- Department of Clinical Chemistry, VU University Medical Center, Amsterdam, Netherlands
During the first trimester of pregnancy fetal trophoblasts invade the maternal decidua, thereby remodeling the maternal spiral arteries. This process of trophoblast invasion is very similar to cancer cell invasion, with multiple signaling pathways shared between the two. Pregnancy-related diseases, e.g., pre-eclampsia, and cancer metastasis start with a decrease or increase in cellular invasion, respectively. Here, we investigate if first trimester placental explants can be used to identify epigenetic factors associated with changes in cellular invasion and their potential use as biomarkers. We show that the outgrowth potential of first trimester explants significantly correlates with promoter methylation of PRKCDBP and MMP2, two genes known to be differentially methylated in both placenta and cancer. The increase in methylation percentage of placental cells coincides with an increase in invasion potential. Subsequently, as a non-invasive marker must be detectable in blood, plasma samples of pregnant and non-pregnant women were analyzed. The MMP2 promoter showed high methylation levels in non-pregnant plasma samples, which decreased in pregnant plasma samples which also contain placental DNA. The decrease in methylated plasma DNA during pregnancy is most likely due to the fractional increase in unmethylated placental DNA. This suggests that the level of unmethylated DNA has the potential to be used as an invasion marker, where higher levels of unmethylated DNA indicate a lower invasion potential of trophoblasts. These proof of principle data provide evidence that human first trimester placental explants are an excellent ex vivo model system to identify (epigenetic) factors and thus potential biomarkers associated with changes in cellular invasion, e.g., to detect pregnancy-related diseases or cancer metastasis. To identify novel biomarkers the next step is to correlate naturally occurring variation in invasion potential to changes in (epigenetic) factors by genome-wide approaches such as massively parallel sequencing.
Introduction
During the first trimester of pregnancy fetal trophoblasts invade the maternal decidua. By invading the decidua the maternal spiral arteries are remodeled by the invasive trophoblasts. This process of trophoblast invasion is very similar to cancer cell invasion, with multiple signaling pathways shared between the two (Ferretti et al., 2007; Holtan et al., 2009), giving rise to, e.g., migration and invasion of stroma and arteries, epithelial-mesenchymal transition, heterotypic interactions, and the shedding of particles into the circulation. Interestingly, multiple pregnancy complications, i.e., IUGR (intra-uterine growth restriction) and pre-eclampsia, show a reduction in trophoblast invasion during first trimester (Steegers et al., 2010).
The fact that trophoblast invasion and cancer cell invasion share many signaling pathways makes the first trimester placenta an excellent model to study factors involved in either increased or decreased invasion potential.
The aim of this study is to investigate if the naturally occurring variation in invasion potential of human first trimester placental explants correlates to changes in epigenetic factors. In the future, this set up can be used as an ex vivo model system to identify factors by genome-wide approaches (e.g., massively parallel sequencing), and thus potential biomarkers associated with changes in cellular invasion, e.g., to detect pregnancy-related diseases or cancer metastasis.
Previously, we identified the STOX1 Y153H SNP to be a susceptibility allele for pre-eclampsia in Dutch females (Van Dijk et al., 2005). When the invasion potential of first trimester placental explants was established by measuring the outgrowth of the explants a significant reduction in invasion potential was found in the placentas homozygous for the Y153H allele (Van Dijk et al., 2010). This finding showed that genetic differences can give rise to measurable changes in outgrowth potential. The study described here investigates if also epigenetic differences can be detected in first trimester placentas with different invasion potentials. When this is the case, not only genetic and protein or mRNA expression differences can function as potential factors, but also epigenetic differences, increasing the identification of novel biomarker panels by genome-wide approaches.
For testing the use of first trimester epigenetic differences in relation to invasion potential the methylation levels of 10 genes were chosen to study. Eight well-known tumor suppressor genes (TP73, RASSF5, RASSF1, APC, DAB2IP, PRKCDBP, WT1, MORF4L1) were chosen which all were described by Grigoriu et al. (2011) to be differentially methylated in first trimester placenta. From the extensive list of proteins and pathways shared by cancer cell invasion and trophoblast invasion, e.g., the WNT signaling pathway, integrins, enzymes, growth factors, and their receptors (see for overviews Ferretti et al., 2007; Holtan et al., 2009), two matrix metalloproteinases were chosen to study, MMP9 and MMP2. Both are found to be expressed in the invasive trophoblast as well as malignant tumors (Ferretti et al., 2007; Holtan et al., 2009).
Materials and Methods
Placental DNA Samples
Placental DNA used (n = 32) originates from placental samples with established outgrowth potential as described previously (Van Dijk et al., 2010). In short, small fragments of placental villi from 6 to 9 week gestation were dissected from placenta, placed on Matrigel and cultured at 3% O2 and 37°C in serum free DMEM/F12 media. Simultaneously, small fragments of the same placentas were kept for DNA isolation. At day 2 and day 4 pictures were taken to measure changes in outgrowth. The explant outgrowth potentials were measured by taking the difference of the distance the extravillous trophoblasts migrated between day 2 and day 4. From each placenta at least three explants were grown, explants that did not show any growth were left out of analyses because they were either not viable or were not properly dissected. For the current study only DNA of placentas were used that had at least two explants with growth.
Plasma DNA Samples
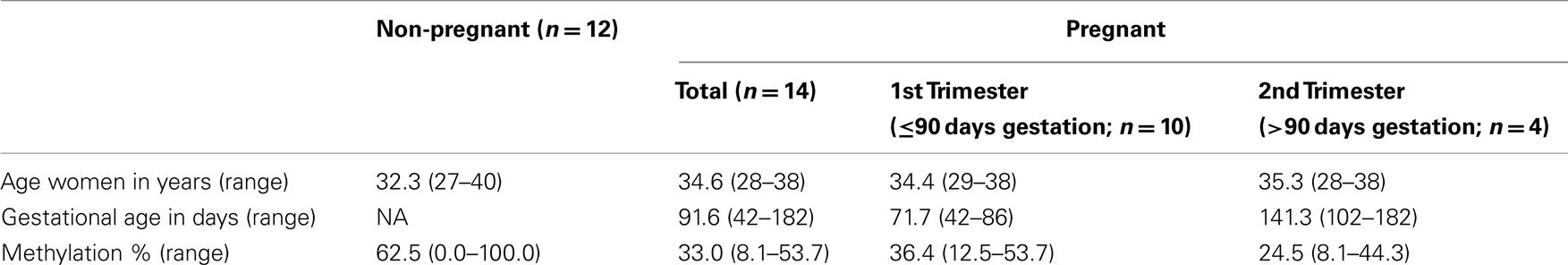
Simultaneous isolation of DNA and RNA from plasma samples was performed as described previously (Oudejans et al., 2003) with modifications (Go et al., 2007). Pregnant sample collection was approved by the ethics committee of the VU University Medical Center. Informed consent was obtained from all women. Isolated DNA samples (n = 26) were precipitated and concentrated to be able to use the DNA amount isolated from 1 ml plasma (∼20 ng) in each reaction. Characteristics of the plasma samples analyzed can be found in Table 1.
DNA Methylation Assessment
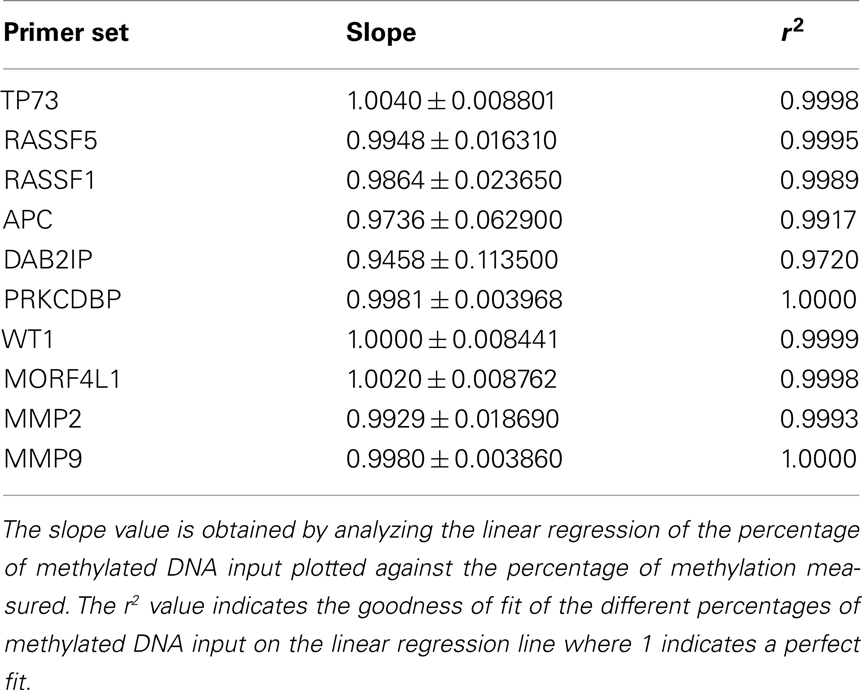
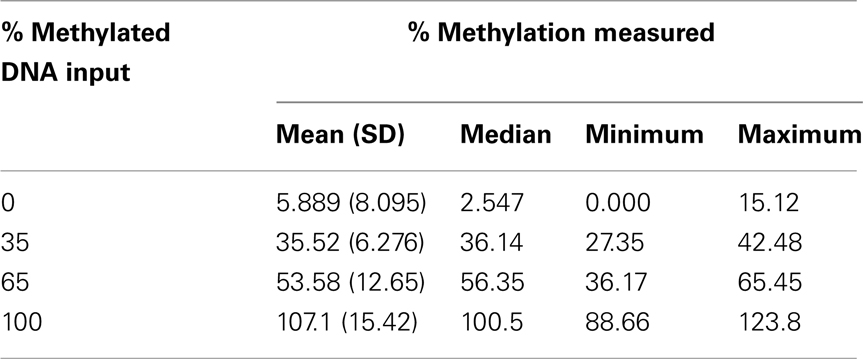
The level of DNA methylation was measured using the OneStep qMethyl kit (Zymo Research). For use with this assay, 10 primer pairs were designed within CpG rich (promoter) regions according to the OneStep qMethyl kit guidelines. Primer sequences can be found in Table 2. In brief, to measure methylation percentages 20 ng DNA (or DNA isolated from 1 ml plasma) in duplicate is placed in a test and a reference reaction mix including primers. The samples are placed in an ABI7300 with the following protocol: 2 h 37°C; 10 min 95°C; 50 cycles 30 s 95°C, 1 min 54°C, 1 min 72°C followed by an dissociation stage to check specificity of the PCR products. The Ct values obtained are used to calculate ΔCt values [Ct(test) – Ct(reference)]. From this methylation percentages are calculated using Methylation% = 100 × 2−ΔCt. This calculation is similar as calculating fold differences in a standard quantitative PCR assay, except that the fold differences are now expressed as percentages by multiplying the fold difference by 100. For example, a “fold difference” of 0.125 (which represents a ΔCt value of 3) using this method gives a methylation percentage of 12.5%, while a ΔCt of 0 (no difference between reference and test reaction) gives a fold difference of 1 indicating 100% methylation. Each primer set used was tested for reaction efficiency by measuring dilutions of fully methylated and unmethylated DNA samples provided with the kit (Table 3). Secondly, calibrator samples consisting of 0, 35, 65, and 100% methylated DNA were run in each experiment. From these the inter-assay and intra-assay reproducibility was investigated, shown in Tables 4 and 5, respectively. From this it can be concluded that at low percentages the errors are smaller, and the assay therefore more sensitive, than at higher levels of methylation.

Table 5. Intra-assay reproducibility showing the positive and negative errors of percentage methylation measured for the four methylated DNA input controls in two independent assays.
Data Analysis
Statistical analysis of the obtained data was carried out with the GraphPad Prism program. In here, linear regression analysis was performed for Figures 1A,B and 2B and Table 3. The p values of the linear regression lines represent the testing of the null hypothesis that the slope is zero, where the p value is calculated from an F test. Data in Tables 1, 4, and 5 and Figure 2A were obtained calculating the mean, median, SD, and SE of the mean of the measurements.
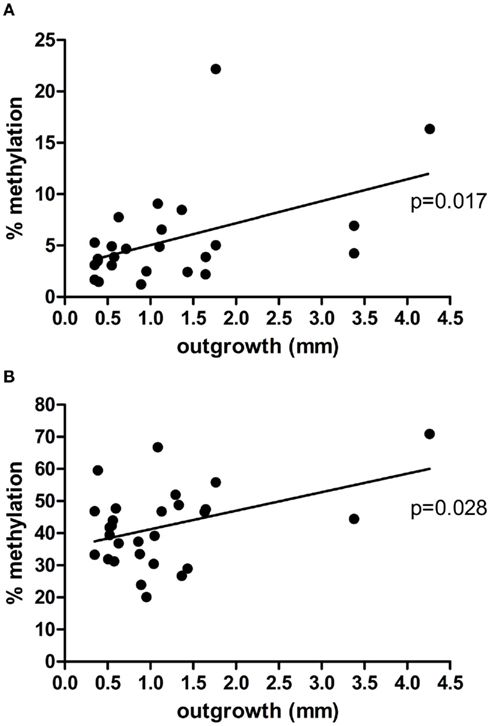
Figure 1. Correlation between placental explant outgrowth and PRKCDBP (A) and MMP2 (B) promoter methylation in first trimester placenta.
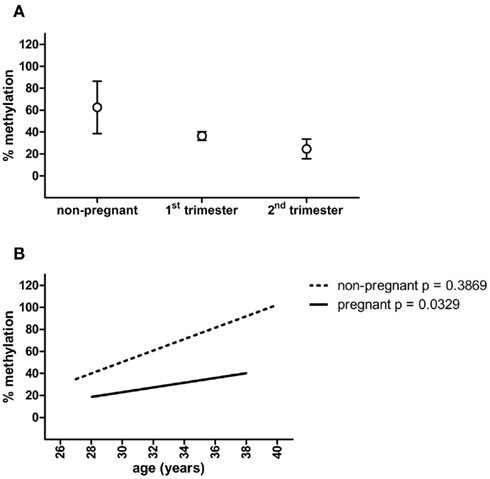
Figure 2. MMP2 promoter methylation in non-pregnant, first trimester and second trimester plasma samples (A), and linear regression of MMP2 methylation percentages plotted against the age of the women (B). Circles represent mean ± SEM.
Results
Outgrowth Correlates with DNA Methylation
DNA samples of first trimester placentas with known outgrowth potential were used to study the epigenetic status of the promoters of two matrix metalloproteinases (MMP9, MMP2) known to be involved in both placentation and cancer (Ferretti et al., 2007), and eight tumor suppressor genes (TP73, RASSF5, RASSF1, APC, DAB2IP, PRKCDBP, WT1, MORF4L1) known to show partial DNA methylation in first trimester placentas (Grigoriu et al., 2011). From the 10 genes chosen to study, PRKCDBP and MMP2 showed a significant linear regression, p = 0.017 and p = 0.028, respectively, where the increase in methylation percentage coincided with an increase in invasion potential (Figure 1). The remaining eight genes did not give significant changes in methylation level upon changes in outgrowth potential (data not shown).
Non-Invasive Biomarker Detection in Blood
To function as a non-invasive marker an (epigenetic) factor must be detectable in blood. Therefore, methylation of PRKCDBP and MMP2 was tested in pregnant plasma and compared to non-pregnant plasma samples. PRKCDBP showed low levels of methylation in both pregnant and non-pregnant samples (data not shown). The MMP2 promoter showed high methylation levels in non-pregnant plasma samples, which decreased in first trimester samples and further decreased in second trimester (Figure 2A; Table 1). Plotting the methylation percentages of the same samples versus the age of the women showed a significant linear regression in pregnant women (p = 0.0329), while the linear regression was not significant in non-pregnant women (Figure 2B). In this graph first and second trimester women were combined as the age range of the second trimester women was not evenly distributed, see also Table 1.
Discussion
The aim of this study was to investigate if the naturally occurring variation in invasion potential of human first trimester placental explants correlates to changes in epigenetic factors. Previously, we showed that genetic differences can give rise to changes in outgrowth potential, as shown for the STOX1 Y153H SNP (Van Dijk et al., 2010). In this study, we describe the use of first trimester placental explants to identify epigenetic differences associated with changes in cellular invasion. Increase in methylation of PRKCDBP and MMP2 in placental cells showed a significant linear regression with increased invasion potential of first trimester placental explants. Increased PRKCDBP methylation levels have frequently been found in several human malignancies, including colorectal, lung, breast, and gastric cancers (Xu et al., 2001; Zöchbauer-Müller et al., 2005; Lee et al., 2008, 2011). Hypermethylation of the promoter region of this gene causes downregulated expression correlating with tumor progression (Lee et al., 2011). This is in accordance with our findings showing increased methylation upon increased outgrowth potential of trophoblasts. Except for the study by Grigoriu et al. (2011) identifying differential methylation of tumor suppressor genes in first trimester placenta by microarray, no other studies have been done on PRKCDBP or MMP2 methylation in the placenta. MMP2 expression and activation levels, however, have been repeatedly found to be increased upon increased trophoblast invasion (Godbole et al., 2011; Onogi et al., 2011; Williams et al., 2011). The literature on increased levels and activation of MMP2 upon tumor progression in various cancer types are numerous, see for a selection of reviews (Levicar et al., 2003; Overall and Dean, 2006; Qian et al., 2011). Furthermore, a decreased methylation status of MMP2 in different cancers types has also been found to be associated with increased tumor invasion (Chernov et al., 2009; Morán et al., 2012). Therefore, although it would have been expected to find a decrease in methylation of the MMP2 promoter, this was not seen in this study. This inconsistency might be a good example of the main difference between trophoblasts and tumorous cells, i.e., the invasion of trophoblasts is tightly regulated in a time and place restricted manner. Methylation of MMP2 upon increased invasion potential can be one of these regulatory mechanisms.
Next, we show that one of the two factors identified can be detected in plasma of both pregnant and non-pregnant women. High MMP2 methylation levels were detected in non-pregnant plasma DNA samples, which decreased in first trimester samples which also contain placental DNA. A further decrease was seen in second trimester samples. The increased amount of unmethylated DNA in pregnant plasma compared to non-pregnant plasma is most likely placental in origin, due to the additional fractional contribution of unmethylated placenta DNA to plasma during pregnancy. The additional fractional increase of unmethylated DNA during the second trimester of pregnancy is consistent with the knowledge that the invasion capacity of the placental trophoblasts also decreases (Zhou et al., 1993). This current methylation pattern seen for MMP2 has been shown before in the SERPINB5 promoter (Chim et al., 2005). SERPINB5 is a well-known tumor suppressor gene in which the promoter furthermore showed downregulated methylation in pre-eclamptic plasma samples. To conclude, the increase in unmethylated DNA during pregnancy suggests that the level of unmethylated DNA has the potential to be used as an invasion marker, where higher levels of unmethylated DNA indicate a lower invasion potential of trophoblasts.
This proof-of-principle study therefore provides evidence that it is possible to use human placental explants as an ex vivo model to identify factors associated with changes in cellular invasion, e.g., to detect pregnancy-related diseases or cancer metastasis. This study shows that not only genetic and protein or mRNA expression differences can function as potential factors, but also epigenetic differences, increasing the identification of novel biomarker panels by genome-wide approaches.
It must be noted that within the current setting MMP2 methylation itself is not usable as a cellular invasion marker. This is due to the high variance of methylation found in non-pregnant samples compared to the pregnant plasma samples, as was shown by the linear regression plots of the methylation percentages versus the age of the women. A linear regression in methylation versus age can be expected when measuring epigenetic effects, however, this was only the case in pregnant samples. The high variance in non-pregnant women might be caused by the observation that plasma samples derived from parous non-pregnant women overall had lower methylation percentages as nulliparous women. However, the amount of parous samples in the non-pregnant group is small (n = 4) and it is not known if the nulliparous women never have been pregnant.
Another issue that needs to be addressed is the method used to measure differences in methylation percentage. The current assay used shows, although within acceptable range at low methylation levels (mean errors around 3%), at higher methylation percentages mean errors up to almost 13%. Therefore, when by genome-wide approaches epigenetic factors are being identified, the method used to measure methylation differences as potential biomarkers should be highly sensitive and accurate at all levels of methylation, which might be achieved by using pyrosequencing or methylation-specific single nucleotide primer extension (MS-SNuPE).
Although MMP2 promoter methylation in the current setting is not usable as a biomarker, it can be envisaged that it does have the potential to be used in combination with other biomarkers yet to be identified. This proof-of-principle study shows that the described method can be used to identify novel biomarkers by correlating naturally occurring variation in invasion potential to changes in (epigenetic) factors by genome-wide approaches such as massively parallel sequencing. Using genome-wide approaches on naturally occurring variation in invasion potential of first trimester placental explants increases the success of identifying primary factors involved in cellular invasion changes. Especially the first trimester placenta is highly informative as this is the period in which trophoblast invasion starts and dysfunction in this process will give complications later in pregnancy. The same holds for cancer cell invasion; when the first signs of metastasis are recognized early, the success of treatment highly increases. As non-invasive early detection markers are still lacking in both pregnancy-related disorders and cancer metastasis new approaches on the identification of novel biomarkers are still urgently needed.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all women to participate in donating blood for this study.
References
Chernov, A. V., Sounni, N. E., Remacle, A. G., and Strongin, A. Y. (2009). Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J. Biol. Chem. 284, 12727–12734.
Chim, S. S., Tong, Y. K., Chiu, R. W., Lau, T. K., Leung, T. N., Chan, L. Y., Oudejans, C. B., Ding, C., and Lo, Y. M. (2005). Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc. Natl. Acad. Sci. U.S.A. 102, 14753–14758.
Ferretti, C., Bruni, L., Dangles-Marie, V., Pecking, A. P., and Bellet, D. (2007). Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 13, 121–141.
Go, A. T., Visser, A., Mulders, M. A., Blankenstein, M. A., Van Vugt, J. M., and Oudejans, C. B. (2007). 44 Single-nucleotide polymorphisms expressed by placental RNA: assessment for use in noninvasive prenatal diagnosis of trisomy 21. Clin. Chem. 53, 2223–2224.
Godbole, G., Suman, P., Gupta, S. K., and Modi, D. (2011). Decidualized endometrial stromal cell derived factors promote trophoblast invasion. Fertil. Steril. 95, 1278–1283.
Grigoriu, A., Ferreira, J. C., Choufani, S., Baczyk, D., Kingdom, J., and Weksberg, R. (2011). Cell specific patterns of methylation in the human placenta. Epigenetics 6, 368–379.
Holtan, S. G., Creedon, D. J., Haluska, P., and Markovic, S. N. (2009). Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 84, 985–1000.
Lee, J. H., Byun, D. S., Lee, M. G., Ryu, B. K., Kang, M. J., Chae, K. S., Lee, K. Y., Kim, H. J., Park, H., and Chi, S. G. (2008). Frequent epigenetic inactivation of hSRBC in gastric cancer and its implication in attenuated p53 response to stresses. Int. J. Cancer 122, 1573–1584.
Lee, J. H., Kang, M. J., Han, H. Y., Lee, M. G., Jeong, S. I., Ryu, B. K., Ha, T. K., Her, N. G., Han, J., Park, S. J., Lee, K. Y., Kim, H. J., and Chi, S. G. (2011). Epigenetic alteration of PRKCDBP in colorectal cancers and its implication in tumor cell resistance to TNFα-induced apoptosis. Clin. Cancer Res. 17, 7551–7562.
Levicar, N., Nuttall, R. K., and Lah, T. T. (2003). Proteases in brain tumour progression. Acta Neurochir. (Wien) 145, 825–838.
Morán, A., Fernández-Marcelo, T., Carro, J., De Juan, C., Pascua, I., Head, J., Gómez, A., Hernando, F., Torres, A. J., Benito, M., and Iniesta, P. (2012). Methylation profiling in non-small cell lung cancer: clinical implications. Int. J. Oncol. 40, 739–746.
Onogi, A., Naruse, K., Sado, T., Tsunemi, T., Shigetomi, H., Noguchi, T., Yamada, Y., Akasaki, M., Oi, H., and Kobayashi, H. (2011). Hypoxia inhibits invasion of extravillous trophoblast cells through reduction of matrix metalloproteinase (MMP)-2 activation in the early first trimester of human pregnancy. Placenta 32, 665–670.
Oudejans, C. B., Go, A. T., Visser, A., Mulders, M. A., Westerman, B. A., Blankenstein, M. A., and Van Vugt, J. M. (2003). Detection of chromosome 21-encoded mRNA of placental origin in maternal plasma. Clin. Chem. 49, 1445–1449.
Overall, C. M., and Dean, R. A. (2006). Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 25, 69–75.
Qian, Q., Wang, Q., Zhan, P., Peng, L., Wei, S. Z., Shi, Y., and Song, Y. (2011). The role of matrix metalloproteinase 2 on the survival of patients with non-small cell lung cancer: a systematic review with meta-analysis. Cancer Invest. 28, 661–669.
Steegers, E. A., Von Dadelszen, P., Duvekot, J. J., and Pijnenborg, R. (2010). Pre-eclampsia. Lancet 376, 631–644.
Van Dijk, M., Mulders, J., Poutsma, A., Könst, A. A., Lachmeijer, A. M., Dekker, G. A., Blankenstein, M. A., and Oudejans, C. B. (2005). Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat. Genet. 37, 514–519.
Van Dijk, M., Van Bezu, J., Van Abel, D., Dunk, C., Blankenstein, M. A., Oudejans, C. B., and Lye, S. J. (2010). The STOX1 genotype associated with pre-eclampsia leads to a reduction of trophoblast invasion by alpha-T-catenin upregulation. Hum. Mol. Genet. 19, 2658–2667.
Williams, P. J., Bulmer, J. N., Innes, B. A., and Broughton Pipkin, F. (2011). Possible roles for folic acid in the regulation of trophoblast invasion and placental development in normal early human pregnancy. Biol. Reprod. 84, 1148–1153.
Xu, X. L., Wu, L. C., Du, F., Davis, A., Peyton, M., Tomizawa, Y., Maitra, A., Tomlinson, G., Gazdar, A. F., Weissman, B. E., Bowcock, A. M., Baer, R., and Minna, J. D. (2001). Inactivation of human SRBC, located within the 11p15.5-p15.4 tumor suppressor region, in breast and lung cancers. Cancer Res. 61, 7943–7949.
Zhou, Y., Damsky, C. H., Chiu, K., Roberts, J. M., and Fisher, S. J. (1993). Preeclampsia is associated with abnormal expression of adhesion molecules by invasive cytotrophoblasts. J. Clin. Invest. 91, 950–960.
Zöchbauer-Müller, S., Fong, K. M., Geradts, J., Xu, X., Seidl, S., End-Pfützenreuter, A., Lang, G., Heller, G., Zielinski, C. C., Gazdar, A. F., and Minna, J. D. (2005). Expression of the candidate tumor suppressor gene hSRBC is frequently lost in primary lung cancers with and without DNA methylation. Oncogene 24, 6249–6255.
Keywords: placenta, pre-eclampsia, cancer metastasis, invasion potential, biomarker
Citation: van Dijk M, Visser A, Posthuma J, Poutsma A and Oudejans CBM (2012) Naturally occurring variation in trophoblast invasion as a source of novel (epigenetic) biomarkers. Front. Gene. 3:22. doi: 10.3389/fgene.2012.00022
Received: 29 November 2011;
Accepted: 28 January 2012;
Published online: 20 February 2012.
Edited by:
Ryan Yuen, The Hospital for Sick Children, CanadaReviewed by:
Nicholas C. Wong, Murdoch Childrens Research Institute, AustraliaRyan Yuen, The Hospital for Sick Children, Canada
Copyright: © 2012 van Dijk, Visser, Posthuma, Poutsma and Oudejans. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Cees B. M. Oudejans, Department of Clinical Chemistry, VU University Medical Center, De Boelelaan 1117, 1081 HV Amsterdam, Netherlands. e-mail:Y2JtLm91ZGVqYW5zQHZ1bWMubmw=
 Marie van Dijk
Marie van Dijk