- 1Institute of Physiology and Basic Medicine, Novosibirsk, Russia
- 2Institute of Medicine and Psychology, Novosibirsk State University, Novosibirsk, Russia
- 3Institute of Cytology and Genetics, Novosibirsk, Russia
- 4Tomsk National Research Medical Center of the Russian Academy of Sciences, Mental Health Research Institute, Tomsk, Russia
- 5Department of Psychotherapy and Psychological Counseling, National Research Tomsk State University, Tomsk, Russia
- 6Institute of Non-Destructive Testing, National Research Tomsk Polytechnic University, Tomsk, Russia
Depressive disorder (DD) is a widespread mental disorder. Although DD is to some extent inherited, the genes contributing to the risk of this disorder and its genetic mechanisms remain poorly understood. A recent large-scale genome-wide association Chinese study revealed a strong association between the SIRT1 gene variants and DD. The aim of this study was to analyze the occurrence of heterozygote carriers and search for rare SNP variants of the SIRT1 gene in a cohort of DD patients as compared with a cohort of randomly selected members of the Russian population. The complete coding sequences of the SIRT1 gene from 1024 DNA samples from the general Russian population and from 244 samples from patients with DD were analyzed using targeted sequencing. Four new genetic variants of the SIRT1 were discovered. While no significant differences in the allele frequencies were found between the DD patients and the general population, differences between the frequencies of homozygote carriers of specific alleles and occurrences of heterozygous were found to be significant for rs2236318 (P < 0.0001), and putatively, rs7896005 (P < 0.05), and rs36107781 (P < 0.05). The study found for the first time that two new SNPs (i.e., 10:69665829 and 10:69665971) along with recently reported ones (rs773025707 and rs34701705), are putatively associated with DD. The revealed DD-associated SIRT1 SNPs might confer susceptibility to this disorder in Russian population of European descent.
Introduction
Depressive disorder (DD or unipolar depression, which includes Major Depressive Disorder [MDD] and dysthymia) is a widespread mental disorder, with heritability as a one of the risk factors. Multiple genes are enganged in development of depression, however, the “genetic architecture” of DD has yet to be elucidated (Peterson et al., 2017).
Recent studies have shown that dysregulation of the function of intracellular proteins has an important role in the pathogenesis of a wide range of mental disorders. The proteins involved in neurobiological processes are believed to be potential targets for pharmacotherapy and/or new markers for diagnostics and the predicted course of affective disorders (Dunman and Violeti, 2013; Losenkov et al., 2014, 2016).
Silent information regulator-1 (SIRT1) deacetylase, a protein with NAD(+)-dependent deacetylation activity, is a member of the sirtuin protein family. The substrates of SIRT1 are histones, transcription factors and some other DNA-binding proteins (Hsu et al., 2016). Deacetylation changes the affinity of proteins to DNA, leading to the alteration of the transcription activity of various genes and, consequently, a variety of phenotypic effects.
Silent information regulator-1 has been shown to mediate various cellular processes including inflammation, mitochondrial biogenesis, cell growth, apoptosis, as well as cell senescence and consequent aging (Donmez and Outeiro, 2013; Wang, 2014; Ng et al., 2015; Tang et al., 2018). The levels of SIRT1 are decreased in both transcriptional and post-transcriptional conditions during aging, accompanied by attenuated mitochondrial biogenesis, an important component of aging-related diseases (Yoshizaki et al., 2010; Rahman and Islam, 2011; Yuan et al., 2016).
Silent information regulator-1 was indexed to be involved into early neuron development, synaptic plasticity, and memory regulation mechanisms (Gao et al., 2010) as well as activate MAO-A in the brain to mediate anxiety and exploratory drive (Libert et al., 2011). It binds to and transcriptionally regulates the gene locus encoding the G protein-coupled receptor GPR50 in the brain, identified as a genetic risk factor for bipolar disorder and DD in women Joerg et al., 2015).
It has been recently evidenced that SIRT1 participates in regulating inflammatory responses via expression of interleukin-6 (Tang et al., 2017). Given that down-regulation of SIRT1 promotes the secretion of IL-6 (Volonte et al., 2015), the engagement of SIRT1 into pathophysiology of DD may be implemented through neuroinflammatory pathways (Ng et al., 2015). Possible roles of mitochondrial dysfunctions and SIRT1 in DD pathogenesis are also being currently under the scope (Kambe and Miyata, 2017).
Silent information regulator-1 is coded by the SIRT1 gene. All of the above on SIRT1 protein as well as recently obtained data on significant down-regulation of the peripheral blood mRNA-based SIRT1 expression in the patients with DD (Luo and Zhang, 2016) strengthen the suggestion that SIRT1 may be regarded as a putative risk gene of DD.
As for the genetic facet of the SIRT1 world, data on genetic variations in SIRT1 and their associations with DD are scarce and contradictory. An association between rs10997875 in SIRT1 gene and DD was found in the Japanese population. Thus, the authors did not find any association between SIRT1 gene and SSRI therapeutic response in DD in the allele/genotype analysis or haplotype analysis (Kishi et al., 2010).
A low-coverage whole-genome analysis of a large cohort of Chinese patients with DD revealed a substantial difference in the frequencies of two SNPs (rs12415800 and rs35936514) between patients with DD and the general population. One of those SNPs (rs12415800) was mapped to the SIRT1 locus (CONVERGE Consortium, 2015). Data from the study of Finnish population evidenced associations of the SIRT1 rs3758391 T allele with DD, odds ratio = 1.19 (Kovanen et al., 2015).
A large study based on the DNA from individuals of European descent participating in the consumer genomics company 23andMe’s optional research initiative revealed 15 new loci associated with DD. Meanwhile, the authors were unable to replicate the strong association for SNPs in the SIRT1 gene found for the Chinese population and only a modest association for rs12415800 was reported (Hyde et al., 2016).
The newest data and meta-analysis on SIRT1 polymorphism and clinical depression in a Han Chinese population did not find any significant difference in allelic distribution of the rs3758391 between controls and patients with DD (Tang et al., 2018). However, the authors evidenced a wide divergence of the T allele frequency of rs3758391 among different ethnic populations (85% in Han Chinese vs. 27% in Caucasians), suggesting that rs3758391 polymorphism could be ethnicity-associated. Finally, the study showed a significant association of SIRT1 expression with rs3758391 in the occipital cortex, involved into pathophysiology of DD. Namely, for this brain region, the carriers of the CC genotype manifested significantly lower levels of SIRT1 expression than those with the TT genotype (Tang et al., 2018).
While there are promising data on SIRT1 as a novel DD risk gene, association of its allelic variants with DD still remains unclear. Moreover, the majority of publications are focused on the analysis of the most frequent allelic variants, whereas potential associations with rare allelic variants ate still unknown. Moreover, the occurrences of heterozygote/homozygote carriers were not analyzed in details. In view of the above, the objective of the study was to identify rare SNP variants of the SIRT1 gene and analyze occurrences of heterozygote/homozygote carriers in a cohort of randomly selected members of the Russian population (Novosibirsk city) in comparison with a cohort of DD patients.
Materials and Methods
Participants
One thousand and twenty-four samples were chosen (according to the list of random numbers generated by SPSS software) from a large collection of genomic DNA obtained from 45 to 69-year-olds of European descent living in Novosibirsk city in 2003–2005 (HAPIEE epidemiological study1). This derived population sample was not analyzed for the presence of psychopathologies.
For the depression sample, DNA was derived from the patients with depression hospitalized in psychiatric units of the Institute of Physiology and Basic Medicine, Novosibirsk and Mental Health Research Institute, Tomsk (a city in close proximity to Novosibirsk) in 2009–2017. The selection criteria included a DSM-IV diagnosis of unipolar depression (MDD or chronic depression) and European ancestry. The diagnoses were assessed by psychiatrists using structured interview – M.I.N.I. (Sheehan et al., 1998; in 71% of cases) or SCID (First et al., 1997; in 29% of cases). All studies were approved by local Ethics Committees, and the signed informed consent included the permission from the patients to perform a genetic analysis of their DNA samples.
The blood was sampled in tubes and frozen at -20°C. Genomic DNA was extracted by the phenol-chloroform method.
NGS Analysis of SIRT1 Gene
Nine exons of the SIRT1 gene were analyzed by targeted sequencing. All exons with their adjacent intron (20–50 bp) were covered by 19 amplicons (225–275 bp). The DNA fragment 5′ adjacent to the SIRT1 gene containing the SNP named rs12415800 was sequenced as well. Primers were designed using AmpliSeq Designer and Primer 3 software. Twenty selected fragments were amplified in two pools. 20 ng of genomic DNA was used for each reaction containing SuperHotTaq DNA polymerase (Bioron, Germany), 3.0 mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each primer. All reactions were performed in 25 μl volume with the following cycling program: 95oC for 2 min, then 25 cycles at 95oC for 15 s, 60oC for 30 s, and 72oC for 40 s.
For each DNA sample, 25 μl of the two PCR reactions were combined and purified by Agencourt AMPure XP (Beckman Coulter, Pasadena, CA, United States). The resulting amplicons were ligated with the barcodes and A/P1 adapters. A and P1 adapters were used in the additional PCR round for the enrichment. Qubit dsDNA HS Assay kit was used for resulting fragments concentration measurement on a Qubit 3.0 fluorimeter (Thermosphere, United States). The lengths of the fragments were determined using a High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, United States) and the 2100 Bioanalyzer (Agilent Technologies). Normalized libraries were used in emulsion PCR with the Ion PGM Hi-Q OT2 kit and sequenced on Ion Torrent PGM (with Ion PGM HI-Q sequencing kit) according to the manufacturer’s instructions. To reduce the number of samples, two samples were pooled for each barcode.
PRINSEQ (Schmieder and Edwards, 2011) was used for quality control, filtering, and trimming the reads. All reads were mapped to the human genome reference GRCh37/hg19 using BWA-MEM v. 0.7.5 (Li and Durbin, 2009). SAM tools (Li et al., 2009) were used for SNVs detection. Selected variants were manually examined with Integrative Genomic Viewer (Robinson et al., 2011).
All SNP variants were named according to the guidelines of the Human Genome Variation Society, HGVS (Den Dunnen and Antonarakis, 2000). Nucleotide and amino acid numbering were based on GenBank reference sequences Nc_000010.10 and NP_036370.2, respectively. If an SNP was found in the pooled sample, samples from the pool were sequenced individually on an ABI 3130 Genetic Analyzer. The most frequent SNPs were individually analyzed by allele-specific real-time PCR.
Statistical analyses of differences in allele frequencies were performed using the Chi-squared test. Hardy-Weinberg equilibrium was calculated using the available web-tool (Rodriguez et al., 2009).
Results
In total, 1024 general population DNA samples and 244 depression patient DNA samples were included into the analysis. The age in the population sample was 53.8 ± 7.0 years (mean ±SD), the male to female ratio was 1:1. The age of depression patients was 49.0 ± 11.4 years, the male to female ratio was 1:4, with most (224) having recurrent MDD (or single episode MDD) in addition to 20 cases of dysthymia. The individuals in both study samples were of European descent.
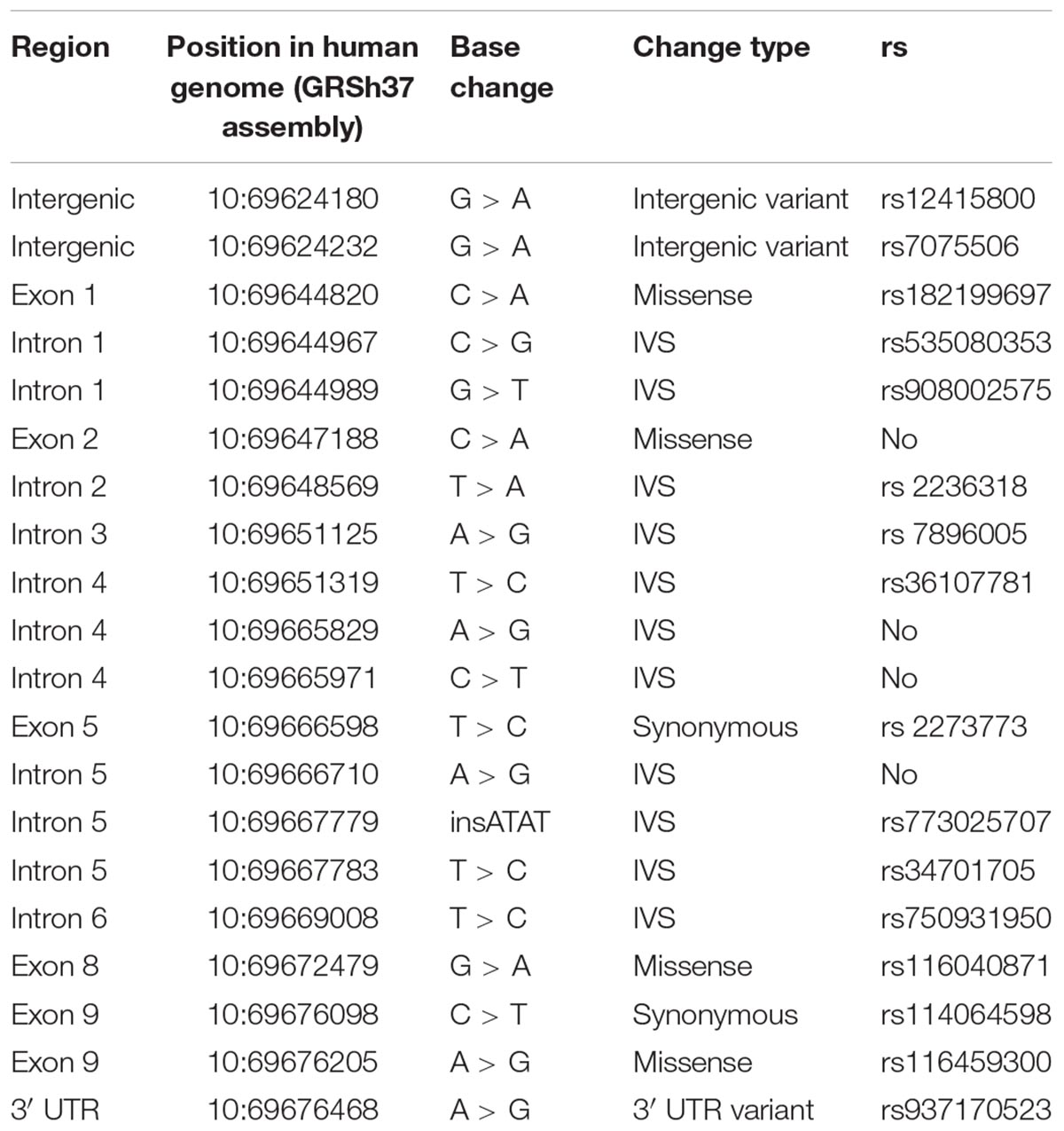
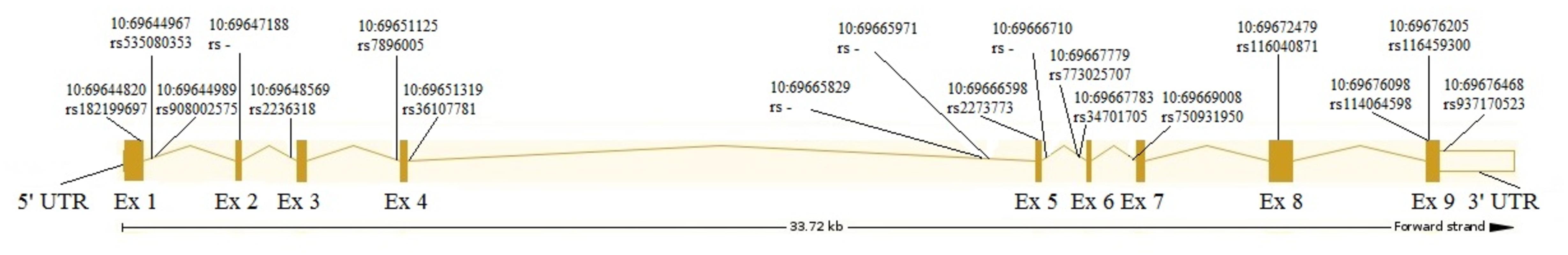
The analysis revealed 20 SNPs in the SIRT1 gene (Table 1 and Figure 1). The frequencies of the 6 most common SNPs are presented in Table 2, and those of the less frequent ( < 1%) SNPs are presented in Table 3. For the latter, only heterozygote carriers were found, which is not surprising for rare SNPs. Six rare SNPs were found only in the general population, four SNPs were found only in DD patients, and four SNPs were found in both groups.
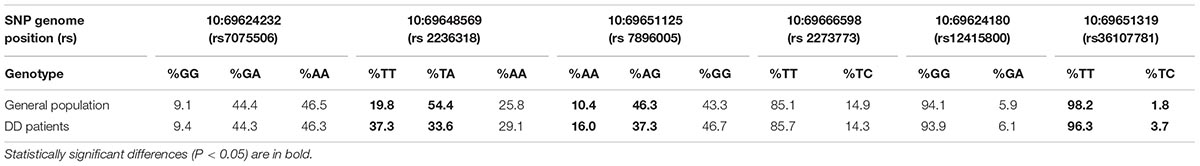
Table 2. The most frequent SIRT1 allele frequencies in Russian population (Ò = 1024) and Depressive Disorder (DD) patients (N = 244).
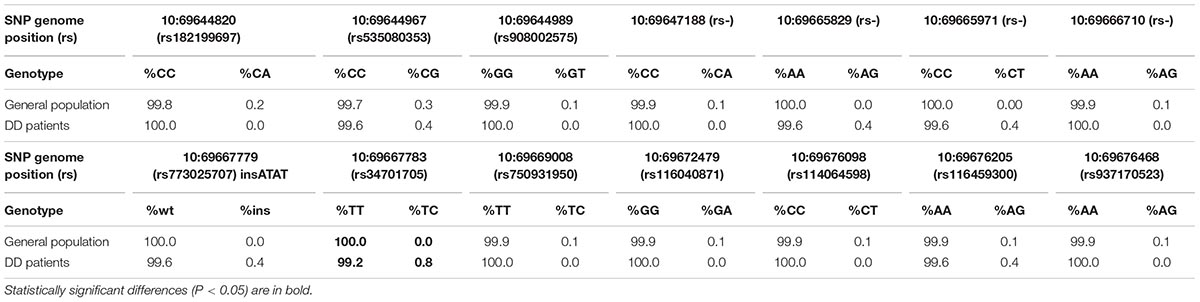
Table 3. The less frequent SIRT1 allele frequencies in Russian population (N = 1024) and Depressive Disorder (DD) patients (N = 244).
Of the 20 SNPs, 4 were identified for the first time; therefore, their rs-numbers are not assigned yet and are not indicated in Table 3. These SNPs were positioned at 10:69647188, 10:69666710, 10:69665829, and 10:69665971.
The occurrences of homozygote carriers for two SNPs – T/T for rs2236318 and A/A for rs7896005 – were significantly higher in DD than in general population (P < 0.001 and P < 0.05, respectively). The occurrences of heterozygote carriers for these two SNPs were significantly lower (P < 0.0001 and P < 0.05, respectively). The occurrences of heterozygote carriers of two additional, less frequent SNPs – rs36107781 and rs34791705 – were significantly higher in DD than in general population (P < 0.05). However, analysis of Hardy-Weinberg equilibrium for these SNPs in the sample of general population revealed equilibrium only for rs2236318 (χ2 = 8.62, P < 0.005).
The allelic frequencies of the SNPs presented in Table 2 are similar to those indicated in the “1000 Genomes” program for European population (data taken from2; Table 4).
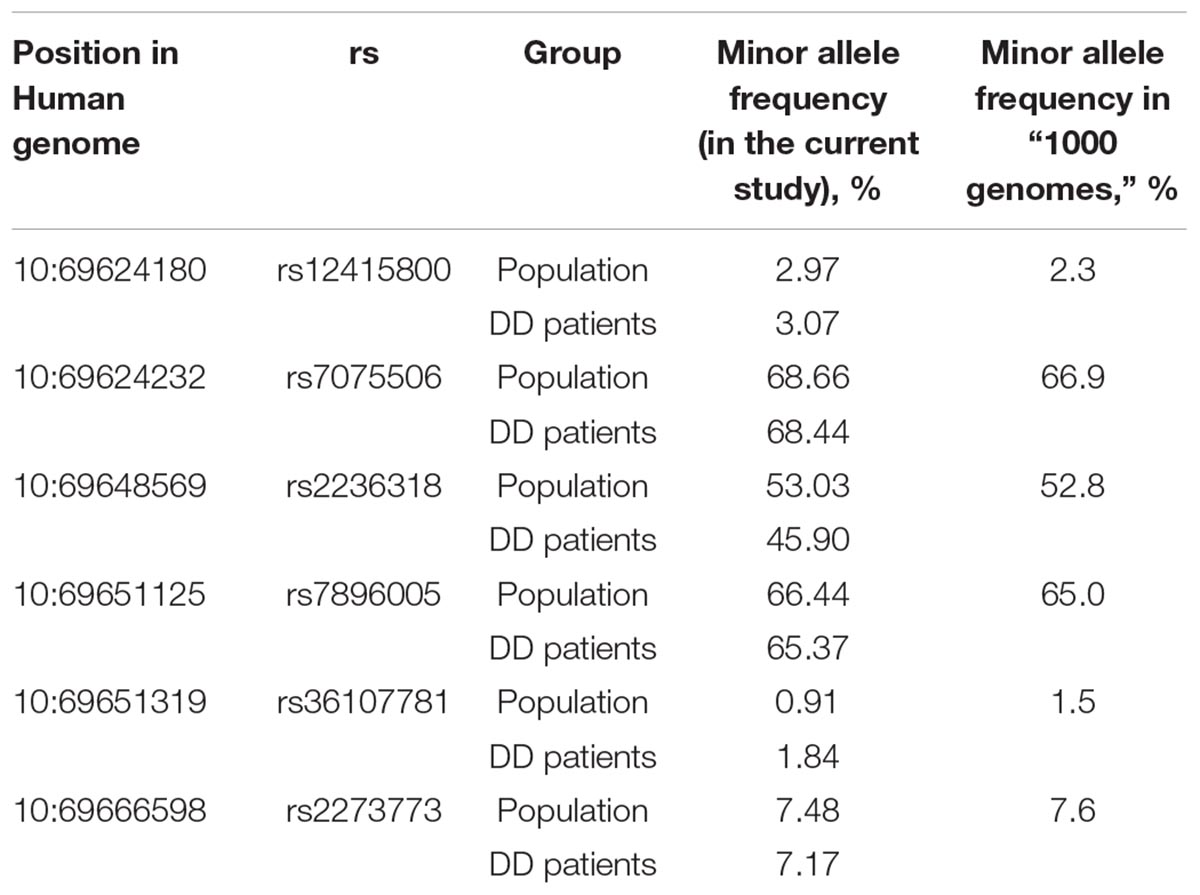
Table 4. Allele frequencies of the most frequent SIRT1 SNPs in general Russian population (N = 1024) and in Depressive Disorder (DD) patients (N = 244) in comparison with the “1000 genomes” study (data for European population).
Discussion
Comprehensive NGS analysis of the SIRT1 structure in the general population DNA samples (N = 1024) and in DD patient DNA samples (N = 244) revealed 20 SNPs (Table 1). Among those SNPs were 4 new genetic variants not described earlier (10:69647188, 10:69665829, 10:69665971, and 10:69666710). Significant differences between these two groups were found in the frequency of heterozygous carriers for rs2236318, rs7896005, rs36107781 and rs34791705 (Tables 2, 3), supported by the Hardy-Weinberg equilibrium in the sample of general population for the first SNP (rs2236318). However, no difference was observed in the allele frequencies (Table 4).
Our results indicate that three frequent SIRT1 genotypes (rs2236318 genotype T/T, rs7896005 genotype A/A and rs36107781 genotype T/C) may be associated with DD, with the rs2236318 genotype T/T identified as having a P-value < 1 × 10-4. The absence of Hardy-Weinberg equilibrium for rs7896005 and rs36107781 in the sample of general population does not allow ascertaining those SNPs that are associated with DD. However, almost perfect Hardy-Weinberg equilibrium for rs2236318 (p < 0.005) and a high prevalence of genotype T/T among DD patients (p < 0.001) demonstrate a strong association of this SNP with DD.
To our knowledge, these associations have been identified for the first time. Two previous studies demonstrated a relationship between DD and SIRT1 rs12415800 allele frequencies in Han Chinese melancholic DD women (CONVERGE Consortium, 2015) and – to a much smaller degree – in an European self-reporting depression sample (Hyde et al., 2016) coded as rs187810158 in that study. In contrast to those studies, we did not find such a difference in rs12415800 (GA heterozygotes 6.1% vs. 5.9%; Table 2). Most likely, it is due to some differences among the samples (ethnicity, gender, difference in diagnostic procedures). Our patients represented both sexes with clinically verified DSM-IV-based diagnoses of depression, mainly ‘classical’ depression, with melancholic features (78%). Moreover, both those studies (CONVERGE Consortium, 2015; Hyde et al., 2016) were focused mainly on the allele frequencies and not on the occurrence of homozygous/heterozygous allele carriers.
Several rare SNPs (2 cases of rs34701705 and 1 case each of rs773025707, 10:69665829, and 10:69665971) were found exclusively in DD patients, which may indicate a possible correlation of these variants with this disorder. Larger cohort studies are needed for verifying these associations.
Most SNPs found in this study are mapped to the introns, although 4 SNPs result in missense mutations: rs182199697 (C > A) results in Pro114Gln, 10:69647188 (C > A) results in Phe148Leu, rs116040871 (G > A) results in Glu536Lys, and rs116459300 (A > G) results in Asn700Ser. Two other SNPs caused synonymous changes (rs114064598 and rs2273773) and one SNP (rs937170523) was mapped to the 3′-UTR (Table 1 and Figure 1).
There are two limitations for this study: (1) while the study suggests an important role for SIRT1 in DD, it should be noted that this disorder represents a polygenic phenotype and that many other loci remain to be discovered beyond SIRT1; (2) the quality of experienced early environment (e.g., parental care, exposure to childhood abuse) was not evaluated in the DD group. Thus, one should keep in mind that the behavioral phenotype of DD featuring worse depressive symptomatology, may putatively be associated with SIRT1 allelic variants. At least, the recent data on a reduction in the levels of the deacetylase SIRT1 in peripheral blood mononuclear cells in this type of DD patients, significantly predicting the extent of behavioral despair, is giving credibility to this suggestion (Lo Iacono et al., 2015).
As the Novosibirsk population is similar to that of Russia, the SIRT1 SNPs frequencies obtained in this study may be generalized to the entire population of European descent in Russia.
Overall, our preliminary findings suggest that the revealed DD-linked SIRT1 SNPs might confer susceptibility to this disorder in Russian population of European descent, thereby extending growing body of data on SIRT1 as a novel DD risk gene.
Ethics Statement
All studies were approved by local Ethics Committees, and the signed informed consent included the permission from the patients to perform a genetic analysis of their DNA samples. The local Ethics Committees are: (1) Committee for Biomedical Ethics of the Institute of Physiology and Basic Medicine, (2) Ethics Committee of the Institute of Preventive and Internal Medicine (now incorporated into the Institute of Cytology and Genetics, Novosibirsk), and (3) Ethics Committee of the Mental Health Research Institute (now incorporated into the Tomsk National Research Medical Center of the Russian Academy of Sciences).
Author Contributions
The study was conceived and supervised by LA, SK, and KD. DNA samples of DD patients were provided by LA, KD, NV, NB, and SI. Population DNA samples were provided by VM and MV. The genetic analysis was done by MA, DB, AG, and SK. The sequence data analysis and statistics were made by MA. The results were discussed and manuscript was written by LA, SK, and KD. All authors approved the final version of the manuscript.
Funding
This work was supported by the Russian Science Foundation grant no. 16-15-00128 (2016–2018) to LA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Maria Lebedinskaya (Novosibirsk), Eugeniya Gadetskaya (Novosibirsk), German Simutkin (Tomsk), and Innokentiy Losenkov (Tomsk) for diagnostics of depressed patients and supervising their DNA sampling, Elena Gornostaeva (Novosibirsk) for bioinformatics genetic analysis.
Footnotes
- ^http://www.ucl.ac.uk/easteurope/hapiee.html
- ^https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes
References
CONVERGE Consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–593. doi: 10.1038/nature14659
Den Dunnen, J. T., and Antonarakis, S. E. (2000). Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 15, 7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N
Donmez, G., and Outeiro, T. F. (2013). SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 5, 344–352. doi: 10.1002/emmm.201302451
Dunman, S. R., and Violeti, B. (2013). Signaling pathways underlying the pathophysiology and treatment of depression: novel mechanisms for rapid-acting agents. Trends Neurosci. 35, 47–56. doi: 10.1016/j.tins.2011.11.004
First, M. G., Spitzer, R. L., Gibbon, M., and Williams, J. B. (1997). Structured Clinical Interview for DSM-IV Axis I Disorders. Clinical Version, Administration Booklet. New York: New York State Psychiatric Institute.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., et al. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109. doi: 10.1038/nature09271
Hsu, W. W., Wu, B., and Liu, W. R. (2016). Sirtuins 1 and 2 are universal histone deacetylases. ACS Chem. Biol. 11, 792–799. doi: 10.1021/acschembio.5b00886
Hyde, C. L., Nagle, M. W., Tian, C., Chen, X., Paciga, C. A., Wendland, J. R., et al. (2016). Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036. doi: 10.1038/ng.3623
Joerg, R., Leheste, J. R., and Torres, G. (2015). Resveratrol: brain effects on SIRT1, GPR50 and photoperiodic signaling. Front. Mol. Neurosci. 8:61. doi: 10.3389/fnmol.2015.00061
Kambe, Y., and Miyata, A. (2017). Possible roles of mitochondrial dysfunctions and SIRT1 in major depressive disorder. Nihon Yakurigaku Zasshi 150, 204–206. doi: 10.1254/fpj.150.204
Kishi, T., Yoshimura, R., Kitajima, T., Okochi, T., Okumura, T., Tsunoka, T., et al. (2010). SIRT1 gene is associated with major depressive disorder in the Japanese population. J. Affect. Disord. 126, 167–173. doi: 10.1016/j.jad.2010.04.003
Kovanen, L., Donner, K., and Partonen, T. (2015). SIRT1 polymorphisms associate with seasonal weight variation, depressive disorders, and diastolic blood pressure in the general population. PLoS One 10:e0141001. doi: 10.1371/journal.pone.0141001
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). 1000 Genome project data processing subgroup. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Libert, S., Pointer, K., Bell, E. L., Das, A., Cohen, D. E., Asara, J. M., et al. (2011). SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell 147, 1459–1472. doi: 10.1016/j.cell.2011.10.054
Lo Iacono, L., Visco-Comandini, F., Valzania, A., Viscomi, M. T., Coviello, M., Giampà, A., et al. (2015). Adversity in childhood and depression: linked through SIRT1. Transl. Psychiatry 5:e629. doi: 10.1038/tp.2015.125
Losenkov, I. S., Ivanova, S. A., Vyalova, N. A., Simutkin, G. G., and Bokhan, N. A. (2014). Proteins of the Akt1/GSK-3 beta signaling pathway in peripheral blood mononuclear cells of patients with affective disorders. Neurochem. J. 31, 240–245.
Losenkov, I. S., Vyalova, N. M., Simutkin, G. G., Bokhan, N. A., and Ivanova, S. A. (2016). An association of AKT1 gene polymorphism with antidepressant treatment response. World J. Biol. Psychiatr. 17, 239–242. doi: 10.3109/15622975.2015.1112921
Luo, X. J., and Zhang, C. (2016). Down-regulation of SIRT1 gene expression in major depressive disorder. Am. J. Psychiatry 173:1046. doi: 10.1176/appi.ajp.2016.16040394
Ng, F., Wijaya, L., and Tang, D. L. (2015). SIRT1 in the brain – connections with aging-associated disorders and lifespan. Front. Cell. Neurosci. 9:64. doi: 10.3389/fncel.2015.00064
Peterson, R. E., Cai, N., Bigdeli, T. B., Li, Y., Reimers, M., Nikulova, A., et al. (2017). The genetic architecture of major depressive disorder in han chinese women. JAMA Psychiatry 74, 162–168. doi: 10.1001/jamapsychiatry.2016.3578
Rahman, S., and Islam, R. (2011). Mammalian Sirt1: insights on its biological functions. Cell Commun. Signal. 9:11. doi: 10.1186/1478-811X-9-11
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Rodriguez, S., Gaunt, T. R., Ian, N. M., and Day, I. N. M. (2009). Hardy-Weinberg equilibrium testing of biological ascertainment for mendelian randomization studies. Am. J. Epidemiol. 169, 505–514. doi: 10.1093/aje/kwn359
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Sheehan, D. V., Lecrubier, Y., Sheehan, K. H., Amorim, P., Janavs, J., Weiller, E., et al. (1998). The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry 59(Suppl. 20), 22–33.
Tang, L., Chen, Q., Meng, Z., Sun, L., Zhu, L., Liu, J., et al. (2017). Suppression of Sirtuin-1 increases IL-6 expression by activation of the Akt pathway during allergic asthma. Cell. Physiol. Biochem. 43, 1950–1960. doi: 10.1159/000484119
Tang, W., Chen, Y., Fang, X., Wang, Y., Fan, W., and Zhang, C. H. (2018). SIRT1 rs3758391 and major depressive disorder: new data and meta-analysis. Neurosci. Bull. 34, 863–866. doi: 10.1007/s12264-018-0235-5
Volonte, D., Zou, H., Bartholomew, J. N., Liu, Z., Morel, P. A., and Galbiati, F. (2015). Oxidative stress-induced inhibition of Sirt1 by caveolin-1 promotes p53-dependent premature senescence and stimulates the secretion of interleukin 6 (IL-6). J. Biol. Chem. 290, 4202–4214. doi: 10.1074/jbc.M114.598268
Wang, Y. (2014). Molecular links between caloric restriction and Sir2/SIRT1 activation. Diabetes Metab. J. 38, 321–329. doi: 10.4093/dmj.2014.38.5.321
Yoshizaki, T., Schenk, S., Imamura, T., Babendure, J. L., Sonoda, N., Bae, E. J., et al. (2010). SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 298, E419–E428. doi: 10.1152/ajpendo.00417.2009
Keywords: SIRT1 gene, SNP, depressive disorders, population, European descent
Citation: Aftanas LI, Anisimenko MS, Berdyugina DA, Garanin AY, Maximov VN, Voevoda MI, Vyalova NM, Bokhan NA, Ivanova SA, Danilenko KV and Kovalenko SP (2019) SIRT1 Allele Frequencies in Depressed Patients of European Descent in Russia. Front. Genet. 9:686. doi: 10.3389/fgene.2018.00686
Received: 01 August 2018; Accepted: 11 December 2018;
Published: 04 January 2019.
Edited by:
Peter Kochunov, University of Maryland, United StatesReviewed by:
Julia Fedotova, Russian Academy of Sciences, RussiaTimo Partonen, National Institute for Health and Welfare, Finland
Copyright © 2019 Aftanas, Anisimenko, Berdyugina, Garanin, Maximov, Voevoda, Vyalova, Bokhan, Ivanova, Danilenko and Kovalenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lyubomir I. Aftanas, bGlhZnRhbmFzQGdtYWlsLmNvbQ==
 Lyubomir I. Aftanas
Lyubomir I. Aftanas Maksim S. Anisimenko1
Maksim S. Anisimenko1 Vladimir N. Maximov
Vladimir N. Maximov Mikhail I. Voevoda
Mikhail I. Voevoda Natalya M. Vyalova
Natalya M. Vyalova Svetlana A. Ivanova
Svetlana A. Ivanova Konstantin V. Danilenko
Konstantin V. Danilenko