearrangement is similar to that of the β chain except that the α chain is composed of only one V (n = ∼70) and one J (n = 61) segment fused again to an invariable constant region. The α and β chains, if recombined successfully, are then co-expressed and assembled with the CD3 chains to form a TCR with specificity that is unique to that cell. Studies have suggested that this mechanism can potentially generate 1012–1015 different possible TCRs (Davis and Bjorkman, 1988). The stochastic generation of the TCR by V, D, J recombination thus constitutes a large, pre-established genotype space for TCR and along with its associated antigenic specificity provides a relatively simple and well-defined fitness landscape of T cell activation. Expression of the successfully rearranged TCR together with the TCR co-receptor CD3 leads to the commitment to the TCRαβ lineage and the expression of CD4 and CD8 co-receptors to create DP thymocytes (Petrie et al., 1990a,b). DP thymocytes expressing TCRαβ move actively within the cortical microenvironment, scanning thymic cortical epithelial cells (cTECs) MHC molecules (Witt et al., 2005). Functional TCRαβ receptors that respond to self peptide–MHC complexes on cTECs are induced to survive and proliferate (Ashton-Rickardt et al., 1993), whereas TCRs defective for MHC binding die from neglect (Sebzda et al., 1994). The selection for functional TCRs within the cortex is known as positive selection. During positive selection, differential kinetics of TCR–MHC interaction drives DP thymocytes to differentiate to become CD4+ or CD8+ single-positive (SP) thymocytes (Sebzda et al., 1994).
Positively selected SP thymocytes then move to the thymic medulla, where they reside for 4–5 days before being licensed to exit the thymus and enter peripheral circulation (Takahama, 2006; McCaughtry et al., 2007). The thymic medulla is a specialized environment where developing SP thymocytes are extensively screened for tolerance to self-antigens, including peripheral tissue-specific antigens (Takahama, 2006). Medullary thymic epithelial cells (mTECs) ectopically express thousands of tissue-restricted antigens (TRAs) (Anderson et al., 2002), a phenomenon unique to mTECs known as promiscuous gene expression (Derbinski et al., 2001). Promiscuous gene expression allows mTECs to present antigens present in all organs within their MHC molecules (Derbinski et al., 2001). In addition to mTECs, the thymic medulla also contains a heterogenous population of thymic dendritic cells (Wu and Shortman, 2005). Two thirds of thymic dendritic cells originate from within the thymus (intrathymic DCs) (Donskoy and Goldschneider, 2003), the remaining third are immigrants from peripheral sites (migratory DCs) (Li et al., 2009). Intrathymic DCs represent a second route to present mTEC-derived antigens to SP thymocytes, which includes secreted molecules, as well as all major subcellular compartments including membrane, cytoplasm, and nuclear antigens (Koble and Kyewski, 2009). Migratory DCs are thought to present both peripheral self antigens as well as potentially presenting innocuous foreign antigens such as those present in gut flora (Klein et al., 2009). Thus promiscuous gene expression by mTECs, combined with antigen presentation by intrathymic and migratory DCs, provides a mirror of self-antigens present throughout the organism. SP thymocytes use their time in the thymic medulla to extensively scan the self antigen-MHC complexes of tMECs, intrathymic DCs, and migratory DCs (Klein et al., 2009). If a cell has too high of an affinity for a self-MHC presented in the thymic medulla, it will trigger activation of the TCR, which during this T-cell developmental stage leads to apoptosis (Daniels et al., 2006). Apoptosis of self-reactive thymocytes in the thymic medulla efficiently removes auto-reactive thymocytes from the T-cell repertoire, a process known as negative selection.
Summary of TCR Activation and Downstream Signaling.
Figure A2 summarizes the primary steps of TCR activation most relevant to our argument. For the purposes of this review, we have not discussed many of the other receptor ligand interactions required for T cell activation (reviewed in Rudolph et al., 2006). However there are several factors that can affect the overall state of T cell activation. With each naïve T cell expressing a unique TCR, T cells must constantly sample MHC molecules if they are to discover the presence of foreign peptide with enough affinity to clear the threshold for activation. On the surface, this would seem like a very rare occurrence. Indeed, experiments using well characterized pathogens with known immunodominant peptides have elegantly shown that the estimated number of cells from a normally derived repertoire, which has sufficient affinity to respond to a particular foreign peptide, is anywhere from 3 × 10−4 to 6 × 10−7 depending on the pathogen (Moon et al., 2007; Alanio et al., 2010). One factor that contributes to appropriate T cell activation in the presence of rare foreign antigens is the length of time the T cell remains engaged with the APC. Studies show that T cell surveying APC loaded with only endogenous peptide display short transient interactions with the APC (Salazar-Fontana and Bierer, 2001). When peptides specific for the TCR are added, the T cell remains engaged to the APC on the order of hours, which ultimately results in T cell activation (Miller et al., 2002, 2004). It is also important to note that other factors can affect how the TCR interacts with the peptide/MHC molecule such as TCR clustering, endocytosis, and degradation of the TCR following engagement (reviewed in Salazar-Fontana and Bierer, 2001; Rudolph et al., 2006; Smith-Garvin et al., 2009).
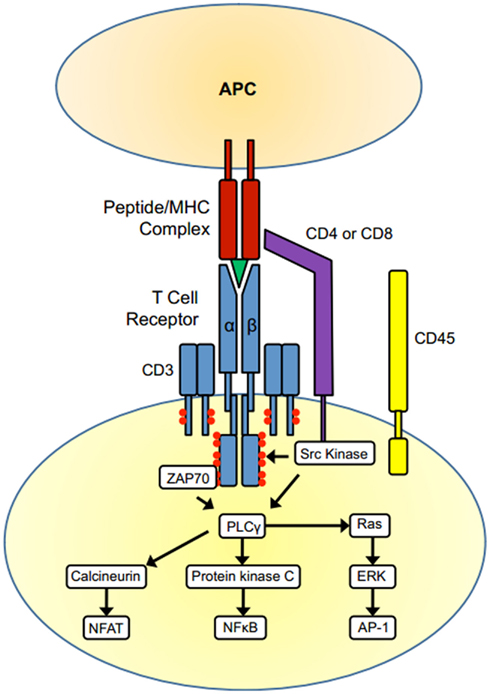
Figure A2. T cell activation. The TCR complex (TCR α and β chains and CD3) and the co-receptors CD4 or CD8 are associated with tyrosine kinases of the Src family. When the TCR binds to an MHC presenting its cognate antigen, the co-receptors CD4/CD8 also bind the MHC molecule on the APC surface. The dephosphorylation of Src kinases by the phosphatase CD45 leads to their activation, and the Src kinases phosphorylate ITAMs within the receptor complex (red circles). ITAM phosphorylation then leads to the recruitment and activation of the kinase Zap70 and the subsequent recruitment of multiple other signaling molecules. One crucial event is activation of the enzyme phospholipase C γ-1, leading to activation of the three main signaling pathways that drive T-cell activation, the Ras-MAP kinase Pathway, the protein kinase C pathway, and calcineurin pathway. These pathways culminate in the activation of the transcription factors NF-κB, NFAT, and AP-1, initiating gene transcription and resulting in the differentiation, proliferation, and effector activity of T-cells.
A Summary of CGV’s Role in T-Cell Activation
In the context of the adaptive immune system, the hide and environment-dependent release of heritable trait differences (CGV) is facilitated by the stochastic recombination of TCR gene segments (source of genetic variation), and the stable positive/negative selection conditions in the thymus. The genome:proteome landscape of T cell receptors contains many distinct proteins that are functionally equivalent for the selection conditions of the thymus, i.e. the landscape contains a high degree of functional neutrality. T cell maturation statistics suggest that this functional neutrality is present in roughly 1–5% of the total TCR G:P landscape. Because selection conditions in the thymus cannot distinguish between these neutral variants, this allows for the accumulation of a genetically diverse repertoire of receptors across the naïve T cell population. However, these T cell receptors retain structural (entropic and thermodynamic) differences that, while hidden in the initial selection context of the thymus, can lead to the expression of highly distinct cell responses (e.g., activation) when T-cells are removed from their initial developmental environment and presented with antigens.
In summary, the stochastic recombination of TCR gene segments, and the positive/negative selection in the thymus combine to establish non-trivial T cell population properties that result in: (1) heritable differences in T-cell receptors that map to the same affinity response for host cells (silent genetic diversity), (2) the exposure of unique affinities for T cells that are presented with foreign antigens (exposure of heritable phenotypic differences), (3) the ability of some of these environment-dependent responses to invoke T-cell activation and subsequent clonal expansion (the discovery of new selected phenotypes) when presented with foreign antigens. Together, these features establish a well-defined role for cryptic genetic variation in an evolving cell population.
References
Alanio, C., Lemaitre, F., Law, H. K., Hasan, M., and Albert, M. L. (2010). Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 115, 3718–3725.
Anderson, M. S., Venanzi, E. S., Klein, L., Chen, Z., Berzins, S. P., Turley, S. J., von Boehmer, H., Bronson, R., Dierich, A., Benoist, C., and Mathis, D. (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401.
Ashton-Rickardt, P. G., Van Kaer, L., Schumacher, T. N., Ploegh, H. L., and Tonegawa, S. (1993). Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell 73, 1041–1049.
Ciliberti, S., Martin, O. C., and Wagner, A. (2007). Innovation and robustness in complex regulatory gene networks. Proc. Natl. Acad. Sci. U.S.A. 104, 13591–13596.
Daniels, M. A., Teixeiro, E., Gill, J., Hausmann, B., Roubaty, D., Holmberg, K., Werlen, G., Hollander, G. A., Gascoigne, N. R., and Palmer, E. (2006). Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729.
Davis, M. M., and Bjorkman, P. J. (1988). T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402.
Derbinski, J., Schulte, A., Kyewski, B., and Klein, L. (2001). Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039.
Donskoy, E., and Goldschneider, I. (2003). Two developmentally distinct populations of dendritic cells inhabit the adult mouse thymus: demonstration by differential importation of hematogenous precursors under steady state conditions. J. Immunol. 170, 3514–3521.
Frank, S. (2007). Maladaptation and the paradox of robustness in evolution. PLoS ONE 2, e1021. doi:10.1371/journal.pone.0001021
Klein, L., Hinterberger, M., Wirnsberger, G., and Kyewski, B. (2009). Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844.
Koble, C., and Kyewski, B. (2009). The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J. Exp. Med. 206, 1505–1513.
Le Douarin, N. M., and Jotereau, F. V. (1975). Tracing of cells of the avian thymus through embryonic life in interspecific chimeras. J. Exp. Med. 142, 17–40.
Li, J., Park, J., Foss, D., and Goldschneider, I. (2009). Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J. Exp. Med. 206, 607–622.
McCaughtry, T. M., Wilken, M. S., and Hogquist, K. A. (2007). Thymic emigration revisited. J. Exp. Med. 204, 2513–2520.
Miller, M. J., Safrina, O., Parker, I., and Cahalan, M. D. (2004). Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J. Exp. Med. 200, 847–856.
Miller, M. J., Wei, S. H., Parker, I., and Cahalan, M. D. (2002). Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296, 1869–1873.
Moon, J. J., Chu, H. H., Pepper, M., McSorley, S. J., Jameson, S. C., Kedl, R. M., and Jenkins, M. K. (2007). Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213.
Nitta, T., Murata, S., Ueno, T., Tanaka, K., and Takahama, Y. (2008). Thymic microenvironments for T-cell repertoire formation. Adv. Immunol. 99, 59–94.
North, R. J. (1969). Cellular kinetics associated with the development of acquired cellular resistance. J. Exp. Med. 130, 299.
Petrie, H. T., Hugo, P., Scollay, R., and Shortman, K. (1990a). Lineage relationships and developmental kinetics of immature thymocytes: CD3, CD4, and CD8 acquisition in vivo and in vitro. J. Exp. Med. 172, 1583–1588.
Petrie, H. T., Pearse, M., Scollay, R., and Shortman, K. (1990b). Development of immature thymocytes: initiation of CD3, CD4, and CD8 acquisition parallels down-regulation of the interleukin 2 receptor alpha chain. Eur. J. Immunol. 20, 2813–2815.
Read, A. F., Day, T., and Huijben, S. (2011). The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 2), :10871.
Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466.
Salazar-Fontana, L. I., and Bierer, B. E. (2001). T-lymphocyte coactivator molecules. Curr. Opin. Hematol. 8, 5–11.
Sebzda, E., Wallace, V. A., Mayer, J., Yeung, R. S., Mak, T. W., and Ohashi, P. S. (1994). Positive and negative thymocyte selection induced by different concentrations of a single peptide. Science 263, 1615–1618.
Smith-Garvin, J. E., Koretzky, G. A., and Jordan, M. S. (2009). T cell activation. Annu. Rev. Immunol. 27, 591–619.
Takahama, Y. (2006). Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6, 127–135.
Whitacre, J. M. (2012). Genetic and environment-induced pathways to innovation: on the possibility of a universal relationship between robustness and adaptation in complex biological systems. Evol. Ecol. 25, 965–975.
Whitacre, J. M., and Bender, A. (2010). Degeneracy: a design principle for achieving robustness and evolvability. J. Theor. Biol. 263, 143–153.
Witt, C. M., Raychaudhuri, S., Schaefer, B., Chakraborty, A. K., and Robey, E. A. (2005). Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 3, e160. doi:10.1371/journal.pbio.0030160
Keywords: cryptic genetic variation, evolution, adaptive immunity, exaptation
Citation: Whitacre JM, Lin J and Harding A (2012) T cell adaptive immunity proceeds through environment-induced adaptation from the exposure of cryptic genetic variation. Front. Gene. 3:5. doi: 10.3389/fgene.2012.00005
Received: 01 December 2011; Paper pending published: 21 December 2011;
Accepted: 06 January 2012; Published online: 09 February 2012.
Edited by:
Firas H. Kobeissy, University of Florida, USAReviewed by:
Ying Xu, West Virginia University, USAKotb Abdelmohsen, National Institutes of Health, USA
Copyright: © 2012 Whitacre, Lin and Harding. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: James M. Whitacre, Computer Science, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK. e-mail:andoaXRhY3JlNzlAZ21haWwuY29t; Angus Harding, Princess Alexandra Hospital, The University of Queensland Diamantina Institute, Woolloongabba, QLD, Australia. e-mail:YS5oYXJkaW5nMUB1cS5lZHUuYXU=


