- UKM Medical Molecular Biology Institute (UMBI), UKM Medical Center, Kuala Lumpur, Malaysia
Circular RNAs (circRNAs) which were once considered as “junk” are now in the spotlight as a potential player in regulating human diseases, especially cancer. With the development of high throughput technologies in recent years, the full potential of circRNAs is being uncovered. CircRNAs possess some unique characteristics and advantageous properties that could benefit medical research and clinical applications. CircRNAs are stable with covalently closed loops that are resistant to ribonucleases, have disease stage-specific expressions and are selectively abundant in different types of tissues. Interestingly, the presence of circRNAs in different types of treatment resistance in human cancers was recently observed with the involvement of a few key pathways. The activation of certain pathways by circRNAs may give new insights to treatment resistance management. The potential usage of circRNAs from this aspect is very much in its infancy stage and has not been fully validated. This mini-review attempts to highlight the possible role of circRNAs as regulators of treatment resistance in human cancers based on its intersection molecules and cancer-related regulatory networks.
Introduction
Increasing evidence has shown that circular RNAs (circRNAs) a form of non-coding RNA is involved in important biological processes and cellular functions (Kristensen et al., 2018). Though the concept of circRNAs first emerged in 1976, it became particularly interesting in recent years when it was found to have regulatory roles in cancer biology. During the early years of discovery, the occurrence of circRNAs was considered as post-transcriptional errors; however, with the advent of various biomedical technologies, the biogenesis and functions of circRNAs have been extensively explored (Wang et al., 2017). As of 2018, there were approximately 30000 circRNAs identified and the numbers are increasing (Si-Tu et al., 2019). There are four categories of circRNAs, exonic circRNA (ecircRNA), circular intronic RNA (ciRNA), exon-intron circRNAs, and intergenic circRNAs (Li Z. et al., 2015; Memczak et al., 2013; Zhang et al., 2013). CircRNAs have covalently closed loops without a free 3′ or 5′ ends (Lasda and Parker, 2014). It is postulated that the formation of circRNAs are via back-splicing where the downstream exons are spliced to upstream exons in reverse order in the primary transcript (Chen and Yang, 2015). Furthermore, several unique properties make circRNAs a promising entity in providing key insights into human diseases. Besides being abundant both in normal and cancer cells, it was also found that circRNAs are specifically expressed at every stage of cell development (Li J. et al., 2015). It was further confirmed that different isoforms of circRNAs from the same gene are expressed differently in different cell types. In several types of cancers such as hepatocellular carcinoma and colorectal cancer, it was noted that the expression level of circRNAs varies according to TNM stage, presence of metastasis and size of tumor (Szabo and Salzman, 2016). Unlike linear RNAs, circRNAs are more stable and are not easily degraded by ribonucleases such as exonuclease or RNase R due to the unexposed 3′ and 5′ terminals (Wang et al., 2017). Moreover, most circRNAs have an average half-life of over 48 h compared to linear mRNA with an average half-life of 10 h, thus making it more available for both research and clinical purposes. In addition to its advantageous properties, studies have found that circRNAs are involved in several biological activities as competing endogenous RNA by sponging miRNAs (Lin ADF and Chen, 2018), RNA binding proteins (RBPs) (Wang et al., 2015) and translating peptides (Granados-Riveron and Aquino-Jarquin, 2016; Du et al., 2017). Of particular interest is the role of circRNAs as miRNA sponge in tumor pathogenesis, and there have been many publications related to this (Wang et al., 2015; Zhang et al., 2017; Kun-Peng et al., 2018). By serving as a miRNA sponge with many binding sites, circRNAs can regulate the expression of miRNA as a competitive inhibitor that suppresses the ability of the miRNA to bind to its target genes. This event can, in turn, increase the levels of the miRNA target causing dysregulation of gene expression and pathological effects on tumor environment (Huang et al., 2015; Palmieri et al., 2018; Zeng et al., 2018). Some of these potential miRNA targets have been reported to function as important regulators of various cellular processes including apoptosis, invasion, migration, and drug resistance in several cancers.
Recently, much evidence was published on the role of circRNAs in disease progression and activation of key pathways like EMT and Wnt (Shen et al., 2019; Wu et al., 2019). Cancers that are gaining popularity like gastric, hepatocellular, lung, and breast are being studied closely with the hope to target the specific circRNAs that are involved in the development of tumor (Shang et al., 2019). Accumulating data on the association between circRNA and tumorigenesis shows promising results. However, little is known about its role in cancer therapy resistance. As therapy resistance remains one of the major clinical hurdles in cancer management, this mini-review aims to explore the potential of circRNAs as a regulator of treatment resistance. We reviewed recent relevant publications focusing on circRNAs in treatment resistance, particularly regarding drug therapy and radiotherapy. We also looked at studies at the network level to explain the relationship of circRNAs with the potential targets and pathways that could influence disease progression.
CircRNA Effects Radiotherapy Receptivity via WNT Pathway
Non-coding RNAs have been linked to tumorigenesis, metastasis, and the development of resistance to treatment (Gong et al., 2014). Radiation therapy is one of the main treatment solutions for esophageal squamous cell carcinoma (ESCC) patients, especially with unresectable esophageal cancer. Unfortunately, radioresistance has been one of the reasons for failed treatments and local tumor recurrence in ESCC (Chen et al., 2017). In a study conducted by Su et al, hsa_circ_001059 and hsa_circ_000167 levels were shown to be dysregulated in radioresistant ESCC cell line as compared to the parental cell line (Su et al., 2016). The analysis showed that circRNA_001059 could sponge to multiple miRNAs including miR-30c-1, miR-30c-2, miR-122, miR-139-3p, miR-339-5p, and miR-1912. In support of this finding, miR-30 and miR-122 were found to be dysregulated in chemoresistant prostate cancer and miR-30 in radiosensitive leukemia cells (Ni et al., 2017; Liamina et al., 2017). These dysregulated circRNAs were mapped to their target genes and were found to be mainly involved in the Wnt signaling pathway and the PI3K/Akt pathway. The crosstalk mechanisms between the Wnt pathway and other non-coding RNAs such as the long non-coding RNA (lncRNAs) have been observed in cancer pathogenesis (Yang et al., 2018) including in breast cancer (Liu et al., 2016; Koval and Katanaev, 2018) and lung cancer (Wan et al., 2016; Zeng et al., 2017). Wnt signaling pathway was shown to be an important regulator in cancer cell responsiveness to radiotherapy (Takebe et al., 2015). The activation of the pathway increases the rate of DNA double-strand break repair and is also involved in breast cancer and colorectal cancer chemotherapy resistance (Flanagan et al., 2015; Pohl et al., 2017). Moreover, Li et al. have found that the expression of circITCH was low in ESCC tissues. It is possible that circITCH could influence the radioresistance of ESCC by inhibiting the target gene via suppression of the Wnt/β signaling pathway as in a previous radioresistance study (Li J. et al., 2015). Besides ESCC, circITCH has also been associated with other cancers as well such as hepatocellular, bladder, lung, and colorectal where it acts as miRNA sponges particularly to miR-7, miR-17 and miR-214 (Li J. et al., 2015; Guo et al., 2018; Shang et al., 2019). Due to the sponging effect, the expression of the protein-coding gene ITCH increases as well (Fang, 2018). It was discovered that the ITCH gene is an important player in tumor formation and responsiveness to chemotherapy (Li J. et al., 2015). Phosphorylation of the mediator protein Dvl2 is required for activation of Wnt signaling and previous research have found that ITCH gene can disrupt Dvl2 phosphorylation and subsequently lead to the inhibition of the signaling pathway (Li J. et al., 2015). It has also been shown that parallel circITCH expression level increases with ITCH mRNA level in colorectal cancer (Huang et al., 2015). Sponging effects of circITCH to miR-7 and miR-20a lead to the downregulation of ITCH via bindings of these miRNAs to the 3'UTR of ITCH, ultimately attenuating the proliferative rate of CRC cell lines (Huang et al., 2015).
Multiple CircRNAs Regulate Chemoresistance via MAPK Pathway
Chemoresistance poses as one of the biggest challenges for cancer patients receiving neoadjuvant, adjuvant or palliative chemotherapy. Similar to radioresistance, the possibility of circRNAs as chemoresistance regulators were recently explored. Evidence on this is still limited but seems promising. It is well known that the Mitogen-Activated Protein Kinase (MAPK) signaling pathway is dysregulated in many cancers such as breast, pancreatic, colon, and leukemia (Li et al., 2014). MAPK pathway is important in controlling cellular activities including but not limited to proliferation, differentiation, apoptosis, survival, inflammation, and influence gene expression. This pathway has also shown potential in regulating treatment resistance involving non-coding RNAs in non-small cell lung cancer and malignant melanoma (Li et al., 2014). A study on EGFR therapy in colorectal cancer showed that global circRNAs were significantly downregulated in mutant Kras cell line, particularly circFAT1 and circARHGAP5 (Dou et al., 2016). Kras functions in activating the MAP3K tier which in turn activates the ERK1/2 cascade while EGFR is an important growth factor which can act as an oncogene by hyper-activating the signaling pathway (Hymowitz and Malek, 2018). In line with this observation, some studies showed that overexpression of EGFR can promote radio and chemotherapy resistance by activating the second and third tiers of the MAPK pathway (Pozzi et al., 2016). The MEK/ERK cascade was shown to be exclusively deactivated when EGFR expression is inhibited. These findings suggest that dysregulation in any tier of this cascade affects the levels of circRNA and may contribute to treatment resistance. However, it is difficult to conclude from this study that the lower levels of circRNA is a regulator of the oncogenic factors or it directly protects the cancer cells from chemoradiation treatment. Thus, a further functional investigation is required to define the role of circRNAs in this instance. Dysregulation of the MAPK pathway was also found in lung adenocarcinoma (LUAD) treatment resistance whereby the increased level of circRNA CCDC66 contributes to the overexpression of EGFR (Joseph et al., 2018). CircCCDC66 acts as a sponge for multiple miRNAs and therefore can have a wider effect on its targeted genes (Weng et al., 2017). Besides the MAPK signaling pathway, HGF-MET pathway was also reported to be involved. The role of HGF-MET pathway in EGFR targeted therapy resistance was first reported in 2007 and since then many studies have shown that increase in its activity causes an elevation in RAS-RAF-MEK-ERK or the MAPK pathways (Ko et al., 2017). Thus, current studies are trying to approach the dual inhibition of MET and EGFR in managing resistance in LUAD patients (Ko et al., 2017). Another notable circRNA that plays a role in MAPK pathway is ciRS-7, which is known to sponge to miR-7 and elevates the expression of target genes (Hansen et al., 2013a; Weng et al., 2017). ciRS-7 is known as a super sponge for miR-7 with more than 70 binding sites and act as an expression inhibitor (Hansen et al., 2013a; Hansen et al., 2013b). MiR-7 which is a well-researched miRNA is known to be a player in the progression of many types of cancers in human and directly targets key oncogenes such as EFGR (Kefas et al., 2008), c-KIT in brain cancer (Tamim et al., 2014), PAX6 in colorectal cancer (Needhamsen et al., 2014) and AKT in hepatocellular carcinoma (Fang et al., 2012). Thus, lower miR-7 level due to the sponging effect of cirS-7 causes an increase in target genes, in this instance EGFR and causes an imbalance to the signaling pathway (Fang et al., 2012).
Several Reported CircRNAs Contribute to Treatment Resistance via PI3K/AKT Pathway
Aberrant fusion-circRNA (f-circRNAs), a new form of circRNA generated by chromosomal translocation, was recently linked to treatment resistance in leukemic cells. Lehmann et al. found that f-circRNAs may potentially contribute to the development of resistance to therapy by protecting the leukemic cells from arsenic trioxide (Lehmann et al., 2001). Arsenic oxide is used to treat newly diagnosed and relapsed leukemic patients where it is known to induce apoptotic and cytotoxic effects in blast cells (Dong et al., 2015). Similarly, another study found that by knocking out the expression of f-circM9, leukemic cells can resume apoptosis and resistance toward Ara-C can be reversed. The team has proved that this type of f-circRNA can contribute to tumorigenesis and resistance to therapy both in vitro and in vivo. Besides the MAPK pathway, the PI3K/AKT signaling pathway was triggered simultaneously by the presence of f-circM9. Similar to MAPK, the PI3K/AKT pathway is another common pathway often associated with cancer. This complex cascade influences cell apoptosis, cell cycle, DNA repair, glucose metabolism, and cell transformation. Mainly it is known to play a critical role in transducing signal between oncogenes and cellular functions (Gao et al., 2017). Inhibition of these two pathways caused drug induced apoptosis, hence the high presence of f-circRNAs in leukemic cells will have a negative effect on treatment response. This could be an indicator of possible treatment failure for leukemic patients (Nanba and Toyooka, 2008).
The hsa_circ_0006528 was found to play a vital role in mediating chemoresistance in breast cancer (Carrle and Bielack, 2006). It was observed that higher expression of circ_0006528 is significantly associated with adriamycin (ADM)-resistance in breast cancers (Miao et al., 2017). Circ_0006528 is derived from exons 2 to 5 of the PRELID gene and is exclusively related to miR-7-5p. Previously, it was reported that low levels of miR-7 contribute to the resistance to chemotherapy, and the findings of this study further supported this (Callaghan et al., 2014). Upregulated circRNAs triggered the MAPK signaling pathway and were shown to regulate the cancer cells response to ADM treatment (Nanba and Toyooka, 2008). PI3K/AKT could also play a part in mediating the drug resistance via regulation of hsa-miR-130b (Miao et al., 2017). Current evidence showed that up-regulation of miRNA-130b mediates chemoresistance and increases the proliferation of breast cancer cells. It also reduces the expression of PTEN target which is a common tumor suppressor gene responsible for cell growth and apoptosis (Di Cristofano and Pandolfi, 2000). It is worth noting that the MAPK and PI3K/AKT pathways are complex and multilayered in nature, thus we could see that different types and levels of circRNAs trigger these pathways with opposing effects. In support of this, activation of AKT pathway was shown to have a better prognosis in luminal breast cancer in contrast to its detrimental effect on the above-mentioned study involving circ_0006528 (Sonnenblick et al., 2019).
PI3K/AKT signaling pathway was also predicted to regulate response to 5-Fluorouracil (5-FU) in colorectal cancer patients. 5-FU, a pyrimidine antagonist, is widely used in managing advanced stage colorectal chemotherapy patients but has a recurrence rate of more than 50% (Yu et al., 2009). Upregulation of hsa_circ_0000504 was observed in 5-FU resistance colorectal cancer, where the circRNA interacts with miR-485-5p which targets STAT3 gene (Xiong et al., 2017). It was demonstrated that by silencing the STAT3 gene, clonogenic survival of cancer cells was significantly reduced (Spitzner et al., 2014). From the KEGG pathway analysis, miR-485-5p was predicted to activate the AKT signaling pathway which is associated with chemoradiotherapy treatment response (Xiong et al., 2017). In another recent study, it was reported that hsa_circ_0004015 was highly expressed in non-small cell lung cancer (NSCLC) which significantly contributes to disease progression and EGFR-Tyrosine Kinase Inhibitors (TKI) resistance (Zhou et al., 2019). TKIs are used as first line drug for NSCLC patients with EGFR mutations. Subsequently, via bioinformatics prediction, miR-1183 and target gene PDPK1 were further studied. It is known that PDPKI a crucial component of the Akt-mTOR pathway, can intervene in cell proliferation and apoptosis (Li et al., 2018). In vitro results concluded that knockdown of hsa_circ_0004015 significantly sensitized NSCLC resistant cell line to TKIs (Zhou et al., 2019). Similarly, another group of investigators also proved the relationship of circRNA in resisting EGF-TKIs treatment by activating AKT/mTOR in EGFR-mutant NSCLC (Cheng et al., 2015)
Interestingly this pathway was also seen activated in cisplatin resistance in gastric cancer (GC) (Huang et al., 2019). Cisplatin is among the main chemotherapy drugs given for GC patients. In this study circRNA AKT3 upregulates PIK3R1 which, in turn increases the treatment resistance via miR-198 suppression and activation of PI3K/AKT signaling pathway. Similarly, cisplatin resistance was also found in human thyroid carcinoma cells in which circRNA EIF6 was studied (Liu et al., 2018). By bioinformatics analysis, miR-144-3p was found to regulate the expression of circRNA EIF6 and Transforming growth factor-α (TGF-α). This growth factor could promote tumor growth via various signaling pathways, such as PI3K/AKT and MEK/VEGF. Besides, a transcription factor known as Forkhead box O (FOXO) was reported to interact with the PI3K/AKT pathway as well and is mostly targeted for cancer treatment therapy (Farhan et al., 2017). In support of this, a group of researchers has found that overexpression of circRNA 0067835 promotes the expression of FOXO3a gene which causes detrimental effect to temporal lobe epilepsy (TLE) (Guo et al., 2018). Researchers discovered that circRNA-0067835 acts as sponge to miR-155 which has binding sites to FOXO3a gene. From these findings, aberrant activation of this pathway not only contributes to the progression of disease but also the cell receptivity to chemotherapy and radiotherapy treatment (Toulany and Rodemann, 2015). This multiplex pathway consisting of many regulatory molecules are seen hyper activated in different types of cancers. Looking in a different lens, this could open a new path for research and therapy management especially in terms of chemoradioresistance.
CircRNA Influences Uptake of Drugs via Drug Transporter Pathway
A group of researchers has found that downregulation of circPVT1 by small inhibiting RNA (siRNA) might weaken resistance to doxorubicin and cisplatin treatment in osteosarcoma (Kun-Peng et al., 2018). Doxorubicin or ADM is an essential component in the treatment of osteosarcoma and cisplatin is the second most commonly used drug given in combination with doxorubicin (Carrle and Bielack, 2006). This study found that cirPVT1 was significantly higher in the chemoresistance group as compared to the normal control group and knockdown of circPVT1 could partly reverse the resistance of doxorubicin and cisplatin (Kun-Peng et al., 2018). Overexpression of circPVT1 is also present in chemoresistant lung cancer as compared to chemosensitive patients. Knockdown of circPVT1 decreased the expression level of a well-known drug transporter gene, ABCB1. From previous discoveries, it was found that the gene promotes chemoresistance via removing the intracellular drugs by P-glycoprotein (P-GP) an ATP-dependent efflux pump that may have to metabolize the drugs in cancer cells (Callaghan et al., 2014).
CircRNA Interrupt Chemotherapy Acceptance by Regulating VEGF Pathway
CircRNA has also been shown to be a potential regulator for gemcitabine resistance in pancreatic cancer (Shao et al., 2018). Gemcitabine, a pro drug used as one of the key drugs in treating pancreatic cancer is able to kill cells with active DNA synthesis and block cell cycle progression at G1/S phase (Plunkett et al., 1991; Plunkett et al., 1995). Researchers found that when chr14:101402109-101464448+ and chr4:52729603-52780244+ were silenced in gemcitabine resistant cell lines, the sensitivity towards the drug was enhanced (Shao et al., 2018). Likewise, when the circRNAs were overexpressed, the gemcitabine resistance towards the cell lines was further intensified. By further exploring the binding site for miRNA, it was shown that miR-145-5p was down-regulated in the tumor tissues and plasma of chemotherapy resistant patients (Shao et al., 2018). MiR-145 is known to be associated with gemcitabine resistance in pancreatic cancer and it is believed that ErbB and VEGF signaling biological pathways were activated (Skrypek et al., 2015; Zhou et al., 2016). The ErbB signaling pathway was reported to be involved in gemcitabine resistance in pancreatic cancer while the VEGF pathway was implicated in the progression of the disease which could also cause drug resistance (Shao et al., 2018). Not much literature is available with regards to VEGF and non-coding RNA in treatment resistance however it is worth to note that there is a link between non-coding RNAs particularly circRNAs, in tumor angiogenesis involving VEGF pathway (Abhinand et al., 2016). As pancreatic cancer is known for its poor prognosis, any avenue that could predict treatment response can ease the burden of unnecessary treatment hassle for patients. Potentially, the circRNAs can be used as biomarkers for planning the treatment of patients’ in a more personalized approach.
CircRNA Causes Treatment Failure via Hypoxia-Inducible Factor-1 Regulatory Pathway
Hypoxia-inducible factor (HIF) pathway is increasingly studied due to its therapeutic potential in disease management. A group of circRNA researchers has joined the bandwagon in exploring the relationship between this pathway and cisplatin resistance in bladder cancer (Su et al., 2019). HIF 1 protein plays an integral part in angiogenesis and responding directly to hypoxia. However, due to this property, the enhancement of this gene can also allow the proliferation of cancer cells. Thus inhibition of this pathway potentially could prevent the metastasis of cancer cells (Ziello et al., 2007). In this study, circELP3 was elevated in hypoxia condition, was found to contribute to treatment resistance in bladder cancer cells (Su et al., 2019). Elevation of circELP3 was also in accordance with the severity of disease in human cancer patients. Stimulatingly, the hypoxia elevated circELP3 was independent to ELP3. In theory, it is expected that the hypoxia-related circRNA is correlated to this angiogenesis pathway however the findings in this study were contradictory. This unexpected finding gives a new perspective to the function of circRNA in regulating cisplatin resistance and further test should be carried out to confirm the independence. Another group of researchers has identified circ_103470 and circ_101102 was dysregulated in endometriosis and HIF-1 may be associated with the pathogenesis (Zhang et al., 2018). However, this was only a profiling study thus the space to explore remains large. Compared to other mentioned pathways, HIF-1 has just commenced. As angiogenesis is one of the key target approaches in cancer study, it is worth to explore this area of the pathway with a combination of circRNA.
Future Directions and Conclusions
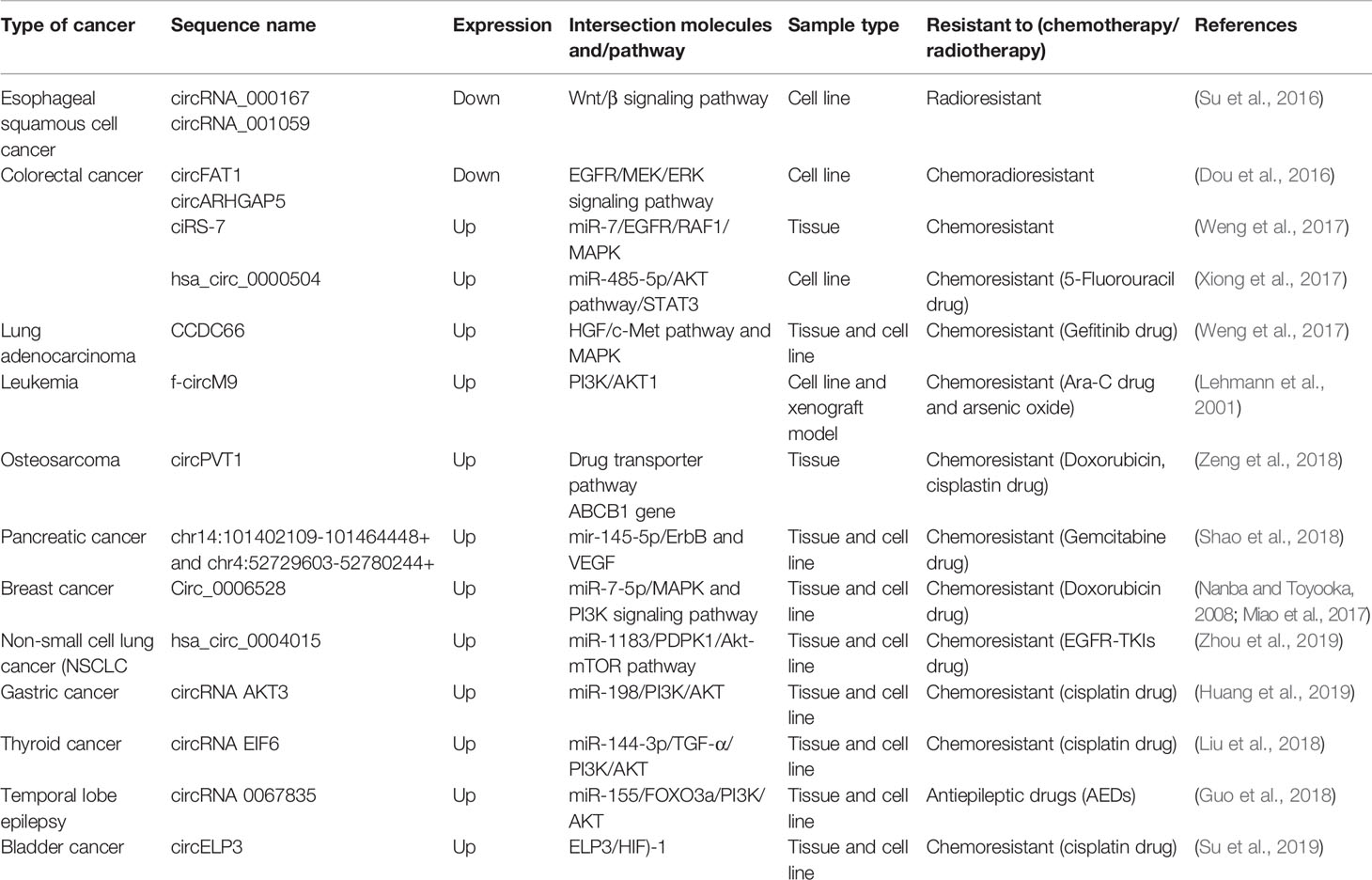
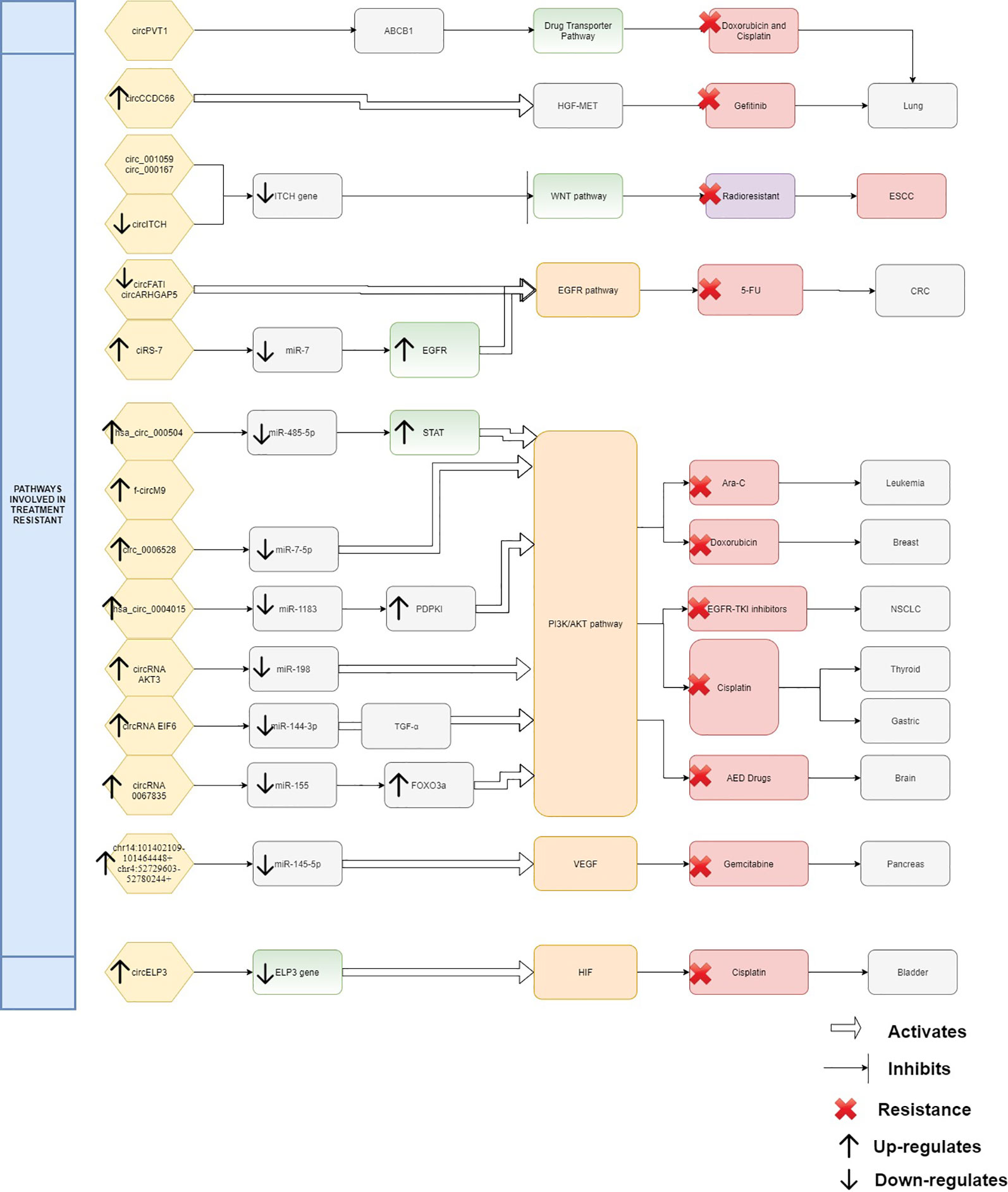
From the evidence compiled as shown in Table 1, it was apparent that there are still many unexplored circRNAs that might be involved in cancer treatment resistance. At this juncture it is difficult to conclude the impact of circRNAs in chemoresistance and radioresistance; however, they have already exhibited enormous potential for further exploration. As treatment resistance in newly diagnosed patients or in recurrence state remains a major challenge, circRNAs could be an ideal marker to predict for treatment failure or even in reversing the resistance. Although the evidence is rather limited and incomplete, it is clear that there are certain interactions between circRNAs and miRNAs which act as contributing factors in disease progression and therapy resistance. By identifying the interactions between circRNA/miRNA/gene/pathway, more opportunities on targeted therapies can be created as shown in Figure 1. In this review, many circRNAs were shown to activate the MAPK pathway and the PI3K signaling pathway. By studying the complex interaction between circRNAs, targeted gene, and the pathways involved, we may be able to detect responses to treatment, early resistance or recurrence in real time. However, analyzing and targeting specific circRNAs persist as a challenge to researchers. Besides than optimizing the feature of circRNA as competing endogenous RNA, limited computational approaches, such as microarray and RNA sequencing, are used for differential expression analysis. Subsequently, inhibition and overexpression of circRNAs are carried out to functionally characterize the targeted circRNAs based on the bioinformatics analysis. These pipelines are rather complex and require tedious preparation for reliable results and remain as a great challenge for novice researchers. It is apparent that there are still gaps to be filled in terms of predicting the target genes and the functional mechanisms in modulating diseases. For validation of the high throughput sequencing, current approach mostly focuses on proving the presence of circRNA by carrying out qPCR and Sanger sequencing. To date, only a few circRNAs have been characterized and validated fully. The unpredictable character of circRNA as a pro-oncogene and anti-tumor molecule is thought-provoking and a limiting factor for immediate clinical application. It is more challenging when the same circRNA exhibit multiple roles in different cancers. Thus, it is difficult to deduce and predict a single role for circRNA. Nonetheless, this shows the flexibility of circRNAs as a biomarker and when manipulated could contribute to prevention or delaying of disease progression. CircRNAs as a predictive marker and potential regulator of treatment resistance is still in the early phase of research and application, nevertheless, the future of circRNAs is certainly promising, and it will be likely that they will become part of the armamentarium to treat cancers in future.
Author Contributions
SJ and NA drafted the article. NA conceived the idea. NM, EH, and RJ provided feedback and critical input.
Funding
This manuscript was funded by the Fundamental Research Grant Scheme (FRGS/1/2017/SKK08/UKM/03/3) awarded by the Ministry of Higher Education Malaysia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abhinand, C. S., Raju, R., Soumya, S. J., Arya, P. S., Sudhakaran, P. R. (2016). VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J. Cell Commun. Signal. 10 (4), 347–354. doi: 10.1007/s12079-016-0352-8
Callaghan, R., Luk, F., Bebawy, M. (2014). Inhibition of the multidrug resistance P-glycoprotein: Time for a change of strategy? Drug Metab. Disposition 42 (4), 623–631. doi: 10.1124/dmd.113.056176
Carrle, D., Bielack, S. S. (2006). Current strategies of chemotherapy in osteosarcoma. Int. Orthopaedics 30 (6), 445–451. doi: 10.1007/s00264-006-0192-x
Chen, L. L., Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12 (4), 381–388. doi: 10.1080/15476286.2015.1020271
Chen, G.-Z., Zhu, H.-C., Dai, W.-S., Zeng, X.-N., Luo, J.-H., Sun, X.-C. (2017). The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J. Thorac. Dis. 9 (3), 849–859. doi: 10.21037/jtd.2017.03.23
Cheng, N., Cai, W., Ren, S, Li, X., Wang, Q., Pan, H., et al. (2015). Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 6 (27), 23582–23593. doi: 10.18632/oncotarget.4361
Di Cristofano, A., Pandolfi, P. P. (2000). The multiple roles of PTEN in tumor suppression. Cell. 100 (4), 387–390. doi: 10.1016/s0092-8674(00)80674-1
Dong, L., Deng, J., Sun, Z. M., Pan, A. P., Xiang, X. J., Zhang, L., et al. (2015). Interference with the β-catenin gene in gastric cancer induces changes to the miRNA expression profile. Tumor Biol. 36 (9), 6973–6983. doi: 10.1007/s13277-015-3415-1
Dou, Y., Cha, D. J., Franklin, J. L., Higginbotham, J. N., Jeppesen, D. K., Weaver, A. M., et al. (2016). Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 6 (1), 37892–37903. doi: 10.1038/srep37982
Du, W. W., Zhang, C., Yang, W., Yong, T., Awan, F. M., Yang, B. B. (2017). Identifying and characterizing circRNA-protein interaction. Theranostics. 7 (17), 4183–4191. doi: 10.7150/thno.21299
Fang, Y., Xue, J. L., Shen, Q., Chen, J., Tian, L. (2012). MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 55 (6), 1852–1862. doi: 10.1002/hep.25576
Fang, Y. (2018). Circular RNAs as novel biomarkers with regulatory potency in human diseases. Futur. Sci. OA 4 (7), FSO314. doi: 10.4155/fsoa-2018-0036
Farhan, M., Wang, H., Gaur, U., Little, P. J., Xu, J., Zheng, W. (2017). FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 13 (7), 815–827. doi: 10.7150/ijbs.20052
Flanagan, L., Lindner, A. U., de Chaumont, C., Kehoe, J., Fay, J., Bacon, O., et al. (2015). BCL2 protein signalling determines acute responses to neoadjuvant chemoradiotherapy in rectal cancer. J. Mol. Med. 93 (3), 315–326. doi: 10.1007/s00109-014-1221-7
Gao, D., Zhang, X., Liu, B., Meng, D., Fang, K., Guo, Z., et al. (2017). Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 9 (9), 1175–1188. doi: 10.2217/epi-2017-0055
Gong, Z., Lin, H. K., Yang, J., Li, J., Yang, L., Le, Y., et al. (2014). Novel insights into the role of microRNA in lung cancer resistance to treatment and targeted therapy. Curr. Cancer Drug Targets. 14 (3), 241–258. doi: 10.2174/1568009614666140305104845
Granados-Riveron, J. T., Aquino-Jarquin, G. (2016). The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta - Gene Regul. Mechanisms. 1859 (10), 1245–1251. doi: 10.1016/j.bbagrm.2016.07.009
Gong, G. H., An, F. M., Wang, Y., Bian, M., Wang, D., Wei, C. X. (2018). Comprehensive circular RNA profiling reveals the regulatory role of the CircRNA-0067835/miR-155 pathway in temporal lobe epilepsy. Cell. Physiol. Biochem. doi: 10.1159/000495589
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., Kjems, J.. (2013a). Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441), 384–388. doi: 10.1038/nature11993
Hansen, T. B., Kjems, J., Damgaard, C. K. (2013b). Circular RNA and miR-7 in Cancer. Cancer Res. 73 (18), 5609–5612. doi: 10.1158/0008-5472.CAN-13-1568
Huang, G., Zhu, H., Shi, Y., Wu, W., Cai, H., Chen, X. (2015). Cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-Catenin Pathway. PloS One. 10 (6), e0131225. doi: 10.1371/journal.pone.0131225
Huang, X., Xu, Z., Li, Z., Zhang, Q., Wang, W., Li, B., et al. (2019). Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer. 18 (71), 1–20. doi: 10.1186/s12943-019-0969-3
Hymowitz, S. G., Malek, S. (2018). Targeting the MAPK Pathway in RAS Mutant Cancers. Cold Spring Harb. Perspect. Med. 8 (11), 1–16. doi: 10.1101/cshperspect.a031492
Joseph, N. A., Chiou, S. H., Lung, Z, Yang, C. L., Lin, T. Y., Chang, H. W., et al. (2018). The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 11 (1), 74–88. doi: 10.1186/s13045-018-0557-9
Kefas, B., Godlewski, J., Comeau, L., Li, Y., Abounader, R., Hawkinson, M., et al. (2008). microRNA-7 inhibits the epidermal growth factor receptor and the akt pathway and is down-regulated in glioblastoma. Cancer Res. 68 (10), 3566–3572. doi: 10.1158/0008-5472.CAN-07-6639
Ko, B., He, T., Gadgeel, S., Halmos, B. (2017). MET/HGF pathway activation as a paradigm of resistance to targeted therapies. Ann. Transl. Med. 5 (1), 4–4. doi: 10.21037/atm.2016.12.09
Koval, A., Katanaev, V. L. (2018). Dramatic dysbalancing of the Wnt pathway in breast cancers. Sci. Rep. 8 (1), 7329–7339. doi: 10.1038/s41598-018-25672-6
Kristensen, L. S., Hansen, T. B., Venø, M. T., Kjems, J. (2018). Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 37 (5), 555–565. doi: 10.1038/onc.2017.361
Kun-Peng, Z., Xiao-Long, M., Chun-Lin, Z. (2018). Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 14 (3), 321–330. doi: 10.7150/ijbs.24360
Lasda, E., Parker, R. (2014). Circular RNAs: diversity of form and function. RNA. 20 (12), –. doi: 10.1261/rna.047126.114
Lehmann, S., Bengtzen, S., Paul, A., Christensson, B., Paul, C. (2001). Effects of arsenic trioxide (As2O3) on leukemic cells from patients with non-M3 acute myelogenous leukemia: studies of cytotoxicity, apoptosis and the pattern of resistance. Eur. J. Haematol. 66 (6), 357–364. doi: 10.1034/j.1600-0609.2001.066006357.x
Li, R., Zhang, L., Jia, L., Duan, Y., Li, Y., Bao, L., et al. (2014). Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 9 (6), e100893. doi: 10.1371/journal.pone.0100893
Li, Z, Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi: 10.1038/nsmb.2959
Li, J., Yang, J., Zhou, P., Le, Y., Zhou, C., Wang, S., et al. (2015). Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 5 (2), 472–480.
Li, F., Zhang, L., Li, W., Deng, J., Zheng, J., An, M., et al. (2015). Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 6 (8), 6001–6013.
Li, D., Mullinax, J. E., Aiken, T., Xin, H., Wiegand, G., Anderson, A., et al. (2018). Loss of PDPK1 abrogates resistance to gemcitabine in label-retaining pancreatic cancer cells. BMC Cancer. 18 (1), 772–787. doi: 10.1186/s12885-018-4690-1
Liamina, D., Saenko, Y., Sibirnyj, W., Khokhlova, A., Saenko, V., Rastorgueva, E., et al. (2017). Radiation-induced changes of microRNA expression profiles in radiosensitive and radioresistant leukemia cell lines with different levels of chromosome abnormalities. Cancers (Basel). 9 (10), 136–152 doi: 10.3390/cancers9100136
Lin ADF, X., Chen, Y. (2018). Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Research 24, 70–78. doi: 10.12659/MSMBR.909737
Liu, H., Wang, G., Yang, L., Qu, J., Yang, Z., Zhou, X. (2016). Knockdown of long non-Coding RNA UCA1 Increases the tamoxifen sensitivity of breast cancer cells through inhibition of wnt/β-catenin pathway. PLoS One. 11 (12), e0168406. doi: 10.1371/journal.pone.0168406
Liu, F., Zhang, J., Qin, L., Yang, Z., Xiong, J., Liu, J., et al. (2018). Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany. NY). 10 (12), 3806–3820. doi: 10.18632/aging.101674
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495 (7441), 333–338. doi: 10.1038/nature11928
Miao, Y., Zheng, W., Li, N., Su, Z., Zhao, L., Zhou, H., et al. (2017). MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci. Rep. 7, 1–12. doi: 10.1038/srep41942
Nanba, K., Toyooka, S. (2008). The allelic distribution of a single nucleotide polymorphism in the PDCD5 gene locus of Japanese non-small cell lung cancer patients. Mol. Med. Rep. 1 (5), 667–671. doi: 10.3892/mmr_00000010
Needhamsen, M., White, R. B., Giles, K. M., Dunlop, S. A., Thomas, M. G. (2014). Regulation of human PAX6 expression by miR-7. Evol. Bioinforma. 10 107–113. doi: 10.4137/EBO.S13739
Ni, J., Bucci, J., Chang, L., Malouf, D., Graham, P., Li, Y. (2017). Targeting microRNAs in prostate cancer radiotherapy. Theranostics 7 (13), 3243–3259. doi: 10.7150/thno.19934
Palmieri, D., Langenberger, D., Dai, Y., Calin, G. A., Dragomir, M. (2018). Circular RNAs in cancer – lessons learned from microRNAs. Front. Oncol. 8 (8). doi: 10.3389/fonc.2018.00179
Plunkett, W., Grindey, G. B., Plunkett, W. (1991). Action of 2',2'-difluorodeoxycytidine on DNA synthesis. Cancer Res. 51 (22), 6110–6117.
Plunkett, W., Huang, P., Xu, Y. Z., Heinemann, V., Grunewald, R., Gandhi, V. (1995). Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin. Oncol. 22 (11), 3–10.
Pohl, S. G., Brook, N., Agostino, M., Arfuso, F., Kumar, A. P., Dharmarajan, A. (2017). Wnt signaling in triple-negative breast cancer. Oncogenesis. 6, e310–e322. doi: 10.1038/oncsis.2017
Pozzi, C., Cuomo, A., Spadoni, I., Magni, E., Silvola, A., Conte, A., et al. (2016). The EGFR-specific antibody cetuximab combined with chemotherapy triggers immunogenic cell death. Nat. Med. 22 (6), 624–631. doi: 10.1038/nm.4078
Shang, Q., Yang, Z., Jia, R., Ge, S. (2019). The novel roles of circRNAs in human cancer. Mol. Cancer 18 (1), 1–10. doi: 10.1186/s12943-018-0934-6
Shao, F., Huang, M., Meng, F., Huang, Q. (2018). Circular RNA Signature Predicts Gemcitabine Resistance of Pancreatic Ductal Adenocarcinoma. Front. Pharmacol. 9, 1–11. doi: 10.3389/fphar.2018.00584
Shen, B., Wang, Z., Li, Z., Song, H., Ding, X.. (2019) Circular RNAs: an emerging landscape in tumor metastasis. Am. J. Cancer Res. 9 (4), 630–643.
Si-Tu, J., Cai, Y, Feng, T., Yang, D., Yuan, S., Yang, X., et al. (2019). Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int. J. Biol. Macromol. 122, 1235–1243. doi: 10.1016/j.ijbiomac.2018.09.076
Skrypek, N., Vasseur, R., Vincent, A., Duchêne, B., Van Seuningen, I., Jonckheere, N. (2015). The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget 6 (13), 10853–10867. doi: 10.18632/oncotarget.3414
Sonnenblick, A., Venet, D., Brohée, S., Pondé, N., Sotiriou, C. (2019). pAKT pathway activation is associated with PIK3CA mutations and good prognosis in luminal breast cancer in contrast to p-mTOR pathway activation. NPJ Breast Cancer. 8 (7), 1–7. doi: 10.1038/s41523-019-0102-1
Spitzner, M., Roesler, B., Bielfeld, C., Emons, G., Gaedcke, J., Wolff, H. A., et al. (2014). STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. Int. J. Cancer. 134 (4), 997–1007. doi: 10.1002/ijc.28429
Su, H., Jin, X., Xie, C., Lin, F., Deng, X., Shen, L., et al. (2016). Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J. Transl. Med. 14 (1), 1–10. doi: 10.1186/s12967-016-0977-7
Su, Y., Yang, W., Jiang, N., Shi, J., Wang, C., Lin, T. (2019). Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int. J. Biol. Sci. 15 (2), 441–452. doi: 10.7150/ijbs.26826
Szabo, L., Salzman, J. (2016). Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692. doi: 10.1038/nrg.2016.114
Takebe, N., Miele, L., Harris, P. J., Jeong, W., Bando, H., Kahn, M., et al. (2015). Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 12 (8), 445–464. doi: 10.1038/nrclinonc.2015.61
Tamim, S., Vo, D. T., Uren, P. J., Qiao, M., Bindewald, E., Kasprzak, W. K., et al. (2014). Genomic analyses reveal broad impact of miR-137 on genes associated with malignant transformation and neuronal differentiation in glioblastoma cells. PLoS One. 9 (1), 85591. doi: 10.1371/journal.pone.0085591
Toulany, M., Rodemann, H. P. (2015). Phosphatidylinositol 3-kinase/Akt signaling as a key mediator of tumor cell responsiveness to radiation. Semin. Cancer Biol. 35, 180–190. doi: 10.1016/j.semcancer.2015.07.003
Wan, L., Zhang, L., Fan, K., Cheng, Z. X., Sun, Q. C., Wang, J. J. (2016). Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. BioMed. Res. Int. 2016, 1–11. doi: 10.1155/2016/1579490
Wang, X., Zhang, Y., Huang, L., Zhang, J., Pan, F., Li, B., et al. (2015). Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int. J. Clin. Exp. Pathol. 8 (12), 16020–16025.
Wang, Y., Mo, Y., Gong, Z., Yang, X., Yang, M., Zhang, S., et al. (2017). Circular RNAs in human cancer. Mol. Cancer. 16 (25), 1–8. doi: 10.1186/s12943-017-0598-7
Weng, W., Wei, Q., Toden, S., Yoshida, K., Nagasaka, T., Fujiwara, T., et al. (2017). Circular RNA ciRS-7 — A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 23 (14), 3918–3928. doi: 10.1158/1078-0432.CCR-16-2541
Wu, J., Qi, X., Liu, L., Hu, X., Liu, J, Yang, J., et al. (2019). Emerging epigenetic regulation of circular RNAs in human cancer. Mol. Ther. - Nucleic Acids 16, 589–596. doi: 10.1016/j.omtn.2019.04.011
Xiong, W., Ai, Y. Q., Li, Y. F., Ye, Q., Chen, Z. T., Qin, J. Y., et al. (2017). Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. BioMed. Res. Int. 2017, 1–8. doi: 10.1155/2017/8421614
Yang, G., Shen, T., Yi, X., Zhang, Z., Tang, C., Wang, L., et al. (2018). Crosstalk between long non-coding RNAs and Wnt/β-catenin signalling in cancer. J. Cell. Mol. Med. 22 (4), 2062–2070.
Yu, Y., Kanwar, S. S., Patel, B. B., Nautiyal, J., Sarkar, F. H., Majumdar, A. P. N. (2009). Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Transl. Oncol. 2 (4), 321–328. doi: 10.1593/tlo.09193
Zeng, Y., Xu, Y., Shu, R., Sun, L., Tian, Y., Shi, C., et al. (2017). Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. Int. J. Mol. Med. 40 (6), 1818–1828. doi: 10.3892/ijmm.2017.3189
Zeng, K., Chen, X., Xu, M., Liu, X., Hu, X., Xu, T., et al. (2018). CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7 article. Cell Death Dis. 9 (4). doi: 10.1038/s41419-018-0454-8
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., et al. (2013). Circular Intronic Long Noncoding RNAs. Mol. Cell. 51 (6), –. doi: 10.1016/j.molcel.2013.08.017
Zhang, Y., Qi, X., Li, G., Li, J., Yu, J., Liu, H., et al. (2017). Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 8 (14), 22936–22943. doi: 10.18632/oncotarget.15288
Zhang, M., Ren, C., Xiao, Y., Xia, X., Fang, X. (2018). Expression profile analysis of circular RNAs in ovarian endometriosis by microarray and bioinformatics. Med. Sci. Monit. 24, 9240–9250. doi: 10.12659/MSM.913885
Zhou, R., Curry, J. M., Roy, L. D., Grover, P., Haider, J., Moore, L. J., et al. (2016). A novel association of neuropilin-1 and MUC1 in pancreatic ductal adenocarcinoma: role in induction of VEGF signaling and angiogenesis. Oncogene. 35 (43), 5608–5618. doi: 10.1038/onc.2015.516
Zhou, Y., Zheng, X., Xu, B., Chen, L., Wang, Q., Deng, H., et al. (2019). Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 508 (2), 527–535. doi: 10.1016/j.bbrc.2018.11.157
Keywords: non-coding RNAs, biomarker, RNA splicing, chemoresistance, radioresistance
Citation: Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R and Abu N (2020) Circular RNAs: Potential Regulators of Treatment Resistance in Human Cancers. Front. Genet. 10:1369. doi: 10.3389/fgene.2019.01369
Received: 31 May 2019; Accepted: 16 December 2019;
Published: 28 January 2020.
Edited by:
Stefano Duga, Humanitas University, ItalyReviewed by:
Marianna Aprile, Italian National Research Council (CNR), ItalyMohammadreza Hajjari, Shahid Chamran University of Ahvaz, Iran
Copyright © 2020 Jeyaraman, Hanif, Ab Mutalib, Jamal and Abu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nadiah Abu, bmFkaWFoLmFidUBwcHVrbS51a20uZWR1Lm15
 Shivapriya Jeyaraman
Shivapriya Jeyaraman Ezanee Azlina Mohamad Hanif
Ezanee Azlina Mohamad Hanif Nurul Syakima Ab Mutalib
Nurul Syakima Ab Mutalib Rahman Jamal
Rahman Jamal Nadiah Abu
Nadiah Abu