- 1Department of Gastroenterology, The People’s Hospital of Liuzhou, Liuzhou, China
- 2Department of Research, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, China
Background: The N-acetyltransferase 1 (NAT1) gene is downregulated in several cancers and associated with patient survival. In this study, we sought to examine the prognostic value and clinical significance of NAT1 methylation in colon adenocarcinoma (COAD).
Methods: Data relating to NAT1 mRNA expression and methylation and clinicopathological features of COAD were extracted from the database of The Cancer Genome Atlas. We compared the mRNA expression and methylation of NAT1 between COAD and normal tissues and performed correlation analysis to assess the association between NAT1 mRNA expression and methylation. Furthermore, we assessed patient survival based on CpG sites in the promoter region of NAT1 and analyzed the association between the NAT1 mRNA expression and CpG site methylation and clinicopathological features. An independent Gene Expression Omnibus (GEO) dataset was used to validate the results.
Results: We found that the expression of NAT1 mRNA was reduced in COAD compared with normal tissues and that mean methylation of the eight CpG sites in the promoter region of NAT1 was higher in COAD tissues than in normal tissues. Furthermore, five CpG sites were demonstrated to be significantly negatively correlated with NAT1 mRNA expression in COAD. Survival analysis indicated that NAT1 mRNA expression and the cg15797286 and cg18509990 sites were associated with the overall survival of COAD patients. Combined survival analysis revealed that combinations of NAT1 mRNA expression with five CpG sites were significantly associated with the overall survival of COAD patients. Both NAT1 mRNA and cg15797286 were associated with the T, N, and clinical stages of COAD. The GEO data indicated that cg15797286 was hypermethylated in recurrent colorectal adenomas.
Conclusions: Methylation of NAT1 is associated with the development of COAD, and may serve as prognostic and treatment biomarkers for COAD.
Introduction
Colorectal cancers (CRCs) are among the most common digestive system malignancies worldwide, of which colon adenocarcinoma (COAD) comprises the majority (Bray et al., 2018). Despite a recent decrease in the incidence of CRC in Western countries, the incidence in those of less than 55 years of age has increased by approximately 2% per year during the past two decades (Siegel et al., 2017). In addition, the burden of CRC in China is considerably higher than that in the USA (Chen et al., 2016). Although numerous studies have examined the mechanisms underlying colorectal carcinogenesis, and despite the fact that certain aspects of the etiology and pathology of CRC are well documented (Baena and Salinas, 2015; Vulcan et al., 2017), many of the associated mechanisms require further elucidation.
DNA methylation is a common epigenetic modification, which may function by silencing gene expression. The aberrant methylation of genes has been demonstrated to play important roles in multiple biological possesses, including DNA repair and apoptosis, and can subsequently lead to carcinogenesis (Saghafinia et al., 2018; Pfeifer, 2018). In addition to environmental factors, the accumulation of multiple genetic and epigenetic alterations or interactions is considered a crucial factor contributing to the pathogenesis of CRC (Jones, 2012; Vogelstein et al., 2013). The aberrant methylation of several genes has been described in previous studies, including that of HOXA2 (Chen et al., 2019) and PBX3 (Sun et al., 2019). The association of gene methylation with CRC has also been documented. For example, aberrant methylation of CDKN2A has been identified in the breast, prostate, renal, and colon cancers, and has been demonstrated to be associated with transcriptional inhibition (Herman et al., 1995). Furthermore, promoter hypermethylation of SFRPs has been shown to inhibit WNT receptor binding, thereby downregulating pathway signaling in CRC (Suzuki et al., 2004), whereas hypermethylation of the hMLH1 promoter has been found to be related to gene inactivation in sporadic CRC (Cunningham et al., 1998). These findings accordingly indicate that the hypermethylation of certain genes is associated with the pathogenesis of CRC. However, the methylation characteristics of many genes in CRC remain to be elucidated.
N-acetyltransferase 1 (NAT1) is a phase II xenobiotic metabolizing enzyme that is expressed in almost all human tissues (Hein et al., 2018). In several cancers, the expression of NAT1 is associated with cell proliferation in vitro and with survival in vivo (Tiang et al., 2011; Minchin and Butcher, 2018). With regard to CRC, although a previous meta-analysis indicated that the NAT1 genotype was not significantly associated with an elevated CRC risk (Liu et al., 2012), a further study has demonstrated that in vivo knock-down of NAT1 in the HT-29 COAD cell line promoted an up-regulation of E-cadherin and cell–cell contact growth inhibition, thereby indicating that NAT1 may be a novel drug target for CRC therapeutics (Tiang et al., 2011). However, little is currently known regarding the methylation of NAT1 in COAD. Therefore, in this study, we aimed to evaluate the prognostic value and clinical significance of NAT1 methylation in COAD using RNA-sequence data obtained from The Cancer Genome Atlas (TCGA) database.
Materials and Methods
Source of Colon Adenocarcinoma-Related N-Acetyltransferase 1 Data
An RNA-seq dataset (Level 3) of COAD was downloaded from the Broad GDAC FireHose website (http://gdac.broadinstitute.org/, accessed June 7, 2019), which includes data for 263 COAD tissues and 30 corresponding adjacent normal colon tissues. We also downloaded DNA methylation profiles for 275 COAD tissues and 39 adjacent normal colon tissues (Illumina Human Methylation 450K) from the Broad GDAC FireHose site. Our use of TCGA data in the present study complied with the TCGA data usage policy.
Data of N-Acetyltransferase 1 mRNA Expression and Methylation of CpG Sites in Cancers
According to data obtained from the MethHC website (http://methhc.mbc.nctu.edu.tw) (Huang et al., 2015), the NAT1 gene has nine variants, each of which has a unique promoter and certain differences with respect to CpG sites (Table S1). However, only the NM_001160179 variant has a significantly higher methylation value (mean beta value > 0.5) in COAD tissues compared with normal tissues (P < 0.001) (Figure S1). We thus selected the NM_001160179 variant of NAT1 for subsequent analysis. We analyzed the expression of NAT1 mRNA in multiple cancers from the TCGA database using the GEPIA websites (http://gepia.cancer-pku.cn/) (Tang et al., 2017), and extracted and visualized the data relating to CpG sites in the NAT1 promoter region using the MethHC website.
Analysis of the Correlation Between N-Acetyltransferase 1 mRNA Expression and Methylation Values
We determined correlations between NAT1 mRNA expression and the mean methylation of NAT1 promoter CpG sites in COAD using Pearson correlation analysis, and subsequently examined the correlation of NAT1 mRNA expression with methylation at each of the promoter CpG sites. A P-value of less than 0.05 was considered indicative of statistical significance.
Analysis of Colon Adenocarcinoma Patient Survival Related to N-Acetyltransferase 1 Gene Expression and CpG Site Methylation
Initially, we applied Kaplan–Meier curve analysis and a Log-Rank test to examine the association between NAT1 mRNA expression and overall survival (OS) in COAD patients, and subsequently performed similar analyses for associations between methylation at each of the NAT1 promoter CpG sites and OS in COAD patients. Finally, we combined data for NAT1 mRNA expression and methylation at each of the CpG sites and analyzed the associations between these combinations and patient survival. Cox regression analysis was employed to assess independent indicators associated with the prognosis of COAD patients. Data were analyzed using R language (version 3.5.1) with a P-value of less than 0.05 indicating statistical significance.
Association Between N-Acetyltransferase 1 CpG Site Methylation and the Clinicopathological Features of Colon Adenocarcinoma
The clinicopathological features of COAD patients, including patient age, gender, TNM stage, and clinical stage, were extracted from the clinical data of COAD patients downloaded from the Broad GDAC FireHose website. The NAT1 mRNA expression and CpG sites that were found to be significantly associated with OS in COAD were used to analyze associations with the clinicopathological features of COAD. Data were analyzed using SPSS version 18.0 software (IBM Inc., Chicago, IL, USA), with a P-value of less than 0.05 indicating statistical significance.
Validation With an Independent Colorectal Cancer Dataset
In order to validate the results obtained for the TCGA dataset, we downloaded an independent dataset of CRC from the Gene Expression Omnibus (GEO) database, and subsequently compared the mean methylation of NAT1 and that of each CpG site in different tissues, with a P-value of less than 0.05 indicating statistical significance.
Results
N-Acetyltransferase 1 mRNA Expression and Methylation in Cancers
The sequences of the NAT1 gene CpG sites are presented in Figure 1A. As shown in Figure 1B, analysis of data from the TCGA database indicated that the expression of NAT1 mRNA was markedly decreased in COAD, kidney chromophobe, and rectum adenocarcinoma tissues compared with the corresponding normal tissues, but increased in those of breast cancer (P < 0.05). The mean methylation of NAT1 and promoter region CpG sites was markedly elevated in COAD tissues compared with that in normal colon tissues, although no significant differences were detected for the gene coding sequence (Figure 1C). The heatmap graph shown in Figure 1D shows the methylation of the eight CpG sites in COAD, which was visualized using the Clustvis web tool (Metsalu and Vilo, 2015). Table 1 lists the names and locations of the eight CpG sites in the promoter region of NAT1.
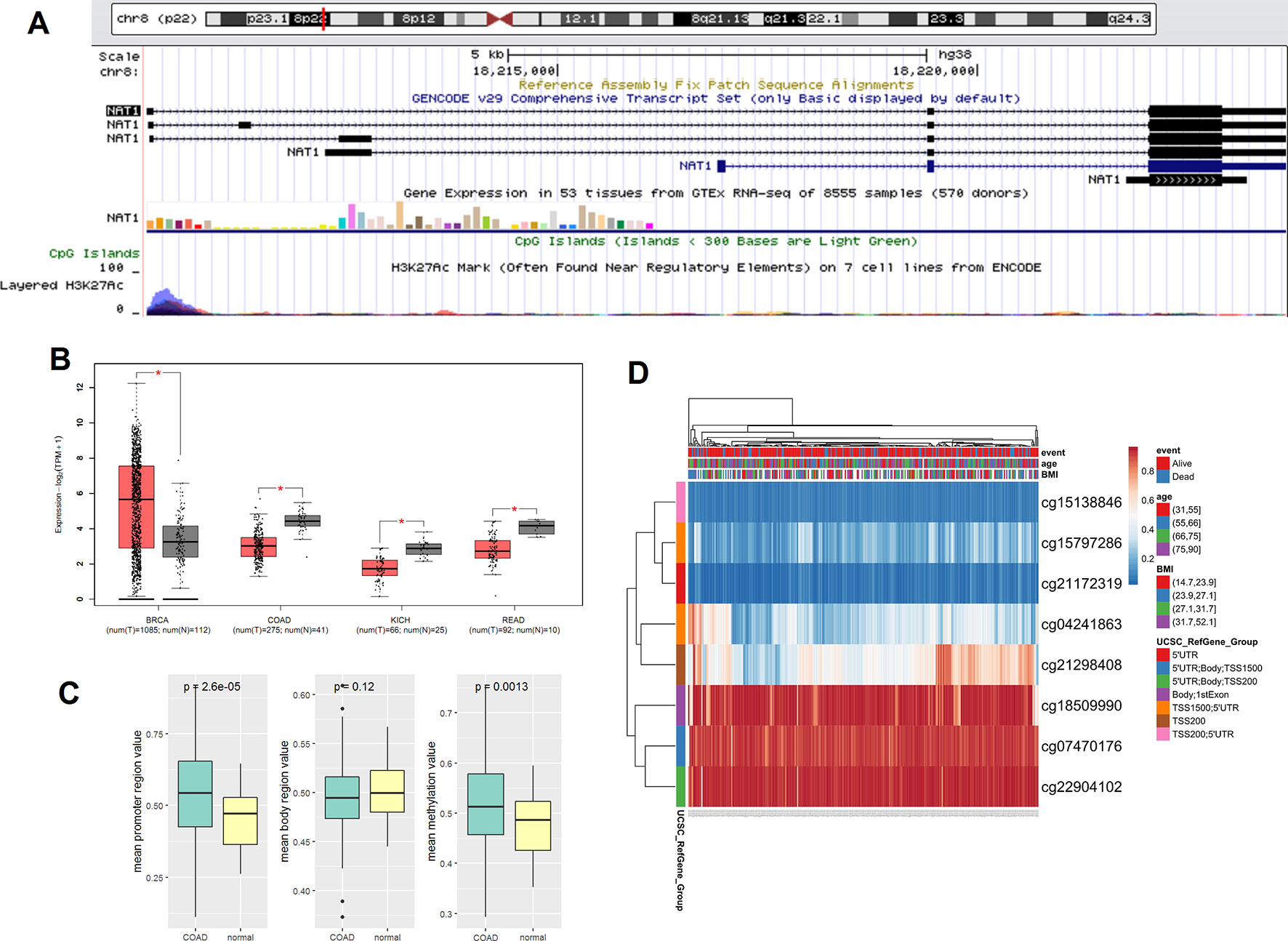
Figure 1 (A) Genomic position and functional annotation of N-acetyltransferase 1 (NAT1) gene from the University of California Santa Cruz genome browser; (B) Comparison of NAT1 mRNA in cancers and corresponding normal tissues; (C) The mean methylation of NAT1 in promoter region of COAD and normal tissues; (D) Heatmap of methylation CpG sites of NAT1 in COAD. *p < 0.05.
Correlation Analysis of N-Acetyltransferase 1 mRNA Expression and Methylation
We initially analyzed the correlation between NAT1 mRNA expression and mean methylation of the promoter region, and accordingly found that NAT1 mRNA expression was significantly negatively correlated with promoter methylation (P < 0.001) (Figure 2A), thereby indicating that methylation of the NAT1 promoter region has a significant influence on the expression of NAT1 mRNA. We subsequently conducted correlation analyses for associations between NAT1 mRNA expression and methylation of each of the eight CpG sites, which revealed that five CpG sites (cg04241863, cg15138846, cg15797286, cg21172319, and cg21298408) were negatively correlated with the expression of NAT1 mRNA in COAD (P < 0.05), whereas cg07470176 was positively correlated with NAT1 mRNA expression (P < 0.05), and the remaining two CpG sites showed no significant correlation (P > 0.05) (Figures 2B–G).
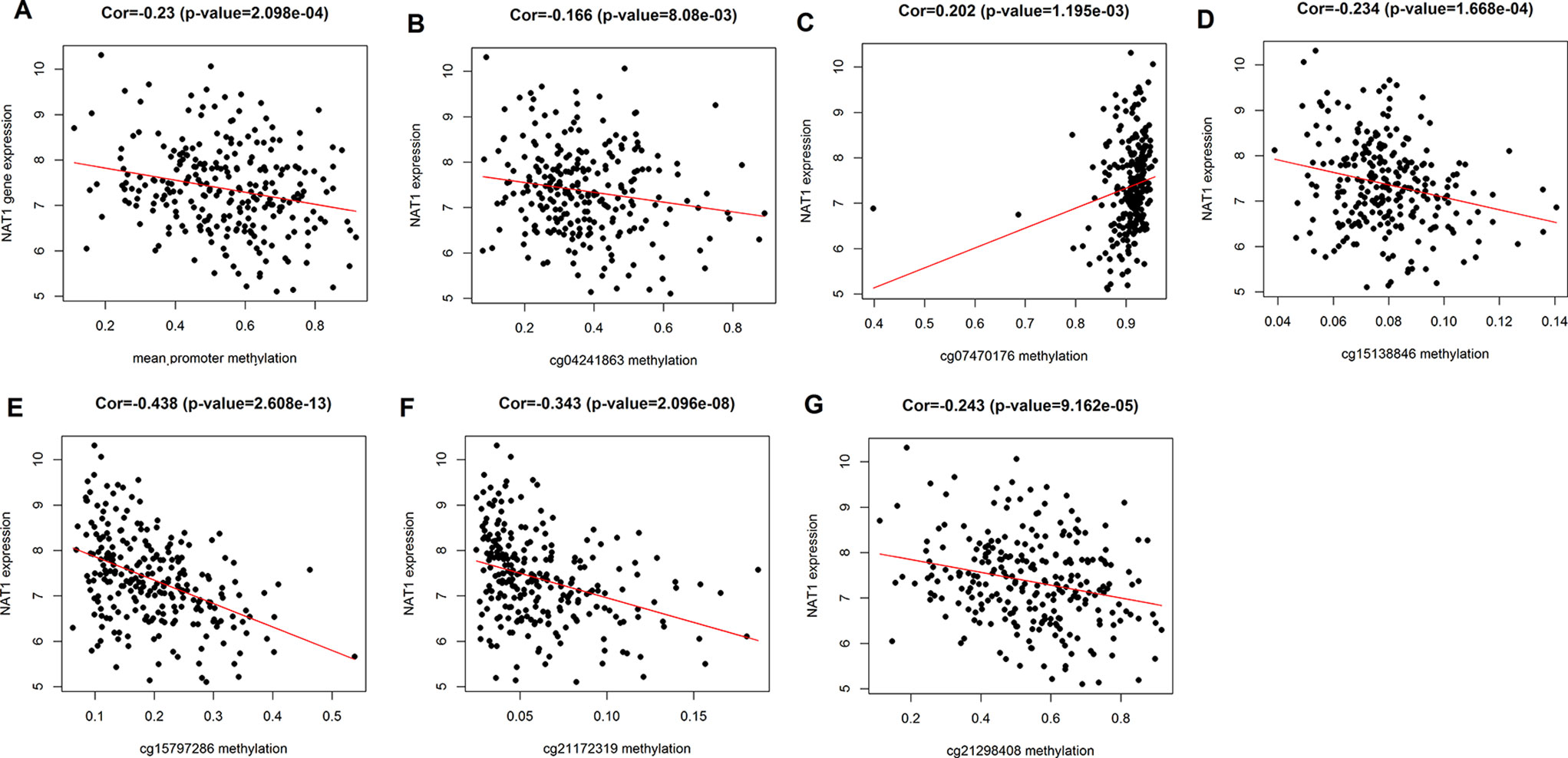
Figure 2 (A) Correlation analysis of N-acetyltransferase 1 (NAT1) mRNA with the mean methylation of promoter region; (B–G) Correlation analysis of NAT1 mRNA with cg04241863, cg07470176, cg15138846, cg15797286, cg21172319, cg21298408.
Analysis of Colon Adenocarcinoma Patient Survival Related to N-Acetyltransferase 1 CpG Site Methylation and Gene Expression
Using the median value as a cut-off, we found that compared with a high expression of NAT1 mRNA, a low expression was associated with a shorter OS in COAD patients. Subsequently, we examined the association between the methylation of individual NAT1 promoter CpG sites and OS, and accordingly found that high methylation of two CpG sites in particular (cg15797286 and cg18509990) was associated with a shorter OS compared with those characterized by low methylation (Figures 3B, C). Consistently, Cox regression analysis revealed that methylation at these two CpG sites independently predicted the prognosis of patients with COAD (P < 0.05) (Table 2).

Figure 3 (A) Kaplan–Meier curve of N-acetyltransferase 1 (NAT1) mRNA expression in COAD; (B) Kaplan–Meier curve of cg15797286 in COAD; (C) Kaplan–Meier curve of cg18509990 in COAD.
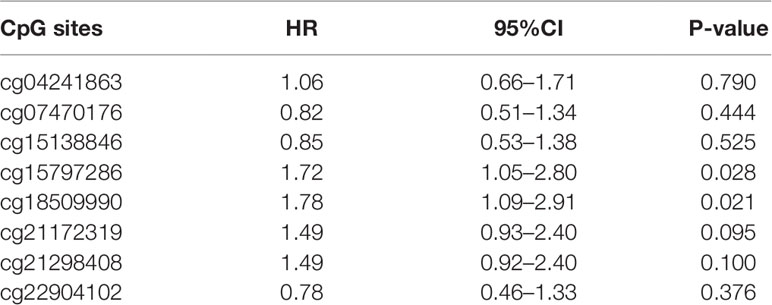
Table 2 N-acetyltransferase 1 (NAT1) CpG sites methylation associated with overall survival in colon adenocarcinoma (COAD).
Analysis of Colon Adenocarcinoma Patient Survival Based on a Combination of N-Acetyltransferase 1 mRNA Expression and CpG Site Methylation
In order to examine further indicators that could be used to predict the prognosis of COAD patients, we performed survival analysis based on combinations of NAT1 mRNA expression and methylation of each CpG site, and accordingly found that seven such combinations were significantly associated with OS in COAD patients (P < 0.05) (Figures 4A–G). Cox regression analysis revealed that six of the seven combinations were independent predictive indicators in patients with COAD (P < 0.05) Table 3.
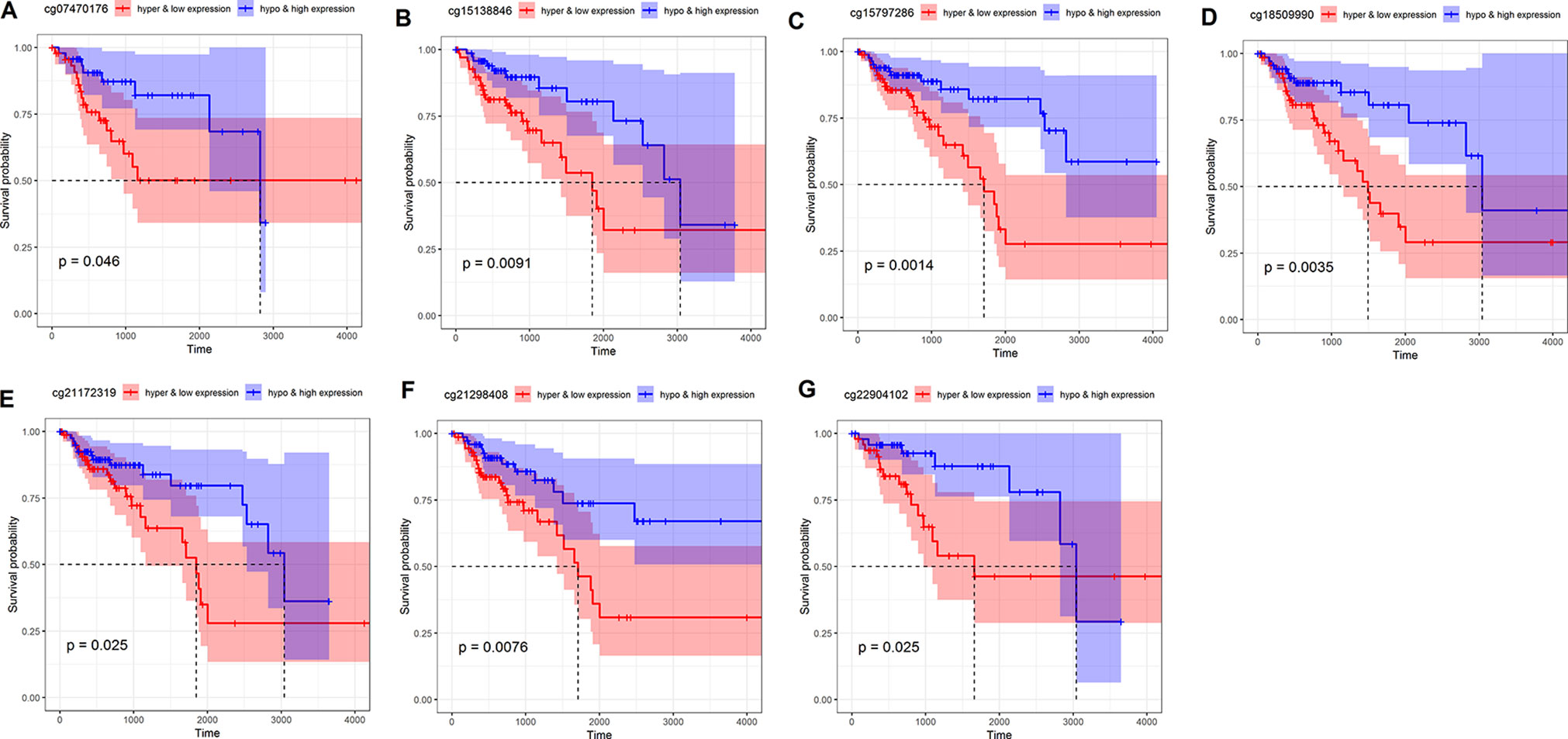
Figure 4 Kaplan–Meier curve of combination of N-acetyltransferase 1 (NAT1) mRNA with CpG sites in COAD. (A) NAT1 mRNA with cg07470176; (B) NAT1 mRNA with cg15138846; (C) NAT1 mRNA with cg15797286; (D) NAT1 mRNA with cg18509990; (E) NAT1 mRNA with cg21172319; (F) NAT1 mRNA with cg21298408; (G) NAT1 mRNA with cg22904102.
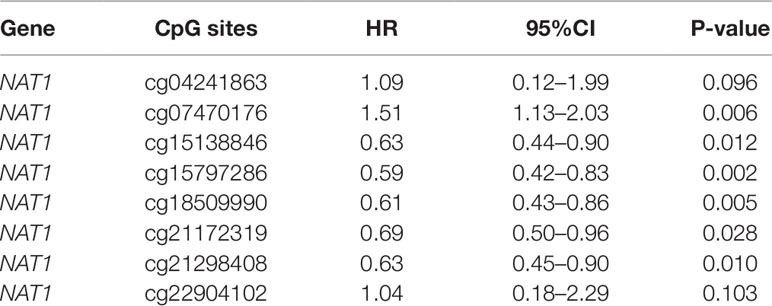
Table 3 Survival analysis of N-acetyltransferase 1 (NAT1) CpG sites combine with mRNA expression in colon adenocarcinoma (COAD).
Association Between N-Acetyltransferase 1 mRNA Expression and CpG Site Methylation and the Clinicopathological Features of Colon Adenocarcinoma
In order to examine the association between NAT1 mRNA expression and CpG site methylation and the clinicopathological features of COAD, we selected NAT1 mRNA and the two CpG sites (cg15797286 and cg18509990) showing significant associations with the OS of COAD patients. We observed that NAT1 mRNA expression was associated with the T stage, N stage, and clinical stage of COAD, and that similar results were obtained for cg15797286 (P < 0.05). In contrast, we detected no associations between cg18509990 and the clinicopathological features of COAD (P > 0.05) (Table 4).
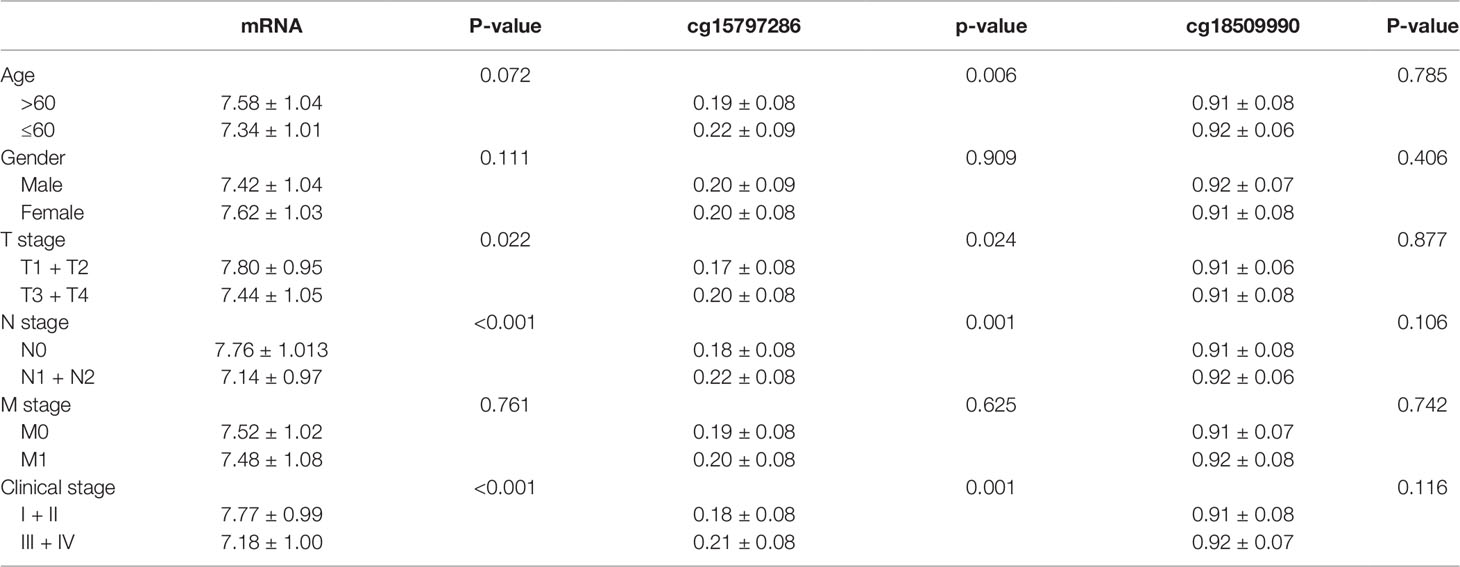
Table 4 Association of N-acetyltransferase 1 (NAT1) mRNA and two CpG sites with clinicopathological features of colon adenocarcinoma (COAD).
Validation of N-Acetyltransferase 1 CpG Site Methylation in Colon Cancer Using a Gene Expression Omnibus Dataset
In order to validate the aforementioned results based on analysis of a TCGA dataset, we downloaded an independent CRC dataset from the GEO database (GSE128067), which included non-recurrent colorectal adenoma (n = 30), recurrent colorectal adenoma (n = 29), and matched pair (n = 10) samples. The results showed that the mean methylation of the eight CpG sites was significantly increased in colorectal adenomas compared with normal tissues. We also found that compared with non-recurrent colorectal adenomas, the cg15797286 and cg21298408 sites in recurrent colorectal adenomas were characterized by higher levels of methylation (Figure 5).
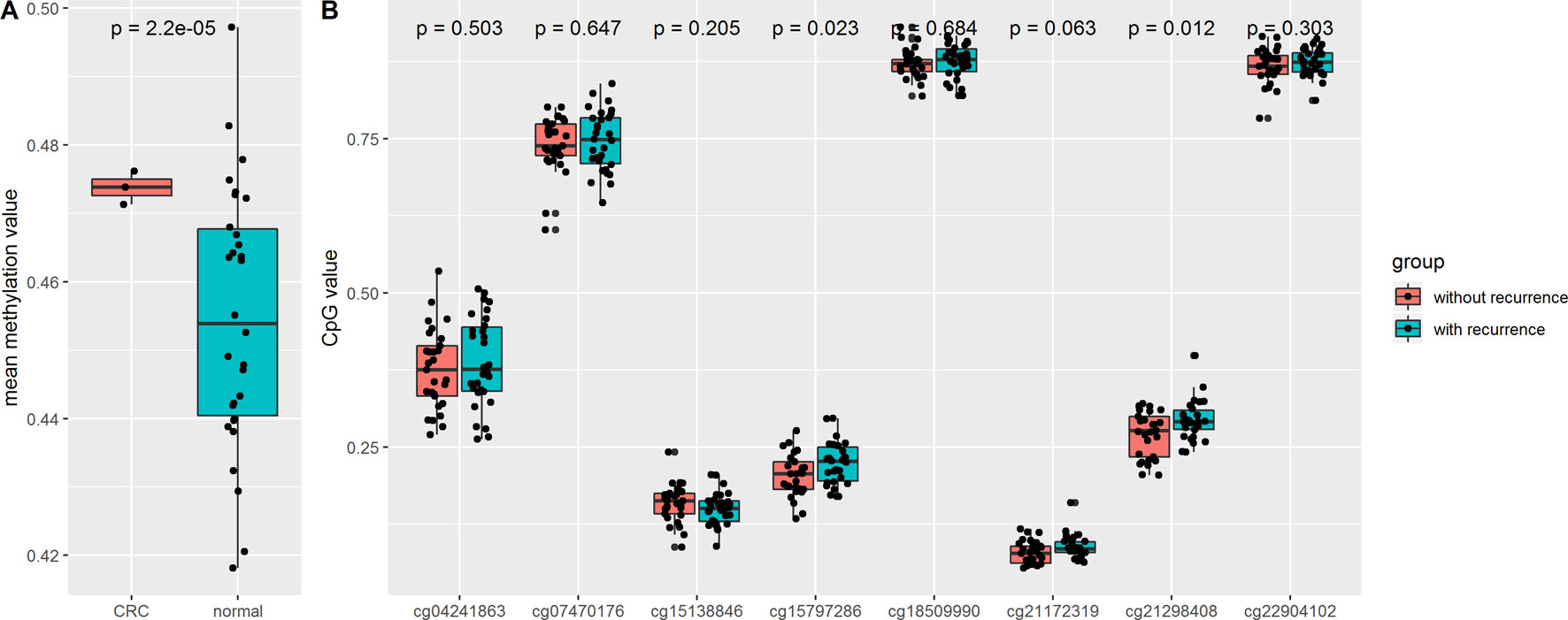
Figure 5 (A) Comparison of mean CpG value of N-acetyltransferase 1 (NAT1) in colorectal adenomas and normal tissues; (B) Comparison of methylation of CpG sites in colorectal adenomas and normal tissues.
Discussion
DNA methylation is a vital type of epigenetic modification that is related to the regulation of gene expression and alternative gene splicing (Jones and Baylin, 2007), and is also implicated in the pathogenesis of numerous diseases, including various cancers (Michalak et al., 2019; Mahmood and Rabbani, 2019). CpG sites are located within or near the promoter regions of genes, and the aberrant methylation of these sites often confers a hypermethylation status and causes gene silencing (Wang et al., 2018). Importantly, DNA methylation is a chemically stable modification and can be detected at relatively low cost, and is therefore regarded as a promising type of non-invasive biomarker that can be used in the diagnosis and prognosis of cancers (Gloss and Samimi, 2014; Chen et al., 2014). To date, studies have identified the methylation of several genes, including Claudin11 (Lee et al., 2016), APC2 (Butcher and Minchin, 2012), and ZNF331 (Liu et al., 2015), that are considered to be potential epigenetic biomarkers for the diagnosis and prognosis of CRC. In addition, a recent study showed that EREG/AREG methylation is associated with the efficacy of anti-EGFR therapy for CRC (Lee et al., 2016).
The NAT1 gene, located on chromosome 8p22, encodes one of two human enzymes that are known to metabolize arylamine- and hydrazine-type drugs (Butcher and Minchin, 2012), and in this regard, a previous study has reported that low NAT1 expression resulted in a distinctly poor response to chemotherapy in breast cancer patients (Minchin and Butcher, 2018). In COAD, NAT1 has been found to be enriched in the caffeine metabolism pathway (Liu et al., 2015), and inhibition of its expression by curcumin has been demonstrated in COAD cells (Chen et al., 2003). In the present study, we found that the expression of NAT1 mRNA was significantly reduced in COAD tissues compared with the corresponding normal tissues, which contrast with observations for the methylation of NAT1. Correlation analysis showed that mean methylation in the promoter region of NAT1 was negatively correlated with NAT1 mRNA expression, suggesting that NAT1 may function as a tumor suppressor gene, and that a high level of methylation could reduce the mRNA expression of this gene in COAD tissues. These findings thus reveal that methylation of NAT1 is associated with the development of COAD, and could thus serve as a potential biomarker for COAD development.
To further evaluate the effect of NAT1 methylation on COAD, we analyzed the correlation between NAT1 mRNA expression and the methylation of promoter CpG sites, and found that methylation of five of the eight CpG sites in the NAT1 promoter was negatively correlated with NAT1 mRNA expression, indicating that these five CpG sites may play regulatory roles in the expression of NAT1 mRNA. We subsequently assessed the prognostic value of all eight CpG sites, and found that only two of the sites were associated with the OS of COAD patients. Given that previous studies have indicated that joint survival analysis based on a combination of data relating to the methylation and mRNA expression of genes can be used to identify candidate prognostic biomarkers (Gao et al., 2018; Lu et al., 2019), we also combined these data in the present study to determine the influence on patient survival. We accordingly found that combinations of NAT1 mRNA expression with six of the eight CpG sites were significantly correlated with the prognosis of COAD patients. Thus, our results indicate that NAT1 mRNA expression, NAT1 methylation, and their combination have potential utility in predicting COAD patient survival.
A number of previous studies have reported the clinical significance of gene methylation in relation to COAD. For example, Ogino et al. (Ogino et al., 2009) investigated the methylation status of 649 COAD patients in two independent cohort studies, and quantified DNA methylation in the CpG island methylator phenotype (CIMP)-specific promoters of eight genes (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1). They found that patients with CIMP-high cancers experienced a significantly low COAD-specific mortality, and that CIMP-high tumors were associated with a significant reduction in COAD-specific mortality, regardless of both MSI and BRAF status. Yagi et al. (Yagi et al., 2010) identified the occurrence of three methylation epigenotypes (high-, intermediate-, and low-methylation) in CRC, and found that the high-methylation epigenotype was correlated with MSI-high and BRAF-mutation(+), whereas KRAS-mutation(+) CRC associated with an intermediate-methylation epigenotype showed worse prognosis. These findings thus provided evidence that gene methylation is associated with the clinical significance and prognosis of COAD.
Nevertheless, although previous studies have demonstrated that the methylation status of several genes is associated with the development of COAD, there is currently limited data available regarding the clinical significance of NAT1 expression and methylation in COAD. In this study, we found that NAT1 mRNA expression was associated with tumor T stage, N stage, and clinical stage, but not M stage. We also evaluated the clinical significance of two CpG sites (cg15797286 and cg18509990), and detected similar associations with respect to cg15797286, although not cg18509990. Interestingly, we observed the trend that the association of NAT1 mRNA expression with each of the evaluated clinicopathological feature was opposite to that of the association of these features with cg15797286 methylation, which further confirmed the correlation between NAT1 mRNA and cg15797286. These results also indicated that NAT1 mRNA expression and cg15797286 methylation are associated with the development of COAD. Moreover, in our analysis of a validated dataset, we found that the methylation of cg15797286 was increased in recurrent CRC, which further verified the association between cg15797286 and the development of COAD. Compared with previous studies that have examined the association of gene methylation with COAD (Ogino et al., 2009; Yagi et al., 2010), in the present study, we focused on a single variant of NAT1, and found that cg15797286 was associated with the clinical stage and survival of COAD patients, which have been reported previously.
To the best of our knowledge, this is the first study that has investigated the prognostic value and clinical significance of NAT1 methylation in COAD. Moreover, we provide the first evidence that the cg15797286 site in the NAT1 promoter region is closely related to the mRNA expression of this gene, as well as to COAD stage and patient survival. Despite these important findings, however, the study does have certain limitations that should be noted. First, the sample size of the validated data obtained from the GEO database was small, and we were unable to distinguish between COAD and rectum adenocarcinoma in the sample data. Thus, we could not verify our results with regard to COAD. Second, due to a lack of survival data for COAD, we were unable to verify the prognostic value of NAT1 methylation in the validated dataset. Third, the biological function of the NAT1 gene and CpG sites and their relationships with COAD remain largely unknown. Consequently, further study of a large COAD cohort is warranted to verify the prognostic value of NAT1 methylation, and in vivo and in vitro experiments are needed in order to elucidate the biological function of NAT1 and its CpG sites with respect to COAD.
Conclusion
In this study, we demonstrate that the NAT1 gene is characterized by a hypermethylated status in COAD, and that the cg15797286 site in the NAT1 promoter is closely related to the development and prognosis of COAD. These findings indicate that the DNA methylation of NAT1 could serve as a prognostic marker and potential therapeutic target for COAD. However, due to the aforementioned limitations, further studies are warranted to validate our results.
Data Availability Statement
Publicly available datasets were used in this study. This data can be found here: http://gdac.broadinstitute.org/.
Author Contributions
Study concept and design: CS, B-LH; Collection and assembly of data: CS, K-ZL, LL and J-LL; Data analysis and interpretation: K-ZL, Y-PT; Manuscript writing and review: All authors.
Funding
This study was partially supported by research funding from the National Natural Science Foundation (No. 81260083), Natural Science Foundation of Guangxi (No. 2018JJA140136), and College Students Innovative Entrepreneurship Project of Guangxi (No. 201910598012; WLXSZX19039).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviation
CRC, colorectal cancer; COAD, colon adenocarcinoma; NAT1, N-acetyltransferase 1; OS, overall survival; APC2, APC regulator of wnt signaling pathway 2; ZNF331, zinc finger protein 331; HOXA2, homeobox protein hox-A2; PBX3, PBX Homeobox 3.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01097/full#supplementary-material
References
Baena, R., Salinas, P. (2015). Diet and colorectal cancer. Maturitas 80 (3), 258–264. doi: 10.1016/j.maturitas.2014.12.017
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68 (6), 394–424. doi: 10.3322/caac.21492
Butcher, N. J., Minchin, R. F. (2012). Arylamine N-acetyltransferase 1: a novel drug target in cancer development. Pharmacol. Rev. 64 (1), 147–165. doi: 10.1124/pr.110.004275
Chen, J. C., Hwang, J. M., Chen, G. W., Tsou, M. F., Hsia, T. C., Chung, J. G. (2003). Curcumin decreases the DNA adduct formation, arylamines N-acetyltransferase activity and gene expression in human colon tumor cells (colo 205). In Vivo 17 (3), 301–309. doi: 10.1016/S0887-2333(03)00020-1
Chen, Z. Y., Zhang, J. L., Yao, H. X., Wang, P. Y., Zhu, J., Wang, W., et al. (2014). Aberrant methylation of the SPARC gene promoter and its clinical implication in gastric cancer. Sci. Rep. 4. doi: 10.1038/srep07035
Chen, D., Song, Y., Zhang, F., Wang, X., Zhu, E., Zhang, X., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66 (2), 115–132. doi: 10.3322/caac.21338
Chen, D., Song, Y., Zhang, F., Wang, X., Zhu, E., Zhang, X., et al. (2019). Genome-wide analysis of lung adenocarcinoma identifies novel prognostic factors and a prognostic score. Front. Genet. 10, 493. doi: 10.3389/fgene.2019.00493
Cunningham, J. M., Christensen, E. R., Tester, D. J., Kim, C. Y., Roche, P. C., Burgart, L. J., et al. (1998). Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 58 (15), 3455–3460.
Gao, C., Zhuang, J., Li, H., Liu, C., Zhou, C., Liu, L., et al. (2018). Exploration of methylation-driven genes for monitoring and prognosis of patients with lung adenocarcinoma. Cancer Cell Int. 18, 194. doi: 10.1186/s12935-018-0691-z
Gloss, B. S., Samimi, G. (2014). Epigenetic biomarkers in epithelial ovarian cancer. Cancer Lett. 342 (2), 257–263. doi: 10.1016/j.canlet.2011.12.036
Hein, D. W., Fakis, G., Boukouvala, S. (2018). Functional expression of human arylamine N-acetyltransferase NAT1*10 and NAT1*11 alleles: a mini review. Pharmacogenet. Genomics 28 (10), 238–244. doi: 10.1097/FPC.0000000000000350v
Herman, J. G., Merlo, A., Mao, L., Lapidus, R. G., Issa, J. P., Davidson, N. E., et al. (1995). Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 55 (20), 4525–4530.
Huang, W. Y., Hsu, S. D., Huang, H. Y., Sun, Y. M., Chou, C. H., Weng, S. L., et al. (2015). MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 43 (Database issue), D856–D861. doi: 10.1093/nar/gku1151
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13 (7), 484–492. doi: 10.1038/nrg3230
Jones, P. A., Baylin, S. B. (2007). The epigenomics of cancer. Cell 128 (4), 683–692. doi: 10.1016/j.cell.2007.01.029
Lee, M. S., McGuffey, E. J., Morris, J. S., Manyam, G., Baladandayuthapani, V., Wei, W., et al. (2016). Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br. J. Cancer 114 (12), 1352–1361. doi: 10.1038/bjc.2016.87
Liu, J., Ding, D., Wang, X., Chen, Y., Li, R., Zhang, Y., et al. (2012). N-acetyltransferase polymorphism and risk of colorectal adenoma and cancer: a pooled analysis of variations from 59 studies. PloS One 7 (8), e42797. doi: 10.1371/journal.pone.0042797
Liu, F., Ji, F., Ji, Y., Jiang, Y., Sun, X1, Lu, Y., et al. (2015). In-depth analysis of the critical genes and pathways in colorectal cancer. Int. J. Mol. Med. 36 (4), 923–930. doi: 10.3892/ijmm.2015.2298
Lu, T., Chen, D., Wang, Y., Sun, X., Li, S., Miao, S., et al. (2019). Identification of DNA methylation-driven genes in esophageal squamous cell carcinoma: a study based on The Cancer Genome Atlas. Cancer Cell Int. 19, 52. doi: 10.1186/s12935-019-0770-9
Mahmood, N., Rabbani, S. A. (2019). DNA Methylation Readers and Cancer: Mechanistic and Therapeutic Applications. Front. Oncol. 9, 489. doi: 10.3389/fonc.2019.00489
Metsalu, T., Vilo, J. (2015). ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43 (W1), W566–W570. doi: 10.1093/nar/gkv468
Michalak, E. M., Burr, M. L., Bannister, A. J., Dawson, M. A. (2019). The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 20 (10), 573–589. doi: 10.1038/s41580-019-0143-1
Minchin, R. F., Butcher, N. J. (2018). Trimodal distribution of arylamine N-acetyltransferase 1 mRNA in breast cancer tumors: association with overall survival and drug resistance. BMC Genomics 19 (1), 513. doi: 10.1186/s12864-018-4894-4
Ogino, S., Nosho, K., Kirkner, G. J., Kawasaki, T., Meyerhardt, J. A., Loda, M., et al. (2009). CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 58 (1), 90–96. doi: 10.1136/gut.2008.155473
Pfeifer, G. P. (2018). Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 19 (4). doi: 10.3390/ijms19041166
Saghafinia, S., Mina, M., Riggi, N., Hanahan, D., Ciriello, G. (2018). Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 251066-1080 (4), e1068. doi: 10.1016/j.celrep.2018.09.082
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barzi, A., et al. (2017). Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67 (3), 177–193. doi: 10.3322/caac.21395
Sun, H., Huang, H., Li, D., Zhang, L., Zhang, Y., Xu, J., et al. (2019). PBX3 hypermethylation in peripheral blood leukocytes predicts better prognosis in colorectal cancer: A propensity score analysis. Cancer Med. 8 (8), 4001–4011. doi: 10.1002/cam4.2321
Suzuki, H., Watkins, D. N., Jair, K. W., Schuebel, K. E., Markowitz, S. D., Chen, W. D., et al. (2004). Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 36 (4), 417–422. doi: 10.1038/ng1330
Tang, Z., Li, C., Kang, B., Gao, G., Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 (W1), W98–W102. doi: 10.1093/nar/gkx247
Tiang, J. M., Butcher, N. J., Cullinane, C., Humbert, P. O., Minchin, R. F. (2011). RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One 6 (2), e17031. doi: 10.1371/journal.pone.0017031
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., Jr., Kinzler, K. W. (2013). Cancer genome landscapes. Science 339 (6127), 1546–1558. doi: 10.1126/science.1235122
Vulcan, A., Manjer, J., Ericson, U., Ohlsson, B. (2017). Intake of different types of red meat, poultry, and fish and incident colorectal cancer in women and men: results from the Malmo Diet and Cancer Study. Food Nutr. Res. 61 (1), 1341810. doi: 10.1080/16546628.2017.1341810
Wang, Q., Wang, G., Liu, C., He, X. (2018). Prognostic value of CpG island methylator phenotype among hepatocellular carcinoma patients: A systematic review and meta-analysis. Int. J. Surg. 54 (Pt A), 92–99. doi: 10.1016/j.ijsu.2018.04.033
Keywords: methylation, N-acetyltransferase 1, colon adenocarcinoma, prognosis, The Cancer Genome Atlas
Citation: Shi C, Xie L-y, Tang Y-p, Long L, Li J-l, Hu B-l and Li K-z (2019) Hypermethylation of N-Acetyltransferase 1 Is a Prognostic Biomarker in Colon Adenocarcinoma. Front. Genet. 10:1097. doi: 10.3389/fgene.2019.01097
Received: 16 July 2019; Accepted: 11 October 2019;
Published: 06 November 2019.
Edited by:
Nejat Dalay, Istanbul University, TurkeyReviewed by:
Naoko Hattori, National Cancer Center Research Institute, JapanSilvia Carolina Galvan, National Autonomous University of Mexico, Mexico
Copyright © 2019 Shi, Xie, Tang, Long, Li, Hu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bang-li Hu, aHViYW5nbGlAZ3htdS5lZHUuY24=; Ke-zhi Li, bGlrZXpoaUBneG11LmVkdS5jbg==
 Cheng Shi
Cheng Shi Li-ye Xie2
Li-ye Xie2 Bang-li Hu
Bang-li Hu