- Waggoner Center for Alcohol and Addiction Research, University of Texas at Austin, Austin, TX, USA
Advances in the fields of genomics and genetics in the last decade have identified a large number of genes that can potentially influence alcohol-drinking behavior in humans as well as animal models. Consequently, the task of identifying efficient molecular targets that could be used to develop effective therapeutics against the disease has become increasingly daunting. One of the reasons for this is the fact that each of the many alcohol-responsive genes only contributes a small effect to the overall mechanism and disease phenotype, as is characteristic of complex traits. Current research trends are hence shifting toward the analysis of gene networks rather than emphasizing individual genes. The discovery of microRNAs and their mechanisms of action on regulation of transcript level and protein translation have made evident the utility of these small non-coding RNA molecules that act as central coordinators of multiple cross-communicating cellular pathways. Cells exploit the fact that a single microRNA can target hundreds of mRNA transcripts and that a single mRNA transcript can be simultaneously targeted by distinct microRNAs, to ensure fine-tuned and/or redundant control over a large number of cellular functions. By the same token, we can use these properties of microRNAs to develop novel, targeted strategies to combat complex disorders. In this review, we will focus on recent discoveries of microRNA signatures in brain of human alcoholics supporting the hypothesis that changes in gene expression and regulation by microRNAs are responsible for long-term neuroadaptations occurring during development of alcoholism. We also discuss insights into the potential modulation of epigenetic regulators by a subset of microRNAs. Taken together, microRNA activity may be controlling many of the cellular mechanisms already known to be involved in the development of alcoholism, and suggests potential targets for the development of novel therapeutic interventions.
Introduction
As highlighted by the World Health Organization: “The harmful use of alcohol is one of the world’s leading health risks. It is a causal factor in more than 60 major types of diseases and injuries and results in approximately 2.5 million deaths each year” (World Health Organization, 2011). Alcoholism, also known as alcohol dependence, is understood as a complex, chronic brain disease with critical biological, behavioral, and socioeconomic components (Leshner, 1997). Heritable factors among the critical biological components are reported to account for about 50% of the risk for alcohol use disorders (AUD), which underscores the impact of genetic elements on disease etiology (Gorini et al., 2011). Nevertheless, the molecular mechanisms underlying genetic adaptations due to excessive alcohol consumption are not fully understood.
The chronic use of alcohol provokes long-term changes in gene and protein expression that allow neurons to adapt through homeostatic alterations in distinct signaling pathways. Among the principal transduction pathways altered by alcohol consumption are those involving receptor tyrosine kinases (which are commonly activated by interaction with growth factors such as EGF, GDNF, BDNF, and insulin among others), serine–threonine kinases (which are mostly intracellular such as PKA, PKC, ERKs, AKT, GSK3, and mTOR, but can also be receptors, e.g., TGFβR), and protein–protein interactions among scaffolding proteins and associated binding partners (examples of scaffolding proteins affected by ethanol are RACK1, PSD95, and Homer2; Ron and Messing, 2011). These signaling pathways are often implicated in the regulation of a variety of transcription factors that consequently affect the expression and activity of many genes (Miranda et al., 2010). Expression profiling studies in postmortem brains of human alcoholics have shown that the transcriptional reprogramming that takes place is brain area-specific and may reflect both preexisting differences in gene expression and alterations in response to alcohol consumption (Mayfield et al., 2008; Gorini et al., 2011). In addition, epigenetic reprogramming, primarily mediated by direct methylation of DNA and acetylation, methylation, and phosphorylation of histone proteins, appears to contribute to the altered gene expression observed in alcoholics and animal models of alcohol consumption (Miranda et al., 2010).
The discovery of microRNAs (miRNAs) and their mechanisms of action is revolutionizing our understanding of gene regulation (Ambros, 2001; Filipowicz et al., 2008). These short (∼17–24 nucleotides long) non-coding RNAs act as post-transcriptional modulators of gene expression by binding to miRNA-recognition elements (MREs) in their target genes. This direct targeting generally results in either suppression of translation or degradation of the targeted mRNA transcript, or both (Filipowicz et al., 2008; Breving and Esquela-Kerscher, 2010). There are instances, however, when miRNAs can increase the expression of a target gene by enhancing mRNA translation (Vasudevan et al., 2007). miRNAs are highly abundant in the brain and play important roles in multiple biological processes such as neuronal differentiation (Cheng et al., 2009), brain development (Fiore et al., 2008), synapse formation and plasticity (Schratt et al., 2006), and neurodegeneration (Schaefer et al., 2007; Bushati and Cohen, 2008). miRNAs also appear to mediate the cellular adaptations induced by exposure to a number of drugs of abuse, including nicotine (Huang and Li, 2008), cocaine (Chandrasekar and Dreyer, 2009), opioids (He et al., 2010), and alcohol (Sathyan et al., 2007; Pietrzykowski et al., 2008; Miranda et al., 2010; Lewohl et al., 2011). In subsequent sections, we discuss major biological processes now known to respond to fluctuations in levels of specific miRNAs, several of which were upregulated in the brain of human alcoholics. We also uncover general patterns of miRNA regulation that may be common to a variety of addiction disorders.
Alcohol-Induced Upregulation of Mirnas in the Human Brain
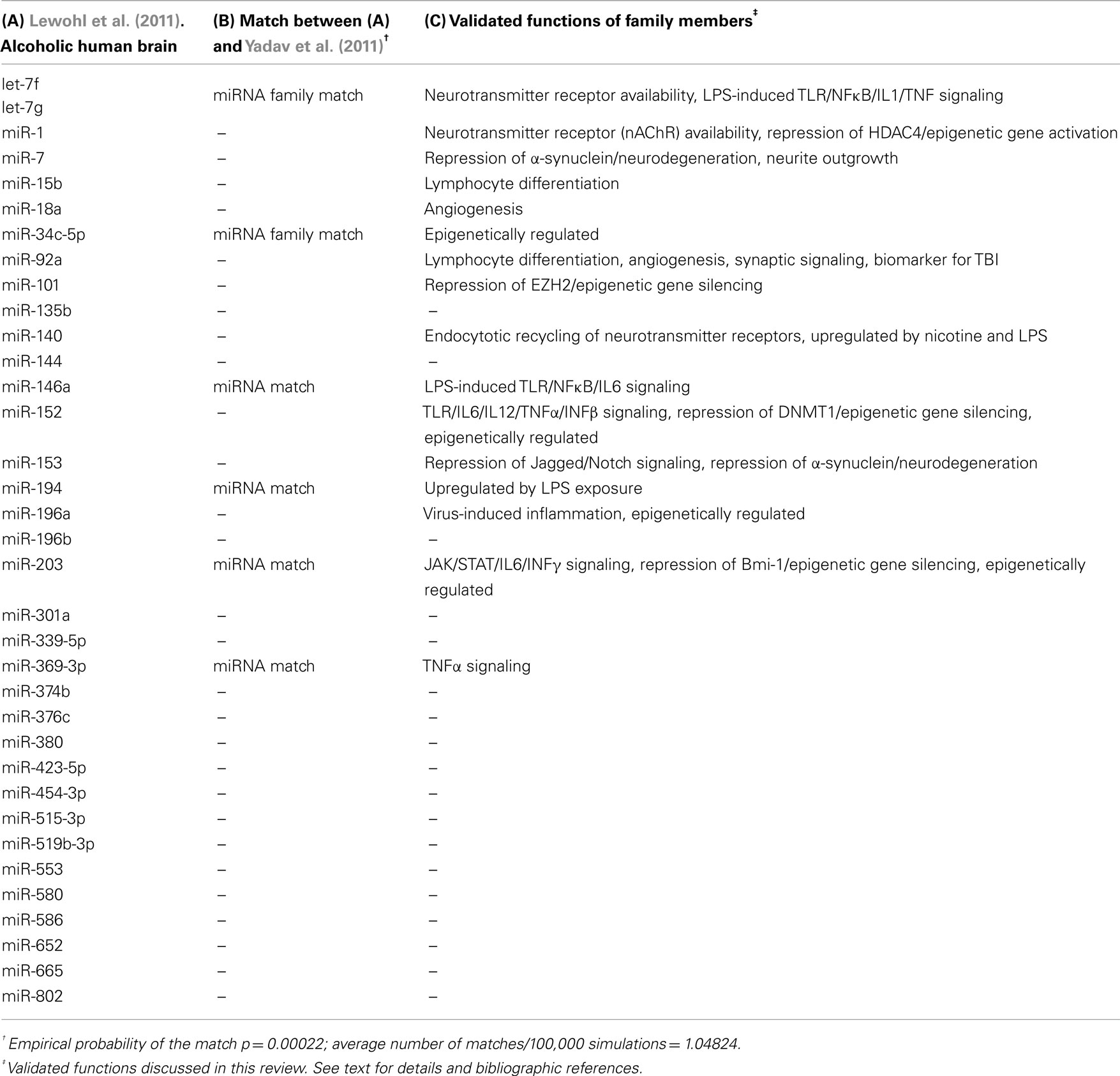
Our group recently published the first comprehensive study describing the impact of alcohol on microRNA levels in the brain of human alcoholics (Lewohl et al., 2011). For this, we conducted miRNA and mRNA microarray studies in prefrontal cortex (PFC) of postmortem human brain samples and developed an integrative statistical analysis highlighting differentially expressed miRNAs that inversely correlated with respective differentially expressed mRNA targets. The report uncovered a majoritarian effect of alcohol on activation of miRNA expression levels and highlighted about 35 human miRNAs that were upregulated in the alcoholic brain samples (Table 1). Interestingly, mRNAs that were predicted to be targeted by this group of upregulated miRNAs were significantly over-represented among the alcohol-downregulated mRNAs, while no such over-representation was detected among the set of alcohol-upregulated mRNAs. This result supports a role for miRNA-dependent inhibition of gene expression in the PFC of human alcoholics. Furthermore, we found that the alcohol-upregulated miRNAs were apparently regulating their putative target genes in a combinatorial fashion (multiple miRNAs targeting the same mRNA). Such a paradigm could be exploited by the cells to either ensure inhibition of the putative targets at any cost (redundancy) or to achieve fine-tuned regulation of specific mRNA expression levels (fine-tuning).
A recent publication by Yadav et al. (2011) described alcohol-induced changes in miRNA expression levels in human-derived neuroblastoma cells. Although the limited model used by these authors is far from reproducing the physiological environment of the alcohol-exposed human brain, their results are informative and corroborate the role of several of the upregulated miRNAs described by our group (Table 1, column A). Interestingly, Yadav and colleagues also found mostly upregulated miRNAs after alcohol exposure. In fact, after we subset their reported differentially expressed miRNAs for those with p ≤ 0.01 (based on our more stringent statistical analyses of only using the lowest p values and reporting adjusted p values to account for multiple testing), only 20 upregulated miRNAs remain statistically significant. Out of these 20 upregulated miRNAs from ethanol-treated cells, six matched upregulated family members in the alcoholic human brain from our studies (miR-369-3p, miR-34c-5p, miR-203, miR-146a, miR-194, and let-7 family members, Table 1, column B). This is statistically highly significant (p = 0.00022), as it is expected that only one miRNA would be common between the two lists by chance (as empirically determined after 100,000 Monte Carlo simulations). This underscores the importance of miRNA upregulation due to alcohol exposure and suggests a consistent involvement of multiple miRNAs in mediating neuroadaptations that could potentially contribute to alcohol dependence.
Cell death-related genes were among the functionally enriched transcripts being targeted by miRNAs in our studies in PFC of human alcoholics (Liu et al., 2006b; Lewohl et al., 2011). A study from the Bakalkin group, also using postmortem PFC of human alcoholics, provides additional support for our findings. These authors found that dysregulation of the cell death machinery by inhibition of the intrinsic apoptotic pathway “may reflect molecular adaptations that counteract alcohol neurotoxicity in cells that survive after many years of alcohol exposure and withdrawal” (Johansson et al., 2008). Although the Yadav group found opposite miRNA-mediated effects inducing neuronal death in cell cultures treated with ethanol for relatively short periods of time (3–72 h), their results do not contradict the neuroadaptive phenomenon we and others observe in chronic alcoholics, but seemingly describe initial effects of acute alcohol exposure as previously documented in animal models (Heaton et al., 1999; Rajgopal et al., 2003; Nowoslawski et al., 2005). Collectively, these findings support the notion that miRNA-mediated neuroadaptations may develop after an initial, also miRNA-mediated, activation of cell death pathways that occurs during the early stages of alcohol abuse.
Our study using postmortem human brains also found that the magnitude of alcohol-related changes in miRNA levels was constrained over a narrow range of only 20–30%, with only a few miRNAs varying outside of this range (Lewohl et al., 2011). We reason that the small changes detected in miRNA, as well as mRNA expression, in PFC of human alcoholics could be due to an increased expression that is localized to a specific cellular compartment, e.g., the neuronal synapse. Such compartmentalized, enhanced differential expression would become diluted as RNA is extracted from total, unfractionated tissue. Alternatively, a larger differential expression may not be compartment-specific but cell type-specific (e.g., differential expression of immune signaling genes in glial cells or differential expression of neuronal genes in specific neuronal subtypes) and similarly become diluted when RNA is extracted from total tissue containing combined cell subpopulations. Both of these possibilities will be discussed further in the following sections.
Human Alcohol-Responsive Mirnas and Neuroinflammation
Inflammation plays critical roles in the pathogenesis of multiple disorders of the central nervous system (CNS; Weiner and Selkoe, 2002; Hunot and Hirsch, 2003) and has recently been implicated as a mechanism of alcoholism-induced neuropathology (Crews et al., 2011; Kelley and Dantzer, 2011; Yakovleva et al., 2011). An important player eliciting activation of immune functions in the CNS in response to alcohol is Toll-like receptor (TLR) 4, which contributes to ethanol-induced activation of nuclear factor kappa B (NFκB) and consequently activates transcription of pro-inflammatory chemokines, cytokines, oxidases, and proteases. TLRs have well-established roles in pathogen detection and initiation of an innate immune response that consequently specifies adaptive responses during infection (O’Neill, 2006).
The role of innate, immune-related genes in alcohol dependency is supported by genetic association studies in humans (Pastor et al., 2000, 2005; Edenberg et al., 2008; Saiz et al., 2009), gene expression microarray studies in postmortem brains of alcoholics (Liu et al., 2006b; Ökvist et al., 2007), and transcriptome meta-analysis in rat brains (Mulligan et al., 2006). Furthermore, recent behavioral studies in mutant mice indicated that deletion of genes involved in aspects of the neuroimmune response to alcohol (B2m, Ctss, Il1rn, Cd14, and Il6) could reduce ethanol consumption in mice (Blednov et al., 2012). Based on these and additional related findings, a hypothesis is developing in part of the alcohol research community that the neurobiology of addiction is due to altered brain signaling driven by immune signaling.
Implications of miRNAs as players in the development of the immune system and the regulation of multiple immune functions are also coming to age (Sonkoly et al., 2008; Belver et al., 2011; O’Neill et al., 2011; Turner et al., 2011). Multiple profiling studies have confirmed the hypothesis that TLR signaling can modulate miRNA expression and this generally depends on NFκB. In all cases described, TLR activation exclusively induces upregulation of miRNAs (O’Neill et al., 2011). Interestingly, this is the same effect (miRNA upregulation) we observe in our own miRNA profiling studies in brain of human alcoholics. In this section, we address a series of recent findings that uncover a potential connection between a group of upregulated miRNAs in postmortem brains of human alcoholics and neuroinflammatory processes triggered by alcohol. We summarize this evidence in a hypothetical model presented in Figure 1.
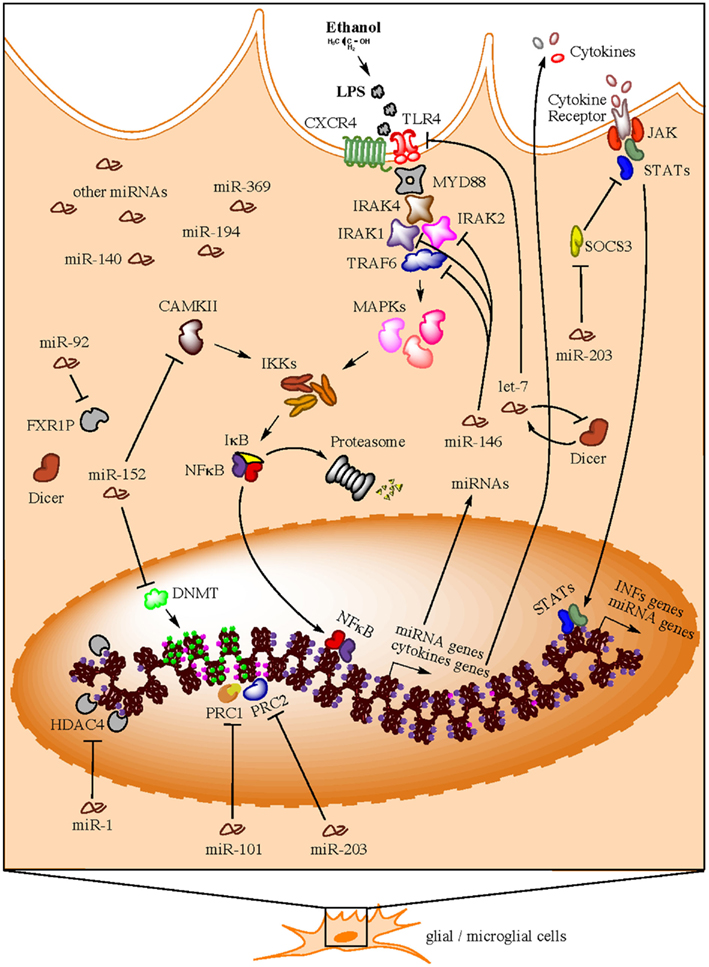
Figure 1. Hypothetical model for neuroimmune-related actions of miRNAs in brain of human alcoholics (e.g., in microglia). Bacterial lipopolysaccharides (LPS) from commensal bacteria, for example, could leak into the bloodstream due to gut leakiness induced by alcohol consumption. A compromised blood brain barrier due to high ethanol concentration in the blood will consequently allow LPS to reach the brain and trigger TLR signaling cascades that activate nuclear factor kappa B (NFκB) and induce the transcription of a variety of pro-inflammatory genes (e.g., cytokines), as well as miRNA genes. Newly synthesized pro-inflammatory factors are secreted to unleash a systemic neuroinflammatory response and to maintain a positive feedback loop in same activated cell. These second round of pro-inflammatory factor production is achieved through the expression of alternative cytokine receptors in the activated cell, which signal back to the nucleus through the JAK/STAT pathway to induce production of additional pro-inflammatory factors (e.g., interferons) and miRNA genes. To avoid over-amplification of these signals and excessive inflammation due to hyper-responsiveness to LPS insult, specific miRNAs (e.g., members of the miR-146, miR-152, and let-7 families) are consequently upregulated in order to suppress TLR4/CXCR4 signaling through inhibition of multiple cascade transducers such as IL1 receptor-associated kinases (IRAKs), TNF receptor-associated factor 6 (TRAF6), and TLR4/CXCR4 themselves, among others. As a compensatory reaction, other miRNAs (e.g., miR-203) that activate the alternative JAK/STAT pathway are also upregulated in an effort to maintain the immune-activated state of the specific cell subtype while promoting a benign, contained inflammatory response. Concomitantly, immune-activated cells implement miRNA-mediated epigenetics mechanisms, such as DNA methylation and histone methylation and/or acetylation) that ensure chromatin modification and global changes in gene expression that allow for long-term homeostatic changes and cellular adaptations under the particular environmental conditions. Expectedly, miRNAs that target epigenetic factors are also activated in order to control and/or fine-tune the ongoing remodeling of the cellular epigenome.
About 30% of the 35 miRNAs upregulated in the PFC of human alcoholics (miR-15, miR-34, miR-92, miR-140, miR-146, miR-152, miR-194, miR-196, miR-203, miR-369, let-7) are modulators of immunity and several of them appear to be involved in regulating TLR signaling. Particularly interesting is the miR-146 family, which is upregulated by bacterial endotoxin lipopolysaccharides (LPS) and controls TLR/NFκB signaling through negative feedback loops involving downregulation of IL1 receptor-associated kinase 1 (IRAK1) and TNF receptor-associated factor 6 (TRAF6) protein levels (Taganov et al., 2006; Zhao et al., 2011). TRAF2 has also being implicated as a miR-146 target (Hou et al., 2009). It was suggested that miR-146 regulatory circuit likely fine-tunes TLR and cytokine signaling, rather than totally abrogating the signal, and that expression of miR-146 may be critical in preventing excess inflammation (Sonkoly et al., 2008). Collectively, IRAK1, IRAK2, and TRAF6 represent important components of the myeloid differentiation primary-response protein 88 (MYD88)-dependent pathway for NFκB activation downstream of TLRs (O’Neill et al., 2011). Correspondingly, several reports have implicated IRAK and TRAF6 transducers in alcohol actions in the brain (Vallés et al., 2004; Guasch et al., 2007; Oliva et al., 2011).
let-7 family members, on the other hand, have been reported to directly target expression of TLR4 and regulate responsiveness to LPS (Chen et al., 2007; Androulidaki et al., 2009) and to form an epigenetic switch together with NFκB and interleukin 6 (IL6) that links inflammation to persistent cell transformation (Iliopoulos et al., 2009). The reported upregulation of let-7 miRNAs in human alcoholics could be understood as necessary to turn off expression of TLRs in glial cells and avoid excessive inflammation due to hyper-responsiveness to LPS insult. This potential regulatory loop could contribute to tolerance and dependency in chronic alcoholics. Furthermore, Blednov and colleagues recently demonstrated that LPS-triggered activation of immune signaling mediated by CD14 [a key component of the LPS-sensing co-clustering complex that also includes Hsp70, Hsp90, CXCR4, GDF5, and TLR4 (Triantafilou and Triantafilou, 2002)] promotes alcohol consumption and alters certain aspects of alcohol reward and aversion in mice (Blednov et al., 2011). Our finding that chemokine receptor CXCR4 is downregulated in brain of human alcoholics and is the most significantly miRNA-over-targeted transcript (Lewohl et al., 2011) supports a role for miRNAs in the regulation of LPS-activated TLR4-transduced immune signaling in chronic alcoholics. CXCR4 has also being recently implicated in opiate-induced hypernociception (White and Wilson, 2010) and polymorphisms in SDF1 (the CXCR4 ligand) associated with several phenotypes, including alcohol consumption (Xiao et al., 2008).
MicroRNA miR-203 is the upregulated miRNA in human alcoholics with the highest over-representation of targets among inversely correlated differentially expressed mRNA transcripts from the same samples (Lewohl et al., 2011). This miRNA has been suggested to regulate IL6 and IFNγ signaling through targeting of the Suppressor of Cytokine Signaling 3 (SOCS3; Sonkoly et al., 2007). SOCS3 is part of a negative feedback loop in cytokine signaling that inhibits the activation of transcription factor STAT3, part of the JAK–STAT signaling cascade that is the basis of the signal transduction mechanism for many cytokine receptors. Upregulation of miR-203 may hence lead to increased and/or longer inflammatory response. Furthermore, miR-203 is predicted to target above mentioned CXCR4 (component of the LPS-sensing complex), which is the alcohol-downregulated mRNA most significantly over-targeted by miRNAs in human alcoholics (Lewohl et al., 2011). These appear to represent compensatory effects that, on one hand, aim at downregulating responsiveness to the LPS insult while, on the other, aim at maintaining required levels of neuroimmune activity. These results support the notion that CXCR4 and miRNAs regulating its expression are important players modulating alcohol actions in the brain of human alcoholics.
The family miR-148/152 was recently reported to inhibit TLR-triggered MHCII expression and functional maturation of dendritic cells (DCs). By targeting CaMKIIα, miR-152 inhibited the production of cytokines, including IL12, IL6, TNFα, and IFNβ, and negatively regulated the innate response (Liu et al., 2010). Coincidently, miR-152 was also upregulated in fetal brain from mice exposed to ethanol during gestation (Wang et al., 2009). Interestingly, Kash et al. (2009) recently found a significant decrease in the total levels of CaMKIIα in the central extended amygdala after ethanol exposure in mice. CaMKIIα expression enhances the extent of desensitization of NR2B-containing NMDA receptors in heterologous cells (Sessoms-Sikes et al., 2005) and regulates ethanol sensitivity of BK channels (Liu et al., 2006a), which suggested the possibility that reduced levels of CaMKIIα could be acting in a general fashion to modulate the ethanol sensitivity of channels in the extended amygdala (Kash et al., 2009). Coincidently, another miRNA, miR-9, also implicated in controlling the activity of the TLR signaling pathway through direct targeting of the NFKB1 gene in human monocytes and neutrophils (Bazzoni et al., 2009; O’Neill et al., 2011), was demonstrated to regulate ethanol sensitivity of BK channels by selectively destabilizing splice variants that contained miR-9 recognition elements, therefore making the BK channels less sensitive to alcohol (Pietrzykowski et al., 2008). We did not detect changes in miR-9 expression in our postmortem human brain study, but such differences among human alcoholics and animal models are plausible due to neurodevelopmental differences between species, stage of alcohol insult, and experimental protocol differences, to name a few. As suggested by O’Neill et al. (2011) miRNAs might also play a role in “controlling the switch from a strong early pro-inflammatory response to the resolution phase of the inflammatory process.” This would also support a case for miRNA and mRNA expression profile discrepancies depending on the stage of the inflammatory response in the particular samples under study.
Evidence is also accumulating for novel mechanisms by which miRNAs can directly regulate mRNAs by facilitating or preventing interaction with RNA-binding proteins. An example of this that also underscores the fact that mRNA stability can be affected by environmental factors, is miR-369-3p (one of the upregulated miRNAs in human alcoholics), which associates directly with the AU-rich elements (AREs) in TNFα mRNA by base pairing and mediates translational activation of TNFα only under growth arrest conditions (Vasudevan and Steitz, 2007; Vasudevan et al., 2007). This effect was dependent on the recruitment of the RNA-binding proteins fragile-X mental retardation-related protein 1 (FXR1) and argonaute 2 (AGo2) and could be reversed when cells were actively proliferating, in which case miR-369-3p would switch roles to represses TNFα. Discovery of such sensitive and complex switch-like regulatory mechanisms emphasizes the extreme fine-tuning capabilities encoded into the biology of miRNAs.
The specific functions of other miRNAs upregulated in human alcoholics, which have also been implicated in immune signaling, remain unknown. Among these are miR-140 and miR-194, both upregulated in in vivo studies in mouse lung after LPS exposure (Moschos et al., 2007); miR-92, which is upregulated in CD4+CD8+ double positive thymocytes in comparison to other stages of T lymphocyte development (Sonkoly et al., 2008); miR-15b, which is upregulated in CD8+ cells when compared with CD4+ T cells or double positive thymocytes (Sonkoly et al., 2008); and miR-196, which have sequence-predicted targets within the hepatitis C virus genomic RNA and is upregulated by antiviral cytokine INFβ (Sonkoly et al., 2008). This highlights the fact that we are just starting to understand the regulatory roles of miRNAs in general. Often, a single miRNA is found to be involved in multiple cellular functions. For example, (1) miR-92a and cluster member miR-18a block angiogenesis when overexpressed in endothelial cells (Bonauer et al., 2009; Doebele et al., 2010), (2) increased miR-92a in plasma levels in patients with traumatic brain injury (TBI) is a good biomarker for the severity of the disease (Redell et al., 2010), and (3) family member miR-92b is involved in synaptic signaling (Ceman and Saugstad, 2011). These diverse functions provide evidence for versatile and complex systemic roles of miRNAs.
Human Alcohol-Responsive Mirnas, Neurotransmitter Signaling, and Synaptic Plasticity
Alcohol, as well as other drugs of abuse, produce long-term changes within the brain reward circuits and these changes are thought to lead to drug tolerance, reward dysfunction, escalation of drug intake, and eventually compulsive use (Russo et al., 2009). Research on the reward circuit has been centered on dopaminergic neurons in the ventral tegmental area (VTA) of the midbrain and their projections to the limbic system, in particular the nucleus accumbens (NAc), dorsal striatum, amygdala, hippocampus, and regions of PFC (Robison and Nestler, 2011). Nevertheless, other neurotransmitter systems seem also to contribute to brain reward responses since animals can still exhibit positive hedonic responses in the absence of dopamine (Hyman et al., 2006). Ethanol as well as opioids, cannabinoids, and nicotine are thought to produce reward partially through non-dopaminergic mechanisms, e.g., μ opioid receptors expressed on NAc neurons, which appear to bypass dopamine inputs from the VTA (Hyman et al., 2006). Studies examining the effect of selective agonist and antagonist drugs have indicated that multiple neurotransmitters, including dopamine, serotonin, acetylcholine, glutamate, GABA, and various peptides, are involved in activation of reward circuits and the production of multiple forms of neuronal plasticity that can convert drug-induced signals into long-term alterations in behavior (Bardo, 1998; Hyman et al., 2006). At the molecular level, complex gene expression mechanisms coordinate long-lasting alterations in dendrite and synapse structure and function. Among these mechanisms, the local control of mRNA translation in neuronal dendrites can account for the tight spatial regulation of plasticity at the level of individual dendrites or spines (Sutton and Schuman, 2006; Schratt, 2009). Recent evidence discussed below indicates that miRNAs play extensive roles in the regulation of these local processes and the development of addictive animal behaviors. We summarize this evidence in a hypothetical model presented in Figure 2.
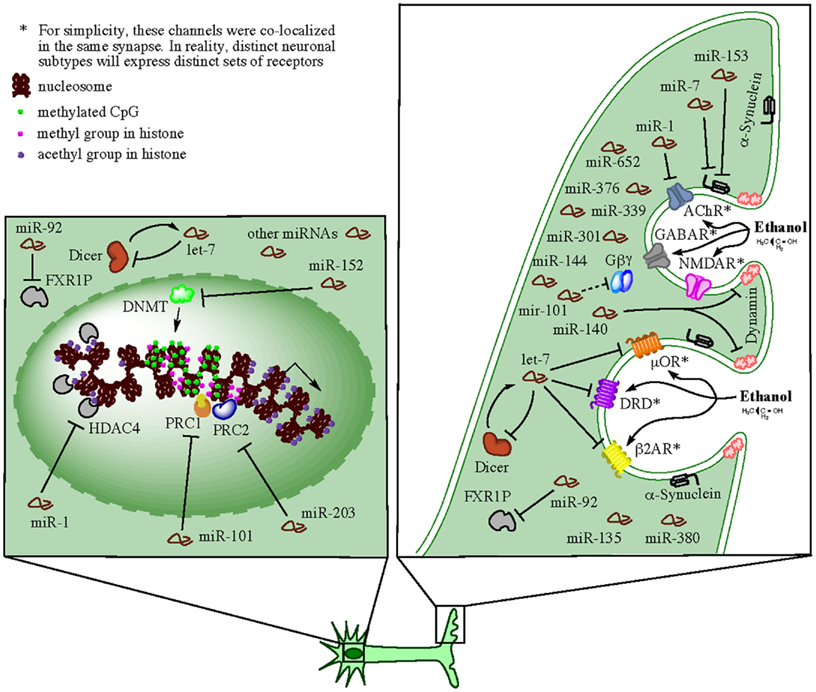
Figure 2. Hypothetical model for synaptic-related actions of miRNAs in brain of human alcoholics (e.g., in neurons). Ethanol can directly or indirectly affect multiple neurotransmitter receptors at the neuronal synapse and activate reward circuits conducive to multiple forms of neuronal plasticity, which in turn, convert the drug-induced signals into long-term alterations in behavior (e.g., alcohol dependency). The receptors affected on a particular synapse depend on the neuronal subtype and the specific subset of receptors expressed. For simplicity, several receptors such as GABAR, NMDAR, AChR, μOR, DRD, and β2AR, were diagramed as co-localized on the same synapse in the cartoon, but this is probably not the case in reality. miRNAs transported to and enriched in the synapses are locally processed by resident miRNA-ribonucleoprotein (miRNP) complexes (containing Dicer and FXR1P among other factors) and exert predominately inhibitory effects on mRNA targets also present at the specific synapses. Members of miRNA families, such as let-7, miR-1, miR-101, miR-140, and several others, downregulate the activity of neurotransmitter receptors by directly targeting respective mRNAs or by interfering with synaptic endocytosis (e.g., through targeting of dynamin and α-synuclein mRNAs). In addition, synaptic miRNAs implement negative feedback loops (through targeting of dicer and FXR1 mRNA, among other miRNA biogenesis-related factors) that auto-regulate their own availability. Such feedback loops ensure a balanced homeostatic control of a variety of synaptic functions. Concomitantly, cells implement miRNA-mediated epigenetics mechanisms, such as DNA methylation and histone methylation and/or acetylation, that ensure chromatin modification and global changes in gene expression that allow for long-term homeostatic changes and cellular adaptations under the particular environmental conditions. Expectedly, miRNAs that target epigenetic factors are also activated in order to control and/or fine-tune the ongoing remodeling of the cellular epigenome.
Members of multiple miRNA families upregulated in the brain of human alcoholics have been collectively found enriched (let-7, miR-1, miR-7, miR-92, miR-135, miR-146, miR-339, miR-376, and miR-380) or depleted (miR-34, miR-101, miR-144, miR-153, miR-301, and miR-652) in rodent synapses (Lugli et al., 2008; Siegel et al., 2009; Eipper-Mains et al., 2011). miR-92 in particular, had been implicated in the regulation of FXR1P, which is an RNA-binding protein found in dendrites and involved in miRNA biogenesis (Ceman and Saugstad, 2011). Lugli et al. (2008) suggested that the miRNAs expressed within dendrites and within dendritic spines were expected to contribute to the regulation of local protein synthesis. Since the main effect of miRNA upregulation in human alcoholic brain is suspected to be downregulation of targeted-gene expression and/or inhibition of protein translation (Lewohl et al., 2011) and blockade of de novo protein synthesis in dendritic spines attenuates formation of long-term memory due to impairment of dendritic growth and remodeling (Schratt, 2009; Eipper-Mains et al., 2011), it might suggest that the observed alcohol upregulation of these synapse-localized miRNAs could have a major impact in the development of new addiction-promoting, long-term memories. Hence, early or accelerated upregulation of alcohol-responsive miRNAs in the brain might preempt development of addiction and could represent a potential therapeutic strategy.
miR-7 and miR-153 have both been recently implicated in the regulation of α-synuclein (Doxakis, 2010). α-synuclein plays a role in activity-dependent maintenance of SNARE complex levels in pre-synaptic terminals (Burre et al., 2010; Greten-Harrison and Polydoro, 2010; Burgoyne and Morgan, 2011) and is involved in dopaminergic neurotransmission and neurodegenerative disorders (Doxakis, 2010). Importantly, there is a large body of evidence linking α-synuclein accumulation to alcohol dependency in humans as well as in rodents (Bönsch et al., 2005; Liang and Carr, 2006; Foroud et al., 2007; Taraskina et al., 2008; Aho et al., 2009). Furthermore, miR-7 has also been implicated in the control of neurite outgrowth in vitro, during neuronal differentiation of human neuroblastoma cell lines (Chen et al., 2010). On the other hand, miR-153 has been implicated in the teratogenic effects of alcohol in mice, although in this case, due to downregulation of the combinatorial function of three miRNAs (miR-21, miR-335, and miR-153). Simultaneous downregulation of these three miRNAs allowed their shared target, Jagged1 (a Notch receptor ligand), to accumulate and induce cell cycle and neuroepithelial maturation (Sathyan et al., 2007).
The Smalheiser group has found that dicer and the Ago homolog eIF2c, both involved in miRNA biogenesis, were expressed in mouse brain synaptic fractions and enriched at post-synaptic densities (PSDs; Lugli et al., 2005; Smalheiser and Lugli, 2009). The findings that NMDA and calcium stimulation of synaptoneurosomes induced calpain-dependent activation of dicer led the authors to propose a model in which (i) glutamate or other neurotransmitter activity at synapses causes a local increase of intracellular calcium levels within the post-synaptic neuron; (ii) this calcium increase needs to be sufficient to activate calpain; (iii) calpain liberates active dicer and eIF2c from the PSDs. Furthermore, the authors postulated that the acute effects of calpain on dicer act as a highly localized, phasic, high-threshold trigger that leads to formation of small RNAs near synapses, which once formed should be relatively long-lasting and subsequently independent of ongoing dicer activity (Lugli et al., 2005). Recent studies in Drosophila and rodents have confirmed the importance of components of the RISC complex at the synapse, which was necessary for the establishment of certain forms of short and long-term memories in these models (Ashraf et al., 2006; Batassa et al., 2010). Remarkably, we found that dicer was downregulated in the PFC of human alcoholics and that, equal to CXCR4, it was the mRNA most significantly over-targeted by upregulated miRNAs (Lewohl et al., 2011), which suggest a relevant role for dicer-miRNA feedback loops in the mediation of and/or neuroadaptation to alcohol actions.
Several of the synaptically modulated miRNA families have been involved in the regulation of neurotransmitter signaling, which is mechanistically related to stimulation of reward circuits and the formation of associative, synapse-specific memories (Hyman et al., 2006). For example, the let-7 family, one of the miRNA families differentially expressed in PFC of human alcoholics, could be modulating the activity of a variety of neurotransmitter receptors. Several members of this family have been reported to regulate receptors such as the μ opioid receptor, the β2-adrenergic receptor, and the dopamine D3 receptor (Pillai, 2005; Chandrasekar and Dreyer, 2009; He et al., 2010). While investigating mechanisms contributing to opioid tolerance in mice, He et al. (2010) identified let-7 family members (let-7a, let-7c, and let-7g were used as representative members) as mediators of translocation and sequestration of μ opioid receptor mRNA into P-bodies, which led to translation repression without affecting the mRNA levels. A similar mechanism for repression of translational initiation of let-7 targets in human cells had previously been reported (Pillai, 2005). The μ opioid receptor is largely distributed along reward circuits, where it mediates the reinforcing activities of morphine and several non-opioid drugs such as alcohol, cannabinoids, and nicotine. The non-opioid drugs act at their own receptors [GABAA and NMDA receptors for alcohol, CB1 and CB2 receptors for cannabinoids, and nicotinic acetylcholine receptor (nAChR) for nicotine] and are likely to induce the release of endogenous opioid peptides that, in turn, activate the μ receptors. This type of receptor seems to function as a convergent molecular gate in the initiation of addictive behaviors (Contet et al., 2004). In addition, Wang et al. (2011) demonstrated that let-7f targeted β2-adrenergic receptor (β2AR) mRNA and repressed expression of the native ADRB2 protein in the human cell line H292. Interestingly, another microRNA that is also upregulated in human alcoholics, miR-15, belongs to one of the only two other groups of miRNAs that have predicted binding sites within the 3′ UTR of the β2AR mRNA (Wang et al., 2011). Recent evidence supports a role for β-adrenergic receptors in retrieval of cocaine-associated memories, mediated by norepinephrine acting at central β-adrenergic receptors in rats (Otis and Mueller, 2011). With this work, Otis and Mueller also demonstrated that propranolol, a commonly prescribed β-blocker, can be used as an adjunct to exposure therapy for the treatment of cocaine addiction. In addition, activation of β2AR also appears to be required for some of the key neurochemical changes that characterize chronic opioid administration in mice (Liang et al., 2007). Yet another let-7 family member, let-7d, was reported to be downregulated by cocaine in rat mesolimbic brain slices, and overexpression of this miRNA in hybrid NG108-15 cells downregulated the dopamine D3 receptor (Chandrasekar and Dreyer, 2009). It is well known that drugs of abuse induce persistent structural and functional changes in the mesolimbic dopaminergic system that lead to high-risk, drug-seeking behaviors, and relapse (Chandrasekar and Dreyer, 2011).
On the other hand, support for an involvement of miR-101 (another miRNA upregulated in the PFC of human alcoholics) in the modulation of GABAergic transmission in response to alcohol consumption can be found in a recent study conducted by the Tabakoff group (Saba et al., 2011). After analyzing gene expression profiles induced by alcohol-drinking in panels of recombinant and isogenic inbred mice, Saba and colleagues found that guanine nucleotide binding protein beta 1 subunit (Gnb1) was the only transcript that changed expression in all groups of high and low alcohol-drinking mice and, in addition, localized within a quantitative trait loci for alcohol consumption. Gnb1 expression level was inversely related to the level of alcohol intake. Based on differential detection of Gnb1 transcript variants, protein levels, and 3′ UTR sequence analysis, the authors suggested that miR-101 could be regulating the amount of Gβ1 protein. Gβ1 protein can initiate a cascade of intracellular signaling events leading to the internalization of GABAA and other receptors (Saba et al., 2011). Although subsequent studies investigating the direct interaction between miR-101 and the Gnb1 transcripts are necessary for validation, we are enthusiastic about the emergence of seemingly interconnected patterns spanning multiple animal species.
Additional evidence for a role of miRNAs in the regulation of endocytotic recycling of neurotransmitter receptors is found in the work of the Li group, who found that miR-140* is upregulated by nicotine treatment and directly binds to dynamin (Dnm1) mRNA to inhibit its expression (Huang and Li, 2008). Dynamin-1 is involved in scission of clathrin-coated vesicles and it is essential for endocytotic synaptic vesicle recycling only during the application of a strong or sustained stimulus when exocytosis of neurotransmitter-filled vesicles is extreme and rapid retrieval of vesicles is required to maintain the synaptic vesicle pool (Ferguson et al., 2007; Etheridge et al., 2009). Proteomic studies of synaptosomal fractions from superior frontal gyrus and occipital cortex of postmortem human brains identified dynamin-1 protein as differentially regulated between alcoholics and controls (decreased expression in alcoholics) and provided evidence for differential alteration of multiple protein isoforms in a brain region-specific manner (Etheridge et al., 2009). Furthermore, our group has demonstrated that dynamin-1 establishes a strong physical interaction with the large conductance voltage- and calcium-activated potassium channel (BKCa), another major player in alcohol actions (Gorini et al., 2010). Interestingly, multiple miRNAs upregulated in human alcoholics, including miR-101, miR-135b, miR-140, miR-203, miR-34c-5p, miR-376, miR-454, miR-519b-3p, miR-580, and miR-586 (Lewohl et al., 2011), are predicted to target dynamin family members, which underscore the role that these proteins and respective targeting-miRNAs play in mediating alcohol actions.
Acetylcholine (ACh) could be another neurotransmitter signaling pathway regulated by miRNAs in human alcoholic brains. miR-1 is a conserved muscle-specific microRNA that regulates aspects of both pre- and post-synaptic function at neuromuscular junctions (Simon et al., 2008). Using C. elegans as model system, Simon et al. (2008) demonstrated that miR-1 regulates the expression level of two nAChR subunits, thereby altering muscle sensitivity to ACh. Alcohol has been reported to enhance the function of naturally expressed α4β2 nAChRs and to inhibit the activity of naturally expressed α7 nAChRs (Narahashi et al., 1999). Furthermore, activation of nAChRs was reported to selectively reduce alcohol consumption in outbred Wistar rats (Dyr et al., 1999), while mutations in transmembrane domains of α2 channel subunits can enhance actions of alcohols on neuronal nicotinic receptors (Borghese et al., 2002).
Taken together, our analysis of potential effects of alcohol-responsive miRNAs on neurotransmitter receptor-mediated signaling indicates that a variety of neurotransmitter-regulated pathways may be simultaneously changing in response to alcohol as well as other drugs of abuse, and that the activity of multiple key miRNA families may be contributing to this particular complex network of interactions. Direct effect of upregulated miRNAs (as in the cohort of human alcoholic studied by Lewohl et al., 2011) over their putative neurotransmitter receptor targets is suggestive of a neuroadaptive response to counteract the drug-induced receptor activation characteristic in multiple types of addiction. On the other hand, since receptor endocytosis contributes to signal termination and desensitization of activated G protein-coupled receptors (Finn and Whistler, 2001; Li and van der Vaart, 2011), miRNA-driven downregulation of dynamin-1, which disrupts neurotransmitter receptor endocytosis, might contribute to the addictive effects of alcohol as well as other drugs of abuse.
Human Alcohol-Responsive Mirnas and Epigenetics
The fact that epigenetic mechanisms underlie adaptation in adult organisms has become clear thanks to an explosion in research into mechanisms by which chromatin and histone modifications impact transcriptional regulation under a variety of environmental insults. These mechanisms have been demonstrated to play a role in drug addiction, as repeated exposure to drugs of abuse induce alterations in histone acetylation, phosphorylation, and methylation levels, as well as DNA methylation levels (Maze and Nestler, 2011; Robison and Nestler, 2011; Wong et al., 2011). In alcoholic patients, a significant increase in global DNA methylation influenced by reduced levels of DNA methyl transferase 3b (DNMT3b) has been reported and a possible subsequent derangement of epigenetic control suggested (Bönsch et al., 2004, 2006). Furthermore, several genomic loci such as nerve growth factor (NGF) and pro-opiomelanocortin gene (POMC) from blood cells and prodynorphin (PDYN) from brain tissue have been found hyper-methylated in human alcoholics (Bönsch et al., 2006; Muschler et al., 2010; Heberlein et al., 2011; Taqi et al., 2011). In this section, we focus on recent studies demonstrating that miRNAs can regulate epigenetic factors and vice versa (Sato et al., 2011), highlighting, in particular, epigenetic-related miRNAs upregulated in the brain of human alcoholics. We summarize this evidence in the hypothetical models presented in Figures 1 and 2 (see nucleus).
miR-101 was reported to specifically repress expression of EZH2, a conserved catalytic subunit within the polycomb repressor complex 2 (PRC2), which is involved in histone methylation and consequent transcriptional silencing (Varambally et al., 2008; Friedman et al., 2009). Overexpression of miR-101 markedly attenuated cell proliferation and apparently configured the histone code of cancer cells to that associated with more benign cellular phenotypes (Varambally et al., 2008). Other miRNAs commonly silenced in a variety of cancers also target factors in the epigenetic pathway; for example, miR-203 targets Bmi-1, a component of the polycomb repressor complex 1 (PRC1) that working together with PRC2, plays an important role in stabilizing epigenetic gene silencing by binding to methylated chromatin (Wellner et al., 2009; Sato et al., 2011), and miR-152 targets DNA methyltransferase DNMT1 (Tsuruta et al., 2011). All these miRNAs have been postulated as tumor suppressors (TS-miRNAs) and all have in common that they specifically downregulate factors involved in altering the epigenetic landscape of cells. These studies and several others (Rodríguez-Paredes and Esteller, 2011; Tsai and Baylin, 2011) demonstrate that when the epigenetic machinery goes awry, cells have increased chances to become malignant and develop cancers. Therefore, it is reasonable to speculate that cells and tissues in non-complicated alcoholics, as well as in any non-complicated group of individuals chronically subjected to environmental stress, should have implemented defense mechanisms such as overexpression of TS-miRNAs in order to avoid early development of malignances.
On the other hand, miR-1 has been reported to repress histone deacetylase 4 (HDAC4) and to promote differentiation of myoblasts (Chen et al., 2006). HDAC4 is highly abundant in mouse brain (Kumar et al., 2005) and its subcellular localization (nuclear or cytoplasmic) in rat hippocampal neurons is specified by neuronal activity (Chawla et al., 2003). Interestingly, Pandey et al. (2008) found that the anxiolytic effects produced by acute alcohol were associated with increased HDAC activity and reduced histone acetylation in the rat amygdala, and suggested that HDAC inhibitors may be potential therapeutic agents in treating alcohol withdrawal symptoms. Nevertheless, somewhat contradictorily, the Nestler group has reported that HDAC inhibition potentiates cocaine’s behavioral effects and that reducing acetylation through viral-mediated overexpression of HDAC4 in the NAc of mice decreased cocaine reward (Kumar et al., 2005). These authors also suggested that stimulation of gene transcription may be the predominant mechanism for cocaine-induced behavioral plasticity. These drug-specific or animal model-specific cellular activities and behaviors warn us about the potential risk of extrapolating results from one model into another, but also underscore the fine-tuning capabilities of these regulatory pathways.
Another subset of miRNA families upregulated in alcoholics, including miR-1, miR-34, miR-152, miR-196, and miR-203, is regulated by epigenetic mechanisms (Sato et al., 2011), which could partly explain how environmental factors could be contributing to disease development. The respective transcriptional units for miR-34b/c and miR-203 both contain CpG islands, and the methylation levels of these CpG islands are inversely correlated with the expression level of the mature miRNA in various cancers, including CNS tumors (Kozaki et al., 2008; Furuta et al., 2010). miR-152 and miR-196a are also silenced by hypermethylation of CpG islands on the respective DNA loci (Hoffman et al., 2009; Tsuruta et al., 2011). It is interesting to note that several of the miRNAs that target factors involved in epigenetic remodeling could be conversely regulated by epigenetic mechanisms, suggesting that miRNAs are elements of epigenetic feedback regulatory loops that fine-tune activity of the pathway.
Conclusion
Given the complex etiology of alcohol abuse disorders and dependence, characterized by brain-wide pathophysiological alterations under continuous crosstalk, we reason that to better understand and treat alcohol abuse, we need to develop organ and system-wide models (even if initially incomplete and speculative to some extent) with the potential to capture the dynamism in such complexity. Once we discover main dynamic relationships conducive to stable states of the disease, we may be able to design strategies to detour or reverse disease progression, akin of chemical reactions transiting through multiple equilibrium states. From the analysis and hypothetical models presented here, the notion emerges that miRNAs are efficiently being upregulated in response to alcohol insult and consequently inducing organ and system-wide homeostatic changes that produce long-term adaptation under the specific cellular environment. Remarkably, miRNAs that localize to and display activity at synapses in neurons are also capable of eliciting distinct and specific activities in other cell types, such as innate immunity-related functions in microglial cells, among others. This underscores the impact that molecular efficiency, signaling crosstalk, and cellular economy plays in the adaptation and evolution of cellular systems, all of which are concepts we have already learnt over the many years of biological research.
Due to the versatility and wide-reaching regulatory power of miRNAs, we expect the future development of novel knockdown and/or overexpression strategies targeting the activity of specific miRNAs, in an effort to prevent or even reverse addiction. Yet, many issues need to be addressed in order to better understand how miRNAs perform their functions and how they could be exploited to combat alcohol abuse. We wonder, for example, whether minor sequence variations in members of miRNA families affect their target specificity and biological function, and whether miRNA family members exist to exert redundant control over specific targets, to fine-tune their post-transcriptional regulation, or to exert a combination of both types of control. We also wonder whether we could unleash the fine-tuning regulatory potential of miRNAs by exploiting the combinatorial capability of miRNA targeting.
On the other hand, the miRNA field would immensely benefit from the development of high throughput technologies and methods for reliable characterization and validation of miRNA targets that are lagging at this time, together with the development of bioinformatic platforms capable of effectively and efficiently integrating the complex combinatorial patterns hidden under the “miRNA code.” Equally thrilling will be the discovery of small molecules that could specifically interact with and modulate the availability of select miRNAs in particular tissues, which would be invaluable to the clinical and therapeutic fields.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jody Mayfield and Dr. Adron Harris for comments on the manuscript. Work in the Mayfield Laboratory is funded by grants from NIAAA (AA020926, AA019382, AA016648, AA012404, AA013518, AA013517). Yury O. Nunez is funded by postdoctoral fellowship under NIH/NIAAA Institutional Research Training Grant 5T32AA007471-24. The authors are grateful to the Australian Brain Donor Programs NSW Tissue Resource Centre for providing alcoholic and control brain tissue for analysis. The Centre is supported by the University of Sydney, National Health and Medical Research Council of Australia, Schizophrenia Research Institute, and NIAAA.
References
Aho, L., Karkola, K., Juusela, J., and Alafuzoff, I. (2009). Heavy alcohol consumption and neuropathological lesions: a post-mortem human study. J. Neurosci. Res. 87, 2786–2792.
Androulidaki, A., Iliopoulos, D., Arranz, A., Doxaki, C., Schworer, S., Zacharioudaki, V., Margioris, A. N., Tsichlis, P. N., and Tsatsanis, C. (2009). The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231.
Ashraf, S. I., McLoon, A. L., Sclarsic, S. M., and Kunes, S. (2006). Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124, 191–205.
Bardo, M. T. (1998). Neuropharmacological mechanisms of drug reward: beyond dopamine in the nucleus accumbens. Crit. Rev. Neurobiol. 12, 37–67.
Batassa, E. M., Costanzi, M., Saraulli, D., Scardigli, R., Barbato, C., Cogoni, C., and Cestari, V. (2010). RISC activity in hippocampus is essential for contextual memory. Neurosci. Lett. 471, 185–188.
Bazzoni, F., Rossato, M., Fabbri, M., Gaudiosi, D., Mirolo, M., Mori, L., Tamassia, N., Mantovani, A., Cassatella, M. A., and Locati, M. (2009). Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. U.S.A. 106, 5282–5287.
Belver, L., Papavasiliou, F. N., and Ramiro, A. R. (2011). MicroRNA control of lymphocyte differentiation and function. Curr. Opin. Immunol. 23, 368–373.
Blednov, Y. A., Benavidez, J. M., Geil, C., Perra, S., Morikawa, H., and Harris, R. A. (2011). Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Behav. Immun. 25(Suppl. 1), S92–S105.
Blednov, Y. A., Ponomarev, I., Geil, C., Bergeson, S., Koob, G. F., and Harris, R. A. (2012). Neuroimmune regulation of alcohol consumption: behavioral validation of genes obtained from genomic studies. Addict. Biol. 17, 108–120.
Bonauer, A., Carmona, G., Iwasaki, M., Mione, M., Koyanagi, M., Fischer, A., Burchfield, J., Fox, H., Doebele, C., Ohtani, K., Chavakis, E., Potente, M., Tjwa, M., Urbich, C., Zeiher, A. M., and Dimmeler, S. (2009). MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324, 1710–1713.
Bönsch, D., Lederer, T., Reulbach, U., Hothorn, T., Kornhuber, J., and Bleich, S. (2005). Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum. Mol. Genet. 14, 967–971.
Bönsch, D., Lenz, B., Fiszer, R., Frieling, H., Kornhuber, J., and Bleich, S. (2006). Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 113, 1299–1304.
Bönsch, D., Lenz, B., Reulbach, U., Kornhuber, J., and Bleich, S. (2004). Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 111, 1611–1616.
Borghese, C. M., Ali, D. N., Bleck, V., and Harris, R. A. (2002). Acetylcholine and alcohol sensitivity of neuronal nicotinic acetylcholine receptors: mutations in transmembrane domains. Alcohol. Clin. Exp. Res. 26, 1764–1772.
Breving, K., and Esquela-Kerscher, A. (2010). The complexities of microRNA regulation: mirandering around the rules. Int. J. Biochem. Cell Biol. 42, 1316–1329.
Burgoyne, R. D., and Morgan, A. (2011). Chaperoning the SNAREs: a role in preventing neurodegeneration. Nat. Cell Biol. 13, 8–9.
Burre, J., Sharma, M., Tsetsenis, T., Buchman, V., Etherton, M. R., and Sudhof, T. C. (2010). α-Synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329, 1663–1667.
Bushati, N., and Cohen, S. M. (2008). MicroRNAs in neurodegeneration. Curr. Opin. Neurobiol. 18, 292–296.
Ceman, S., and Saugstad, J. (2011). MicroRNAs: meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease. Pharmacol. Ther. 130, 26–37.
Chandrasekar, V., and Dreyer, J.-L. (2009). microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42, 350–362.
Chandrasekar, V., and Dreyer, J.-L. (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36, 1149–1164.
Chawla, S., Vanhoutte, P., Arnold, F. J. L., Huang, C. L.-H., and Bading, H. (2003). Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159.
Chen, H., Shalom-Feuerstein, R., Riley, J., Zhang, S.-D., Tucci, P., Agostini, M., Aberdam, D., Knight, R. A., Genchi, G., Nicotera, P., Melino, G., and Vasa-Nicotera, M. (2010). miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem. Biophys. Res. Commun. 394, 921–927.
Chen, J.-F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., Conlon, F. L., and Wang, D. Z. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233.
Chen, X.-M., Splinter, P. L., O’Hara, S. P., and LaRusso, N. F. (2007). A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 282, 28929–28938.
Cheng, L.-C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Contet, C., Kieffer, B. L., and Befort, K. (2004). Mu opioid receptor: a gateway to drug addiction. Curr. Opin. Neurobiol. 14, 370–378.
Crews, F. T., Zou, J., and Qin, L. (2011). Induction of innate immune genes in brain create the neurobiology of addiction. Brain Behav. Immun. 25(Suppl. 1), S4–S12.
Doebele, C., Bonauer, A., Fischer, A., Scholz, A., Reiss, Y., Urbich, C., Hofmann, W.-K., Zeiher, A. M., and Dimmeler, S. (2010). Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115, 4944–4950.
Doxakis, E. (2010). Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 285, 12726–12734.
Dyr, W., Koros, E., Bienkowski, P., and Kostowski, W. (1999). Involvement of nicotinic acetylcholine receptors in the regulation of alcohol drinking in Wistar rats. Alcohol Alcohol. 34, 43–47.
Edenberg, H. J., Xuei, X., Wetherill, L. F., Bierut, L., Bucholz, K., Dick, D. M., Hesselbrock, V., Kuperman, S., Porjesz, B., Schuckit, M. A., Tischfield, J. A., Almasy, L. A., Nurnberger, J. I. Jr., and Foroud, T. (2008). Association of NFKB1, which encodes a subunit of the transcription factor NF-kappaB, with alcohol dependence. Hum. Mol. Genet. 17, 963–970.
Eipper-Mains, J. E., Kiraly, D. D., Palakodeti, D., Mains, R. E., Eipper, B. A., and Graveley, B. R. (2011). microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 17, 1529–1543.
Etheridge, N., Lewohl, J. M., Mayfield, R. D., Harris, R. A., and Dodd, P. R. (2009). Synaptic proteome changes in the superior frontal gyrus and occipital cortex of the alcoholic brain. Proteomics Clin. Appl. 3, 730–742.
Ferguson, S. M., Brasnjo, G., Hayashi, M., Wölfel, M., Collesi, C., Giovedi, S., Raimondi, A., Gong, L. W., Ariel, P., Paradise, S., O’Toole, E., Flavell, R., Cremona, O., Miesenböck, G., Ryan, T. A., and De Camilli, P. (2007). A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science 316, 570–574.
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat. Rev. Genet. 9, 102–114.
Finn, A. K., and Whistler, J. L. (2001). Endocytosis of the mu opioid receptor reduces tolerance and a cellular hallmark of opiate withdrawal. Neuron 32, 829–839.
Fiore, R., Siegel, G., and Schratt, G. (2008). MicroRNA function in neuronal development, plasticity and disease. Biochim. Biophys. Acta 1779, 471–478.
Foroud, T., Wetherill, L. F., Liang, T., Dick, D. M., Hesselbrock, V., Kramer, J., Nurnberger, J., Schuckit, M., Carr, L., Porjesz, B., Xuei, X., and Edenberg, H. J. (2007). Association of alcohol craving with alpha-synuclein (SNCA). Alcohol. Clin. Exp. Res. 31, 537–545.
Friedman, J. M., Liang, G., Liu, C.-C., Wolff, E. M., Tsai, Y. C., Ye, W., Zhou, X., and Jones, P. A. (2009). The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 69, 2623–2629.
Furuta, M., Kozaki, K.-I., Tanaka, S., Arii, S., Imoto, I., and Inazawa, J. (2010). miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31, 766–776.
Gorini, G., Bell, R. L., and Mayfield, R. D. (2011). Molecular targets of alcohol action: translational research for pharmacotherapy development and screening. Prog. Mol. Biol. Transl. Sci. 98, 293–347.
Gorini, G., Ponomareva, O., Shores, K. S., Person, M. D., Harris, R. A., and Mayfield, R. D. (2010). Dynamin-1 co-associates with native mouse brain BKCa channels: proteomics analysis of synaptic protein complexes. FEBS Lett. 584, 845–851.
Greten-Harrison, B., and Polydoro, M. (2010). αβγ-Synuclein triple knockout mice reveal age-dependent neuronal dysfunction. Proc. Natl. Acad. Sci. U.S.A. 107, 19573–19578.
Guasch, R. M., Blanco, A. M., Pérez-Aragó, A., Miñambres, R., Talens-Visconti, R., Peris, B., and Guerri, C. (2007). RhoE participates in the stimulation of the inflammatory response induced by ethanol in astrocytes. Exp. Cell Res. 313, 3779–3788.
He, Y., Yang, C., Kirkmire, C. M., and Wang, Z. J. (2010). Regulation of opioid tolerance by let-7 family microRNA targeting the opioid receptor. J. Neurosci. 30, 10251–10258.
Heaton, M. B., Moore, D. B., Paiva, M., Gibbs, T., and Bernard, O. (1999). Bcl-2 overexpression protects the neonatal cerebellum from ethanol neurotoxicity. Brain Res. 817, 13–18.
Heberlein, A., Muschler, M., Frieling, H., Behr, M., Eberlein, C., Wilhelm, J., Gröschl, M., Kornhuber, J., Bleich, S., and Hillemacher, T. (2011). Epigenetic down regulation of nerve growth factor during alcohol withdrawal. Addict Biol. doi:10.1111/j.1369-1600.2010.00307.x
Hoffman, A. E., Zheng, T., Yi, C., Leaderer, D., Weidhaas, J., Slack, F., Zhang, Y., Paranjape, T., and Zhu, Y. (2009). microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 69, 5970–5977.
Hou, J., Wang, P., Lin, L., Liu, X., Ma, F., An, H., Wang, Z., and Cao, X. (2009). MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183, 2150–2158.
Huang, W., and Li, M. D. (2008). Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1). Int. J. Neuropsychopharmacol. 12, 537.
Hunot, S., and Hirsch, E. C. (2003). Neuroinflammatory processes in Parkinson’s disease. Ann. Neurol. 53(Suppl. 3), S49–S58; discussion S58–S60.
Hyman, S. E., Malenka, R. C., and Nestler, E. J. (2006). Neural mechanisms of addiction: the role of reward-related learning and memory. Annu. Rev. Neurosci. 29, 565–598.
Iliopoulos, D., Hirsch, H. A., and Struhl, K. (2009). An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706.
Johansson, S., Ekström, T. J., Marinova, Z., Ökvist, A., Sheedy, D., Garrick, T., Harper, C., Kuzmin, A., Yakovleva, T., and Bakalkin, G. (2008). Dysregulation of cell death machinery in the prefrontal cortex of human alcoholics. Int. J. Neuropsychopharmacol. 12, 109.
Kash, T. L., Baucum, A. J., Conrad, K. L., Colbran, R. J., and Winder, D. G. (2009). Alcohol exposure alters NMDAR function in the bed nucleus of the stria terminalis. Neuropsychopharmacology 34, 2420–2429.
Kelley, K. W., and Dantzer, R. (2011). Alcoholism and inflammation: neuroimmunology of behavioral and mood disorders. Brain Behav. Immun. 25, S13–S20.
Kozaki, K.-I., Imoto, I., Mogi, S., Omura, K., and Inazawa, J. (2008). Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 68, 2094–2105.
Kumar, A., Choi, K.-H., Renthal, W., Tsankova, N. M., Theobald, D. E. H., Truong, H.-T., Russo, S. J., Laplant, Q., Sasaki, T. S., Whistler, K. N., Neve, R. L., Self, D. W., and Nestler, E. J. (2005). Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314.
Lewohl, J. M., Nunez, Y. O., Dood, P. R., Tiwari, G. R., Harris, R. A., and Mayfield, R. D. (2011). Up-regulation of MicroRNAs in brain of human alcoholics. Alcohol. Clin. Exp. Res. 35, 1928–1937.
Li, M. D., and van der Vaart, A. D. (2011). MicroRNAs in addiction: adaptation’s middlemen. Mol. Psychiatry 16, 1159–1168.
Liang, D.-Y., Shi, X., Li, X., Li, J., and Clark, J. D. (2007). The beta2 adrenergic receptor regulates morphine tolerance and physical dependence. Behav. Brain Res. 181, 118–126.
Liang, T., and Carr, L. G. (2006). Regulation of alpha-synuclein expression in alcohol-preferring and -non preferring rats. J. Neurochem. 99, 470–482.
Liu, J., Asuncion-Chin, M., Liu, P., and Dopico, A. M. (2006a). CaM kinase II phosphorylation of slo Thr107 regulates activity and ethanol responses of BK channels. Nat. Neurosci. 9, 41–49.
Liu, J., Lewohl, J. M., Harris, R. A., Iyer, V. R., Dodd, P. R., Randall, P. K., and Mayfield, R. D. (2006b). Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology 31, 1574–1582.
Liu, X., Zhan, Z., Xu, L., Ma, F., Li, D., Guo, Z., Li, N., and Cao, X. (2010). MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J. Immunol. 185, 7244–7251.
Lugli, G., Larson, J., Martone, M. E., Jones, Y., and Smalheiser, N. R. (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905.
Lugli, G., Torvik, V. I., Larson, J., and Smalheiser, N. R. (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 106, 650–661.
Mayfield, R. D., Harris, R. A., and Schuckit, M. A. (2008). Genetic factors influencing alcohol dependence. Br. J. Pharmacol. 154, 275–287.
Maze, I., and Nestler, E. J. (2011). The epigenetic landscape of addiction. Ann. N. Y. Acad. Sci. 1216, 99–113.
Miranda, R. C., Pietrzykowski, A. Z., Tang, Y., Sathyan, P., Mayfield, D., Keshavarzian, A., Sampson, W., and Hereld, D. (2010). MicroRNAs: master regulators of ethanol abuse and toxicity. Alcohol. Clin. Exp. Res. 34, 575–587.
Moschos, S. A., Williams, A. E., Perry, M. M., Birrell, M. A., Belvisi, M. G., and Lindsay, M. A. (2007). Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 8, 240. doi:10.1186/1471-2164-8-240
Mulligan, M. K., Ponomarev, I., Hitzemann, R. J., Belknap, J. K., Tabakoff, B., Harris, R. A., Crabbe, J. C., Blednov, Y. A., Grahame, N. J., Phillips, T. J., Finn, D. A., Hoffman, P. L., Iyer, V. R., Koob, G. F., and Bergeson, S. E. (2006). Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl. Acad. Sci. U.S.A. 103, 6368–6373.
Muschler, M. A. N., Hillemacher, T., Kraus, C., Kornhuber, J., Bleich, S., and Frieling, H. (2010). DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J. Neural Transm. 117, 513–519.
Narahashi, T., Aistrup, G., and Marszalec, W. (1999). Neuronal nicotinic acetylcholine receptors: a new target site of ethanol. Neurochemistry 35, 131–141.
Nowoslawski, L., Klocke, B. J., and Roth, K. A. (2005). Molecular regulation of acute ethanol-induced neuron apoptosis. J. Neuropathol. Exp. Neurol. 64, 490–497.
Ökvist, A., Johansson, S., Kuzmin, A., Bazov, I., Merino-Martinez, R., Ponomarev, I., Mayfield, R. D., Harris, R. A., Sheedy, D., Garrick, T., Harper, C., Hurd, Y. L., Terenius, L., Ekström, T. J., Bakalkin, G., and Yakovleva, T. (2007). Neuroadaptations in human chronic alcoholics: dysregulation of the NF-kappaB system. PLoS ONE 2, e930. doi:10.1371/journal.pone.0000930
Oliva, J., Bardag-Gorce, F., Li, J., French, B. A., and French, S. W. (2011). S-adenosylmethionine prevents the up regulation of Toll-like receptor (TLR) signaling caused by chronic ethanol feeding in rats. Exp. Mol. Pathol. 90, 239–243.
O’Neill, L. A., Sheedy, F. J., and McCoy, C. E. (2011). MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175.
O’Neill, L. A. J. (2006). How Toll-like receptors signal: what we know and what we don’t know. Curr. Opin. Immunol. 18, 3–9.
Otis, J. M., and Mueller, D. (2011). Inhibition of β-adrenergic receptors induces a persistent deficit in retrieval of a cocaine-associated memory providing protection against reinstatement. Neuropsychopharmacology 36, 1912–1920.
Pandey, S. C., Ugale, R., Zhang, H., Tang, L., and Prakash, A. (2008). Brain chromatin remodeling: a novel mechanism of alcoholism. J. Neurosci. 28, 3729–3737.
Pastor, I. J., Laso, F. J., Avila, J. J., Rodríguez, R. E., and González-Sarmiento, R. (2000). Polymorphism in the interleukin-1 receptor antagonist gene is associated with alcoholism in Spanish men. Alcohol. Clin. Exp. Res. 24, 1479–1482.
Pastor, I. J., Laso, F. J., Romero, A., and González-Sarmiento, R. (2005). Interleukin-1 gene cluster polymorphisms and alcoholism in Spanish men. Alcohol Alcohol. 40, 181–186.
Pietrzykowski, A. Z., Friesen, R. M., Martin, G. E., Puig, S. I., Nowak, C. L., Wynne, P. M., Siegelmann, H. T., and Treistman, S. N. (2008). Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59, 274–287.
Pillai, R. S. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576.
Rajgopal, Y., Chetty, C. S., and Vemuri, M. C. (2003). Differential modulation of apoptosis-associated proteins by ethanol in rat cerebral cortex and cerebellum. Eur. J. Pharmacol. 470, 117–124.
Redell, J. B., Moore, A. N., Ward, N. H., Hergenroeder, G. W., and Dash, P. K. (2010). Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 27, 2147–2156.
Robison, A. J., and Nestler, E. J. (2011). Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637.
Rodríguez-Paredes, M., and Esteller, M. (2011). Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339.
Ron, D., and Messing, R. O. (2011). Signaling pathways mediating alcohol effects. Curr. Top. Behav. Neurosci. doi:10.1007/7854_2011_161. [Epub ahead of print].
Russo, S. J., Mazei-Robison, M. S., Ables, J. L., and Nestler, E. J. (2009). Neurotrophic factors and structural plasticity in addiction. Neuropharmacology 56(Suppl. 1), 73–82.
Saba, L. M., Bennett, B., Hoffman, P. L., Barcomb, K., Ishii, T., Kechris, K., and Tabakoff, B. (2011). A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology 60, 1269–1280.
Saiz, P. A., Garcia-Portilla, M. P., Florez, G., Corcoran, P., Arango, C., Morales, B., Leza, J. C., Alvarez, S., Díaz, E. M., Alvarez, V., Coto, E., Nogueiras, L., and Bobes, J. (2009). Polymorphisms of the IL-1 gene complex are associated with alcohol dependence in Spanish Caucasians: data from an association study. Alcohol. Clin. Exp. Res. 33, 2147–2153.
Sathyan, P., Golden, H. B., and Miranda, R. C. (2007). Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 27, 8546–8557.
Sato, F., Tsuchiya, S., Meltzer, S. J., and Shimizu, K. (2011). MicroRNAs and epigenetics. FEBS J. 278, 1598–1609.
Schaefer, A., O’Carroll, D., Tan, C. L., Hillman, D., Sugimori, M., Llinas, R., and Greengard, P. (2007). Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 204, 1553–1558.
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., and Greenberg, M. E. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289.
Sessoms-Sikes, S., Honse, Y., Lovinger, D. M., and Colbran, R. J. (2005). CaMKIIalpha enhances the desensitization of NR2B-containing NMDA receptors by an autophosphorylation-dependent mechanism. Mol. Cell. Neurosci. 29, 139–147.
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., Khudayberdiev, S., Leuschner, P. F., Busch, C. J., Kane, C., Hübel, K., Dekker, F., Hedberg, C., Rengarajan, B., Drepper, C., Waldmann, H., Kauppinen, S., Greenberg, M. E., Draguhn, A., Rehmsmeier, M., Martinez, J., and Schratt, G. M. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716.
Simon, D. J., Madison, J. M., Conery, A. L., Thompson-Peer, K. L., Soskis, M., Ruvkun, G. B., Kaplan, J. M., and Kim, J. K. (2008). The MicroRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133, 903–915.
Smalheiser, N. R., and Lugli, G. (2009). microRNA regulation of synaptic plasticity. Neuromolecular Med. 11, 133–140.
Sonkoly, E., Ståhle, M., and Pivarcsi, A. (2008). MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18, 131–140.
Sonkoly, E., Wei, T., Janson, P. C. J., Sääf, A., Lundeberg, L., Tengvall-Linder, M., Norstedt, G., Alenius, H., Homey, B., Scheynius, A., Ståhle, M., and Pivarcsi, A. (2007). MicroRNAs: novel regulators involved in the pathogenesis of psoriasis. PLoS ONE 2, e610. doi:10.1371/journal.pone.0000610
Sutton, M. A., and Schuman, E. M. (2006). Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58.
Taganov, K. D., Boldin, M. P., Chang, K.-J., and Baltimore, D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486.
Taqi, M. M., Bazov, I., Watanabe, H., Sheedy, D., Harper, C., Alkass, K., Druid, H., Wentzel, P., Nyberg, F., Yakovleva, T., and Bakalkin, G. (2011). Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict. Biol. 16, 499–509.
Taraskina, A. E., Filimonov, V. A., Kozlovskaya, Y. A., Morozova, M. N., Gaschin, D. V., and Schwarzman, A. L. (2008). High level of alpha-synuclein mRNA in peripheral lymphocytes of patients with alcohol dependence syndrome. Bull. Exp. Biol. Med. 146, 609–611.
Triantafilou, M., and Triantafilou, K. (2002). Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 23, 301–304.
Tsai, H.-C., and Baylin, S. B. (2011). Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 21, 502–517.
Tsuruta, T., Kozaki, K.-I., Uesugi, A., Furuta, M., Hirasawa, A., Imoto, I., Susumu, N., Aoki, D., and Inazawa, J. (2011). miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 71, 6450–6462.
Turner, M. L., Schnorfeil, F. M., and Brocker, T. (2011). MicroRNAs regulate dendritic cell differentiation and function. J. Immunol. 187, 3911–3917.
Vallés, S. L., Blanco, A. M., Pascual, M., and Guerri, C. (2004). Chronic ethanol treatment enhances inflammatory mediators and cell death in the brain and in astrocytes. Brain Pathol. 14, 365–371.
Varambally, S., Cao, Q., Mani, R.-S., Shankar, S., Wang, X., Ateeq, B., Laxman, B., Cao, X., Jing, X., Ramnarayanan, K., Brenner, J. C., Yu, J., Kim, J. H., Han, B., Tan, P., Kumar-Sinha, C., Lonigro, R. J., Palanisamy, N., Maher, C. A., and Chinnaiyan, A. M. (2008). Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699.
Vasudevan, S., and Steitz, J. A. (2007). AU-rich-element-mediated upregulation of translation by FXR1 and argonaute 2. Cell 128, 1105–1118.
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934.
Wang, L.-L., Zhang, Z., Li, Q., Yang, R., Pei, X., Xu, Y., Wang, J., Zhou, S. F., and Li, Y. (2009). Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum. Reprod. 24, 562–579.
Wang, W. C. H., Juan, A. H., Panebra, A., and Liggett, S. B. (2011). MicroRNA let-7 establishes expression of β2-adrenergic receptors and dynamically down-regulates agonist-promoted down-regulation. Proc. Natl. Acad. Sci. U.S.A. 108, 6246–6251.
Weiner, H. L., and Selkoe, D. J. (2002). Inflammation and therapeutic vaccination in CNS diseases. Nature 420, 879–884.
Wellner, U., Schubert, J., Burk, U. C., Schmalhofer, O., Zhu, F., Sonntag, A., Waldvogel, B., Vannier, C., Darling, D., zur Hausen, A., Brunton, V. G., Morton, J., Sansom, O., Schüler, J., Stemmler, M. P., Herzberger, C., Hopt, U., Keck, T., Brabletz, S., and Brabletz, T. (2009). The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495.
White, F., and Wilson, N. (2010). Opiate-induced hypernociception and chemokine receptors. Neuropharmacology 58, 35–37.
Wong, C. C. Y., Mill, J., and Fernandes, C. (2011). Drugs and addiction: an introduction to epigenetics. Addiction 106, 480–489.
World Health Organization. (2011). Global Status Report on Alcohol and Health (Geneva: WHO Press), 1–85.
Xiao, Q., Ye, S., Oberhollenzer, F., Mayr, A., Jahangiri, M., Willeit, J., Kiechl, S., and Xu, Q. (2008). SDF1 gene variation is associated with circulating SDF1alpha level and endothelial progenitor cell number: the Bruneck Study. PLoS ONE 3, e4061. doi:10.1371/journal.pone.0004061
Yadav, S., Pandey, A., Shukla, A., Talwelkar, S. S., Kumar, A., Pant, A. B., and Parmar, D. (2011). miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J. Biol. Chem. 286, 37347–37357.
Yakovleva, T., Bazov, I., Watanabe, H., Hauser, K. F., and Bakalkin, G. (2011). Transcriptional control of maladaptive and protective responses in alcoholics: a role of the NF-κB system. Brain Behav. Immun. 25(Suppl. 1), S29–S38.
Keywords: human postmortem brain, alcohol dependence, alcoholism, gene expression, miRNA, non-coding RNA
Citation: Nunez YO and Mayfield RD (2012) Understanding alcoholism through microRNA signatures in brains of human alcoholics. Front. Gene. 3:43. doi: 10.3389/fgene.2012.00043
Received: 07 February 2012; Accepted: 09 March 2012;
Published online: 03 April 2012.
Edited by:
Matthew Reilly, National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism, USAReviewed by:
Walter Lukiw, Louisiana State University, USAZhaohui Wang, University of Texas Southwestern Medical Center, USA
Copyright: © 2012 Nunez and Mayfield. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: R. Dayne Mayfield, Waggoner Center for Alcohol and Addiction Research, University of Texas at Austin, Austin, TX 78712, USA. e-mail:ZGF5bmUubWF5ZmllbGRAbWFpbC51dGV4YXMuZWR1