Abstract
Circadian rhythms produce a biological measure of the time of day. In plants, circadian regulation forms an essential adaptation to the fluctuating environment. Most of our knowledge of the molecular aspects of circadian regulation in plants is derived from laboratory experiments that are performed under controlled conditions. However, it is emerging that the circadian clock has complex roles in the coordination of the transcriptome under natural conditions, in both naturally occurring populations of plants and in crop species. In this review, we consider recent insights into circadian regulation under natural conditions. We examine how circadian regulation is integrated with the acute responses of plants to the daily and seasonally fluctuating environment that also presents environmental stresses, in order to coordinate the transcriptome and dynamically adapt plants to their continuously changing environment.
Introduction
The Earth rotates on its axis approximately every 24 h. This causes daily cycles in a variety of environmental conditions, such as the presence, spectrum and direction of light, ambient temperature, relative humidity, and the activity of herbivores. Furthermore, the axial tilt of the Earth causes seasonal changes in the photoperiod and other environmental parameters. The daily environmental cycles appear to have driven the evolution of circadian rhythms. Circadian rhythms are biological cycles that have a period of about 24 h and persist in the absence of external cues. A key feature of circadian rhythms is that they define the daily phase of biological processes, and therefore organize the daily timing of the transcriptome so that cellular processes occur at appropriate times of day and in a coordinated manner (Harmer et al., 2000). Circadian regulation increases the performance and fitness of higher plants, including crops (Green et al., 2002; Dodd et al., 2005; Turner et al., 2005; Graf et al., 2010; Izawa et al., 2011; Müller et al., 2015).
The molecular functioning of plant circadian rhythms and the circadian organization of the transcriptome has been investigated predominantly under laboratory conditions. However, circadian rhythms are thought to have evolved as an adaptation to naturally fluctuating environments, and an understanding of their importance for ecosystems and field-grown crops requires knowledge of circadian regulation under natural conditions. Here, we explore the recent expansion of understanding of molecular and cellular aspects of circadian regulation under natural conditions, with a focus on the regulation of the transcriptome.
Features of Circadian Rhythms
The circadian system in plants is often considered to include several conceptual components. First, input or entrainment pathways adjust the phase of the circadian oscillator to match the phase of the environment by changing the circadian phase in response to specific environmental cues known as zeitgebers. Second, the circadian oscillator is a molecular network that produces an estimate of the time of day that, in plants, consists of an interconnected network of genes and proteins arranged in feedback loops. Third, output mechanisms communicate the measure of the time of day generated by the circadian oscillator to clock-controlled processes in the cell. Finally, signaling pathways that entrain the circadian clock and others that participate in environmental responses are, themselves, circadian regulated so that the magnitude of their response to a defined stimulus depends upon the time of day. This aspect of circadian regulation is known as the circadian “gating” of signal transduction (Hotta et al., 2007). Next, we summarize key features of circadian regulation in Arabidopsis thaliana, and the reader is referred to other review articles for greater depth (Nagel and Kay, 2012; Hsu and Harmer, 2014; Huang and Nusinow, 2016; Millar, 2016). There are differences in circadian clock architecture between Arabidopsis and crop species, which are summarized in a number of excellent articles (Song et al., 2010; Bendix et al., 2015; Nakamichi, 2015). Key differences between Arabidopsis and crops for consideration include effects of polyploidy, genome duplication and hybrid vigor (Ermolaeva et al., 2003; Ni et al., 2009; Hotta et al., 2013; Liu et al., 2014), allelic diversity (Xie et al., 2015), and species- and variety-specific alterations in the relationship between the circadian clock and control of flowering time (Turner et al., 2005; Cockram et al., 2007; Song et al., 2015).
Mechanisms of Circadian Regulation in Plants
Entrainment is the process whereby the phase of the circadian oscillator is adjusted to match the daily phase of the environment. This is crucial to allow the oscillator to provide a faithful biological estimate of the time of day. In Arabidopsis, entrainment occurs in response to light, temperature, and metabolic cues (Somers et al., 1998; Salomé and McClung, 2005; Kim et al., 2007; Haydon et al., 2013). The entrainment mechanism involves the phytochrome, cryptochrome, and other LOV-domain photoreceptors such as ZEITLUPE (ZTL) (Somers et al., 1998; Kim et al., 2007), temperature responses mediated by PRR7, PRR9, and ELF3 (Salomé and McClung, 2005; Thines and Harmon, 2010), and metabolite signaling by sugar-sensing protein kinases such as SNF1-RELATED PROTEIN KINASE1.1 (KIN10/AKIN10/SnRK1.1) and also PHYTOCHROME INTERACTING FACTOR (PIF) proteins (Haydon et al., 2013; Shin et al., 2017; Shor et al., 2017; Frank et al., 2018). Entrainment in Arabidopsis generally takes the form of parametric entrainment, whereby the pace of the oscillator is transiently accelerated or decelerated in order to adjust the circadian phase (Covington et al., 2001; Michael et al., 2003; Rémi et al., 2010). This contrasts nonparametric entrainment, which involves an instantaneous reset of the circadian phase. Effects upon the circadian oscillator of factors such as relative humidity (Mwimba et al., 2018), phytohormones (Hanano et al., 2006) and ROS (Lai et al., 2012) might allow these factors to entrain the circadian oscillator, although whether they are genuinely zeitgebers would benefit from further assessment by formal approaches such as the construction of phase response curves.
The circadian oscillator is formed from transcription/translation loops that are connected primarily by repressive feedback (Hsu and Harmer, 2014), augmented by post-translational and epigenetic mechanisms. Over 20 circadian clock-related genes have been identified in Arabidopsis, with homologs of these present in other plants including crops. CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are MYB-like transcription factors forming part of a core feedback loop within the circadian oscillator (Schaffer et al., 1998; Wang and Tobin, 1998). CCA1 and LHY transcript abundance peaks in the morning and decreases throughout the day. CCA1 and LHY repress the expression of the evening-phased TIMING OF CAB EXPRESSION1 (TOC1) by binding to its promoter (Harmer et al., 2000; Lu et al., 2009; Yakir et al., 2009). Inversely, TOC1 regulates CCA1 and LHY (Alabadı́ et al., 2001) by binding to their promoters to repress their expression (Gendron et al., 2012; Huang et al., 2012). CCA1 and LHY also repress other evening-phased genes, including GIGANTEA (GI), LUXa ARRHYTHMO (LUX), BROTHER OF LUX ARRHYTHMO (BOA), EARLY FLOWERING3 (ELF3), and ELF4 (Dai et al., 2011; Lau et al., 2011; Lu et al., 2012; Nagel and Kay, 2012). Furthermore, CCA1 and LHY appear to repress day-phased PRR9 and PRR7 in a time of day-dependent manner (Kamioka et al., 2016; Adams et al., 2018). PRR9 reaches peak abundance after dawn, followed by PRR7, and these act as CCA1 and LHY repressors along with their homolog PRR5. Furthermore, PRR-family proteins repress transcription of the morning-phased REVEILLE8 (RVE8), which is within the same protein family as CCA1 and LHY (Rawat et al., 2009; Nakamichi et al., 2010; Farinas and Mas, 2011; Nakamichi et al., 2012; Farré and Liu, 2013). In contrast to the morning RVE8 transcript peak, RVE8 protein levels peak during the afternoon and RVE8 appears to positively regulate hundreds of evening-phased genes by binding to evening element (EE) promoter motifs to induce their transcription (Rawat et al., 2011; Hsu et al., 2013). RVE4 and RVE6 are close homologs to RVE8 that might function redundantly with RVE8 (Hsu et al., 2013). A further component of the circadian oscillator is the evening complex (EC), incorporating LUX, ELF3, and ELF4. Within this, ELF3 and ELF4 negatively regulate the morning gene PRR9 and induce CCA1 expression by interacting with BOA (Dai et al., 2011; Dixon et al., 2011; Helfer et al., 2011; Nusinow et al., 2011; Herrero et al., 2012).
Circadian-regulated cis elements within gene promoters appear to couple the circadian oscillator with the rhythmic regulation of the transcriptome. Therefore, in addition to forming parts of the circadian oscillator, circadian oscillator components are positioned within output pathways to provide transcriptional regulation of circadian-regulated genes. There is a rich set of circadian-regulated cis elements within the promoters of the Arabidopsis genome, and these confer distinct circadian phases to distinct subsets of transcripts (Harmer et al., 2000; Covington and Harmer, 2007). For example, the circadian oscillator components CCA1 and LHY are thought to bind to the promoters of at least 439 (CCA1) and 722 genes (LHY), respectively (Nagel et al., 2015; Kamioka et al., 2016; Adams et al., 2018). Similarly, interaction between circadian oscillator components and other signaling proteins, such as the PIF proteins involved in environmental signaling, can change their promoter-binding activity and allow the integration of circadian and environmental cues (Martín et al., 2018).
Environmental Differences Between Laboratory and Field Conditions Will Impact Circadian Regulation
Laboratory investigations of plant circadian rhythms commonly involve the entrainment of plants to square-wave light/dark cycles in the presence of uniform temperature conditions (Figure 1A). The plants are transferred subsequently to constant light and temperature conditions for investigation of the rhythmic process under question (Figure 1B). Under permissive constant conditions, the circadian-regulated process will “free run” and its characteristics (e.g., circadian period, phase, damping) can be evaluated and quantified (Figure 1B). Plants can be exposed to a variety of other types of entrainment conditions, such as temperature fluctuations, which are usually also imposed with square-wave patterns. For this analysis, it is common to discard data collected during the first 24 h of constant conditions because this often includes transient responses to the final dawn that are not representative of the subsequent free-running rhythm (Figure 1B) (Dodd et al., 2014).
Figure 1
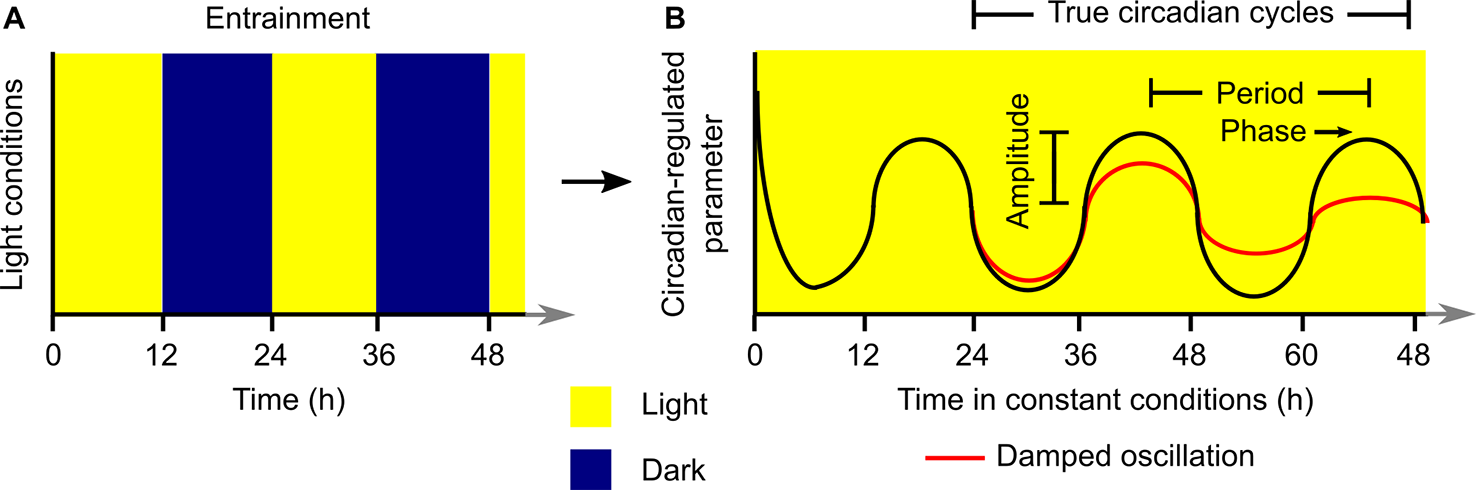
Generalized structure for execution of an experiment to investigate properties of a circadian rhythm under laboratory conditions. The plant is (A) entrained to a square-wave light/dark (or warm/cold) cycle and subsequently (B) transferred to constant conditions where the circadian-regulated feature of the plant will free-run. Analysis of the resultant rhythm (B) with a variety of quantitative tools can be used to extract information about the circadian period, phase, and amplitude. Data collected during the first 24 h of constant conditions is not considered to be a true circadian cycle, and these data are often discarded prior to quantitative analysis of free-running rhythms.
These structured environmental conditions are important for investigating the architecture and function of the circadian oscillator, and the downstream circadian regulation of transcription, metabolism, development, and physiology. However, plants growing under natural conditions are unlikely to experience conditions of continuous light, darkness, constant temperature or square-wave light/dark cycles (Figures 2A–E). There are also differences in environmental conditions caused by the weather, which are superimposed on the 24-h cycle. For example, the temperature can differ between successive days (e.g., Figures 2B, E). Therefore, understanding the contribution of the circadian oscillator to temporal structures in plants growing under natural conditions requires knowledge of the functioning of the circadian oscillator under either controlled conditions that fluctuate in a manner more representative of natural environments, or under naturally occurring field conditions (Figures 2B–E). Key differences between commonly used laboratory conditions (Figure 2A) and those experienced by plants in nature include gradual changes in irradiance at dawn and dusk, unpredictable changes in light and temperature conditions due to the weather (Figures 2A, B). There are also progressive seasonal changes in photoperiod, irradiance, and temperature (Figures 2B–E). While there have been comparisons of differences between plants grown in growth chambers and field conditions (Limpens et al., 2012; Poorter et al., 2016), these studies have not considered the impacts upon circadian regulation of these differences. Because a number of environmental parameters that differ between chamber and field conditions are key entrainment cues for the clock, there are likely to be fascinating differences in circadian function under field conditions that add depth to our understanding of plant circadian regulation. Here, we focus on the interactions between circadian and abiotic environmental cues that regulate the transcriptome under natural conditions.
Figure 2

Examples of differences in environmental parameters that will impact circadian rhythms of plants in growth chamber and field conditions. (A) Typical entrainment program used in the laboratory to study rhythmic processes, characterized by a 12-h light/12-h dark square-wave cycle of illumination combined with constant temperature conditions. (B–E) Light and temperature conditions in a naturally-fluctuating environment, monitored for 24 h during four cardinal points of the year. In this case, the environment is characterized by gradual daily changes in light and temperature conditions, seasonal differences in the temperature and light environment, and short-term variations caused by altering weather conditions. Data were collected during 2018–2019 at a field site equipped with a meteorological station, Taka-cho, Nishiwaki, Hyogo Prefecture, Japan (35° 05’ N, 134° 54’ E) (Kudoh et al., 2018; Nagano et al., 2019).
Effects of Light Conditions in Natural Environments Upon Plant Circadian Regulation
Plants in natural environments experience fluctuating light levels during the course of the day due to changes in cloud cover and shading by neighbouring plants (Figures 2B–E). These fluctuations affect the intensity as well as the wavelengths of light that are incident upon plants. This is likely to alter entrainment, because increases in light intensity shorten the circadian period in Arabidopsis (Somers et al., 1998). Both red and blue light provide entraining inputs to the circadian clock (Millar et al., 1995; Somers et al., 1998). In Arabidopsis, red light is sensed by the phytochromes phyA-E, while blue light is sensed by the cryptochromes cry1 and cry2. However, because plants are normally exposed to white light, combined information from these and other photoreceptors will influence the circadian oscillator (Oakenfull and Davis, 2017). In the laboratory, continuous far red light causes a shorter circadian period, so it is possible that under natural conditions the ratio of red to far red light (R:FR), which is altered by shade, the threat of shade, and the angle of incident light, will influence circadian regulation (Wenden et al., 2011). However, there is little information concerning the effect of R:FR more representative of natural environments upon the circadian oscillator. In addition to the phototransduction pathways that act upon the circadian oscillator through the photoreceptors, photosynthetic light harvesting leads to the production of photosynthetic sugars and reactive oxygen species (ROS). Photosynthetic sugars can entrain the circadian oscillator (Haydon et al., 2013; Frank et al., 2018) and there is evidence that ROS can adjust circadian oscillator function (Lai et al., 2012), so a fascinating area for future investigation would be to identify the relevance of these mechanisms that couple metabolism with circadian regulation under natural conditions.
Under square-wave environmental fluctuations, the transition from dark to light and vice versa is instantaneous, while under natural conditions plants experience a gradual increase and decrease in light intensity at dawn and dusk, respectively (Figure 1). A comparison of plant growth under square-wave and naturally fluctuating light environments identified that plants grown under fluctuating light have thinner leaves, decreased light absorption by leaves, and a decrease in carbon assimilation compared with plants grown under square-wave light/dark cycles (Vialet-Chabrand et al., 2017). In sugar beet, a gradual increase in light intensity caused more efficient carbon flow within plants compared with sudden transitions between light and dark (Fondy et al., 1989). These differences in physiology could potentially reflect effects of altered transcriptome dynamics between plants grown under natural and laboratory conditions of light.
Effects of Temperature Conditions in Natural Environments Upon Plant Circadian Regulation
Under natural conditions, in addition to the day/night temperature changes, there are temperature fluctuations during and between days depending on the weather conditions and season. Experiments with field-grown rice suggest that the circadian clock is robust to fluctuating environments, although the accumulation of individual transcripts is affected by temperature (Matsuzaki et al., 2015). The circadian period is relatively robust to temperature fluctuations within a specific range, typically between 12°C and 27°C, which is known as temperature compensation of the circadian clock (Gould et al., 2006). Laboratory studies have identified that PRR7, PRR9, an interaction between GI and CCA1/LHY, and FLC have roles in temperature compensation (Salomé and McClung, 2005; Edwards et al., 2006; Gould et al., 2006; Salomé et al., 2010). QTL analysis has also suggested that ZTL and GI contribute to this response (Edwards et al., 2005). Modeling suggests that blue light signaling might participate in temperature compensation through the cryptochrome photoreceptors (Gould et al., 2013). Formal quantification of temperature compensation within the circadian oscillator requires analysis of the properties of circadian rhythms under free-running conditions. However, plants under field conditions experience a greater range of varying environmental parameters in comparison to the very structured environmental conditions used to study temperature compensation in the laboratory. It is possible that responses of the circadian oscillator to the range of environmental parameters presented by field conditions provides an opportunity to investigate the environmental parameter space within which circadian rhythms remain temperature compensated.
The difference in temperature between day and night can also act as an entrainment cue. The circadian clock can be entrained by temperature fluctuations as small as 4°C (Gould et al., 2006). Daily temperature fluctuations with a warmer day and cooler night (e.g., cycles of 12 h at 31°C, 12 h at 20°C, known as thermocycles) can drive daily oscillations of 12% and 8% of genes in rice and poplar under constant light, respectively (Filichkin et al., 2011). Although many laboratory experiments investigating circadian rhythms do not incorporate day/night temperature fluctuations, such a difference can affect circadian regulation (Mizuno et al., 2014) and be essential for some aspects of plant performance. For example, a 1°C increase in the temperature during the night can decrease rice grain yield by 10% (Peng et al., 2004). Furthermore, warm night temperatures disrupt rhythmically expressed genes, particularly those genes driven by thermocyles (Desai et al., 2019), which has impacts upon productivity.
Interactions Between Abiotic Stress Responses and Circadian Regulation Will Shape the Transcriptome Under Natural Conditions
Responses to low temperature and circadian regulation are linked intrinsically. The C-REPEAT/DRE BINDING FACTOR (CBF) proteins are major regulators of cold acclimation (Thomashow et al., 2001). CBF1, 2, and 3 transcripts undergo circadian regulation in Arabidopsis under ambient temperature conditions, with transcript abundance peaking 4 h after subjective dawn (Harmer et al., 2000; Fowler et al., 2005). The circadian clock has two main effects on the CBF pathway; not only is there circadian regulation of basal CBF transcript accumulation under ambient temperatures, but there is also circadian gating of cold-induced accumulation of CBF transcripts. In this context, low temperature treatment causes greater CBF transcript accumulation 4 h after subjective dawn compared with other times of day (Fowler et al., 2005). CCA1 and LHY positively regulate the CBF-pathway because circadian cycling of CBF genes is absent from the cca1 lhy double mutants, which also have reduced freezing tolerance (Dong et al., 2011). Interestingly, a low red/far-red ratio of light causes greater CBF transcript accumulation in Arabidopsis in a manner that depends upon the time of day, suggesting that light quality might influence cold acclimation in a circadian-dependent manner (Franklin and Whitelam, 2007). There is also a seasonal dimension to this circadian regulation, with plants grown under short days at ambient temperatures having greater daily amplitudes of the rhythm of CBF transcript abundance (Lee and Thomashow, 2012). This is thought to increase freezing tolerance of plants grown under short photoperiods compared with long photoperiods (Lee and Thomashow, 2012). Furthermore, under long day conditions the CBF pathway is negatively regulated by PIF4, PIF7 and PHYB (Kidokoro et al., 2009; Lee and Thomashow, 2012). This suggests that fluctuating light and temperature, as well as photoperiod, will also impact the involvement of the circadian oscillator in the transcriptomic responses of plants to abiotic stress.
Cold acclimation responses differ between plants acclimated at constant temperatures under laboratory conditions compared with the variable temperature conditions in natural environments. For example, cold acclimation of plants in the field arises from multiple peaks of CBF gene expression that are concurrent with temperature decreases during the night (Hiraki et al., 2018). This contrasts the single peak of CBF expression during the daytime reported in plants acclimated at a constant temperature of 2°C in a growth chamber (Hiraki et al., 2018). The single or double peaks of CBF expression correlate with multiple Ca2+ signals induced by fluctuating temperatures, which is consistent with the notion that Ca2+ signals integrate temperature fluctuations into specific signatures that inform the expression of cold-induced genes (Kiegle et al., 2000; Hiraki et al., 2018). Photoperiod and light intensity also inform daily oscillations of cytosolic free calcium ([Ca2+]cyt) within the plant (Love et al., 2004). It is possible that under natural conditions, convergence between [Ca2+]cyt signals induced by light, temperature, circadian regulation, and other perturbations produce specific transcriptional dynamics within the circadian oscillator that have downstream effects upon the transcriptome.
When investigating molecular aspects of plant abiotic stress responses using laboratory experiments, control experimental conditions are necessary but might lead to artefacts in the interpretation of transcriptomic responses to the environment under natural conditions. For example, in the context of low temperature responses, plants in the laboratory are not usually exposed to daily fluctuating temperatures before exposure to cold. Furthermore, cold temperature treatments are usually applied rapidly, with plants transferred from ambient to cold temperatures within a matter of minutes, whereas this is unlikely to occur under natural conditions (e.g., Figure 2E). Under natural conditions these previous environmental fluctuations, combined with longer term epigenetic storage of environmental information (Aikawa et al., 2010), might modify the transcriptomic response to current environments such that interpretations of transcriptional responses from laboratory conditions might not translate directly into transcriptomic responses under natural conditions.
Contribution of Alternative Splicing to the Rhythmic Transcriptome Under Natural Conditions
An essential mechanism of transcriptome and proteome regulation in plants is alternative splicing (AS). This is the process whereby more than one transcript can be generated from a single precursor mRNA (pre-mRNA) through the use of alternative splice sites in response to developmental cues, environmental cues, and stresses (Reddy et al., 2013). These alternative transcripts can contain premature termination codons (PTCs) that cause targeting by the nonsense-mediated decay (NMD) pathway, therefore regulating the amount of coding mRNA present (Hug et al., 2016). AS can also produce proteins with altered length or domain arrangements, impacting their function or stability, so expanding the proteome diversity (Syed et al., 2012). It is thought that approximately 61% of intron-containing genes undergo AS in Arabidopsis (Marquez et al., 2012). AS occurs extensively across core genes of the circadian oscillator (Romanowski and Yanovsky, 2015), with mutations in AS genes leading to improper processing of clock genes. For example, a mutation in the STIPL1 and PRMT5 genes, which participate in the regulation of AS, alters the accumulation of circadian clock transcripts due to less efficient splicing of PRR9 and causing a longer circadian period (Sanchez et al., 2010; Jones et al., 2012).
Recent analysis of the Arabidopsis transcriptome highlights a massive and rapid differential AS in response to cold temperature exposure, including a substantial proportion of circadian clock associated genes (Calixto et al., 2018). Splicing patterns of CCA1 transcripts alter in response to biotic and abiotic cues such as pathogen exposure, cold and high light, causing the accumulation of the long intron-retaining CCA1 isoform as well as a phase delay (Filichkin et al., 2010; Filichkin et al., 2015). Upon transfer to 4°C, the abundance of functional Arabidopsis LHY protein was reduced due to retention of the first intron in the LHY 5’-UTR, resulting in NMD (James et al., 2012a). AS of the myb-domain transcription factor RVE8 as well as barley orthologs of LHY and PRR7 also occurs in response to low temperature (James et al., 2012b; Calixto et al., 2016). Furthermore, a recent study showed that in sugarcane under field conditions, AS events of clock genes relates closely to temperature fluctuations, with splice forms expressed more highly at low temperatures (Dantas et al., 2019a).
There is also evidence that AS might contribute to the temperature compensation of circadian regulation (James et al., 2012a; James et al., 2012b). In Arabidopsis, the spliceosomal assembly factor GEMIN2 attenuates the effects of temperature on circadian period by regulating AS events. Due to differences in the splicing of clock genes in GEMIN2 mutants exposed to low temperatures for 24 h (Schlaen et al., 2015), it has been suggested that AS acts to continuously adjust the circadian clock in fluctuating temperatures under natural conditions (Dantas et al., 2019a). This means that a natural environment with fluctuating light and temperature conditions may drive alterations in AS that lead to distinct transcriptome and proteome profiles depending on the environmental conditions.
Circadian Regulation of the Transcriptome in Crops Under Natural Conditions
An important aim of plant sciences research is to contribute to the development of the next generation of crop varieties through breeding, genetic modification, and gene editing. The incredibly pervasive influence of circadian regulation upon plant metabolism and physiology means that the circadian oscillator affects traits of agricultural importance, with a large number of oscillator components being identified as important determinants of agricultural traits (Bendix et al., 2015; Nakamichi, 2015). Translation of laboratory research concerning circadian rhythms into information of agricultural benefit requires an understanding of circadian regulation in crops under both artificial and natural environments. Understanding circadian regulation in crops provides opportunities to alter the latitudinal range of cultivation (Müller et al., 2015; Nakamichi, 2015), intercept developmental and signaling processes including photoperiodism (Turner et al., 2005; Nakamichi, 2015) and stress responses (Fowler et al., 2005; Bieniawska et al., 2008; Nakamichi et al., 2009; Seo et al., 2012; Nakamichi et al., 2016), manipulate plant defences or pollination biology (Roden and Ingle, 2009; Goodspeed et al., 2012; Yon et al., 2017a; Yon et al., 2017b), and fuse chemical biology and circadian biology to develop and optimize agrochemical use (Belbin et al., 2019; Uehara et al., 2019). On the other hand, plant breeding that causes inadvertent alterations in circadian clock function could have deleterious consequences for crop performance.
Daily programs of transcriptional regulation have been investigated in several crop species growing under natural conditions; particularly rice, sugarcane, and pineapple (Izawa et al., 2011; Nagano et al., 2012; Matsuzaki et al., 2015; Ming et al., 2015; Dantas et al., 2019a; Dantas et al., 2019b). Fewer studies have directly tested roles for circadian clock components in crops under field conditions. In one study, field-grown rice plants harboring a null mutation of GIGANTEA were found to have altered flowering time, stomatal conductance, fertility and grain weight compared with the background line, with the GIGANTEA mutation altering the expression patterns of about 75% of the transcriptome (Izawa et al., 2011). Furthermore, under field conditions this GIGANTEA mutant has altered accumulation of transcripts associated with GA and auxin signaling, and potential alterations in ABA and jasmonate signaling (Itoh and Izawa, 2011). When considered as a whole, the circadian oscillator of field-grown rice appears to be relatively robust to environmental fluctuations, even though individual genes have transient responses to environmental changes (Matsuzaki et al., 2015).
Tobacco is another important crop where roles for circadian clock genes have been investigated in wild relatives. Manipulations of the circadian oscillator of a wild tobacco species (Nicotiana attenuata) have provided insights into roles for circadian regulation under naturally fluctuating conditions, with a focus upon ecology and ecophysiology. For example, antisense-mediated misexpression of NaLHY and NaTOC1 identified potential roles for the circadian oscillator gating photosynthetic responses to light under field conditions (Joo et al., 2017). The N. attenuata circadian oscillator also appears to contribute to the rate of flower opening and flower angle (Yon et al., 2016). Although circadian regulation modified flower selection by moths under laboratory conditions (Yon et al., 2016), these findings did not extrapolate with statistical significance to field conditions (Yon et al., 2017b). This illustrates how roles for circadian regulation can differ between field and laboratory conditions, and the need to extrapolate cautiously from the laboratory to field.
The major seed crop sunflower also gains benefits from circadian regulation. Sunflower plants track the sun such that they point eastwards around dawn and west at dusk (Vandenbrink et al., 2014). Disruption of this solar tracking in pot-grown plants in the field, by rotating pots daily so that the plant orientation was incorrect, reduced biomass and leaf area compared with correctly orientated controls (Atamian et al., 2016). In addition, correctly orientated sunflower heads received more pollinator visits than incorrectly orientated sunflowers. This is in part due to greater solar warming of the flower heads of correctly orientated plants (Atamian et al., 2016). It will be fascinating in future to identify which sunflower homologs of circadian oscillator components drive the differential growth response that underlies this mechanism (Atamian et al., 2016).
Quantitative genetics is providing valuable insights into the relationship between circadian regulation and phenotypic characteristics under laboratory and field conditions. For example, laboratory experiments have demonstrated that allelic variation in GIGANTEA leads to variations in the circadian period in B. rapa (Xie et al., 2015). In addition, experiments using a recombinant inbred line population of B. rapa revealed a correlation between circadian regulation, stomatal conductance, and CO2 assimilation, but not with reporters for water use efficiency (Edwards et al., 2012). This could suggest that circadian regulation does not contribute to drought responses associated with gas exchange traits (Edwards et al., 2012), although we note that water use efficiency under well-watered conditions is not necessarily an indicator of drought tolerance (Blum, 2009). In a separate study, the QTL that determine characteristics of circadian regulation (e.g., period and phase) were investigated in a RIL population of Arabidopsis (Rubin et al., 2019). Seedlings of the RIL population were entrained under field conditions at several times of year, and circadian rhythms were subsequently investigated in the laboratory using bioluminescence imaging. Intriguingly, this identified that the QTL determining the circadian phase differed between each season of entrainment (Rubin et al., 2019). While these population genetics studies did not investigate the dynamics of the entire transcriptome under field conditions, in future, it could be informative to apply circadian transcriptome analysis protocols to RIL populations under natural or laboratory conditions to better understand the mechanistic relationship between QTLs identified, changes in circadian regulation, and physiological outcomes.
A key challenge for investigations of rhythmic transcriptional regulation in crop species under natural conditions is the complexity of some crop genomes, particularly due to polyploidy. While sequencing technologies combined with analytical tools such as HomeoRoq (Akama et al., 2014) can allow quantification of transcripts derived from distinct gene homeologs in polyploids, it is also the case that the level of replication and/or sampling frequency required for timecourse studies can be high. When combined with the sequencing depth that is required to distinguish gene homeologs, sequencing cost constraints might arise.
Perspectives
Naturally fluctuating conditions impose an extraordinarily complex set of modifications to the interactions between the circadian oscillator and its outputs (Figure 3). There are multiple positions within the circadian system where signals communicating environmental information will modify the functioning of the circadian system (Figure 3). This includes the entrainment mechanisms, direct effects of the environment upon the circadian oscillator, and circadian gating of environmental signaling and response pathways (Figure 3). There are also direct effects of environmental cues upon circadian-regulated transcripts that are downstream of the circadian clock, such that circadian and environmental inputs are integrated separately by the transcripts (Figure 3). Furthermore, cascades of transcriptional regulation having a hierarchical organization will provide multiple additional positions for the entry of environmental cues into the circadian-regulated transcriptional cascades (Figure 3). This will lead to a continuous and dynamic adjustment of both the circadian oscillator and its outputs (Webb et al., 2019) through the action of these multiple signal inputs, with this dynamic adjustment itself altering in response to a naturally fluctuating environment. We suggest that there is future scope to extend beyond the application of transcriptional analysis under field conditions to daily cycles of integrated “omics” measures such as epigenetic states, protein abundance, and post-translational modifications and fluctuations of key primary and secondary metabolites. Furthermore, additional investigation of mutants and gene-edited plants grown outdoors in the context of circadian regulation is likely to be exceedingly informative. Advances in growth chamber technology such as fully programmable solar simulation will allow systematic investigation of the contributions of specific complex environmental parameters to circadian regulation and its outputs, thereby helping to integrate our interpretations of results from field and laboratory experiments. An interesting challenge arising from such investigations in naturally occurring plant populations is that the study species will occupy different developmental stages in distinct seasons, adding to the complexity of comparing circadian regulation between, for example, the rosette and cauline leaves of model brassica species. There is clear evidence that statistical modeling can help to unravel these complexities of daily and seasonal transcriptional programs under natural conditions (Aikawa et al., 2010; Nagano et al., 2012; Satake et al., 2013; Hepworth et al., 2018; Nagano et al., 2019). Such approaches could be invaluable to future plant sciences research as they have the potential to predict effects of future climates upon circadian function and resultant impacts upon crop and ecosystem performance.
Figure 3
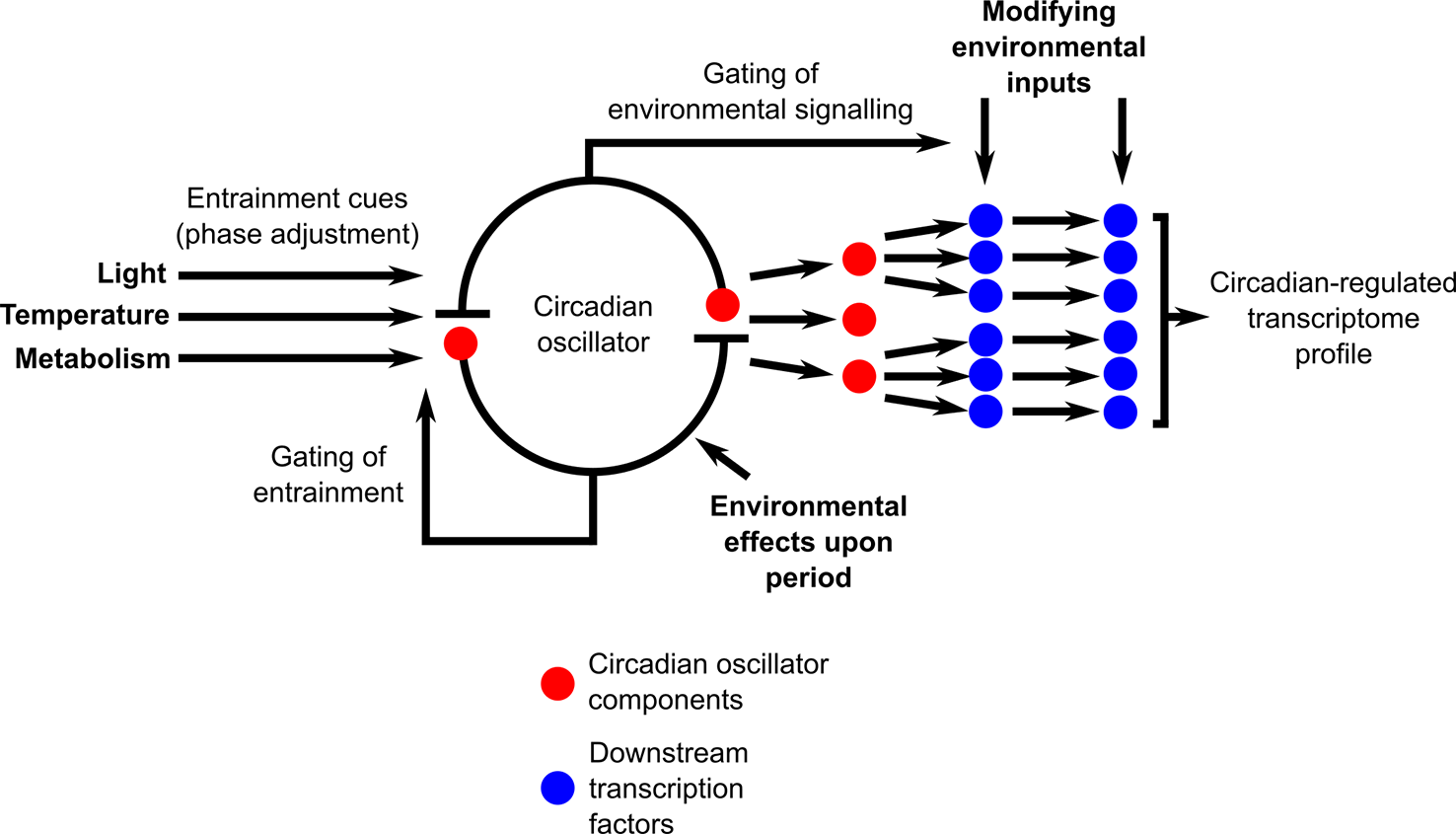
Circadian-regulated processes receive multiple environmental inputs, leading to complex interactions between circadian regulation and environmental conditions upon the transcriptome under natural conditions. Diagram conceptualizes the circadian system as a core circadian oscillator that is entrained by environmental cues and that produces output timing signals that regulate the transcriptome through a transcription factor cascade. Bold-face text indicates potential sites of environmental modification of circadian regulation, with environmental cues acting upon both oscillator inputs, oscillator function, oscillator outputs, and upon the sensitivity of environmental signaling through the process of circadian gating. In this conceptual scheme, circadian oscillator components that are transcriptional regulators (red circles) also function as the initial output step from the circadian oscillator that regulates key downstream transcription factors. In Arabidopsis, this role is fulfilled by oscillator components such as CCA1, LHY, the PRR, GI, and the evening complex (Nakamichi et al., 2012; Liu et al., 2013; Nagel et al., 2015; Liu et al., 2016; Ezer et al., 2017; Adams et al., 2018; Nohales et al., 2019).
Funding
The authors thank the BBSRC (BB/J014400/1), The Leverhulme Trust (RPG-2018-216), Consejo Nacional de Ciencia y Tecnología Mexico (Conacyt), the University of Bristol, and the Bristol Centre for Agricultural Innovation for funding.
Statements
Author contributions
PP and AD conceived and planned the article. PP, TM, DC-C, CG, AY, HK and AD wrote the article. TM and HK collected field environmental condition data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Adams S. Grundy J. Veflingstad S. R. Dyer N. P. Hannah M. A. Ott S. et al . (2018). Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol.220 (3), 893–907. doi: 10.1111/nph.15415
2
Aikawa S. Kobayashi M. J. Satake A. Shimizu K. K. Kudoh H. (2010). Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc. Natl. Acad. Sci.107 (25), 11632. doi: 10.1073/pnas.0914293107
3
Akama S. Shimizu-Inatsugi R. Shimizu K. K. Sese J. (2014). Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res.42 (6), e46–e46. doi: 10.1093/nar/gkt1376
4
Alabadı́ D. Oyama T. Yanovsky M. J. Harmon F. G. Más P. Kay S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science293 (5531), 880. doi: 10.1126/science.1061320
5
Atamian H. S. Creux N. M. Brown E. A. Garner A. G. Blackman B. K. Harmer S. L. (2016). Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science353 (6299), 587. doi: 10.1126/science.aaf9793
6
Belbin F. E. Hall G. J. Jackson A. B. Schanschieff F. E. Archibald G. Formstone C. et al . (2019). Plant circadian rhythms regulate the effectiveness of a glyphosate-based herbicide. Nat. Commun.10 (1), 3704–3704. doi: 10.1038/s41467-019-11709-5
7
Bendix C. Marshall C. M. Harmon F. G. (2015). Circadian clock genes universally control key agricultural traits. Mol. Plant8 (8), 1135–1152. doi: 10.1016/j.molp.2015.03.003
8
Bieniawska Z. Espinoza C. Schlereth A. Sulpice R. Hincha D. K. Hannah M. A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol.147 (1), 263–279. doi: 10.1104/pp.108.118059
9
Blum A. (2009). Effective use of water (EUW) and not water-use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Res.112 (2), 119–123. doi: 10.1016/j.fcr.2009.03.009
10
Calixto C. P. G. Simpson C. G. Waugh R. Brown J. W. S. (2016). Alternative splicing of barley clock genes in response to low temperature. PloS One11 (12), e0168028–e0168028. doi: 10.1371/journal.pone.0168028
11
Calixto C. P. G. Guo W. James A. B. Tzioutziou N. A. Entizne J. C. Panter P. E. et al . (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell30 (7), 1424. doi: 10.1105/tpc.18.00177
12
Cockram J. Jones H. Leigh F. J. O’Sullivan D. Powell W. Laurie D. A. et al . (2007). Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot.58 (6), 1231–1244. doi: 10.1093/jxb/erm042
13
Covington M. F. Harmer S. L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PloS Biol.5 (8), e222. doi: 10.1371/journal.pbio.0050222
14
Covington M. F. Panda S. Liu X. L. Strayer C. A. Wagner D. R. Kay S. A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell13 (6), 1305. doi: 10.1105/TPC.000561
15
Dai S. Wei X. Pei L. Thompson R. L. Liu Y. Heard J. E. et al . (2011). BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell23 (3), 961. doi: 10.1105/tpc.111.084293
16
Dantas L. L. B. Calixto C. P. G. Dourado M. M. Carneiro M. S. Brown J. W. S. Hotta C. T. (2019a). Alternative splicing of circadian clock genes is temperature regulated in field-grown sugarcane. bioRxiv. doi: 10.1101/718890
17
Dantas L. L. B. de Lima N. O. Alves-Lima C. Nishiyama M. Y. Carneiro M. S. Souza G. M. et al . (2019b). Rhythms of transcription in field-grown sugarcane are highly organ specific. bioRxiv. doi: 10.1101/607002
18
Desai J. S. Lawas L. M. F. Valente A. M. Leman A. R. Grinevich D. O. Jagadish S. V. K. et al . (2019). Warm nights disrupt global transcriptional rhythms in field-grown rice panicles. bioRxiv. doi: 10.1101/702183
19
Dixon L. E. Knox K. Kozma-Bognar L. Southern M. M. Pokhilko A. Millar A. J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol.21 (2), 120–125. doi: 10.1016/j.cub.2010.12.013
20
Dodd A. N. Salathia N. Hall A. Kévei E. Tóth R. Nagy F. et al . (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science309 (5734), 630. doi: 10.1126/science.1115581
21
Dodd A. N. Dalchau N. Gardner M. J. Baek S. J. Webb A. A. R. (2014). The circadian clock has transient plasticity of period and is required for timing of nocturnal processes in Arabidopsis. New Phytol.201, 168–179. doi: 10.1111/nph.12489
22
Dong M. A. Farré E. M. Thomashow M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the c-repeat binding factor (cbf) pathway in Arabidopsis. Proc. Natl. Acad. Sci.108 (17), 7241. doi: 10.1073/pnas.1103741108
23
Edwards K. D. Lynn J. R. Gyula P. Nagy F. Millar A. J. (2005). Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics170 (1), 387. doi: 10.1534/genetics.104.035238
24
Edwards K. D. Anderson P. E. Hall A. Salathia N. S. Locke J. C. W. Lynn J. R. et al . (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell18 (3), 639–650. doi: 10.1105/tpc.105.038315
25
Edwards C. E. Ewers B. E. McClung C. R. Lou P. Weinig C. (2012). Quantitative variation in water-use efficiency across water regimes and its relationship with circadian, vegetative, reproductive, and leaf gas-exchange traits. Mol. Plant5 (3), 653–668. doi: 10.1093/mp/sss004
26
Ermolaeva M. D. Wu M. Eisen J. A. Salzberg S. L. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol.51 (6), 859–866. doi: 10.1023/A:1023001130337
27
Ezer D. Jung J.-H. Lan H. Biswas S. Gregoire L. Box M. S. et al . (2017). The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants3, 17087. doi: 10.1038/nplants.2017.87
28
Farinas B. Mas P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J.66 (2), 318–329. doi: 10.1111/j.1365-313X.2011.04484.x
29
Farré E. M. Liu T. (2013). The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. In Plant Biol.16 (5), 621–629. doi: 10.1016/j.pbi.2013.06.015
30
Filichkin S. A. Priest H. D. Givan S. A. Shen R. Bryant D. W. Fox S. E. et al . (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res.20 (1), 45–58. doi: 10.1101/gr.093302.109
31
Filichkin S. A. Breton G. Priest H. D. Dharmawardhana P. Jaiswal P. Fox S. E. et al . (2011). Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PloS One6 (6), e16907. doi: 10.1371/journal.pone.0016907
32
Filichkin S. A. Cumbie J. S. Dharmawardhana P. Jaiswal P. Chang J. H. Palusa S. G. et al . (2015). Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant8 (2), 207–227. doi: 10.1016/j.molp.2014.10.011
33
Fondy B. R. Geiger D. R. Servaites J. C. (1989). Photosynthesis, carbohydrate metabolism, and export in Beta vulgaris L. and Phaseolus vulgaris L. during square and sinusoidal light regimes. Plant Physiol.89, 396–402. doi: 10.1104/pp.89.2.39
34
Fowler S. G. Cook D. Thomashow M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol.137 (3), 961. doi: 10.1104/pp.104.058354
35
Frank A. Matiolli C. C. Viana A. J. C. Hearn T. J. Kusakina J. Belbin F. E. et al . (2018). Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63 . Curr. Biol.28 (16), 2597–2606.e2596. doi: 10.1016/j.cub.2018.05.092
36
Franklin K. A. Whitelam G. C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet.39 (11), 1410–1413. doi: 10.1038/ng.2007.3
37
Gendron J. M. Pruneda-Paz J. L. Doherty C. J. Gross A. M. Kang S. E. Kay S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci.109 (8), 3167. doi: 10.1073/pnas.1200355109
38
Goodspeed D. Chehab E. W. Min-Venditti A. Braam J. Covington M. F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci.109 (12), 4674. doi: 10.1073/pnas.1116368109
39
Gould P. D. Locke J. C. W. Larue C. Southern M. M. Davis S. J. Hanano S. et al . (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell18 (5), 1177. doi: 10.1105/tpc.105.039990
40
Gould P. D. Ugarte N. Domijan M. Costa M. Foreman J. MacGregor D. et al . (2013). Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol.9 (1), 650. doi: 10.1038/msb.2013.7
41
Graf A. Schlereth A. Stitt M. Smith A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci.107 (20), 9458. doi: 10.1073/pnas.0914299107
42
Green R. M. Tingay S. Wang Z.-Y. Tobin E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol.129 (2), 576. doi: 10.1104/pp.004374
43
Hanano S. Domagalska M. A. Nagy F. Davis S. J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes to Cells11 (12), 1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x
44
Harmer S. L. Hogenesch J. B. Straume M. Chang H.-S. Han B. Zhu T. et al . (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science290 (5499), 2110. doi: 10.1126/science.290.5499.2110
45
Haydon M. J. Mielczarek O. Robertson F. C. Hubbard K. E. Webb A. A. R. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature502, 689. doi: 10.1038/nature12603
46
Helfer A. Nusinow D. A. Chow B. Y. Gehrke A. R. Bulyk M. L. Kay S. A. (2011). LUX Arrhythmo encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol.21 (2), 126–133. doi: 10.1016/j.cub.2010.12.021
47
Hepworth J. Antoniou-Kourounioti R. L. Bloomer R. H. Selga C. Berggren K. Cox D. et al . (2018). Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat. Commun.9 (1), 639. doi: 10.1038/s41467-018-03065-7
48
Herrero E. Kolmos E. Bujdoso N. Yuan Y. Wang M. Berns M. C. et al . (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell24 (2), 428. doi: 10.1105/tpc.111.093807
49
Hiraki H. Uemura M. Kawamura Y. (2018). Calcium signaling-linked CBF/DREB1 gene expression was induced depending on the temperature fluctuation in the field: views from the natural condition of cold acclimation. Plant Cell Physiol.60 (2), 303–317. doi: 10.1093/pcp/pcy210
50
Hotta C. T. Gardner M. J. Hubbard K. E. Baek S. J. Dalchau N. Suhita D. et al . (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ.30 (3), 333–349. doi: 10.1111/j.1365-3040.2006.01627.x
51
Hotta C. T. Nishiyama M. Y.Jr., and SouzaG. M. (2013). Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PloS One8 (8), e71847. doi: 10.1371/journal.pone.0071847
52
Hsu P. Y. Harmer S. L. (2014). Wheels within wheels: the plant circadian system. Trends In Plant Sci.19 (4), 240–249. doi: 10.1016/j.tplants.2013.11.007
53
Hsu P. Y. Devisetty U. K. Harmer S. L. (2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife2, e00473. doi: 10.7554/eLife.00473
54
Huang H. Nusinow D. A. (2016). Into the evening: complex interactions in the Arabidopsis circadian clock. Trends In Genet.32 (10), 674–686. doi: 10.1016/j.tig.2016.08.002
55
Huang W. Pérez-García P. Pokhilko A. Millar A. J. Antoshechkin I. Riechmann J. L. et al . (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science336 (6077), 75. doi: 10.1126/science.1219075
56
Hug N. Longman D. Cáceres J. F. (2016). Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res.44 (4), 1483–1495. doi: 10.1093/nar/gkw010
57
Itoh H. Izawa T. (2011). A study of phytohormone biosynthetic gene expression using a circadian clock-related mutant in rice. Plant Signaling Behav.6 (12), 1932–1936. doi: 10.4161/psb.6.12.18207
58
Izawa T. Mihara M. Suzuki Y. Gupta M. Itoh H. Nagano A. J. et al . (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell23 (5), 1741. doi: 10.1105/tpc.111.083238
59
James A. B. Syed N. H. Bordage S. Marshall J. Nimmo G. A. Jenkins G. I. et al . (2012a). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell24 (3), 961–981. doi: 10.1105/tpc.111.093948
60
James A. B. Syed N. H. Brown J. W. S. Nimmo H. G. (2012b). Thermoplasticity in the plant circadian clock: how plants tell the time-perature. Plant Signaling Behav.7 (10), 1219–1223. doi: 10.4161/psb.21491
61
Jones M. A. Williams B. A. McNicol J. Simpson C. G. Brown J. W. S. Harmer S. L. (2012). Mutation of Arabidopsis Spliceosomal Timekeeper Locus1 causes circadian clock defects. Plant Cell24 (10), 4066. doi: 10.1105/tpc.112.104828
62
Joo Y. Fragoso V. Yon F. Baldwin I. T. Kim S.-G. (2017). Circadian clock component, LHY, tells a plant when to respond photosynthetically to light in nature. J. Integr. Plant Biol.59 (8), 572–587. doi: 10.1111/jipb.12547
63
Kamioka M. Takao S. Suzuki T. Taki K. Higashiyama T. Kinoshita T. et al . (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell28 (3), 696. doi: 10.1105/tpc.15.00737
64
Kidokoro S. Maruyama K. Nakashima K. Imura Y. Narusaka Y. Shinwari Z. K. et al . (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol.151 (4), 2046. doi: 10.1104/pp.109.147033
65
Kiegle E. Moore C. A. Haseloff J. Tester M. A. Knight M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J.23 (2), 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
66
Kim W.-Y. Fujiwara S. Suh S.-S. Kim J. Kim Y. Han L. et al . (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature449, 356. doi: 10.1038/nature06132
67
Kudoh H. Honjo M. N. Nishio H. Sugisaka J. (2018). “The long-term “in natura” study sites of Arabidopsis halleri for plant transcription and epigenetic modification analyses in natural environments,” in Plant Transcription Factors: Methods and Protocols. Ed. YamaguchiN. (New York, NY: Springer New York), 41–57.
68
Lai A. G. Doherty C. J. Mueller-Roeber B. Kay S. A. Schippers J. H. M. Dijkwel P. P. (2012). Circadian Clock-Associated 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci.109 (42), 17129. doi: 10.1073/pnas.1209148109
69
Lau On S. Huang X. Charron J.-B. Lee J.-H. Li G. Deng X. W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell43 (5), 703–712. doi: 10.1016/j.molcel.2011.07.013
70
Lee C.-M. Thomashow M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci.109 (37), 15054. doi: 10.1073/pnas.1211295109
71
Limpens J. Granath G. Aerts R. Heijmans M. M. P. D. Sheppard L. J. Bragazza L. et al . (2012). Glasshouse vs field experiments: do they yield ecologically similar results for assessing N impacts on peat mosses? New Phytol.195 (2), 408–418. doi: 10.1111/j.1469-8137.2012.04157.x
72
Liu T. Carlsson J. Takeuchi T. Newton L. Farré E. M. (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7 . Plant J.76 (1), 101–114. doi: 10.1111/tpj.12276
73
Liu S. Liu Y. Yang X. Tong C. Edwards D. Parkin I. A. P. et al . (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun.5 (1), 3930. doi: 10.1038/ncomms4930
74
Liu T. L. Newton L. Liu M.-J. Shiu S.-H. Farré E. M. (2016). A G-Box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol.170 (1), 528. doi: 10.1104/pp.15.01562
75
Love J. Dodd A. N. Webb A. A. R. (2004). Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell16 (4), 956. doi: 10.1105/tpc.020214
76
Lu S. X. Knowles S. M. Andronis C. Ong M. S. Tobin E. M. (2009). Circadian Clock Associated1 And Late Elongated Hypocotyl function synergistically in the circadian clock of Arabidopsis. Plant Physiol.150 (2), 834. doi: 10.1104/pp.108.133272
77
Lu S. X. Webb C. J. Knowles S. M. Kim S. H. J. Wang Z. Tobin E. M. (2012). CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol.158 (2), 1079. doi: 10.1104/pp.111.189670
78
Marquez Y. Brown J. W. S. Simpson C. Barta A. Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res.22 (6), 1184–1195. doi: 10.1101/gr.134106.111
79
Martín G. Rovira A. Veciana N. Soy J. Toledo-Ortiz G. Gommers C. M. M. et al . (2018). Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol.28 (2), 311–318.e315. doi: 10.1016/j.cub.2017.12.021
80
Matsuzaki J. Kawahara Y. Izawa T. (2015). Punctual transcriptional regulation by the rice circadian clock under fluctuating field conditions. Plant Cell27 (3), 633. doi: 10.1105/tpc.114.135582
81
Michael T. P. Salomé P. A. McClung C. R. (2003). Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci.100 (11), 6878. doi: 10.1073/pnas.1131995100
82
Millar A. J. Straume M. Chory J. Chua N. H. Kay S. A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science267 (5201), 1163. doi: 10.1126/science.7855596
83
Millar A. J. (2016). The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol.67 (1), 595–618. doi: 10.1146/annurev-arplant-043014-115619
84
Ming R. VanBuren R. Wai C. M. Tang H. Schatz M. C. Bowers J. E. et al . (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet.47 (12), 1435–1442. doi: 10.1038/ng.3435
85
Mizuno T. Kitayama M. Oka H. Tsubouchi M. Takayama C. Nomoto Y. et al . (2014). The EC night-time repressor plays a crucial role in modulating circadian clock transcriptional circuitry by conservatively double-checking both warm-night and night-time-light signals in a synergistic manner in Arabidopsis thaliana. Plant Cell Physiol.55 (12), 2139–2151. doi: 10.1093/pcp/pcu144
86
Müller N. A. Wijnen C. L. Srinivasan A. Ryngajllo M. Ofner I. Lin T. et al . (2015). Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet.48, 89. doi: 10.1038/ng.3447 http://www.nature.com/articles/ng.3447-supplementary-information.
87
Mwimba M. Karapetyan S. Liu L. Marqués J. McGinnis E. M. Buchler N. E. et al . (2018). Daily humidity oscillation regulates the circadian clock to influence plant physiology. Nat. Commun.9 (1), 4290–4290. doi: 10.1038/s41467-018-06692-2
88
Nagano A. J. Sato Y. Mihara M. Antonio B. A. Motoyama R. Itoh H. et al . (2012). Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell151 (6), 1358–1369. doi: 10.1016/j.cell.2012.10.048
89
Nagano A. J. Kawagoe T. Sugisaka J. Honjo M. N. Iwayama K. Kudoh H. (2019). Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nat. Plants5 (1), 74–83. doi: 10.1038/s41477-018-0338-z
90
Nagel D. H. Kay S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol.22 (16), R648–R657. doi: 10.1016/j.cub.2012.07.025
91
Nagel D. H. Doherty C. J. Pruneda-Paz J. L. Schmitz R. J. Ecker J. R. Kay S. A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci.112 (34), E4802. doi: 10.1073/pnas.1513609112
92
Nakamichi N. Kusano M. Fukushima A. Kita M. Ito S. Yamashino T. et al . (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol.50 (3), 447–462. doi: 10.1093/pcp/pcp004
93
Nakamichi N. Kiba T. Henriques R. Mizuno T. Chua N.-H. Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell22 (3), 594. doi: 10.1105/tpc.109.072892
94
Nakamichi N. Kiba T. Kamioka M. Suzuki T. Yamashino T. Higashiyama T. et al . (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci.109 (42), 17123. doi: 10.1073/pnas.1205156109
95
Nakamichi N. Takao S. Kudo T. Kiba T. Wang Y. Kinoshita T. et al . (2016). Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol.57 (5), 1085–1097. doi: 10.1093/pcp/pcw057
96
Nakamichi N. (2015). Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol.56 (4), 594–604. doi: 10.1093/pcp/pcu181
97
Ni Z. Kim E.-D. Ha M. Lackey E. Liu J. Zhang Y. et al . (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature457 (7227), 327–331. doi: 10.1038/nature07523
98
Nohales M. A. Liu W. Duffy T. Nozue K. Sawa M. Pruneda-Paz J. L. et al . (2019). Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Dev. Cell49 (6), 840–851.e848. doi: 10.1016/j.devcel.2019.04.030
99
Nusinow D. A. Helfer A. Hamilton E. E. King J. J. Imaizumi T. Schultz T. F. et al . (2011). The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature475 (7356), 398–402. doi: 10.1038/nature10182
100
Oakenfull R. J. Davis S. J. (2017). Shining a light on the Arabidopsis circadian clock. Plant Cell Environ.40 (11), 2571–2585. doi: 10.1111/pce.13033
101
Peng S. Huang J. Sheehy J. E. Laza R. C. Visperas R. M. Zhong X. et al . (2004). Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. United States America101 (27), 9971. doi: 10.1073/pnas.0403720101
102
Poorter H. Fiorani F. Pieruschka R. Wojciechowski T. van der Putten W. H. Kleyer M. et al . (2016). Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol.212 (4), 838–855. doi: 10.1111/nph.14243
103
Rémi J. Merrow M. Roenneberg T. (2010). A circadian surface of entrainment: Varying T, τ, and photoperiod in Neurospora crassa. J. Biol. Rhythms25 (5), 318–328. doi: 10.1177/0748730410379081
104
Rawat R. Schwartz J. Jones M. A. Sairanen I. Cheng Y. Andersson C. R. et al . (2009). REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci.106 (39), 16883. doi: 10.1073/pnas.0813035106
105
Rawat R. Takahashi N. Hsu P. Y. Jones M. A. Schwartz J. Salemi M. R. et al . (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PloS Genet.7 (3), e1001350. doi: 10.1371/journal.pgen.1001350
106
Reddy A. S. N. Marquez Y. Kalyna M. Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell25 (10), 3657. doi: 10.1105/tpc.113.117523
107
Roden L. C. Ingle R. A. (2009). Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell21 (9), 2546. doi: 10.1105/tpc.109.069922
108
Romanowski A. Yanovsky M. J. (2015). Circadian rhythms and post-transcriptional regulation in higher plants. Front. In Plant Sci.6, 437–437. doi: 10.3389/fpls.2015.00437
109
Rubin M. J. Brock M. T. Davis S. J. Weinig C. (2019). QTL underlying circadian clock parameters under seasonally variable field settings in Arabidopsis thaliana. (Bethesda Md.)9 (4), 1131–1139. doi: 10.1534/g3.118.200770
110
Salomé P. A. McClung C. R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell17 (3), 791. doi: 10.1105/tpc.104.029504
111
Salomé P. A. Weigel D. McClung C. R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell22 (11), 3650. doi: 10.1105/tpc.110.079087
112
Sanchez S. E. Petrillo E. Beckwith E. J. Zhang X. Rugnone M. L. Hernando C. E. et al . (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature468 (7320), 112–116. doi: 10.1038/nature09470
113
Satake A. Kawagoe T. Saburi Y. Chiba Y. Sakurai G. Kudoh H. (2013). Forecasting flowering phenology under climate warming by modelling the regulatory dynamics of flowering-time genes. Nat. Commun.4 (1), 2303. doi: 10.1038/ncomms3303
114
Schaffer R. Ramsay N. Samach A. Corden S. Putterill J. Carré I. A. et al . (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell93 (7), 1219–1229. doi: 10.1016/S0092-8674(00)81465-8
115
Schlaen R. G. Mancini E. Sanchez S. E. Perez-Santángelo S. Rugnone M. L. Simpson C. G. et al . (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. United States America112 (30), 9382–9387. doi: 10.1073/pnas.1504541112
116
Seo P. J. Park M.-J. Lim M.-H. Kim S.-G. Lee M. Baldwin I. T. et al . (2012). A self-regulatory circuit of Circadian Clock-Associated1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell24 (6), 2427. doi: 10.1105/tpc.112.098723
117
Shin J. Sánchez-Villarreal A. Davis A. M. , Du, S.-x., BerendzenK. W.KonczC.et al. (2017). The metabolic sensor AKIN10 modulates the Arabidopsis circadian clock in a light-dependent manner. Plant Cell Environ.40 (7), 997–1008. doi: 10.1111/pce.12903
118
Shor E. Paik I. Kangisser S. Green R. Huq E. (2017). Phytochrome Interacting Factors mediate metabolic control of the circadian system in Arabidopsis. New Phytol.215 (1), 217–228. doi: 10.1111/nph.14579
119
Somers D. E. Devlin P. F. Kay S. A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science282 (5393), 1488. doi: 10.1126/science.282.5393.1488
120
Song Y. H. Ito S. Imaizumi T. (2010). Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. In Plant Biol.13 (5), 594–603. doi: 10.1016/j.pbi.2010.05.004
121
Song Y. H. Shim J. S. Kinmonth-Schultz H. A. Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol.66 (1), 441–464. doi: 10.1146/annurev-arplant-043014-115555
122
Syed N. H. Kalyna M. Marquez Y. Barta A. Brown J. W. S. (2012). Alternative splicing in plants–coming of age. Trends In Plant Sci.17 (10), 616–623. doi: 10.1016/j.tplants.2012.06.001
123
Thines B. Harmon F. G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci.107 (7), 3257–3262. doi: 10.1073/pnas.0911006107
124
Thomashow M. F. Gilmour S. J. Stockinger E. J. Jaglo-Ottosen K. R. Zarka D. G. (2001). Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol. Plantarum112 (2), 171–175. doi: 10.1034/j.1399-3054.2001.1120204.x
125
Turner A. Beales J. Faure S. Dunford R. P. Laurie D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science310 (5750), 1031. doi: 10.1126/science.1117619
126
Uehara T. N. Mizutani Y. Kuwata K. Hirota T. Sato A. Mizoi J. et al . (2019). Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci.116 (23), 11528. doi: 10.1073/pnas.1903357116
127
Vandenbrink J. P. Brown E. A. Harmer S. L. Blackman B. K. (2014). Turning heads: the biology of solar tracking in sunflower. Plant Sci.224, 20–26. doi: 10.1016/j.plantsci.2014.04.006
128
Vialet-Chabrand S. Matthews J. S. A. Simkin A. J. Raines C. A. Lawson T. (2017). Importance of fluctuations in light on plant photosynthetic acclimation. Plant Physiol.173 (4), 2163. doi: 10.1104/pp.16.01767
129
Wang Z.-Y. Tobin E. M. (1998). Constitutive expression of the Circadian Clock Associated 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell93 (7), 1207–1217. doi: 10.1016/S0092-8674(00)81464-6
130
Webb A. A. R. Seki M. Satake A. Caldana C. (2019). Continuous dynamic adjustment of the plant circadian oscillator. Nat. Commun.10 (1), 550. doi: 10.1038/s41467-019-08398-5
131
Wenden B. Kozma-Bognár L. Edwards K. D. Hall A. J. W. Locke J. C. W. Millar A. J. (2011). Light inputs shape the Arabidopsis circadian system. Plant J.66 (3), 480–491. doi: 10.1111/j.1365-313X.2011.04505.x
132
Xie Q. Lou P. Hermand V. Aman R. Park H. J. Yun D.-J. et al . (2015). Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci.112 (12), 3829. doi: 10.1073/pnas.1421803112
133
Yakir E. Hilman D. Kron I. Hassidim M. Melamed-Book N. Green R. M. (2009). Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol.150 (2), 844. doi: 10.1104/pp.109.137414
134
Yon F. Joo Y. Cortés Llorca L. Rothe E. Baldwin I. T. Kim S.-G. (2016). Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol.209 (3), 1058–1066. doi: 10.1111/nph.13681
135
Yon F. Kessler D. Joo Y. Cortés Llorca L. Kim S.-G. Baldwin I. T. (2017a). Fitness consequences of altering floral circadian oscillations for Nicotiana attenuata. J. Integr. Plant Biol.59 (3), 180–189. doi: 10.1111/jipb.12511
136
Yon F. Kessler D. Joo Y. Kim S.-G. Baldwin I. T. (2017b). Fitness consequences of a clock pollinator filter in Nicotiana attenuata flowers in nature. J. Integr. Plant Biol.59 (11), 805–809. doi: 10.1111/jipb.12579
Summary
Keywords
circadian rhythms, plant sciences, transcriptomics, environmental signaling, signal integration, phenology, Arabidopsis
Citation
Panter PE, Muranaka T, Cuitun-Coronado D, Graham CA, Yochikawa A, Kudoh H and Dodd AN (2019) Circadian Regulation of the Plant Transcriptome Under Natural Conditions. Front. Genet. 10:1239. doi: 10.3389/fgene.2019.01239
Received
10 October 2019
Accepted
08 November 2019
Published
29 November 2019
Volume
10 - 2019
Edited by
Kenji Fukushima, Julius Maximilian University of Würzburg, Germany
Reviewed by
Norihito Nakamichi, Nagoya University, Japan; Matt Jones, University of Glasgow, United Kingdom
Updates
Copyright
© 2019 Panter, Muranaka, Cuitun-Coronado, Graham, Yochikawa, Kudoh and Dodd.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antony N. Dodd, antony.dodd@jic.ac.uk
This article was submitted to Evolutionary and Population Genetics, a section of the journal Frontiers in Genetics
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.