- Department of Computational Molecular Biology, Max-Planck-Institute for Molecular Genetics, Berlin, Germany
A commentary on
Definition of a consensus binding site for p53
by el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W., and Vogelstein, B. (1992). Nat. Genet. 1, 45–49.
Repeated regions of the genome harbor more functional information than commonly assumed. Two decades ago, a highly influential paper describing the consensus binding site for the key transcription factor p53 was published in Nature Genetics by el-Deiry et al. (1992). Recently, it has been observed that many p53 binding sites are species-specific (Jegga et al., 2008), suggesting a remarkable flexibility in the p53 gene regulatory network. Consistent with this idea, several recent studies have reported the existence of p53 binding sites in sequences of primate-specific interspersed repeats, including retroviral long terminal repeats (LTRs; Wang et al., 2007), short (Alus;Zemojtel et al., 2009; Cui et al., 2011), and long interspersed nuclear elements (LINEs; Harris et al., 2009), highlighting the role of transposition in the emergence of cis-regulatory elements (Feschotte, 2008; Schmidt et al., 2012).
We reanalyzed the 20 genomic sequences originally used by el-Deiry et al. (1992) to construct the p53 consensus binding motif. All of these binding sites could be uniquely mapped to the reference human genome sequence (hg19). Strikingly, as many as seven of these binding sites (∼35%) reside in one of three repeat classes: LTR, LINE, and DNA transposons (Table 1). Interestingly, this small set includes members of the primate-specific LTR10 and MER61 families, which were previously identified as enriched in copies with a functional p53 site (Wang et al., 2007). Additionally, we found that one of the early reported p53 binding sites is composed of a low-complexity repeat.
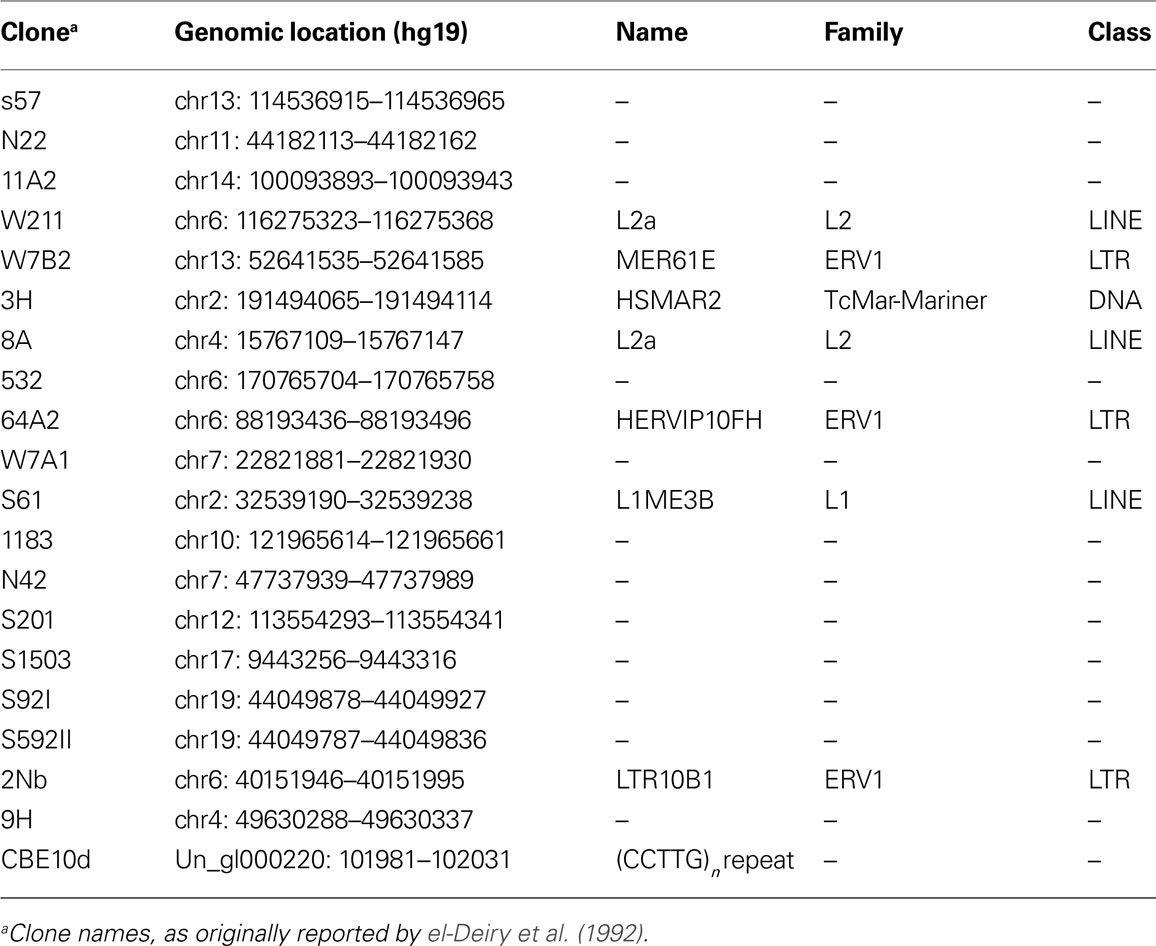
Table 1. Characterization of 20 sequences reported by el-Deiry et al. (1992).
In summary, the original work of el-Deiry et al. (1992) published 20 years ago already contained evidence for the involvement of transposable elements in spreading species-specific p53 binding sites. This raised the question: how many more gems are hidden in previously generated data sets?
References
Cui, F., Sirotin, M. V., and Zhurkin, V. B. (2011). Impact of Alu repeats on the evolution of human p53 binding sites. Biol. Direct 6, 2.
el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W., and Vogelstein, B. (1992). Definition of a consensus binding site for p53. Nat. Genet. 1, 45–49.
Feschotte, C. (2008). Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405.
Harris, C. R., Dewan, A., Zupnick, A., Normart, R., Gabriel, A., Prives, C., Levine, A. J., and Hoh, J. (2009). p53 responsive elements in human retrotransposons. Oncogene 28, 3857–3865.
Jegga, A. G., Inga, A., Menendez, D., Aronow, B. J., and Resnick, M. A. (2008). Functional evolution of the p53 regulatory network through its target response elements. Proc. Natl. Acad. Sci. U.S.A. 105, 944–949.
Schmidt, D., Schwalie, P. C., Wilson, M. D., Ballester, B., Gonçalves, A., Kutter, C., Brown, G. D., Marshall, A., Flicek, P., and Odom, D. T. (2012). Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148, 335–348.
Wang, T., Zeng, J., Lowe, C. B., Sellers, R. G., Salama, S. R., Yang, M., Burgess, S. M., Brachmann, R. K., and Haussler, D. (2007). Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U.S.A. 104, 18613–18618.
Citation: Zemojtel T and Vingron M (2012) P53 binding sites in transposons. Front. Gene. 3:40. doi: 10.3389/fgene.2012.00040
Received: 22 February 2012; Accepted: 01 March 2012;
Published online: 20 March 2012.
Copyright: © 2012 Zemojtel and Vingron. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence:emVtb2p0ZWxAbW9sZ2VuLm1wZy5kZQ==
