Long non-coding RNAs (LncRNAs) are RNAs with more than 200 nucleotides and are mostly transcribed by RNA polymerase II from different regions across the genome. They are currently known as key regulators of cellular function through different mechanisms such as epigenetic regulation, miRNA sponging, and modulating of proteins and enzyme cofactors (Kurokawa, 2011; Nie et al., 2012; Flynn and Chang, 2014; Birgani et al., 2017; Marchese et al., 2017). By this way, they are implicated in development pathways (Amaral and Mattick, 2008). Different lncRNAs such as HOTAIR can play their important roles by changing the chromatin states of the genome (Mercer and Mattick, 2013). Rinn et al. introduced this RNA as a spliced and polyadenylated RNA with 2,158 nucleotides (Hajjari et al., 2013). HOTAIR, as one of the featured lncRNAs, is located between HOXC11 and HOXC12 on chromosome 12q13.3. HOTAIR forms stem-loop structures which bind to histone modification complexes lysine-specific demethylase 1 (LSD1) and Polycomb Repressive Complex2 (PRC2) in order to recruit them on specific target genes. This RNA interacts with Polycomb repressive Complex2 (PRC2) and has a lot of targets such as HOXD. By this way, PRC2 can repress the desired genes leading into increased growth, proliferation, survival, metastasis, invasion, and drug resistance in some cancer cells (Rinn et al., 2007; He et al., 2011; Davidovich et al., 2013; Hajjari et al., 2014; Martens-Uzunova et al., 2014; Zhao et al., 2014). So, different studies have indicated the dysregulation of HOTAIR in different types of cancers in recent years (Gupta et al., 2010; Kogo et al., 2011; Yang et al., 2011; Niinuma et al., 2012; Hajjari et al., 2013; Kim et al., 2013; Li et al., 2013).
In recent studies, there are some reports indicating the role of HOTAIR SNPs which make it a significant cancer susceptibility locus and provide high risk for some cancers (Qi et al., 2016), like breast (Bayram et al., 2015, 2016; Yan et al., 2015), gastric (Pan et al., 2016; Tian et al., 2016), cervical (Guo et al., 2016; Qiu et al., 2016), papillary thyroid carcinoma (Zhu et al., 2016), osteosarcoma (Zhou et al., 2016), prostate (Taheri et al., 2017), ovarian (Wu et al., 2016; Qiu et al., 2017), and colorectal cancers (Xue et al., 2014). This is an interesting point because these SNPs may have effect on gene expression, function, and regulators of epigenome (Hajjari and Rahnama, 2017). Therefore, we think that more studies on these SNPs can reveal the potential of these SNPs for considering them as markers of progression and diagnosis of different cancers.
Figure 1 shows the locations of these SNPs within HOTAIR gene. Herein, we present different SNPs to highlight their potential for further studies.
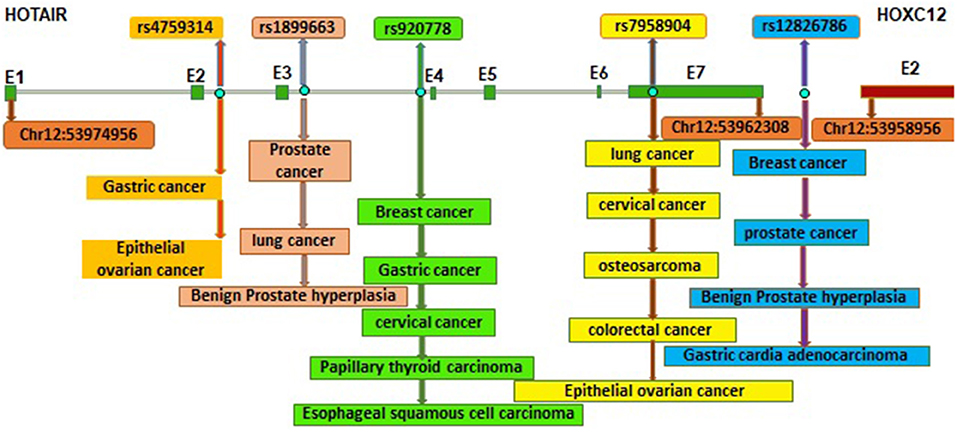
Figure 1. Locations of different SNPs within HOTAIR gene and their association with different types of cancer (E: Exon, exons of HOTAIR, and HOXC12 are shown by green and red boxes). Genomic positions are based on the UCSC Genome browser on Human Dec. 2013 (GRCh38/hg38) assembly.
There are some reports indicating the association between HOTAIR rs12826786 SNP which is located between HOTAIR and HOXC12. The increased risk for some cancers such as breast (BC) (Bayram et al., 2016), gastric adenocarcinoma (GCA) (Guo et al., 2015), prostate cancer (PC), and benign prostate hyperplasia (BPH) (Taheri et al., 2017) has been reported. For instance, women who are carriers of this polymorphism, have an increased risk of BC in both codominant and recessive inheritance models (Bayram et al., 2016). With regard to the location of this SNP, it seems that this SNP has effect on the regulation of HOTAIR gene in the cell. So, the analysis of HOTAIR dysregulation and its correlation with this SNP can be proposed in different types of cancers in different population.
rs920778 is another polymorphism which is located in the intronic enhancer of HOTAIR gene. TT genotype of this SNP has been found to affect the gene expression and make the risk for various cancers (Bayram et al., 2015) such as gastric (Pan et al., 2016), esophageal squamous cell carcinoma (Zhang et al., 2014), cervical (Qiu et al., 2016), and papillary thyroid carcinoma (Zhu et al., 2016). In addition, CC genotype of this SNP might be a cause of breast cancer in both codominant and recessive inheritance genetic models (Bayram et al., 2015).
There are some studies reporting the association between the dysregulation of HOTAIR and rs920778. HOTAIR up-regulation has been suggested as a result of rs920778 in gastric cancer (Xu et al., 2013; Pan et al., 2016). Also, the aberrant expression of HOTAIR in esophageal squamous cell carcinoma seems to be the result of a specific allele of rs920778 (Gupta et al., 2010; Zhang et al., 2014; Dai et al., 2017). Furthermore, there is higher expression of HOTAIR in female papillary thyroid carcinoma tissues because of a specific genetic polymorphism of this gene (Zhu et al., 2016).
Another SNP annotated as rs4759314 is also located in a promoter region in one of the introns of HOTAIR. It is of noted that AG/GG genotypes of the rs4759314 were associated with gastric cancer risk. The expression effects of heterozygotes individuals with G allele were more than homozygotes in the patients in co-dominant models (Du et al., 2015). However, in a controversial report, the HOTAIR gene expression found to be higher in ovarian cancer patients with AG/AA genotypes of rs4759314 (Wu et al., 2016).
Another SNP located in the intronic region of HOTAIR is rs1899663. Due to its location in a putative regulatory element, it seems that this SNP can affect gene expression and regulation. There are some association between HOTAIR rs1899663 T allele and BPH (Benign prostate hyperplasia) patients. Also, The rs1899663 is associated with prostate cancer risk in co-dominant, dominant and recessive inheritance models. Researchers have reported that this SNP changes the affinity for binding of PAX-4, SPZ1, and ZFP281 transcription factors which can alter the HOTAIR gene expression level (Taheri et al., 2017).
Among the SNPs in HOTAIR gene, one named “rs7958904” is an exonic polymorphism. So, it seems that HOTAIR rs7958904 polymorphism can affect the secondary structure of HOTAIR.
It is of noted that CC genotypes of HOTAIR rs7958904 has been reported to be associated with decreased osteosarcoma (Zhou et al., 2016), EOC (Wu et al., 2016), and colorectal cancers risk (Xue et al., 2014). In an study on osteosarcoma patients classified by age, gender, and tumor locations, it was shown that CC genotypes of the HOTAIR rs7958904 can reduce osteosarcoma risk as well as HOTAIR expression level (Zhou et al., 2016). However, cervical cancer patients with CC genotypes of this SNP had higher HOTAIR expression (Jin et al., 2017). Furthermore, with regard to the up-regulation of HOTAIR in lung cancer (Jiang et al., 2017) the SNP has been reported as a region to be associated with chemotherapy response in lung cancer patients through effect on HOTAIR function or expression (Xue et al., 2014; Gong et al., 2016).
HOTAIR have abnormal expression in the different human cancers. Different studies have revealed the cellular and molecular mechanisms in which HOTAIR is involved (Hajjari and Salavaty, 2015; Gong et al., 2016). Recently, some studies indicating the potential role of SNPs of HOTAIR in cancer susceptibility have been published. However, these studies are mostly derived from Asian population. Also, there are some controversial results on this field of study. With regard to the importance of HOTAIR regulation and function, more experiments on different populations, and ethnics are expected to reveal the importance of HOTAIR polymorphisms. Other polymorphisms in HOTAIR gene such Indel and CNV may be considered in future. However, the association between these SNPs and regulation/structure of HOTAIR has to be checked in various cancers. Also, we believe that whole genome sequencing projects can help to find the relation between the SNPs of this RNA with other SNPs in different cancers in future.
Author Contributions
MH designed and wrote the manuscript. SR worked on gathering the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Shahid Chamran university of Ahvaz for supporting our study.
References
Amaral, P. P., and Mattick, J. S. (2008). Noncoding RNA in development. Mammalian Genome. 19, 454–492. doi: 10.1007/s00335-008-9136-7
Bayram, S., Sümbül, A. T., Batmaci, C. Y., and Genç, A. (2015). Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumor Biol. 36, 3863–3870. doi: 10.1007/s13277-014-3028-0
Bayram, S., Sümbül, A. T., and Dadaş, E. (2016). A functional HOTAIR rs12826786 C> T polymorphism is associated with breast cancer susceptibility and poor clinicopathological characteristics in a Turkish population: a hospital-based case–control study. Tumor Biol. 37, 5577–5584. doi: 10.1007/s13277-015-4430-y
Birgani, M. T., Hajjari, M., Shahrisa, A., Khoshnevisan, A., Shoja, Z., Motahari, P., et al. (2017). Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol. Oncol. Res. 24, 329–37. doi: 10.1007/s12253-017-0241-3
Dai, W., Ko, J. M. Y., Choi, S. S. A., Yu, Z., Ning, L., Zheng, H., et al. (2017). Whole-exome sequencing reveals critical genes underlying metastasis in esophageal squamous cell carcinoma. J. Pathol. 242, 500–510. doi: 10.1002/path.4925
Davidovich, C., Zheng, L., Goodrich, K. J., and Cech, T. R. (2013). Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257. doi: 10.1038/nsmb.2679
Du, M., Wang, W., Jin, H., Wang, Q., Ge, Y., Lu, J., et al. (2015). The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 6, 31255–31262. doi: 10.18632/oncotarget.5158
Flynn, R. A., and Chang, H. Y. (2014). Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 14, 752–761. doi: 10.1016/j.stem.2014.05.014
Gong, W.-J., Yin, J.-Y., Li, X.-P., Fang, C., Xiao, D., Zhang, W., et al. (2016). Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumor Biol. 37, 8349–8358. doi: 10.1007/s13277-015-4497-5
Guo, L., Lu, X., Zheng, L., Liu, X., and Hu, M. (2016). Association of long non-coding RNA HOTAIR polymorphisms with cervical cancer risk in a Chinese population. PLoS ONE. 11:e0160039. doi: 10.1371/journal.pone.0160039
Guo, W., Dong, Z., Bai, Y., Guo, Y., Shen, S., Kuang, G., et al. (2015). Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumor Biol. 36, 2845–2854. doi: 10.1007/s13277-014-2912-y
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464, 1071–1076. doi: 10.1038/nature08975
Hajjari, M., Behmanesh, M., Sadeghizadeh, M., and Zeinoddini, M. (2013). Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med. Oncol. 30:670. doi: 10.1007/s12032-013-0670-0
Hajjari, M., Khoshnevisan, A., and Shin, Y. K. (2014). Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumor Biol. 35, 10645–10663. doi: 10.1007/s13277-014-2636-z
Hajjari, M., and Rahnama, S. (2017). HOTAIR long non-coding RNA: characterizing the locus features by the in silico approaches. Genomics Inform. 15, 170–177. doi: 10.5808/GI.2017.15.4.170
Hajjari, M., and Salavaty, A. (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12, 1–9. doi: 10.7497/j.issn.2095-3941.2015.0006
He, S., Liu, S., and Zhu, H. (2011). The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evolu. Biol. 11:102. doi: 10.1186/1471-2148-11-102
Jiang, C., Yang, Y., Yang, Y., Guo, L., Huang, J., Liu, X., et al. (2017). LncRNA-HOTAIR affects tumorigenesis and metastasis of non-small cell lung cancer by up-regulating miR-613. Oncol. Res. 26, 725–734. doi: 10.3727/096504017X15119467381615
Jin, H., Lu, X., Ni, J., Sun, J., Gu, B., Ding, B., et al. (2017). HOTAIR rs7958904 polymorphism is associated with increased cervical cancer risk in a Chinese population. Sci. Rep. 7:3144. doi: 10.1038/s41598-017-03174-1
Kim, K., Jutooru, I., Chadalapaka, G., Johnson, G., Frank, J., Burghardt, R., et al. (2013). HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 32, 1616–1625. doi: 10.1038/onc.2012.193
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326. doi: 10.1158/0008-5472.CAN-11-1021
Kurokawa, R. (2011). Long noncoding RNA as a regulator for transcription. Prog. Mol. Subcell. Biol. 51, 29–41. doi: 10.1007/978-3-642-16502-3_2
Li, D., Feng, J., Wu, T., Wang, Y., Sun, Y., Ren, J., et al. (2013). Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am. J. Pathol. 182, 64–70. doi: 10.1016/j.ajpath.2012.08.042
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18:206. doi: 10.1186/s13059-017-1348-2
Martens-Uzunova, E. S., Böttcher, R., Croce, C. M., Jenster, G., Visakorpi, T., and Calin, G. A. (2014). Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 65, 1140–1151. doi: 10.1016/j.eururo.2013.12.003
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Nie, L., Wu, H.-J., Hsu, J.-M., Chang, S.-S., LaBaff, A. M., Li, C.-W., et al. (2012). Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 4, 127–150.
Niinuma, T., Suzuki, H., Nojima, M., Nosho, K., Yamamoto, H., Takamaru, H., et al. (2012). Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72, 1126–36. doi: 10.1158/0008-5472.CAN-11-1803
Pan, W., Liu, L., Wei, J., Ge, Y., Zhang, J., Chen, H., et al. (2016). A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol. Carcinogen. 55, 90–96. doi: 10.1002/mc.22261
Qi, Q., Wang, J., Huang, B., Chen, A., Li, G., Li, X., et al. (2016). Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget. 7, 38775–38784. doi: 10.18632/oncotarget.9608
Qiu, H., Liu, Q., Li, J., Wang, X., Wang, Y., Yuan, Z., et al. (2016). Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. Apmis. 124, 567–573. doi: 10.1111/apm.12550
Qiu, H., Wang, X., Guo, R., Liu, Q., Wang, Y., Yuan, Z., et al. (2017). HOTAIR rs920778 polymorphism is associated with ovarian cancer susceptibility and poor prognosis in a Chinese population. Future Oncol. 13, 347–355. doi: 10.2217/fon-2016-0290
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Taheri, M., Habibi, M., Noroozi, R., Rakhshan, A., Sarrafzadeh, S., Sayad, A., et al. (2017). HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 613, 20–24. doi: 10.1016/j.gene.2017.02.031
Tian, T., Li, C., Xiao, J., Shen, Y., Lu, Y., Jiang, L., et al. (2016). Quantitative assessment of the polymorphisms in the HOTAIR lncRNA and cancer risk: a meta-analysis of 8 case-control studies. PLoS ONE. 11:e0152296. doi: 10.1371/journal.pone.0152296
Wu, H., Shang, X., Shi, Y., Yang, Z., Zhao, J., Yang, M., et al. (2016). Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget. 7, 41047–41052. doi: 10.18632/oncotarget.8535
Xu, Z.-Y., Yu, Q.-M., Du, Y.-A., Yang, L.-T., Dong, R.-Z., Huang, L., et al. (2013). Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 9, 587–597. doi: 10.7150/ijbs.6339
Xue, Y., Gu, D., Ma, G., Zhu, L., Hua, Q., Chu, H., et al. (2014). Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 30, 303–310. doi: 10.1093/mutage/geu076
Yan, R., Cao, J., Song, C., Chen, Y., Wu, Z., Wang, K., et al. (2015). Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol. 39, 978–985. doi: 10.1016/j.canep.2015.10.025
Yang, Z., Zhou, L., Wu, L.-M., Lai, M.-C., Xie, H.-Y., Zhang, F., et al. (2011). Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18, 1243–1250. doi: 10.1245/s10434-011-1581-y
Zhang, X., Zhou, L., Fu, G., Sun, F., Shi, J., Wei, J., et al. (2014). The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 35, 2062–2067. doi: 10.1093/carcin/bgu103
Zhao, W., An, Y., Liang, Y., and Xie, X. (2014). Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 18, 1930–1936.
Zhou, Q., Chen, F., Fei, Z., Zhao, J., Liang, Y., Pan, W., et al. (2016). Genetic variants of lncRNA HOTAIR contribute to the risk of osteosarcoma. Oncotarget. 7, 19928–19934. doi: 10.18632/oncotarget.7957
Keywords: HOTAIR, lncRNA, cancer, SNP, polymorphism
Citation: Hajjari M and Rahnama S (2019) Association Between SNPs of Long Non-coding RNA HOTAIR and Risk of Different Cancers. Front. Genet. 10:113. doi: 10.3389/fgene.2019.00113
Received: 22 May 2018; Accepted: 30 January 2019;
Published: 28 February 2019.
Edited by:
Chandan Kumar, University of Michigan Health System, United StatesReviewed by:
Venugopal Thayanithy, University of Minnesota, United StatesCopyright © 2019 Hajjari and Rahnama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammadreza Hajjari, TW9oYW1hZC5oYWphcmlAZ21haWwuY29t
 Mohammadreza Hajjari
Mohammadreza Hajjari Saghar Rahnama
Saghar Rahnama