- 1Department of Genetics, Shahid Chamran University of Ahvaz, Ahvaz, Iran
- 2Department of Genetics, School of Biological Sciences, Tarbiat Modares University, Tehran, Iran
- 3Laboaratory of Molecular Pathology and Cancer Genomics, Department of Pharmacy, Seoul National University College of Pharmacy, Seoul, Korea
A commentary on
The beginning of the road for non-coding RNAs in normal hematopoiesis and hematologic malignancies
by Heuston, E. F., Lemon, K. T., and Arceci, R. J. (2011). Front. Genet. 2:94. doi: 10.3389/fgene.2011.00094
The human genome encodes ~20,000 proteins; however, protein-coding genes represent <2% of the total genome (Gibb et al., 2011). Since this discovery, several studies have pointed out that at least 90% of the genome is actively transcribed (Birney et al., 2007; Costa, 2010). This field provoked a spirited debate, which is evident from the number of original articles, reviews, letter to the editors, commentaries, and rebuttals.
Many long non-coding RNAs (lncRNAs), ranging from 0.2 to ~100 kilobases (kb) in length, are transcribed from the genome. LncRNAs affect various cellular functions such as gene regulation, genomic imprinting, RNA maturation, and translation (Wang and Chang, 2011). In addition, the differential expression of lncRNAs has recently been linked to carcinogenesis, including gastric cancer (Hajjari et al., 2013). In recent reports, lncRNAs have been attributed to oncogenic and/or tumor suppressor roles (Reviewed in Gibb et al., 2011; Qiu et al., 2013). Taken together, these reports support the possible involvement of lncRNAs in the initiation and/or progression of breast, colorectal, liver, lung, and gastric cancers (Hajjari and Khoshnevisan, 2013). However, little is known about the potential role of lncRNAs in leukemia. In fact, there is a paucity of reports on the characterization of lncRNAs in this type of cancer.
In the December 2011 edition of Frontiers in Genetics, Heuston et al. (2011) published a review on non-coding RNAs in hematologic malignancies. They highlighted the potential roles for non-coding RNAs including microRNAs and lncRNAs in the development of acute and chronic leukemia. While this is the only review that described the expression patterns of ncRNAs in leukemia, it may provide a foundation for future research in this field.
Heuston et al., referred to several reports on lncRNAs such as ANRIL, lncRNA-P21, MEG3, Dleu2, HOTAIRM1, EGO, and lncRNA-a7 in leukemia, highlighting the potential benefit of research on lncRNAs for the development of much needed diagnostic, prognostic, and therapeutic targets (Heuston et al., 2011). In addition, based on the previous studies, Heuston et al., proposed to study more about the functional role of MEG3 in leukemia. In our opinion, this review provides an overview of the published reports on the subject and represents a suitable guide for researchers involved in the field of molecular pathology of leukemia.
To test if any prior evidence on the potential role of lncRNAs in leukemia progression existed, we queried the Oncomine Database (www.oncomine.org) for five lncRNAs: HOTAIR, ANRIL, MEG3, H19, and UCA1. All five lncRNAs were among the most cited lncRNAs associated with a potential role in cancer.
We found that two lncRNAs including MEG3 and H19 are up-regulated in acute myeloid leukemia (AML) cancer compared to normal blood cells (Fold change > 1.5, P-value < 0.01). As presented in Table 1, the results showed that H19, UCA1, and MEG3 are up-regulated in French-American-British (FAB) classification subtypes M3, M2, and M3, respectively. The expression of H19 is up-regulated in AML compared to acute lymphoblastic leukemia (ALL). In conclusion, taken together, the review from Heuston et al. and our data shed lights on the roles of lncRNAs in different types of leukemia and may prove useful for the development of molecular diagnosis and targeted therapy for leukemia.
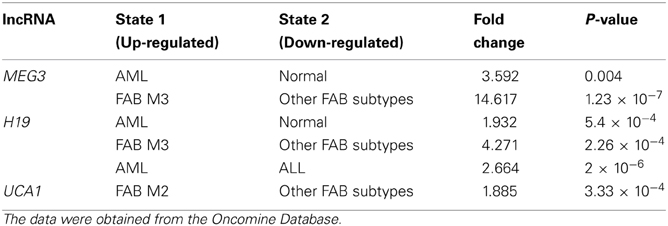
Table 1. Expression level of the lncRNAs in Leukemia (the results are expressed as fold change between cancer and normal tissues or cancer subtypes).
References
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Costa, F. F. (2010). Non-coding RNAs: meet thy masters. Bioessays 32, 599–608. doi: 10.1002/bies.200900112
Gibb, E. A., Brown, C. J., and Lam, W. L. (2011). The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10, 38. doi: 10.1186/1476-4598-10-38
Hajjari, M., Behmanesh, M., Sadeghizadeh, M., and Zeinoddini, M. (2013). Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med. Oncol. 30, 670. doi: 10.1007/s12032-013-0670-0
Hajjari, M., and Khoshnevisan, A. (2013). Potential long non-coding RNAs to be considered as biomarkers or therapeutic targets in gastric cancer. Front. Genet. 4:210. doi: 10.3389/fgene.2013.00210
Heuston, E. F., Lemon, K. T., and Arceci, R. J. (2011). The beginning of the road for non-coding RNAs in normal hematopoiesis and hematologic malignancies. Front. Genet. 2:94. doi: 10.3389/fgene.2011.00094
Qiu, M. T., Hu, J. W., Yin, R., and Xu. L. (2013). Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 34, 613–620. doi: 10.1007/s13277-013-0658-6
Keywords: long non-coding RNA (lncRNA), expression, translational research, leukemia, carcinogenesis
Citation: Hajjari M, Khoshnevisan A and Shin YK (2013) Long non-coding RNAs in hematologic malignancies: road to translational research. Front. Genet. 4:250. doi: 10.3389/fgene.2013.00250
Received: 22 October 2013; Accepted: 02 November 2013;
Published online: 20 November 2013.
Edited by:
Alistair R. Forrest, RIKEN Omics Science Center, JapanCopyright © 2013 Hajjari, Khoshnevisan and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:eWtlZXNoaW5Ac251LmFjLmty
 Mohammadreza Hajjari
Mohammadreza Hajjari Atefeh Khoshnevisan
Atefeh Khoshnevisan Young Kee Shin
Young Kee Shin