- 1South Texas Diabetes and Obesity Institute, Regional Academic Health Center, University of Texas at San Antonio, Harlingen, TX, USA
- 2Department of Nutrition, University of North Carolina at Chapel Hill, Kannapolis, NC, USA
- 3University of North Carolina Nutrition Research Institute, University of North Carolina at Chapel Hill, Kannapolis, NC, USA
- 4Department of Genetics, Texas Biomedical Research Institute, San Antonio, TX, USA
- 5Department of Biochemistry, University of New Mexico School of Medicine, Albuquerque, NM, USA
- 6Dialysis Clinic, Inc., Albuquerque, NM, USA
- 7Arbor Research Collaborative for Health, Ann Arbor, MI, USA
- 8Southwest National Primate Research Center, San Antonio, TX, USA
- 9Department of Medicine, Division of Nephrology, University of New Mexico School of Medicine, Albuquerque, NM, USA
The objective of this study is to identify genetic factors associated with chronic kidney disease (CKD) and related cardiometabolic phenotypes among participants of the Genetics of Kidney Disease in Zuni Indians study. The study was conducted as a community-based participatory research project in the Zuni Indians, a small endogamous tribe in rural New Mexico. We recruited 998 members from 28 extended multigenerational families, ascertained through probands with CKD who had at least one sibling with CKD. We used the Illumina Infinium Human1M-Duo version 3.0 BeadChips to type 1.1 million single nucleotide polymorphisms (SNPs). Prevalence estimates for CKD, hyperuricemia, diabetes, and hypertension were 24%, 30%, 17% and 34%, respectively. We found a significant (p < 1.58 × 10-7) association for a SNP in a novel gene for serum creatinine (PTPLAD2). We replicated significant associations for genes with serum uric acid (SLC2A9), triglyceride levels (APOA1, BUD13, ZNF259), and total cholesterol (PVRL2). We found novel suggestive associations (p < 1.58 × 10-6) for SNPs in genes with systolic (OLFML2B), and diastolic blood pressure (NFIA). We identified a series of genes associated with CKD and related cardiometabolic phenotypes among Zuni Indians, a population with a high prevalence of kidney disease. Illuminating genetic variations that modulate the risk for these disorders may ultimately provide a basis for novel preventive strategies and therapeutic interventions.
Introduction
The Zuni Indians are experiencing interrelated epidemics of chronic kidney disease (CKD) and related features of the cardiometabolic syndrome including obesity, diabetes, dyslipidemia, and hypertension that are intermediate phenotypes for CKD (Stidley et al., 2002; Shah et al., 2003; Scavini et al., 2007; MacCluer et al., 2010). Ethnicity also influences the risk for the development of CKD and related phenotypes (Johnson et al., 2009). Genetic studies, including candidate gene and genome-wide association studies (GWAS), have been conducted to elucidate the effects of specific genes on the variation in CKD and cardiometabolic risk factors. These include studies conducted in Caucasians (Hwang et al., 2007; Parsa et al., 2013), African Americans (Edwards et al., 2008; Willer et al., 2013; Bidulescu et al., 2014), Asians (Yamakawa-Kobayashi et al., 2012; Willer et al., 2013), Mexican Americans (Farook et al., 2013; Thameem et al., 2013), Pima Indians (Bian et al., 2013; Hanson et al., 2013, 2014), and in the 13 American Indian tribes participating in the Strong Heart Family Study (Franceschini et al., 2013; Voruganti et al., 2014). To decrease the burden of kidney disease and related intermediate phenotypes in the Zuni Pueblo, we established the Zuni Kidney Project (ZKP) in partnership with the Indian Health Service, University of New Mexico Health Sciences Center, Texas Biomedical Research Institute and Dialysis Clinic, Inc. (DCI; Stidley et al., 2002).
The Zuni Indians reside in the Zuni Pueblo, located in McKinley County, NM, USA. The population was 6,302 in the 2010 US Census and 97% of inhabitants were American Indians (Zuni Pueblo Quick Facts). Emigration and immigration rates are low and therefore the population is relatively endogamous. The majority of adults work as artisans, making jewelry and fetishes. The ZKP previously conducted a population-based, cross-sectional survey that reported high prevalence estimates, age-and sex-adjusted to the Zuni population, for decreased estimated glomerular filtration rate (eGFR; Scavini et al., 2007), albuminuria (Shah et al., 2003), and hematuria (Tentori et al., 2003). Prevalence estimates for albuminuria and hematuria were higher for diabetic than non-diabetic participants (Shah et al., 2003). The prevalence of end-stage renal disease (ESRD) among the Zuni Indians, adjusted for age and gender, was 20.0-, 4.4-, and 5.6-fold higher than that among European– and African–Americans and the composite estimate for all American Indians (Shah et al., 2003).
Recently the ZKP conducted the Genetics of Kidney Disease in Zuni Indians (GKDZI) Study to explore the hypothesis that genetic factors modulate susceptibility to CKD and related phenotypes. Studies of extended families, such as GKDZI, offer advantages over studies of sib pairs or unrelated individuals for gene discovery since they have enhanced statistical power, are more homogenous and allow for greater genotyping quality control (Laird and Lange, 2008). The current manuscript presents the results of a GWAS in extended, multigenerational families of Zuni Indians.
Materials and Methods
Study Design
We conducted a GWAS in extended families of Zuni Indians. The study cohort consisted of 30 extended families, of which 28 were multigenerational. The families were ascertained through probands with kidney disease, who had at least one sibling with kidney disease. The Institutional Review Boards of all participating institutions and the Zuni Tribal Council approved the protocol. All participants gave written informed consent.
Setting
The study was conducted in the Zuni Pueblo. Recruitment occurred from February 2005 through May 2009. Data were collected from February 2005 through June 2009.
Participants
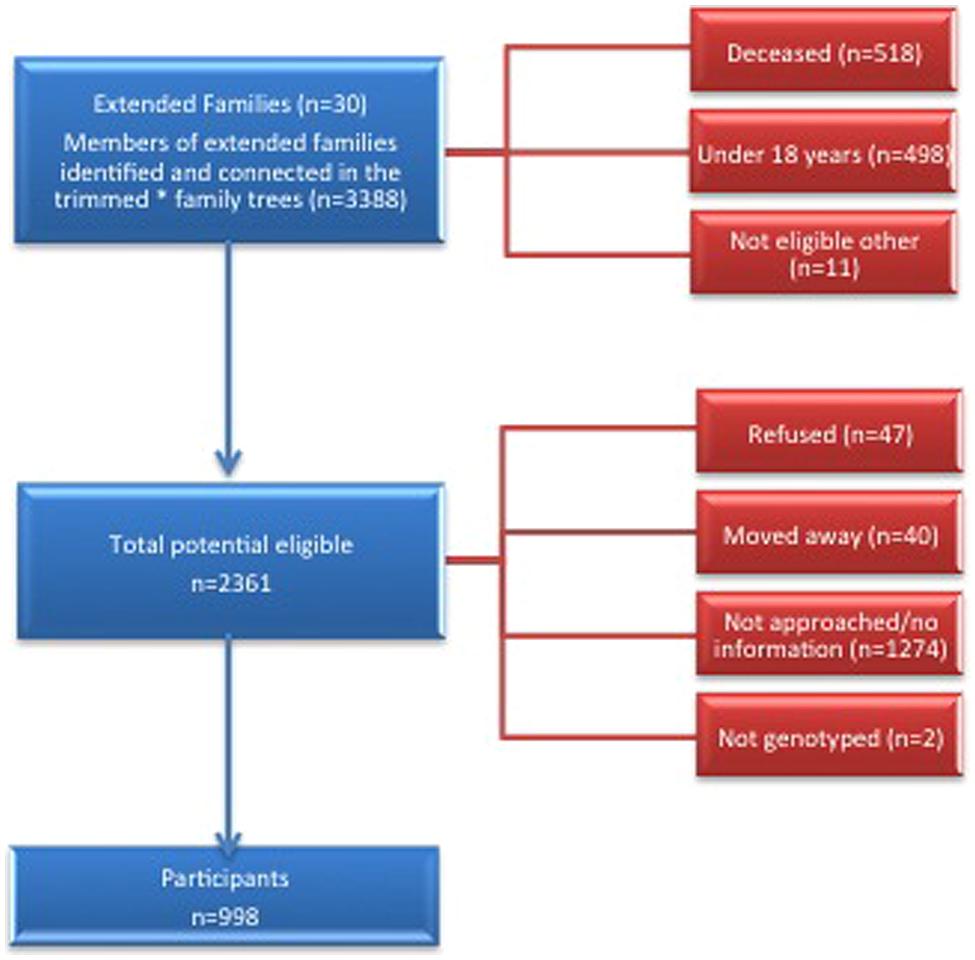
We conducted a cross-sectional study of extended families ascertained through probands with CKD who had at least one sibling with CKD. Potential probands were identified from the ZKP’s previous population-based study of kidney disease, which estimated the prevalence of incipient [15%, (13.1–16.9%)] and overt [4.7% (3.6–5.8%)] albuminuria among 1483 participants (Shah et al., 2003). Eligibility criteria for probands and affected siblings included age ≥18 years, a urine albumin to creatinine ratio (UACR) ≥0.2 in at least two of three urine samples or a reduced serum creatinine-based eGFR, modified for American Indians (Shara et al., 2012) using the Chronic Renal Insufficiency Cohort (CRIC) criteria (Feldman et al., 2003). We invited all first-degree (parents, siblings, and offspring), second-degree (aunts, uncles, nieces, nephews, grandparents, and grandchildren) and third-degree (first cousins, great aunts, great uncles, etc.) relatives of probands and their spouses over 18 years of age to participate. See the consort diagram for details of the recruitment process (Figure 1). We used PEDSYS for data entry, quality control, report generation, and preparation of data files for statistical genetic analysis (Dyke, 1994).
Phenotypic Variables
A random blood sample was drawn from each participant. Blood was drawn for chemistry profile, hemoglobin A1c (HbA1c), and serum creatinine and serum uric acid (SUA). We also measured serum triglycerides, HDL-, LDL-, and total serum cholesterol. We considered a participant to have diabetes if they met ≥1 of the following conditions: (1) history of diabetes, (2) random plasma glucose level ≥200 mg/dL (American Diabetes Association, 2012), (3) HbA1c ≥6.5% (American Diabetes Association, 2012), (4) receiving diabetes medication(s). Three urine samples were collected on separate days from each participant. A participant was considered to have CKD if UACR ≥0.2 in ≥2 of 3 urine samples or if the eGFR was reduced. We also measured blood pressure and calculated body mass index (BMI). Participants were classified as hypertensive if they met ≥1 of the following conditions: (1) history of hypertension; (2) SBP or DBP ≥140 and ≥90 mm Hg, respectively (73–75), or (3) receiving antihypertensive medication(s).
Genotypic Variables
DNA samples were obtained from peripheral blood mononuclear cells. We conducted genome-wide genotyping for 998 participants using the Illumina Human1M-Duo V3.0 BeadChips (Illumina, San Diego, CA, USA) that contain ~1.1 million single nucleotide polymorphism (SNP) assays. Illumina included sample-dependent and independent controls on their chips to test for accuracy of the procedure. Genotype calls were obtained after scanning on the Illumina BeadStation 500GX and analysis using the GenomeStudio software.
Genotyping Quality Control
Specific SNPs were removed from analysis if they had call rates <95% (4,867 SNPs), deviated from Hardy–Weinberg equilibrium (15), were mono-allelic (136,917) or had rare alleles occurring in fewer than five individuals (85,397). SNP genotypes were checked for Mendelian consistency using the program SimWalk (Sobel and Lange, 1996). The estimates of allele frequencies and their SE were obtained by a maximum likelihood estimation method that accounts for pedigree structure using the Sequential Oligogenic Linkage Analysis Routines (SOLAR; http://solar.txbiomedgenetics.org), version 4.3 (Almasy and Blangero, 1998), a program package that is used for association analysis, linkage analysis, transmission disequilibrium tests, and other statistical genetic analyses. Linkage disequilibrium, taking relatedness into account, was also calculated using SOLAR. Missing genotypes were imputed from pedigree data using MERLIN version 1.1.2 (Abecasis et al., 2002).
Reducing Bias in Biological Samples
Reducing bias in UACR
To minimize classification bias, we obtained three urine samples from each participant. The median interval between urine collections was 2 days. We classified albuminuria and hematuria using the mode of three urine samples. UACR was classified as normal (<0.03), incipient (0.03–0.19), or overt (≥0.20). If all three samples were discordant, we used the median value. Urine albumin was measured using nephelometry (Liu et al., 2011; Nicol et al., 2011).
Reducing bias in eGFR
We used the four-variable Modification of Diet in Renal Disease (MDRD) Study equation, modified for use in American Indians to estimate GFR based on a single serum sample (Shah et al., 2003; Scavini et al., 2007). Limitations of this equation include limited validation data in American Indians and the lack of calibration of the serum creatinine assay. Serum creatinine levels are influenced by muscle mass. We recognize that the CKD-EPI equation may out-perform the MDRD equation among people with near normal kidney function (Levey et al., 2009). Unfortunately, however, there are few data on the performance of the CKD-EPI equation among American Indians. We categorized eGFR using the National Kidney Foundation’s (2002) Kidney Disease Outcomes Quality Initiative (KDOQI; K/DOQI guidelines) and the CRIC age-specific criteria (Feldman et al., 2003). Hyperuricemia was defined as SUA >6 mg/dl in women and SUA >7 mg/dl in men.
Genome-wide association analysis (GWA analysis)
Measured genotype analyses were performed using SOLAR version 4.3 (Almasy and Blangero, 1998). The number of SNPs included in the GWA analysis was 884,161. All phenotypes were transformed by inverse normalization to meet assumptions of normality. We obtained residuals using linear regression models adjusted for age, sex, their interactions and higher order terms. Our subjects were ascertained for CKD. To adjust for ascertainment bias, we took a conservative approach by computing likelihood for pedigrees incorporating the CKD phenotype as an additional covariate for kidney function phenotypes (eGFR, UACR, and serum creatinine; Farook et al., 2012). Additional covariates included hypertension and diabetic status. Individuals excluded from analysis included those taking diabetes medications for analysis of HbA1c, antihypertensive medications for analysis of SBP and DBP, and statins for analysis of lipid traits (triglycerides, total-, HDL-, and LDL-cholesterol).
Each SNP genotype was converted in MERLIN version 1.1.2 (Abecasis et al., 2002) to a covariate measure equal to 0, 1, or 2 copies of the minor allele (or, for missing genotypes, the weighted covariate based on imputation). These covariates were included in the variance-components mixed models for measured genotype analyses (Boerwinkle et al., 1986) versus null models that incorporated the random effect of kinship and fixed effects such as age, sex, their interaction and higher order terms. For the initial GWA screen, we tested each SNP covariate independently as a one degree of freedom likelihood ratio test. An adjusted alpha value for significance, using a Moskvina–Schmidt calculation (Moskvina and Schmidt, 2008) based on the effective number of independent SNPs given LD (n = 323,965 SNPs) in the Zuni families, provided the adjusted genome-wide significant and genome-wide suggestive thresholds of 1.58 × 10-7 and 1.58 × 10-6, respectively. We performed the quantitative transmission disequilibrium test (QTDT) to test for population stratification (Havill et al., 2005). The power calculations were implemented in SOLAR 4.3.
Results
Study Participants
The descriptive characteristics of the GKDZI participants for the variables included in the GWAS are presented in Table 1. The mean age was 37.1 ± 13.6 years and 52% were males. Nearly 19% of the participants were diabetic, 34% were hypertensive, 30% had hyperuricemia, and 24% had CKD at the time of the GKDZI clinic exam. The GWAS included 998 individuals with available DNA samples. Genotype distributions of all significantly associated SNPs conformed to the Hardy–Weinberg equilibrium. Population stratification was not significant as per the QTDT and therefore did not confound our associations.
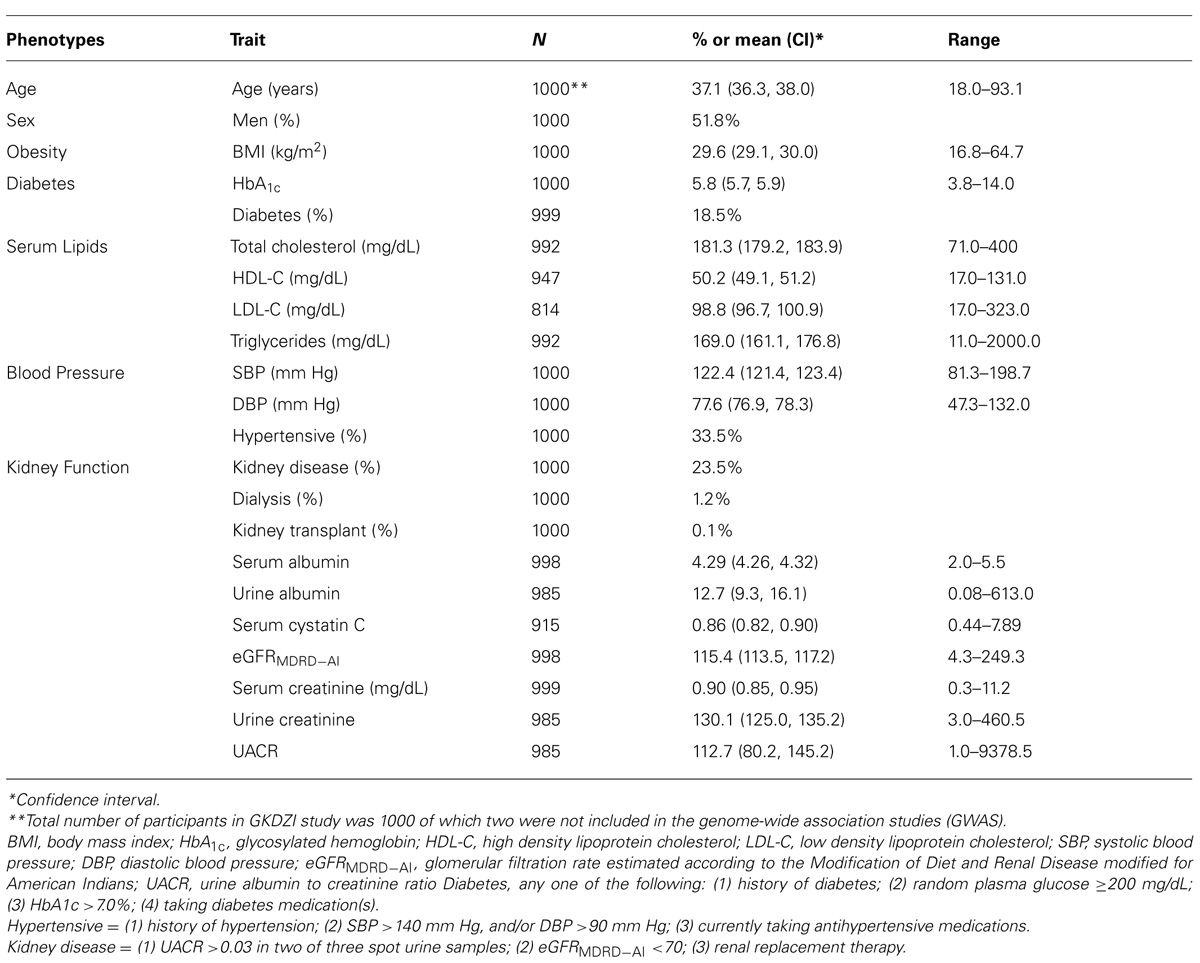
TABLE 1. Characteristics of traits related to kidney disease, diabetes, and CVD in GKDZI participants.
Genome-Wide Association Analysis
Kidney traits
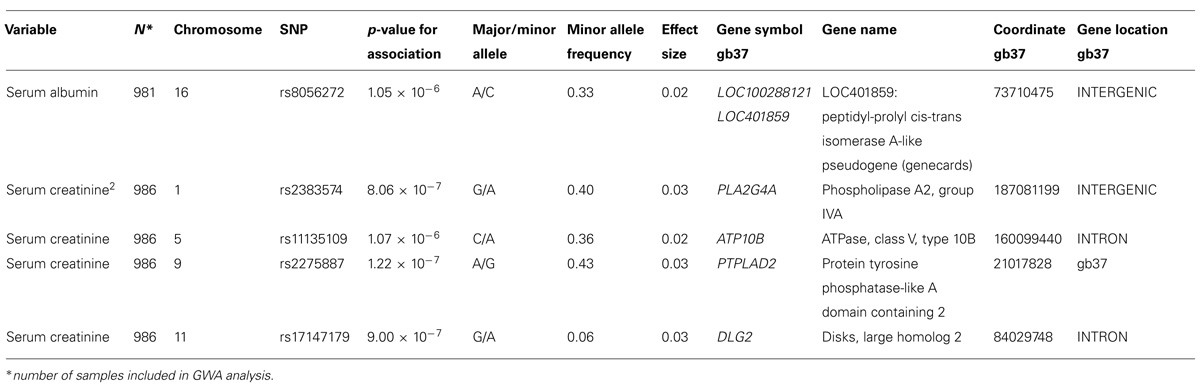
A genome-wide significant association was identified for serum creatinine (Table 2). An intronic variant (minor allele G) in the protein tyrosine phosphatase-like A domain containing 2 (PTPLAD2) gene on Chromosome 9 was significantly associated (p = 1.2 × 10-7) with increased serum creatinine concentrations, with an effect size (residual phenotypic variance that is contributed by the minor allele of the SNP) of 3.0%. Evidence of suggestive association was found for serum creatinine with phospholipase A2 group 4a (PLA2G4A), ATPase, Class V, type 10B (ATP10B), and disks, large homolog 2 (Drosophila; DLG2; Table 2). However, we did not find any significant or suggestive associations for eGFR or the urine to albumin creatinine ratio (UACR). In addition, we found significant associations of SUA with several SNPs in solute carrier family 2, member 9 (SLC2A9) gene (rs6449213, rs938555, rs16890979, rs12499857, rs734553, rs6832439, rs13125646, rs13131257, rs13145758, and rs9998811; Figure 2). Minor alleles of most of these SNPs (shown in detail in Table 3) were associated with lower SUA levels.
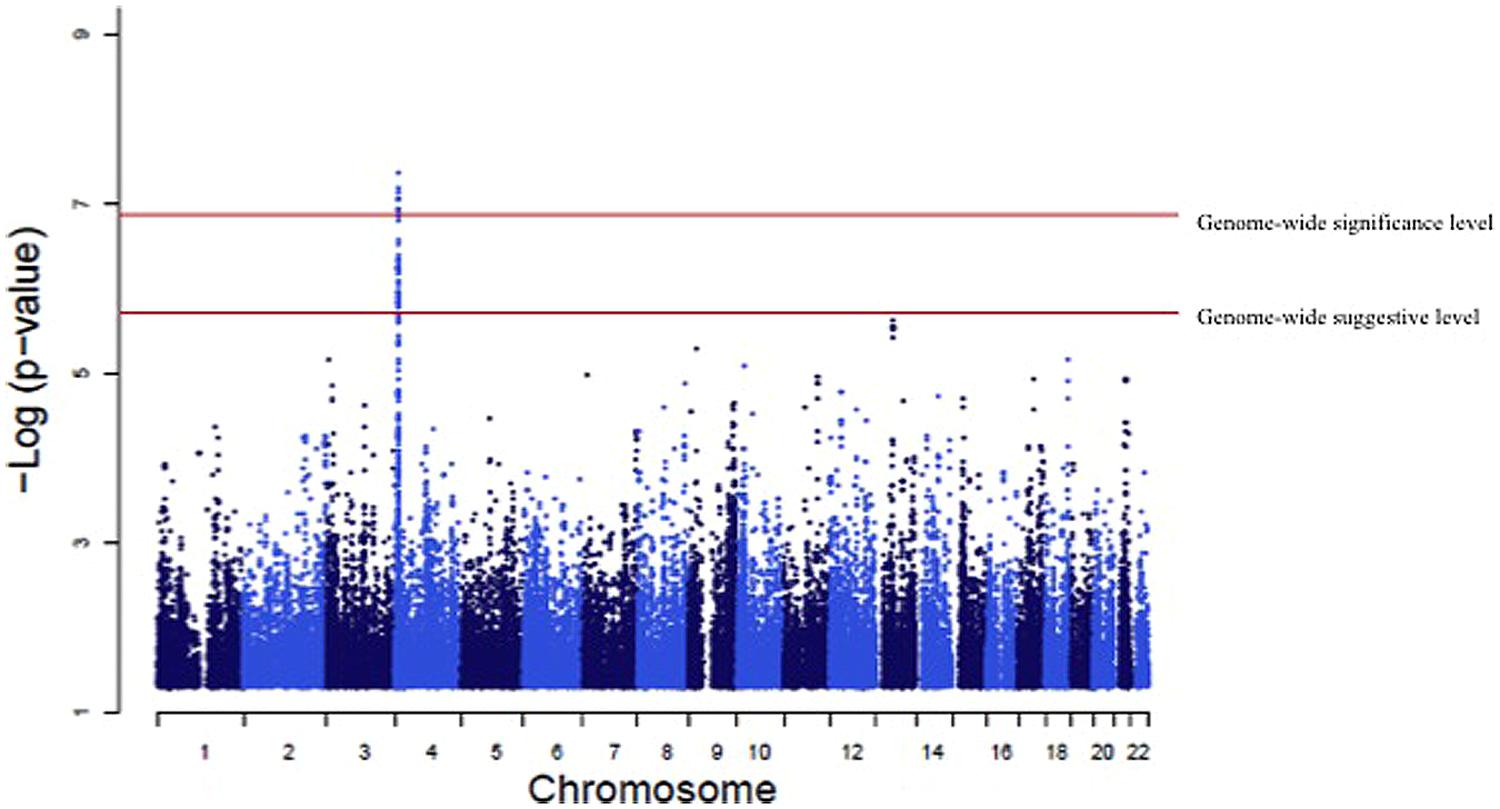
FIGURE 2. Genome-wide association of serum uric acid (SUA) levels. Manhattan Plot for the results of the genome wide association analysis with SUA levels. The genome-wide distribution of p-values for each of the SUA associated single nucleotide polymorphisms (SNPs) is shown. The adjusted genome-wide significant and genome-wide suggestive thresholds were set at 1.58 × 10-7 and 1.58 × 10-6, respectively. The x axis represents the genomic position of SLC2A9 SNPs; the y axis shows the -log10 p-value. There were significant associations with 10 SLC2A9 SNPs (rs6449213, rs938555, rs16890979, rs12499857, rs734553, rs6832439, rs13125646, rs13131257, rs13145758, and rs9998811).
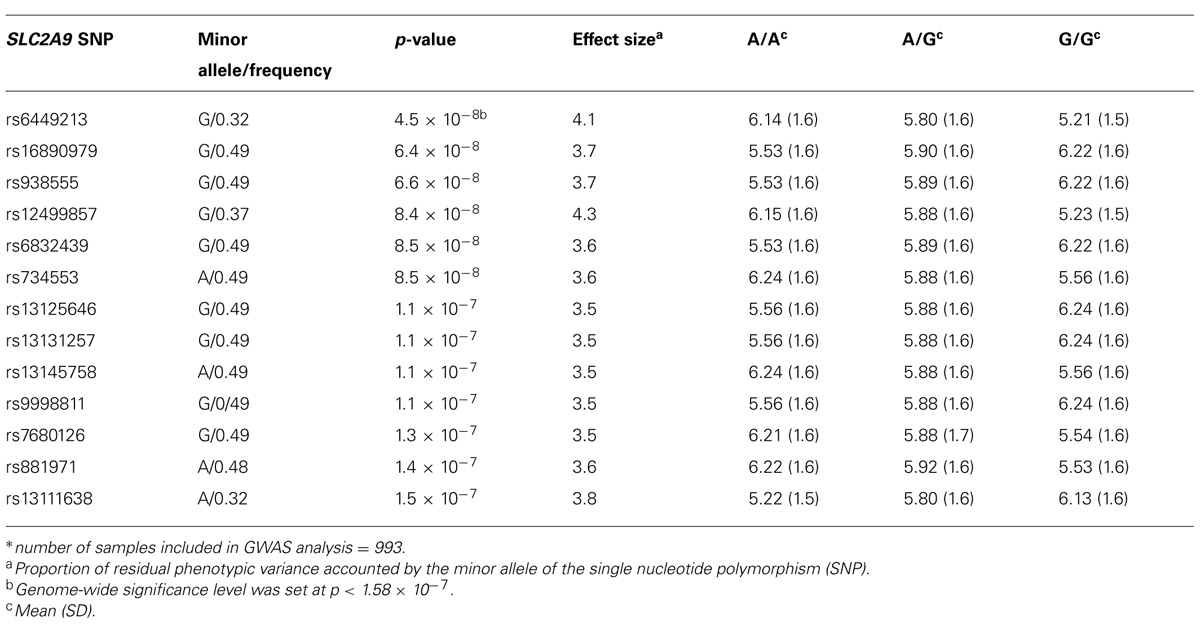
TABLE 3. Significant associations and genotype-specific means of serum uric acid (SUA) levels* (mg/dl).
Lipid phenotypes
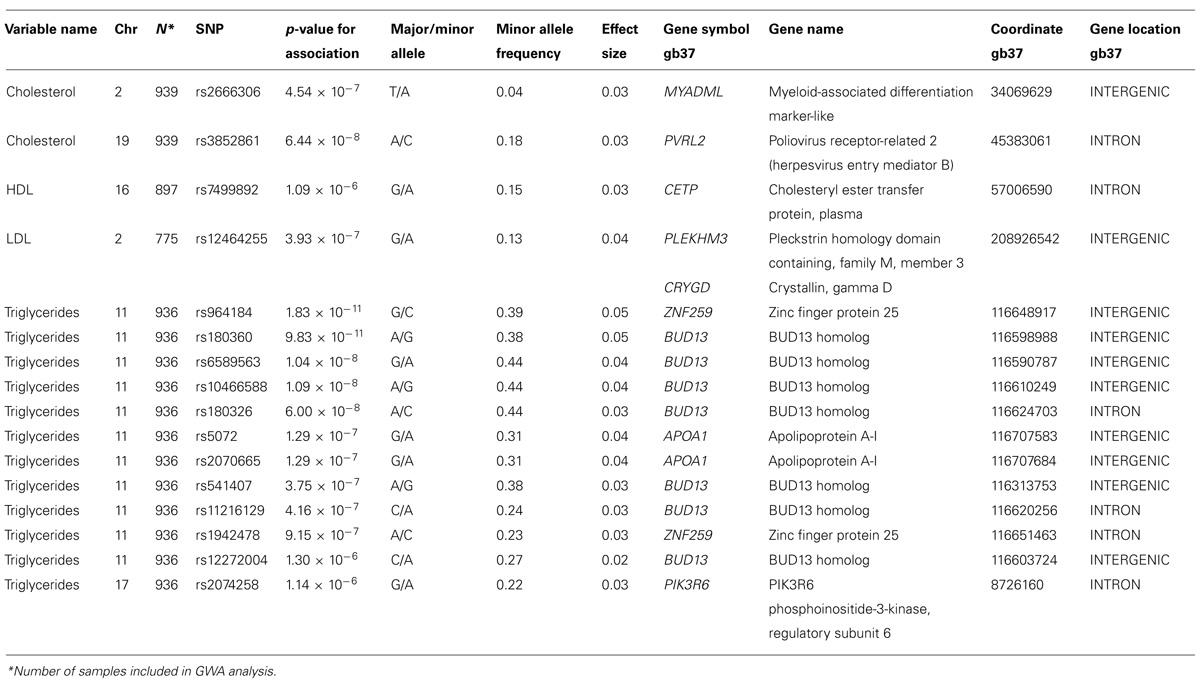
We analyzed the levels of four lipid phenotypes, e.g., triglycerides, high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, and total cholesterol in the GWAS. The strongest association was found for triglycerides for SNPs near the zinc finger protein 259 (ZNF259), apolipoprotein A-1 (APOA1), and BUD13 homolog (BUD13) genes on Chromosome 11 (Table 4). Triglycerides were significantly associated (p = 1.83 × 10-11 to 6.00 × 10-8) with four SNPs near genes and one intronic SNP in BUD13 whose mean effect size ranged from 3.2 to 4.4% (Table 4). All minor alleles of SNPs (effect sizes ranging between 2.3 and 4.8%) except rs180360 (effect size = 4.9%) were associated with increased triglycerides. Two of the SNPs near BUD13 (rs10466588, rs6589563) were in complete LD. Two associated SNPs near the APOA1 gene were also in complete LD. The mean effect size for the two SNPs was 3.6%. Figure 3 provides a Manhattan Plot for the results of the genome-wide association analysis with triglyceride levels. The minor allele (C) of an intronic SNP (rs3852861) in the poliovirus receptor-related 2 (PVRL2) gene on Chromosome 19 was significantly associated (p = 6.44 × 10-8) with increased total cholesterol. The mean effect size was 3.4% (Table 4). We also found evidence of suggestive associations for triglycerides, HDL-, LDL-, and total cholesterol on Chromosomes 17, 16, 2, and 2, respectively.
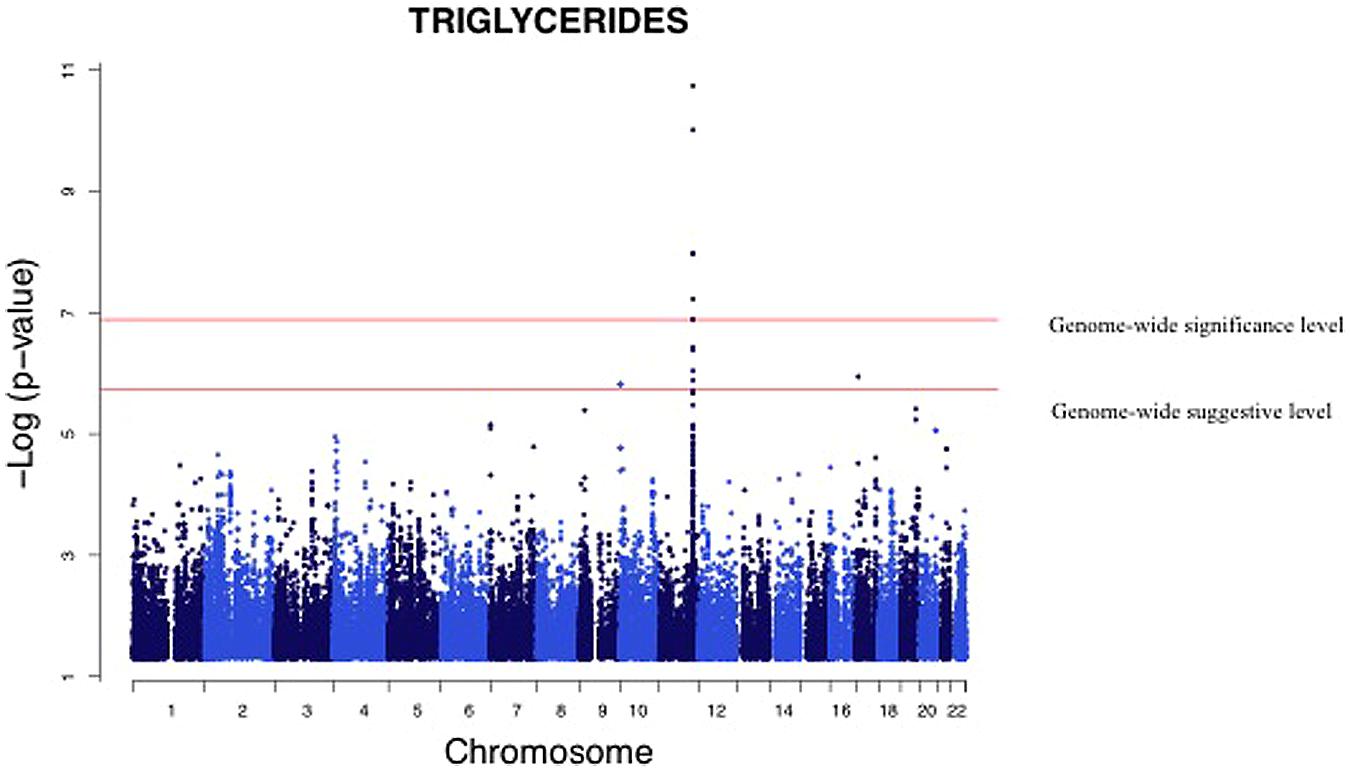
FIGURE 3. Genome-wide association analysis of serum triglycerides. Manhattan Plot for the results of the genome wide association analysis with serum triglyceride levels. The genome-wide distribution of p-values for each of the serum triglyceride associated SNPs is shown. The adjusted genome-wide significant and genome-wide suggestive thresholds were set at 1.58 × 10-7 and 1.58 × 10-6, respectively. The x axis represents the genomic position of the triglyceride associated SNPs; the y axis shows the -log10 p-value. The strongest association was for SNPs near the zinc finger protein 259 (ZNF259), apolipoprotein A-1 (APOA1), and BUD 13 homolog (BUD13) genes on Chromosome 11.
Blood pressure
Although GWA analysis of systolic (SBP) and diastolic blood pressure (DBP) yielded no significant associations, several exhibited evidence of suggestive associations (Data not shown). There were associations with two SNPs near the olfactomedin-like 2B (OLFML2B) gene on Chromosome 1 that approached significance (p = 9.68 × 10-7). The average effect size was 3.6% and was associated with increases in SBP. There was one intronic SNP in nuclear factor I/A (NFIA) on Chromosome 1 that showed evidence of suggestive association (p = 1.23 × 10-6) with decreased DBP.
Discussion
The most significant findings of the first GWAS in Zuni Indians were the strong associations of PTPLAD2, SLC2A9, PVRL2, and BUD13 with serum levels of creatinine, uric acid, total cholesterol and triglycerides, respectively. Although, GWA analysis of BMI, SBP, DBP, and HbA1c provided no significant associations, some traits approached significance and several exhibited evidence of suggestive association.
We identified a novel significant association of an intronic variant in the PTPLAD2 gene on Chromosome 9 with an increased serum creatinine concentration. This gene is part of very long chain fatty acid dehydratase HACD family and has a key role in the dehydration step of the very long chain fatty acid metabolism (Ikeda et al., 2008). Also implicated in tumor suppression (Zuni Pueblo QuickFacts from the US Census Bureau, 2014), this gene has not been previously reported to be associated with serum creatinine. We also found association, albeit suggestive, between serum creatinine and PLA2G4A, ATP10B, and DLG2 SNPs. Their role in kidney function is not clear, except that in the kidney, cytosolic phospholipase A2 seems to play a role in GFR, vascular tone and water transport (Downey et al., 2001).
The strong association between SUA levels and SLC2A9 SNPs is a replication and confirmation of these associations in several populations. Most of these studies were conducted in European populations (Doring et al., 2008; Li et al., 2008; Vitart et al., 2008) as well as in Asian (Tabara et al., 2010; Guan et al., 2011), African American (Dehghan et al., 2008; Rule et al., 2011; Tin et al., 2011) and Mexican American populations (Voruganti et al., 2013). The effect sizes or the proportion of residual variance in a phenotype that is explained by the minor allele of the SNP ranged between 3.5 and 4.3% in this study which is similar to what has been reported by these studies. Similar results were found in a candidate gene study in American Indians where only seven SLC2A9 SNPs were genotyped (Voruganti et al., 2014). In addition, Caulfield et al. (2008) not only confirmed this association in six different cohorts of European ancestry but showed that SLC2A9 can exchange glucose for urate in the process of secretion of urate into the urine in functional studies.
Hyperuricemia is associated with hypertension (Johnson et al., 2005), CKD (Kim et al., 2014), insulin resistance (Cirillo et al., 2006), and cardiovascular disease (Puddu et al., 2012), although causality has not been established. SLC2A9 was originally identified as glucose transporter 9 (GLUT9). However, it facilitates electrogenic transport of both hexoses and uric acid in the proximal tubule (Witkowska et al., 2012). There are two forms, SLC2A9a and SLC2A9b, which are expressed in the basolateral and apical membranes of the proximal tubule, respectively. Their amino acid sequences are identical, except that SLC2A9b has a shorter and modified N-terminus. Both forms are active in urate transport in the proximal tubule (Kimura et al., 2014). Kidney function and SUA are interrelated (Kang et al., 2002). The anti-hypertensive drug losartan lowers SUA (Burnier et al., 1996; Wurzner et al., 2001; Hamada et al., 2002) and confers long-term protection of kidney function (Brenner et al., 2001). A recent GWAS conducted in Mexican Americans, reported a nominal association between UACR and SLC2A9 SNPs (Voruganti et al., 2013). We found nominal associations between SLC2A9 SNPs and kidney function phenotypes (Data not shown). Our results related to kidney function phenotypes replicate results of studies conducted in Mexican Americans and other American Indian tribes (Voruganti et al., 2013, 2014). However, our study is different from others as the participating individuals in our study were ascertained for CKD.
Total serum cholesterol was significantly associated with an intronic SNP (rs3852861) in the PVRL2 gene on Chromosome 19. PVRL2 is located 17 kb downstream from the apolipoprotein E (APOE) gene and has also been associated with severity of multiple sclerosis (Evangelou et al., 1999; Schmidt et al., 2002), late-onset Alzheimer’s disease (Corder et al., 1993), and peripheral T-cell lymphomas (Liestol et al., 2000). A study of Caucasian patients with coronary artery disease found a relationship between homozygosity of the A allele in a polymorphism of the PVRL2/PRR2 gene and premature cardiovascular disease (Freitas et al., 2002). The authors suggested that this finding could be related to viral association or linkage disequilibrium between PRR2 and nearby (17 kb centromeric) apolipoprotein E (APOE; Willer et al., 2008). This gene was also associated with LDL cholesterol in a Caucasian population although the chromosomal region is not the same (Talmud et al., 2009). We also found evidence of suggestive association of cholesterol esterase transfer protein (CETP) with HDL cholesterol which is a replication of several studies (Feitosa et al., 2014; Singaraja et al., 2014; Walia et al., 2014).
The association of triglycerides with four SNPs near and one SNP in the BUD13/ZNF259 region replicates results observed in a Mexican cohort (Weissglas-Volkov et al., 2010), a meta-analysis of individuals of European descent (Schunkert et al., 2011), a Finnish cohort (Kristiansson et al., 2012) and Asian Indians (Braun et al., 2012). The minor allele frequency for rs964184 is higher among Zuni Indians (39%) than among Mexicans (27%), Asian Indians (15%) or Whites (12%). The ZNF259/BUD13 associations with triglyceride levels have been reported in GWAS in the Framingham Study (Suchindran et al., 2010), which also showed an association with lipoprotein-associated phospholipase A2 (Lp-PLA2), a risk factor and possible therapeutic locus for CVD. Similarly, ZNF259 was significantly associated with Lp-PLA2 activity in a meta-analysis of five population-based studies (Grallert et al., 2012). ZNF259 codes for ZPR1, a zinc (as well as some other metals) binding protein, which may play a role in signal transfer from cell cytoplasm to the nucleus and cell proliferation (Galcheva-Gargova et al., 1998). This region of the ZNF259/BUD13, APOA1/C3/A4/A5 genes has also been associated with coronary artery disease (Waterworth et al., 2010).
In addition, we also found some novel, albeit suggestive, associations of various SNPs with cardiometabolic phenotypes such as SNPs near OLFML2B and NOS1AP with blood pressure phenotypes. Although these SNPs have not been associated with blood pressure before, they have been associated with the Short QT-Syndrome among individuals from the UK and North America (Eijgelsheim et al., 2009; Nolte et al., 2009). Similarly, variants in the NFIA gene, which encodes the nuclear factor 1 family of transcription factors have been associated with QRS duration in individuals of European descent (Sotoodehnia et al., 2010).
Strengths and Limitations
The strengths of our study include a dense GWAS using an Illumina chip that was state of the art at the time the study was conducted. Conducting the study in extended families from a relatively endogamous population increased our statistical power and minimized potential population stratification. Furthermore, we utilized state of the art programs for conducting genetic analyses (SOLAR, MERLIN). All study staff members working in Zuni were Zuni, which enhanced community acceptance of performing genetic studies in the Pueblo. Our study also had some significant limitations. We did not perform direct measurements of GFR. The serum creatinine assay was performed in a clinical laboratory and not standardized. The GFR estimating equation has not been validated in Zuni Indians. Kidney biopsies were not performed and may have led to misclassification. We did not account for all possible genetic X environmental interactions. In addition, we did not have positive controls. However, several of our significant loci have been previously identified in individuals without kidney disease.
Conclusion
The results of the GKDZI study support our hypothesis that genetic factors significantly influence susceptibility to CKD and related cardiometabolic phenotypes among Zuni Indians.
Conflict of Interest Statement
Dr. Philip G. Zager is an employee of both the University of New Mexico and Dialysis Clinic Inc. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the contribution and support of the Zuni Governors and Tribal Council and sincerely thank the Zuni people for welcoming us in their homes. We thank Dr. Andrew Narva and Dr. James Whitfield and the Strong Heart Study investigators for their support. Power calculations were conducted by Dr. Jack W. Kent, Jr. Excellent technical support was provided by Maria Del Pillar Villegas, Vanessa Ayala, Amuche Ezeilo, Leslie Firkins, and Serena Cumber. Finally, the authors thank the National Institute of Diabetes and Digestive and Kidney Diseases (Dr. Rebekah Rasooly) and the Advisory Committee for their guidance.
Support
This study was supported in part by grants DK066660-03 and DK57300-05 from the National Institutes of Health (NIH); 5M01RR00997 from the University of New Mexico Clinical Research Center; and P30 ES-012072 from the National Institute of Environmental Health Sciences, and Dialysis Clinic Inc. At the Texas Biomedical Research Institute, these studies were conducted in facilities constructed with support from the Research Facilities Improvement Program grant C06 RR013556 from the National Center for Research Resources, NIH. The AT&T Genomics Computing Center supercomputing facilities used for statistical genetic analyses were supported in part by a gift from the SBC Foundation. The statistical genetics computer package, SOLAR, is supported by grant R01 MH059490 from the National Institute of Mental Health.
Financial Disclosures
Philip G. Zager is an employee of both the University of New Mexico Health Sciences Center and Dialysis Clinic Inc. (DCI). Susan S. Paine is and Arlene Bobelu was a DCI employee. The remaining authors declare that they have no relevant financial interests. NIDDK appointed an independent Data Safety Monitoring Board, which had input into study design. Dialysis Clinic Inc., other than Philip G. Zager, Susan S. Paine, and Arlene Bobelu had no input into study design.
References
Abecasis, G. R., Cherny, S. S., Cookson, W. O., and Cardon, L. R. (2002). Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101. doi: 10.1038/ng786
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Almasy, L., and Blangero, J. (1998). Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211. doi: 10.1086/301844
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
American Diabetes Association. (2012). Standards of medical care in diabetes– (2012). Diabetes Care 35, S11–S63. doi: 10.2337/dc12-s011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bian, L., Traurig, M., Hanson, R. L., Marinelarena, A., Kobes, S., Muller, Y. L.,et al. (2013). MAP2K3 is associated with body mass index in American Indians and Caucasians and may mediate hypothalamic inflammation. Hum. Mol. Genet. 22, 4438–4449. doi: 10.1093/hmg/ddt291
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bidulescu, A., Choudhry, S., Musani, S. K., Buxbaum, S. G., Liu, J., Rotimi, C. N.,et al. (2014). Associations of adiponectin with individual European ancestry in African Americans: the Jackson heart study. Front. Genet. 5:22. doi: 10.3389/fgene.2014.00022
Boerwinkle, E., Chakraborty, R., and Sing, C. F. (1986). The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann. Hum. Genet. 50, 181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Braun, T. R., Been, L. F., Singhal, A., Worsham, J., Ralhan, S., Wander, G. S.,et al. (2012). A replication study of GWAS-derived lipid genes in Asian Indians: the chromosomal region 11q23.3 harbors loci contributing to triglycerides. PLoS ONE 7:e37056. doi: 10.1371/journal.pone.0037056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenner, B. M., Cooper, M. E., de Zeeuw, D., Keane, W. F., Mitch, W. E., Remuzzi, G.,et al. (2001). Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med. 345, 861–869. doi: 10.1056/NEJMoa011161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burnier, M., Roch-Ramel, F., and Brunner, H. R. (1996). Renal effects of angiotensin II receptor blockade in normotensive subjects. Kidney Int. 49, 1787–1790. doi: 10.1038/ki.1996.268
Caulfield, M. J., Munroe, P. B., O’Neill, D., Witkowska, K., Charchar, F. J., Doblado, M.,et al. (2008). SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 5:e197. doi: 10.1371/journal.pmed.0050197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cirillo, P., Sato, W., Reungjui, S., Heinig, M., Gersch, M., Sautin, Y.,et al. (2006). Uric acid, the metabolic syndrome, and renal disease. J. Am. Soc. Nephrol. 17, S165–S168. doi: 10.1681/ASN.2006080909
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corder, E. H., Saunders, A. M., Strittmatter, W. J., Schmechel, D. E., Gaskell, P. C., Small, G. W.,et al. (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. doi: 10.1126/science.8346443
Dehghan, A., Kottgen, A., Yang, Q., Hwang, S. J., Kao, W. L., Rivadeneira, F.,et al. (2008). Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet 372, 1953–1961. doi: 10.1016/S0140-6736(08)61343-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doring, A., Gieger, C., Mehta, D., Gohlke, H., Prokisch, H., Coassin, S.,et al. (2008). SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40, 430–436. doi: 10.1038/ng.107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Downey, P., Sapirstein, A., O’Leary, E., Sun, T. X., Brown, D., and Bonventre, J. V. (2001). Renal concentrating defect in mice lacking group IV cytosolic phospholipase A(2). Am. J. Physiol. Renal Physiol. 280, F607–F618.
Dyke, B. (1994). PEDSYS, a Pedigree Data Management System User’s Manual. Population Genetics Laboratory Technical Report, No. 2, 2nd Edn. San Antonio, TX: Southwest Foundation for Biomedical Research, 226.
Edwards, K. L., Hutter, C. M., Wan, J. Y., Kim, H., and Monks, S. A. (2008). Genome-wide linkage scan for the metabolic syndrome: the GENNID study. Obesity (Silver Spring) 16, 1596–1601. doi: 10.1038/oby.2008.236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eijgelsheim, M., Newton-Cheh, C., Aarnoudse, A. L., Van Noord, C., Wittleman, J. C., Hofman, A.,et al. (2009). Genetic variation in NOS1AP is associated with sudden cardiac death: evidence from the Rotterdam study. Hum. Mol. Genet. 18, 4213–4218. doi: 10.1093/hmg/ddp356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Evangelou, N., Jackson, M., Beeson, D., and Palace, J. (1999). Association of the APOE epsilon4 allele with disease activity in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 67, 203–205. doi: 10.1136/jnnp.67.2.203
Farook, V. S., Coletta, D. K., Puppala, S., Schneider, J., Chittoor, G., Hu, S. L.,et al. (2013). Linkage of type 2 diabetes on chromosome 9p24 in Mexican Americans: additional evidence from the Veterans Administration Genetic Epidemiology Study (VAGES). Hum. Hered. 76, 36–46. doi: 10.1159/000354849
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farook, V. S., Puppala, S., Schneider, J., Fowler, S. P., Chittoor, G., Dyer, T. D.,et al. (2012). Metabolic syndrome is linked to chromosome 7q21 and associated with genetic variants in CD36 and GNAT3 in Mexican Americans. Obesity (Silver Spring) 20, 2083–2092. doi: 10.1038/oby.2012.74
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feitosa, M. F., Wojczynski, M. K., Straka, R., Kammerer, C. M., Lee, J. H., Kraja, A. T.,et al. (2014). Genetic analysis of long-lived families reveals novel variants influencing high density-lipoprotein cholesterol. Front. Genet. 5:159. doi: 10.3389/fgene.2014.00159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feldman, H. I., Appel, L. J., Chertow, G. M., Cifelli, D., Cizman, B., Daugirdas, J.,et al. (2003). The Chronic Renal Insufficiency Cohort (CRIC) study: design and methods. J. Am. Soc. Nephrol. 14, S148–S153. doi: 10.1097/01.ASN.0000070149.78399.CE
Franceschini, N., Haack, K., Goring, H. H., Voruganti, V. S., Laston, S., Almasy, L.,et al. (2013). Epidemiology and genetic determinants of progressive deterioration of glycaemia in American Indians: the strong heart family study. Diabetologia 56, 2194–2202. doi: 10.1007/s00125-013-2988-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Freitas, E. M., Phan, T. C., Herbison, C. E., Christiansen, F. T., Taylor, R. R., and Van Bockxmeer, F. M. (2002). The poliovirus receptor related 2 (PRR2) and apolipoprotein E genes and coronary heart disease. J. Cardiovasc. Risk 9, 59–65. doi: 10.1097/00043798-200202000-00009
Galcheva-Gargova, Z., Gangwani, L., Konstantinov, K. N., Mikrut, M., Theroux, S. J., Enoch, T.,et al. (1998). The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell 9, 2963–2971. doi: 10.1091/mbc.9.10.2963
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grallert, H., Dupuis, J., Bis, J. C., Dehghan, A., Barbalic, M., Baumert, J.,et al. (2012). Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: meta-analysis of genome-wide association studies from five community-based studies. Eur. Heart J. 33, 238–251. doi: 10.1093/eurheartj/ehr372
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guan, M., Zhou, D., Ma, W., Chen, Y., Zhang, J., and Zou, H. (2011). Association of an intronic SNP of SLC2A9 gene with serum uric acid levels in the Chinese male Han population by high-resolution melting method. Clin. Rheumatol. 30, 29–35. doi: 10.1007/s10067-010-1597-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hamada, T., Hisatome, I., Kinugasa, Y., Matsubara, K., Shimizu, H., Tanaka, H.,et al. (2002). Effect of the angiotensin II receptor antagonist losartan on uric acid and oxypurine metabolism in healthy subjects. Intern. Med. 41, 793–797. doi: 10.2169/internalmedicine.41.793
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanson, R. L., Guo, T., Muller, Y. L., Fleming, J., Knowler, W. C., Kobes, S.,et al. (2013). Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes 62, 2984–2991. doi: 10.2337/db12-1767
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanson, R. L., Muller, Y. L., Kobes, S., Guo, T., Bian, L., Ossowski, V.,et al. (2014). A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes 3, 369–376. doi: 10.2337/db13-0416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Havill, L. M., Dyer, T. D., Richardson, D. K., Mahaney, M. C., and Blangero, J. (2005). The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 6:S91. doi: 10.1186/1471-2156-6-S1-S91
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hwang, S. J., Yang, Q., Meigs, J. B., Pearce, E. N., and Fox, C. S. (2007). A genome-wide association for kidney function and endocrine-related traits in the NHLBI’s framingham heart study. BMC Med. Genet. 8:S10. doi: 10.1186/1471-2350-8-S1-S10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ikeda, M., Kanao, Y., Yamanaka, M., Sakuraba, H., Mizutani, Y., Igarashi, Y.,et al. (2008). Characterization of four mammalian 3-hydroxyacyl-CoA dehydratases involved in very long-chain fatty acid synthesis. FEBS Lett. 582, 2435–2440. doi: 10.1016/j.febslet.2008.06.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, R. J., Feig, D. I., Herrera-Acosta, J., and Kang, D. H. (2005). Resurrection of uric acid as a causal risk factor in essential hypertension. Hypertension 45, 18–20. doi: 10.1161/01.HYP.0000150785.39055.e8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, W. D., Kroon, J. J., Greenway, F. L., Bouchard, C., Ryan, D., and Katzmarzyk, P. T. (2009). Prevalence of risk factors for metabolic syndrome in adolescents: National Health and Nutrition Examination Survey (NHANES), 2001-2006. Arch. Pediatr. Adolesc. Med. 163, 371–377. doi: 10.1001/archpediatrics.2009.3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kang, D. H., Nakagawa, T., Feng, L., Watanabe, S., Han, L., Mazzali, M.,et al. (2002). A role for uric acid in the progression of renal disease. J. Am. Soc. Nephrol. 13, 2888–2897. doi: 10.1097/01.ASN.0000034910.58454.FD
Kim, W. J., Kim, S. S., Bae, M. J., Yi, S., Jeon, Y. K., Kim, B. H.,et al. (2014). High-normal serum uric acid predicts the development of chronic kidney disease in patients with type 2 diabetes mellitus and preserved kidney function. J. Diabetes Complications 28, 130–134. doi: 10.1016/j.jdiacomp.2013.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kimura, T., Takahashi, M., Yan, K., and Sakurai, H. (2014). Expression of SLC2A9 isoforms in the kidney and their localization in polarized epithelial cells. PLoS ONE 9:e84996. doi: 10.1371/journal.pone.0084996
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kristiansson, K., Perola, M., Tikkanen, E., Kettunen, J., Surakka, I., Havulinna, A. S.,et al. (2012). Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ. Cardiovasc. Genet. 5, 242–249. doi: 10.1161/CIRCGENETICS.111.961482
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laird, N. M., and Lange, C. (2008). Family-based methods for linkage and association analysis. Adv. Genet. 60, 219–252. doi: 10.1016/S0065-2660(07)00410-5
Levey, A. S., Stevens, L. A., Schmid, C. H., Zhang, Y. L., Castro, A. F. III, Feldman, H. I.,et al. (2009). A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 150, 604–612. doi: 10.7326/0003-4819-150-9-200905050-00006
Li, H., Pascal, V., Martin, M. P., Carrington, M., and Anderson, S. K. (2008). Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 4:e1000254. doi: 10.1371/journal.pgen.1000254
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liestol, K., Kvittingen, E. A., Rootwelt, H., Dunlop, O., Goplen, A. K., Pedersen, J. C.,et al. (2000). Association between apolipoprotein E genotypes and cancer risk in patients with acquired immunodeficiency syndrome. Cancer Detect. Prev. 24, 496–499.
Liu, R., Li, G., Cui, X. F., Zhang, D. L., Yang, Q. H., Mu, X. Y.,et al. (2011). Methodological evaluation and comparison of five urinary albumin measurements. J. Clin. Lab. Anal. 25, 324–329. doi: 10.1002/jcla.20477
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
MacCluer, J. W., Scavini, M., Shah, V. O., Cole, S. A., Laston, S. L., Voruganti, V. S.,et al. (2010). Heritability of measures of kidney disease among Zuni Indians: the Zuni Kidney Project. Am. J. Kidney Dis. 56, 289–302. doi: 10.1053/j.ajkd.2010.03.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moskvina, V., and Schmidt, K. M. (2008). On multiple-testing correction in genome-wide association studies. Genet. Epidemiol. 32, 567–573. doi: 10.1002/gepi.20331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
National Kidney Foundation. (2002). K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis. 39, S1–S266.
Nicol, J. L., Hoy, W. E., Su, Q., Atkins, R. C., and Polkinghome, K. R. (2011). Reproducibility of urinary albumin assays by immunonephelometry after long-term storage at -70 degrees C. Am. J. Kidney Dis. 58, 685–687. doi: 10.1053/j.ajkd.2011.06.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nolte, I. M., Wallace, C., Newhouse, S. J., Waggott, D., Fu, J., Soranzo, N.,et al. (2009). Common genetic variation near the Phospholamban gene is associated with cardiac repolarisation: meta-analysis of three genome-wide association studies. PLoS ONE 4:e6138. doi: 10.1371/journal.pone.0006138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parsa, A., Fuchsberger, C., Kottgen, A., O’Seaghdha, C. M., Pattaro, C., de Andrade, M.,et al. (2013). Common variants in Mendelian kidney disease genes and their association with renal function. J. Am. Soc. Nephrol. 24, 2105–2117. doi: 10.1681/ASN.2012100983
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puddu, P., Puddu, G. M., Cravero, E., Vizioli, L., and Muscari, A. (2012). Relationships among hyperuricemia, endothelial dysfunction and cardiovascular disease: molecular mechanisms and clinical implications. J. Cardiol. 59, 235–242. doi: 10.1016/j.jjcc.2012.01.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rule, A. D., de Andrade, M., Matsumoto, M., Mosley, T. H., Kardia, S., and Turner, S. T. (2011). Association between SLC2A9 transporter gene variants and uric acid phenotypes in African American and white families. Rheumatology (Oxford) 50, 871–878. doi: 10.1093/rheumatology/keq425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scavini, M., Stidley, C. A., Paine, S. S., Shah, V. O., Tentori, F., Bobelu, A.,et al. (2007). The burden of chronic kidney disease among the Zuni Indians: the Zuni Kidney Project. Clin. J. Am. Soc. Nephrol. 2, 509–516. doi: 10.2215/CJN.02780806
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmidt, S., Barcellos, L. F., DeSombre, K., Rimmier, J. B., Lincoln, R. R., Bucher, P.,et al. (2002). Association of polymorphisms in the apolipoprotein E region with susceptibility to and progression of multiple sclerosis. Am. J. Hum. Genet. 70, 708–717. doi: 10.1086/339269
Schunkert, H., Konig, I. R., Kathiresan, S., Reilly, M. P., Assimes, T. L., Holm, H.,et al. (2011). Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338. doi: 10.1038/ng.784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, V. O., Scavini, M., Stidley, C. A., Tentori, F., Welty, T. K., Kessler, D. S.,et al. (2003). Epidemic of diabetic and nondiabetic renal disease among the Zuni Indians: the Zuni Kidney Project. J. Am. Soc. Nephrol. 14, 1320–1329. doi: 10.1097/01.ASN.0000059920.00228.A0
Shara, N. M., Wang, H., Mete, M., Al-Balha, Y. R., Azalddin, N., Lee, E. T.,et al. (2012). Estimated GFR and incident cardiovascular disease events in American Indians: the strong heart study. Am. J. Kidney Dis. 60, 795–803. doi: 10.1053/j.ajkd.2012.06.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singaraja, R. R., Tietjen, I., Hovingh, G. K., Franchini, P. L., Radomski, C., Wong, K.,et al. (2014). Identification of four novel genes contributing to familial elevated plasma HDL cholesterol in humans. J. Lipid Res. 55, 1693–1701. doi: 10.1194/jlr.M048710
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sobel, E., and Lange, K. (1996). Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58, 1323–1337.
Sotoodehnia, N., Isaacs, A., de Bakker, P. I., Dorr, M., Newton-Choh, C., Nolte, I. M.,et al. (2010). Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076. doi: 10.1038/ng.716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stidley, C. A., Shah, V. O., Narva, A. S., Dalton, D., MacCluer, J. W., Bolbelu, A.,et al. (2002). A population-based, cross-sectional survey of the Zuni Pueblo: a collaborative approach to an epidemic of kidney disease. Am. J. Kidney Dis. 39, 358–368. doi: 10.1053/ajkd.2002.30557
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suchindran, S., Rivedal, D., Guyton, J. R., Milledge, T., Gao, X., Benjamin, A.,et al. (2010). Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham heart study. PLoS Genet. 6:e1000928. doi: 10.1371/journal.pgen.1000928
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tabara, Y., Kohara, K., Kawamoto, R., Hiura, Y., Nishimura, K., Morisaki, T.,et al. (2010). Association of four genetic loci with uric acid levels and reduced renal function: the J-SHIPP Suita study. Am. J. Nephrol. 32, 279–286. doi: 10.1159/000318943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Talmud, P. J., Drenos, F., Shah, S., Shah, T., Palmen, J., Verzilli, C.,et al. (2009). Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am. J. Hum. Genet. 85, 628–642. doi: 10.1016/j.ajhg.2009.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tentori, F., Stidley, C. A., Scavini, M., Shah, V. O., Narva, A. S., Paine, S.,et al. (2003). Prevalence of hematuria among Zuni Indians with and without diabetes: the Zuni Kidney Project. Am. J. Kidney Dis. 41, 1195–1204. doi: 10.1016/S0272-6386(03)00351-2
Thameem, F., Kawalit, I. A., Adler, S. G., and Abboud, H. E. (2013). Susceptibility gene search for nephropathy and related traits in Mexican-Americans. Mol. Biol. Rep. 40, 5769–5779. doi: 10.1007/s11033-013-2680-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tin, A., Woodward, O. M., Kao, W. H., Liu, C. T., Lu, X., Nalls, M. A.,et al. (2011). Genome-wide association study for serum urate concentrations and gout among African Americans identifies genomic risk loci and a novel URAT1 loss-of-function allele. Hum. Mol. Genet. 20, 4056–4068. doi: 10.1093/hmg/ddr307
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vitart, V., Rudan, I., Hayward, C., Gray, N. K., Floyd, J., Palmer, C. N.,et al. (2008). SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet. 40, 437–442. doi: 10.1038/ng.106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Voruganti, V. S., Franceschini, N., Haack, K., Laston, S., MacCluer, J. W., Umans, J. G.,et al. (2014). Replication of the effect of SLC2A9 genetic variation on serum uric acid levels in American Indians. Eur. J. Hum. Genet. 22, 938–943. doi: 10.1038/ejhg.2013.264
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Voruganti, V. S., Kent, J. W. Jr., and Debnath, S., Cole, S. A., Haack, K., Goring, H. H.,et al. (2013). Genome-wide association analysis confirms and extends the association of SLC2A9 with serum uric acid levels to Mexican Americans. Front. Genet. 4:279. doi: 10.3389/fgene.2013.00279
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walia, G. K., Gupta, V., Aggarwal, A., Asghar, M., Dudbridge, F., Timpson, N.,et al. (2014). Association of common genetic variants with lipid traits in the Indian population. PLoS ONE 9:e101688. doi: 10.1371/journal.pone.0101688
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waterworth, D. M., Ricketts, S. L., Song, K., Chen, L., Zhao, J. H., Ripatti, S.,et al. (2010). Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 30, 2264–2276. doi: 10.1161/ATVBAHA.109.201020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weissglas-Volkov, D., Aguilar-Salinas, C. A., Sinsheimer, J. S., Riba, L., Huertas-Vazquez, A., Ordonez-Sanchez, M. L.,et al. (2010). Investigation of variants identified in caucasian genome-wide association studies for plasma high-density lipoprotein cholesterol and triglycerides levels in Mexican dyslipidemic study samples. Circ. Cardiovasc. Genet. 3, 31–38. doi: 10.1161/CIRCGENETICS.109.908004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Willer, C. J., Sanna, S., Jackson, A. U., Scuteri, A., Bonnycastle, L. L., Clarke, R.,et al. (2008). Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169. doi: 10.1038/ng.76
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Willer, C. J., Schmidt, E. M., Sengupta, S., Peloso, G. M., Gustafsson, S., Kanoni, S.,et al. (2013). Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283. doi: 10.1038/ng.2797
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Witkowska, K., Smith, K. M., Yao, S. Y., Ng, A. M., O’Neill, D., Karpinski, E.,et al. (2012). Human SLC2A9a and SLC2A9b isoforms mediate electrogenic transport of urate with different characteristics in the presence of hexoses. Am. J. Physiol. Renal Physiol. 303, F527–F539. doi: 10.1152/ajprenal.00134.2012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wurzner, G., Gerster, J. C., Chiolero, A., Maillard, M., Fallab-Stubi, C. L., Brunner, H. R.,et al. (2001). Comparative effects of losartan and irbesartan on serum uric acid in hypertensive patients with hyperuricaemia and gout. J. Hypertens. 19, 1855–1860. doi: 10.1097/00004872-200110000-00021
Yamakawa-Kobayashi, K., Natsume, M., Aoki, S., Nakano, S., Inamori, T., Kasezewa, N.,et al. (2012). The combined effect of the T2DM susceptibility genes is an important risk factor for T2DM in non-obese Japanese: a population based case-control study. BMC Med. Genet. 13:11. doi: 10.1186/1471-2350-13-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, S., Wang, Z., Zhang, Z., Wang, J., Li, Y., Yao, L.,et al. (2014). PTPLAD2 is a tumor suppressor in esophageal squamous cell carcinogenesis. FEBS Lett. 588, 981–989. doi: 10.1016/j.febslet.2014.01.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zuni Pueblo QuickFacts from the US Census Bureau. (2014). US Census Bureau. State and County QuickFacts. Available at: http://quickfacts.census.gov/qfd/index.html
Keywords: single nucleotide polymorphisms, association, kidney function, serum uric acid, triglycerides
Citation: Laston SL, Voruganti VS, Haack K, Shah VO, Bobelu A, Bobelu J, Ghahate D, Harford AM, Paine SS, Tentori F, Cole SA, MacCluer JW, Comuzzie AG and Zager PG (2015) Genetics of kidney disease and related cardiometabolic phenotypes in Zuni Indians: the Zuni Kidney Project. Front. Genet. 6:6. doi: 10.3389/fgene.2015.00006
Received: 16 October 2014; Accepted: 08 January 2015;
Published online: 30 January 2015.
Edited by:
Karen T. Cuenco, Genentech, USAReviewed by:
Yiran Guo, Children’s Hospital of Philadelphia, USASarah Buxbaum, Jackson State University, USA
Copyright © 2015 Laston, Voruganti, Haack, Shah, Bobelu, Bobelu, Ghahate, Harford, Paine, Tentori, Cole, MacCluer, Comuzzie and Zager. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip G. Zager, Department of Medicine, Division of Nephrology, University of New Mexico School of Medicine, ACC5, Albuquerque, NM 87131-5271, USA e-mail:cHphZ0B1bm0uZWR1
†These authors have contributed equally to this work.
 Sandra L. Laston
Sandra L. Laston V. Saroja Voruganti
V. Saroja Voruganti Karin Haack
Karin Haack Vallabh O. Shah
Vallabh O. Shah Arlene Bobelu5
Arlene Bobelu5 Shelley A. Cole
Shelley A. Cole Anthony G. Comuzzie
Anthony G. Comuzzie Philip G. Zager
Philip G. Zager