- College of Plant Protection, Nanjing Agricultural University, State and Local Joint Engineering Research Center of Green Pesticide Invention and Application, Nanjing, China
Transmembrane channel-like (TMC) genes encode a family of evolutionarily conserved membrane proteins. Mutations in the TMC1 and TMC2 cause deafness in humans and mice. However, their functions in insects are is still not well known. Here we cloned three tmc genes, Nltmc3, Nltmc5, and Nltmc7 from brown planthoppers. The predicted amino acid sequences showed high identity with other species homologs and have the characteristic eight or nine transmembrane domains and TMC domain architecture. We detected these three genes in all developmental stages and examined tissues. Interestingly, we found Nltmc3 was highly expressed in the female reproductive organ especially in the oviduct. RNAi-mediated silencing of Nltmc3 substantially decreased the egg-laying number and impaired ovary development. Our results indicate that Nltmc3 has an essential role in the ovary development of brown planthoppers.
Introduction
Transmembrane channel-like (TMC) proteins have been identified from insects to mammals (Keresztes et al., 2003; Kurima et al., 2003; Guo et al., 2016). Eight TMC proteins were presented in vertebrates including humans and mice (Keresztes et al., 2003; Kurima et al., 2003). They can be grouped into three subfamilies A, B, and C, in terms of their sequence homology and similarities of the genomic structures of their respective genes (Keresztes et al., 2003). The TMC protein subfamily A consists of three proteins, TMC1, TMC2, and TMC3; subfamily B contains two proteins, TMC5 and TMC6; And subfamily C include three members, TMC4, TMC7, and TMC8 (Keresztes et al., 2003; Kurima et al., 2003). In Caenorhabditis elegans, two tmc genes have been cloned (Chatzigeorgiou et al., 2013). However, the Drosophila genome only encodes one tmc homolog (Guo et al., 2016). All tmc genes are strongly predicted to encode proteins with at least six transmembrane domains and a novel conserved CWETXVGQEly(K/R)LtvXD amino-acid sequence motif that termed as TMC domain (Keresztes et al., 2003; Kurima et al., 2003).
TMC1 and TMC2, first identified in deaf human patients, are essential for hearing in mice (Kawashima et al., 2015). TMC1 and TMC2 are necessary for the mechano-transduction currents of hair cells (Pan et al., 2013, 2018). Recent studies have showed that TMC1 and TMC2 are pore-forming subunits of mechanosensory transduction channels (Pan et al., 2018; Jia et al., 2019). The tmc1 gene in C. elegans was reported to encode a sodium-sensitive cation channel and participates in nociceptive neuron-mediated alkaline and salt chemo-sensation (Chatzigeorgiou et al., 2013; Zhang et al., 2015; Wang et al., 2016). Recent studies showed that TMC proteins in nematodes modulate egg laying and membrane excitability through a background leak conductance (Yue et al., 2018). In Drosophila, the tmc gene was involved in proprioception, food texture detection and egg-laying texture discrimination (Guo et al., 2016; Zhang et al., 2016; Wu et al., 2019).
The brown planthopper (BPH), Nilaparvata lugens (Stål), (Hemiptera: Delphacidae), is a serious pest on rice in China. It has cause loss of rice production more than $300 million annually in Asia (Min et al., 2014). Chemical insecticides are mainly used for BPH control. However, due to the large scale and intensive use of insecticides, BPH has evolved high levels of resistance to many of the major classes of insecticide (Wu et al., 2018). Hence, it is urgent to find new insecticide targets to develop novel insecticides. Although tmc genes have been characterized in mice, nematodes, and fruit flies, few studies have been performed to investigate functions of tmc genes in other insects. In this study, we characterized the tmc gene family of the BPH. We found that three tmc genes were present in the genome of BPH. The expression of these three tmc genes were investigated and we found that silencing of Nltmc3 gene, which is homology of tmc gene in Drosophila and C. elegans, impairs the egg-laying and ovary development in BPH.
Materials and Methods
Insects
Nilaparvata lugens was collected from a rice field at the Plant Protection Station of Jiangpu County (Jiangsu, China). They were reared on Taichung Native 1 (TN1) rice seedlings in the laboratory. The rearing conditions were 27 ± 1°C, with 70 ± 10% relative humidity and a 16 h:8 h (Light:Dark) photoperiod.
Identification and Cloning of Nltmc Genes
The amino acid sequence of D. melanogaster TMC protein was used to screen against N. lugens genomic and transcriptomic databases for identification of its homologs in N. lugens. Open reading frames (ORFs) were predicted with EditSeq (version 5.02, DNAstar, Madison, WI, United States).
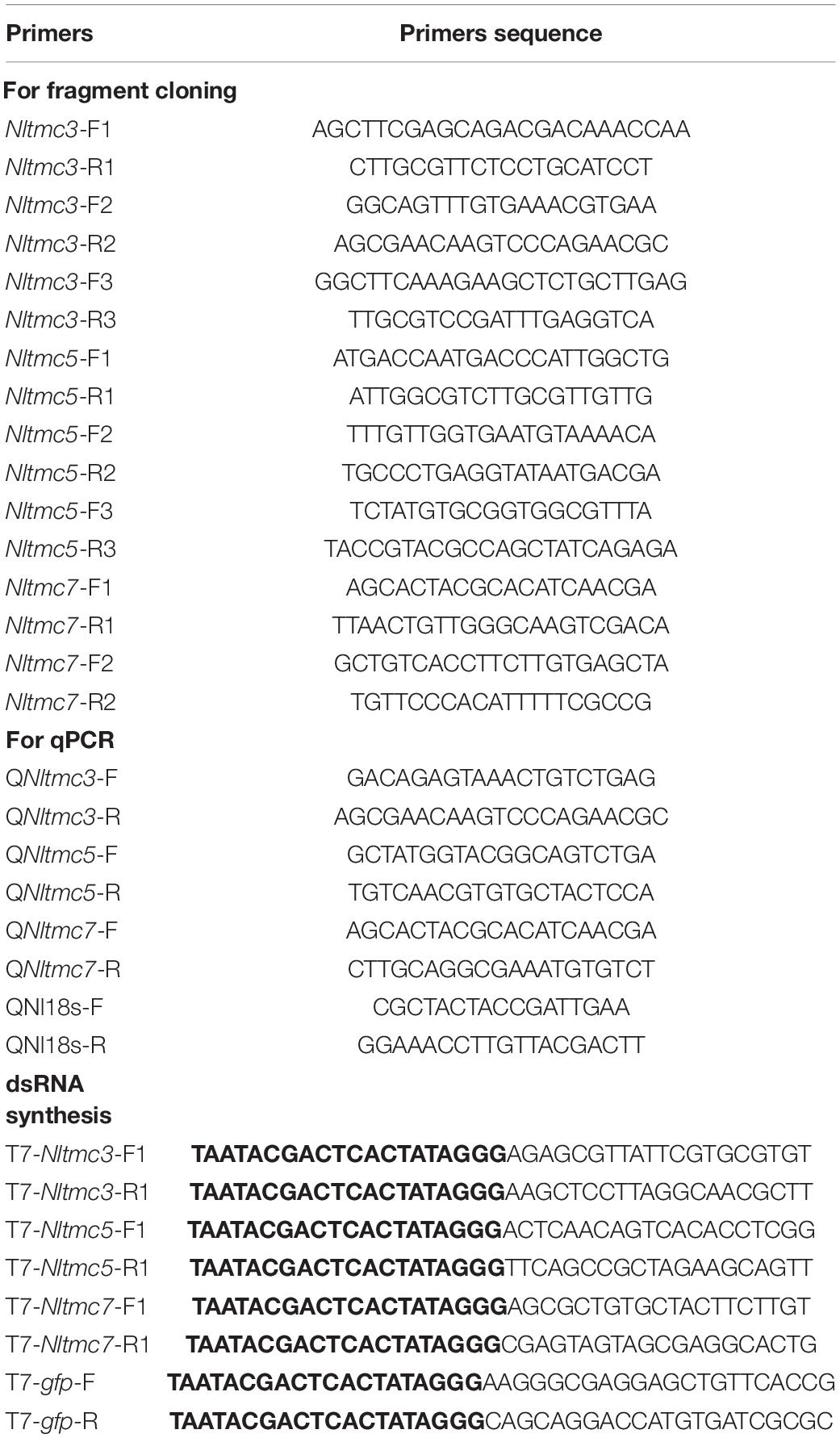
Total RNA was isolated from whole insects using the TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. Residual genomic DNA was removed by RQ1 RNase-Free DNase (Promega). Single-stranded cDNA was synthesized from the total RNA with M-MLV reverse transcriptase and oligo (dT)18 (BioTeke, Beijing, China). The forward primer Nltmc-comp-F and the reverse primer Nltmc-comp-R were used to amplify the full-length or fragment gene by means of PCR on cDNA from adult N. lugens using TransTaq HiFi DNA Polymerase (TransGen Biotech, Beijing, China). The purified PCR products were sub-cloned into pGEM®-T easy vector (Promega, Madison, WI) and then sequenced using the 3730 XL DNA analyzer (Applied Biosystems, Carlsbad, CA, United States. The primers corresponding to each gene are listed in Table 1.
Sequence Analysis and Phylogenetic Tree Construction
The exon and intron architectures of Nltmc genes were predicted based on the alignments of putative cDNA against their corresponding genomic sequences in Spidey1, and then structured on the website of GSDS v2.02 (Hu et al., 2015). The transmembrane segments and topology of NlTMC proteins were predicted by TMHMM v2.03. Multiple alignments of the complete amino acid sequences were performed with Clustal Omega4. Phylogenetic tree was constructed using MEGA 5.2.2 software with the Maximum Likelihood method and bootstrapped with 1,000 replications. The branch support values are expressed as percentages. The accession numbers of the sequences used in the phylogenetic analysis are listed in Table 2.
Table 2. Accession numbers of amino acid sequences used in the phylogenetic and sequence alignment analysis.
Gene Expression Profile Analysis
Developmental stage samples were collected from eggs (n = 100∼120), first-instar (n = 80), second-instar (n = 60), third-instar (n = 230), fourth-instar (n = 15∼20), fifth-instar nymphs (n = 15), and adults of both sexes and wing forms: brachypterous female (BF), macropterous female (MF), brachypterous male (BM), and macropterous male adults (MM) (n = 10). Eggs were collected at 4 days (central development stage) after the oviposition since the egg stage is 6–7 days. Nymphs were collected every 24 h from the beginning of each instar until molting and all adults were collected 4 days after eclosion.
Different tissue samples including head, wing, gut, Malpighian tubule (MT), female reproductive organ (FRO), ovary (OA), oviduct (OU), copulatory pouch (CP), and spermatheca (SE) were dissected from brachypterous female adults collected 4 days after eclosion. The first-strand cDNA was synthesized with HiScript® II Q RT SuperMix for qPCR (+ gDNA wiper) kit (Vazyme, Nanjing, China) using an oligo(dT)18 primer and 500 ng total RNA template in a 10 μl reaction, following the instructions.
Real-time qPCRs were employed to investigate relative expression of Nltmc genes in the various samples using the UltraSYBR Mixture (with ROX) Kit (CWBIO, Beijing, China). The PCR was performed in 20 μl reaction including 4 μl of 10-fold diluted cDNA, 1 μl of each primer (10 μM), 10 μl 2× UltraSYBR Mixture, and 6 μl RNase-free water. The standard two-step PCR cycle conditions were as follows: 95°C for 10 min, and then 40 cycles of amplification consisting of 95°C for 15 s, 60°C for 40 s, followed by melting curve analysis. Pairs of gene-specific primers used for real-time qPCR were designed using the Primer Premier 5 Software (Table 1). The relative quantification of Nltmc was calculated according to the comparative 2–ΔΔCT method (Livak and Schmittgen, 2001).
Double-Stranded RNA (dsRNA) Preparation and Injection
The fragment coding sequence of Nltmc genes and green fluorescent protein (gfp) were amplified by PCR using specific primers conjugated with the T7 RNA polymerase (Table 1). PCR-generated DNA templates were then used to synthesize dsRNA, which contains T7 promoter sequences at each end. We used a MEGAscript T7 transcription kit (Ambion, Austin, TX, United States) to produce the specific dsRNA of each gene as the manufacturer’s instruction. The quality and size of the dsRNA products were verified by 1% agarose gel electrophoresis. Thirty fully mated female adults (collected 4 days after eclosion) were injected with approximately 50 nl of purified dsRNA (5,000 ng/μl) via mesothorax and were reared with rice seedlings. A set of 6–10 insects at 3 days after injection was selected to verify dsRNA knockdown efficiency by qRT-PCR. The remaining individuals were used for observations of eggs laid and female survival. Four to six biological replications were performed.
Egg-Laying, Survival Assay and Quantification of Egg Number
For egg-laying assay, RNAi injected females (fully mated) were transferred vials with fresh rice seedlings. Number of laid eggs were counted under a stereomicroscope (Zeiss) after 3 days. The female survival was recorded 8 days after injection of dsRNA. At least four to six vials per treatment were observed. Ovaries of mated females were prepared under the stereomicroscope (Zeiss). Pictures of ovaries were taken using a light microscope with a digital video camera (Zeiss, ProgRes 3008 mF, Jenoptik, Jena, Germany). The number of eggs per ovary were measured and counted.
Statistics
Experimental data was analyzed using GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA, United States). The two-tailed unpaired Student’s t-test or one-way analysis of variance (ANOVA) of Duncan’s multi-range test were used to test the differences between two or more than two normal distribution data.
Results
Sequence Analysis of tmc Gene Family in N. lugens
We identified three tmc genes in the genome and transcriptome database of N. lugens. These three tmc genes were cloned by PCR and then confirmed by DNA sequencing (Table 1). One fragment and two full-lengths of different cDNA clones were obtained. These sequences were designated Nltmc3 (GenBank accession number: MT576068), Nltmc5 (MT576067), and Nltmc7 (MT576069) according to their similarity to other invertebrate and vertebrate tmc genes (Keresztes et al., 2003; Kurima et al., 2003).
We cloned the fragment of Nltmc3 gene that consists of 4,476-bp cDNA encoding 1,492 amino acids. We tried to clone the full-length of this gene using 5′-RACE and 3′-RACE technology. Unfortunately, we did not get the positive results. We then cloned the full-length of Nltmc5 and Nltmc7 gene. The complete ORF of Nltmc5 and Nltmc7 encodes 692 and 780 amino acids, respectively. Exon-intron organization was analyzed by comparing cloned cDNAs and the corresponding genomic sequence, revealing that Nltmc3 is located on scaffold 754 and scaffold 3202, Nltmc5 is located on scaffold 943, and Nltmc7 is located on scaffold 2298 (Figure 1). TMHMM2.0 strongly predicts the presence of eight or nine transmembrane-spanning domains in each of the TMC proteins. They all encode a conserved TMC domain that share the completely conserved amino acid triplet C (cysteine) – W (tryptophan) – E (glutamic acid), predicted to be located on the extracellular loop upstream of TM6 (Figures 2–4) (Keresztes et al., 2003). Amino-acid sequence comparisons between the NlTMC proteins and other species TMC proteins show high overall amino acid similarity at the transmembrane region and TMC domain (Figures 2–4). The encoded protein of NlTMC3, similar with CeTMC1, CeTMC2, and DmTMC, has large ORFs. BLASTP analyses of protein sequence alignment showed that NlTMC3 had 60, 66, and 66%, sequence similarity with the TMC proteins of Drosophila melanogaster and C. elegans. Interestingly, we found two internal repeats between TM5 and TM6 in the Nltmc3 gene (Figure 2). In mammals, eight TMC proteins can be grouped into three subfamilies A, B, and C, based on sequence homology (Keresztes et al., 2003). Phylogenetic tree comparison showed that NlTMC3 clustered with MmTMC1, MmTMC2, MmMTC3, CeTMC1, CeTMC2, and DmTMC, which belongs to A subfamily. NlTMC5 is assembled in a group that contains MmTMC5 and MmTMC6, which belongs to B subfamily. And NlTMC7 clustered with MmTMC7 and HsTMC7 that belongs to C subfamily (Figure 5).
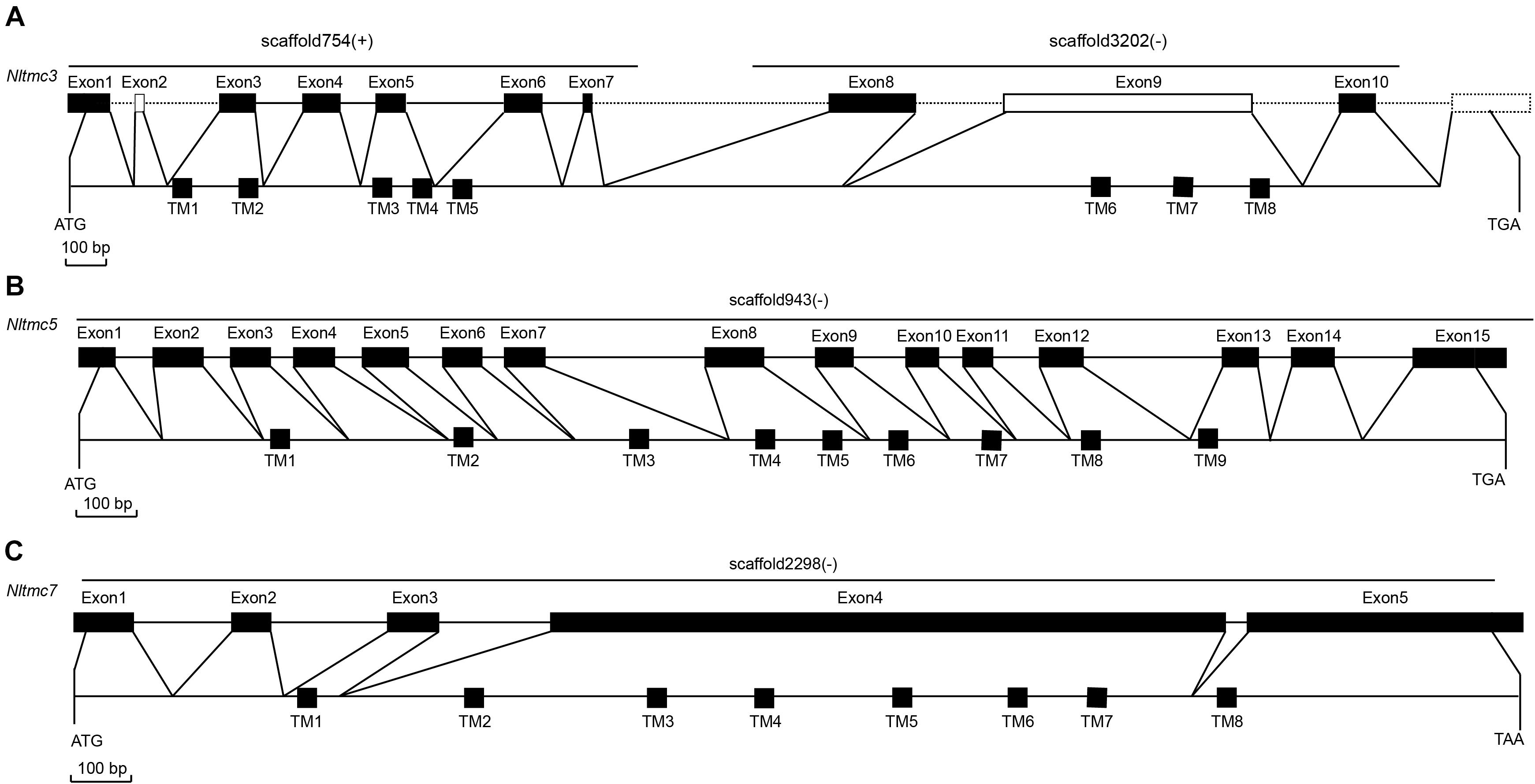
Figure 1. Genomic structures of Nltmc3 (A), Nltmc5 (B), and Nltmc7 (C) genes and the domain structures of encoded proteins. The exon–intron organization of Nltmc genes was determined by sequence comparison between genomic sequence and putative cDNA sequence. The top line of every gene represents the original genomic scaffold sequences. The predicted start codon (ATG), stop codon (TAG or TAA) and the scaffold of gene locus (“ + ” represent the same orientation with scaffold; “ – ” represent the reverse orientation with scaffold) are also shown in the corresponding positions. The bottom line represents the full-length sequence of the transcript. The transmembrane regions are indicated by the black squares. In the predicted topologies of the receptors, the transmembrane regions are indicated as TM1-8 or TM1-9.

Figure 2. Amino acid sequence alignment of NlTMC3 and orthologs genes from Caenorhabditis elegans (CeTMC1: NP_508221.3; CeTMC2: NP_001335510.1), and Drosophila melanogaster (DmTMC: NP_001303362.1). The amino acid position is shown on the right. Identical residues between orthologs sequences are shown as white characters against the black background, and conservative substitutions shown as shading. Black lines represent the transmembrane domain (TM), white squares represent the internal repeats (IRs), and red line represents TMC domain.
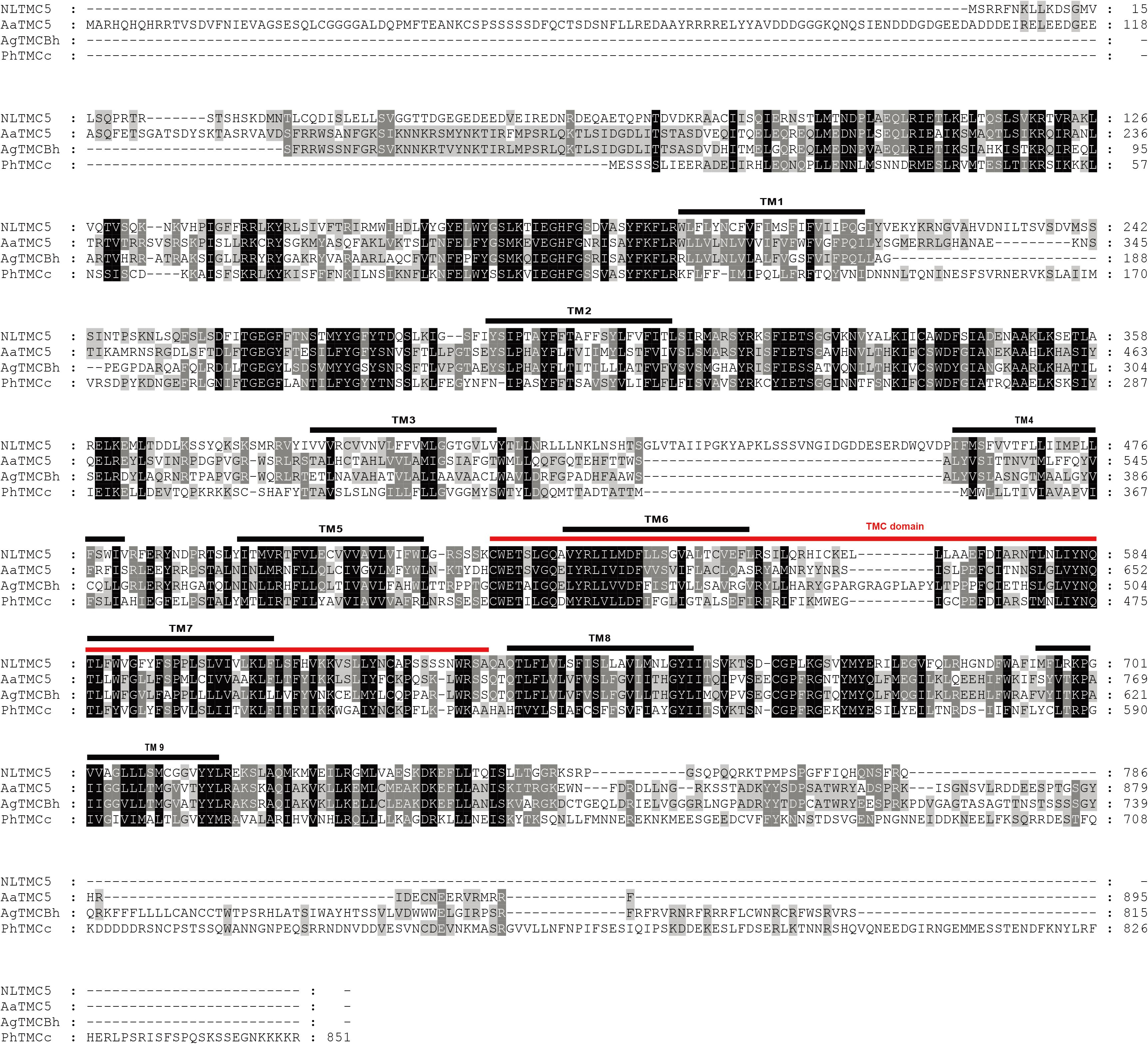
Figure 3. Amino acid sequence alignment of NlTMC5 and orthologs genes from Aedes aegypti (XP_021695152.1), Anopheles gambiae (XP_310494.4), and Pediculus humanus corporis (XP_002427541.1). The amino acid position is shown on the right. Identical residues between orthologs sequences are shown as white characters against the black background, and conservative substitutions shown as shading. Black lines represent the transmembrane domain (TM), and red line represents TMC domain.
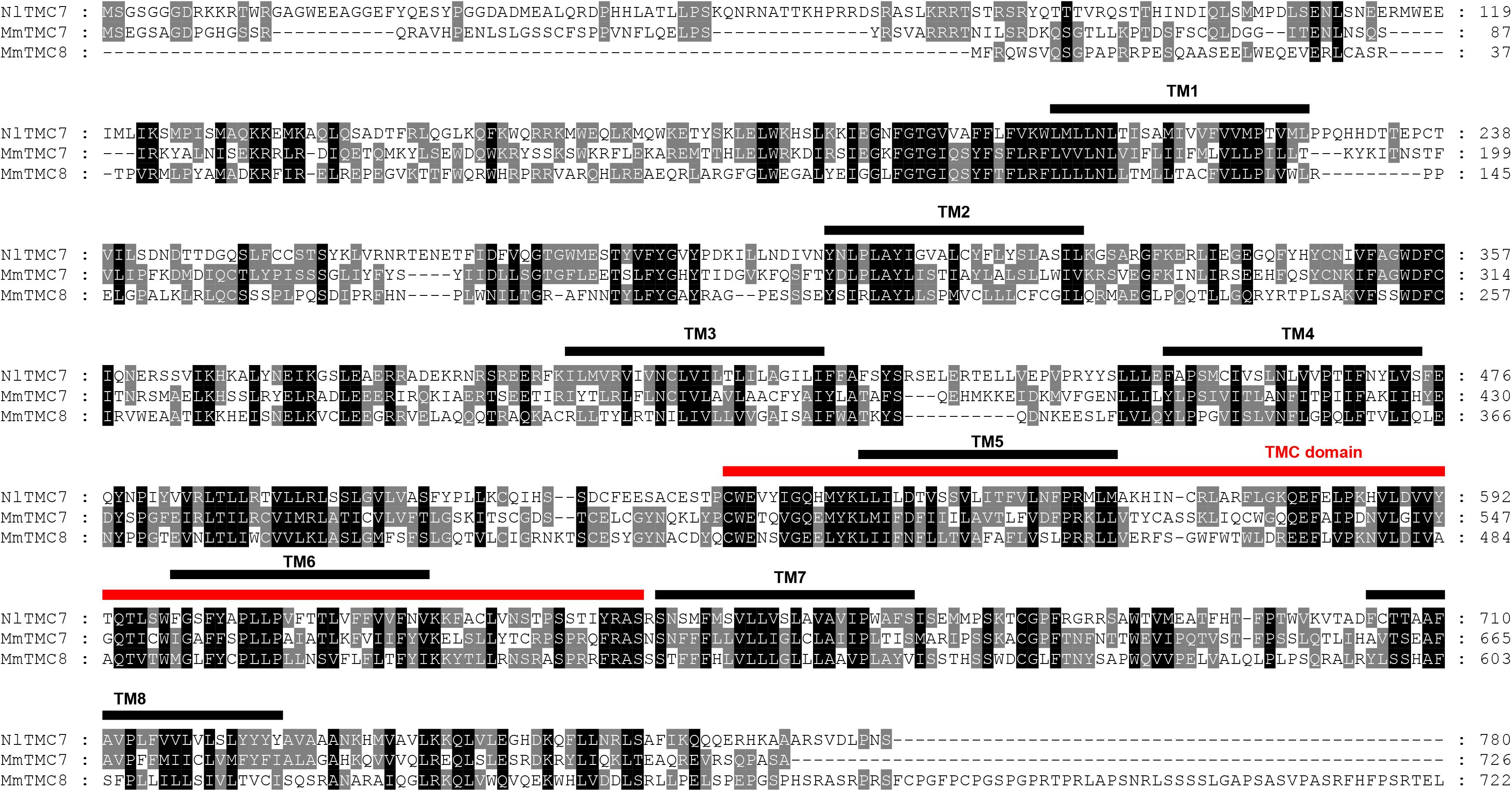
Figure 4. Amino acid sequence alignment of NlTMC7 and orthologs genes from Mus musculus (MmTMC7: NP_766064.2; MmTMC8: NP_001182017.1). The amino acid position is shown on the right. Identical residues between orthologs sequences are shown as white characters against the black background, and conservative substitutions shown as shading. Black lines represent the transmembrane domain (TM), and red line represents TMC domain.
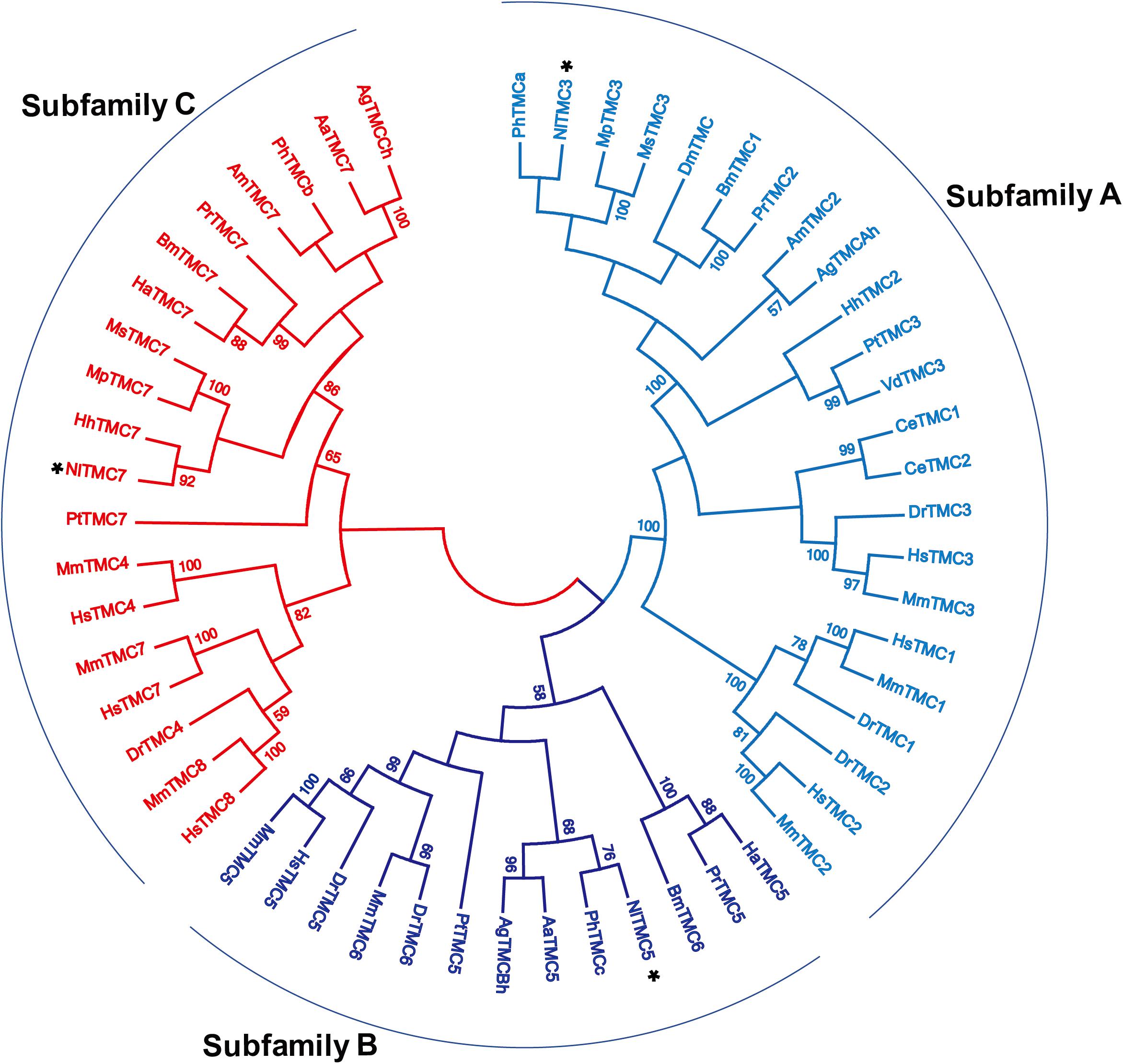
Figure 5. Phylogenetic analysis of three NlTMCs and various TMC proteins. Maximum likelihood tree was constructed by MEGA software. The numbers at the nodes of the branches represent the percentage of bootstrap support (1,000 replications) for each branch. The gene names followed by their GenBank accession numbers are listed in Table 2. Aa, Aedes aegypti; Ag, Anopheles gambiae; Am, Apis mellifera; Bm, Bombyx mori; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Dr, Danio rerio; Ha, Helicoverpa armigera; Hh, Halyomorpha halys; Hs, Homo sapiens; Mm, Mus musculus; Mp, Myzus persicae; Ms, Melanaphis sacchari; Pr, Pieris rapae; Pt, Parasteatoda tepidariorum; Vd, Varroa destructor; Ph, Pediculus humanus corporis.
Developmental and Tissue-Specific Expression Patterns of Nltmc Genes
The relative expression level of three Nltmc genes in different developmental stages and tissues were measured by qPCR (Figure 6). The results showed that the expression levels of the Nltmc genes varied between the developmental stages including egg, 1st–5th instar nymph, and 4-day old adults (MM, MF, BM, and BF). Among them, Nltmc3 and Nltmc7 were highly expressed in the nymphs compared with other developmental stages. Nltmc5 was more highly expressed in MM and MF than BM and BF adults indicated that Nltmc5 might involve in the wing polymorphism in BPH (Figures 6A,D,G).
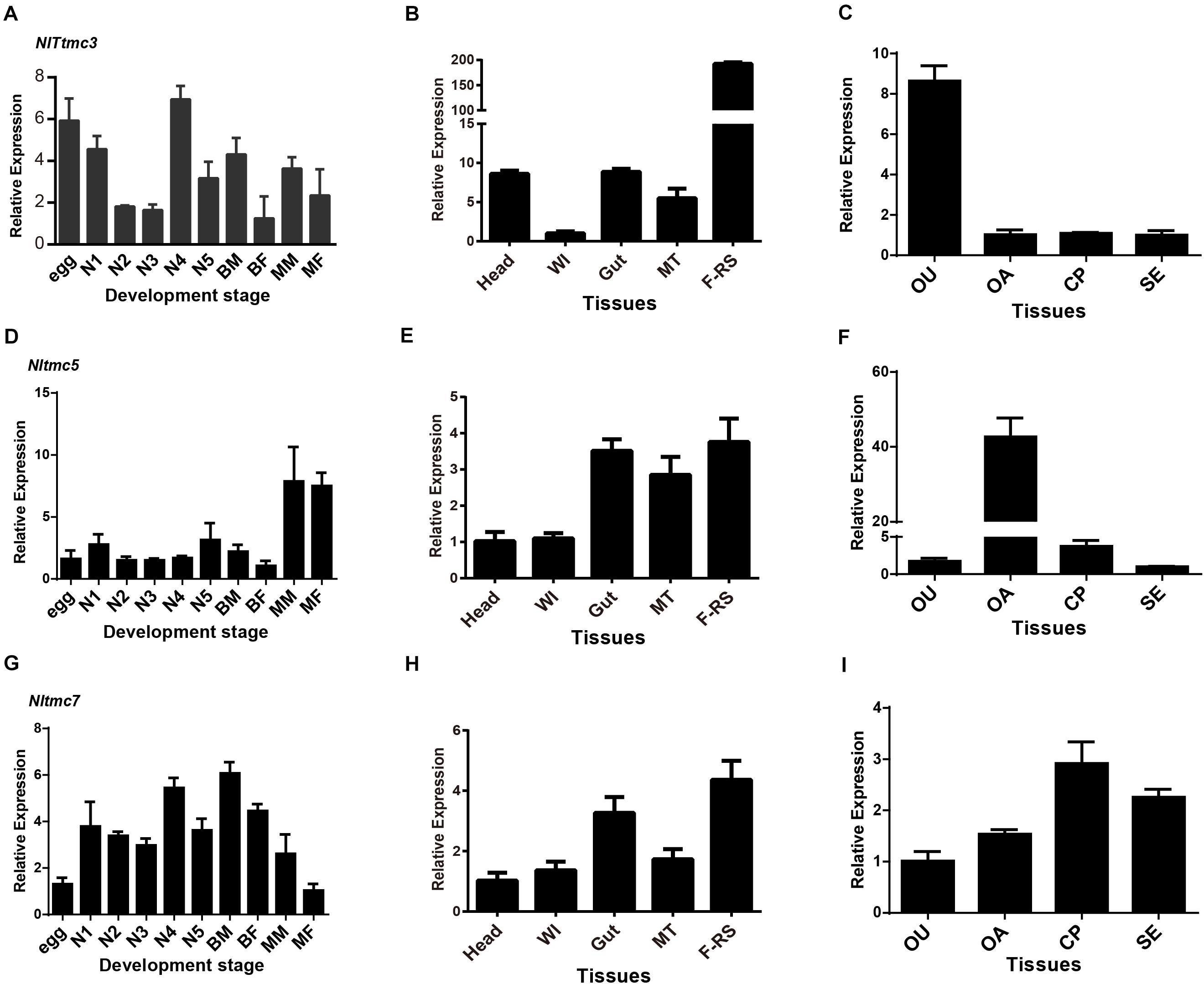
Figure 6. The expression patterns of three Nltmc genes. (A,D,G) Expression patterns of three Nltmc genes at different developmental stages including egg, 1st to 5th instar nymphs, and adults of MF (macropterous female), BF (brachypterous female), MM (macropterous male), and BM (brachypterous male). (B,E,H) Expression patterns of three Nltmc genes in various tissues including head, wing (WI), gut, Malpighian tubule (MT), and female reproductive organ (FRO). (C,F,I) Expression patterns of three Nltmc gene in the four female reproductive organ regions including the oviduct (OU), ovary (OA), copulatory pouch (CP), and the spermatheca (SE). Data are expressed as the mean ± s.e.m. (n > 3).
We further investigated the relative expression level of three Nltmc genes in various female adult tissues, including the head, wing, gut, Malpighian tubules (MT) and FRO using qPCR method. In the examined tissues, all Nltmc genes were mostly expressed in reproductive organs compared with other tissues (Figures 6B,E,H). This indicated that these genes could be involved in reproduction in the BPH. We next examined the expression pattern of three Nltmc genes within the FRO (Figures 6C,F,I). Interestingly, Nltmc3 was highly expressed in the oviduct (OU). While, Nltmc5 was the most expressed in the ovary (OA). And Nltmc7 was almost expressed equally in the four examined tissues (Figures 6C,F,I).
Silencing of Nltmc3 Affects Egg-Laying of N. lugens
Next, we tested whether Nltmc genes are involved in the egg laying of N. lugens. Using RNAi technology, we silenced all of the Nltmc genes in the N. lugens (Figures 7A–C). The dsRNA-injection did not negatively affect the survival of N. lugens (Figure 7D). However, the dsNltmc3-injected planthoppers showed the decreased eggs (Figure 7E). While silencing Nltmc5 and Nltmc7 had little impact on the egg-laying rate of BPH (Figure 7E).
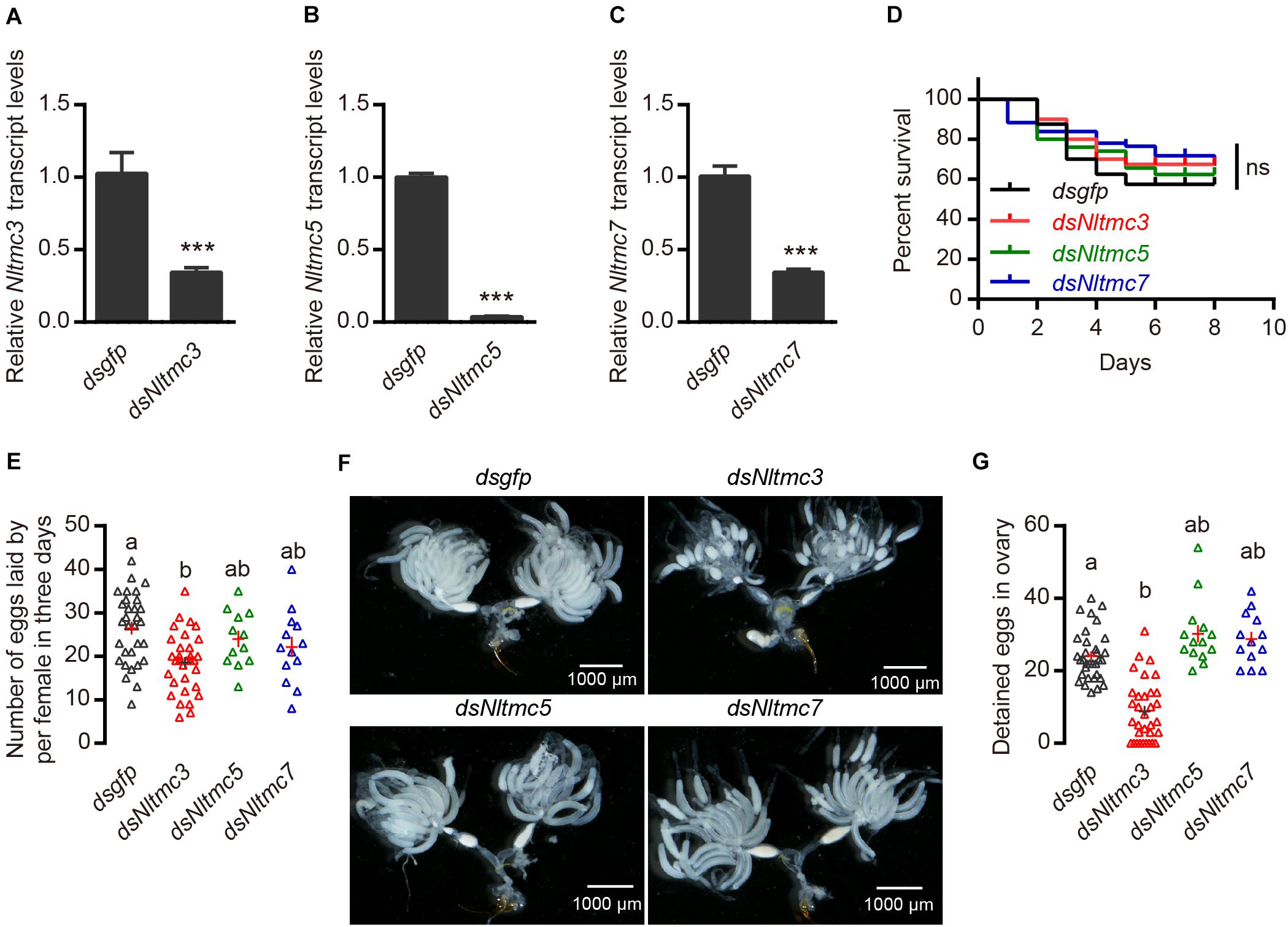
Figure 7. RNAi-mediated silencing of Nltmc3 gene reduce eggs laid of female brown planthopper. (A–C) Downregulation of three Nltmc genes using Nltmc-RNAi leads to a reduction in mRNA expression level. (D) No significant difference was observed in survival between dsNltmc and dsgfp. (E) Silencing of Nltmc3 gene by dsNltmc3 resulted in a significant reduction of eggs laid. While silence of Nltmc5 and Nltmc7 gene have no impacts on egg-laying of brown planthopper. (F) The effect of RNAi on the ovary development in the dsgfp (control), dsNltmc3, dsNltmc5, and dsNltmc7 groups. Scale bar = 1,000 μm. (G) The detained eggs in ovary measured at 4 days old females injected with dsRNA for Nltmc3, Nltmc5, Nltmc7, or gfp. Groups that share at least one letter (e.g., a vs ab) are statistically indistinguishable, and groups that have different letters (e.g., a vs b) are statistically different. One-way ANOVA followed by Tukey’s multiple comparisons test, p < 0.05. All data are presented as means ± s.e.m.
Silencing of Nltmc3 Leads to Undeveloped Ovaries of N. lugens
Next, we investigated the underlying mechanism of Nltmc3 involved in the egg laying of N. lugens. We did not observe any developmental defects of BPH after Nltmc3 gene silencing (data not shown). We then examined ovary development in females at 7-day-adulthood (3-day after injection of dsRNA). For dsgfp-injected females, ovaries were fully developed (Figure 7F). By contrast, ovaries of dsNltmc3-injected females were small and poorly developed (Figure 7F). In the Nltmc3-RNAi planthoppers, we observed less detained eggs per ovary (Figure 7G). However, silencing Nltmc5 and Nltmc7 has no impact on the ovarian development in the BPH (Figures 7F,G).
Discussion
Eight tmc genes were cloned in vertebrate (Keresztes et al., 2003; Kurima et al., 2003). All TMC proteins are strongly predicted to encode at least six conserved transmembrane domains and a conserved TMC domain (Kurima et al., 2003). In insects, the tmc gene was only cloned and investigated in Drosophila (Guo et al., 2016; Zhang et al., 2016). There is only one tmc gene in the Drosophila genome (Guo et al., 2016). And two tmc genes were found in the C. elegans genome (Jia et al., 2019). They all belongs to subfamily A tmc gene family. In this study, we identified three tmc genes in the BPH genome, suggestive of diverse separation of the tmc genes in different species. They can be sub-divided into three subfamilies A, B and C. There are three tmc genes in BPH. Three TMC proteins in BPH were well clustered with other species’ TMC proteins. They are homologs with Myzus persicae tmc3, Aedes aegypti tmc5, and Myzus persicae tmc7, respectively, thus, we used homologs genes’ names for the three tmc genes of the BPH. Interestingly, we found that the Nltmc3 gene was highly expressed in the FRO. The RNAi-based functional analysis indicated that Nltmc3 was involved in female fecundity and ovary development.
NlTMC3 protein exhibits sequence conservation with TMC subfamily A members in other species including Drosophila and C. elegans, in the putative transmembrane domains (Figure 2). NlTMC3 is much larger than its nematode or mouse homologs. A similar result was also found in the fruit fly (Guo et al., 2016). Besides this, we discovered one internal repeat between TM5 and TM6 (Figure 2). It is of interest to determine whether this repeat has any physiological meanings in the future. We found two other tmc genes, Nltmc5 and Nltmc7, which belong to subfamily B and C, respectively. From our phylogenetic analysis, we also found their homology gene in silkworm, mosquito and honeybee (Figure 5). However, these two subfamily genes were lost in the genome of Drosophila and C. elegans.
We also examined the distribution pattern of three Nltmc genes. The results revealed a ubiquitous expression of Nltmc genes in all developmental stages and examined tissues, indicating the possibility of a vast array of physiological functions for Nltmc genes. tmc1 and tmc2 are components of the mechano-transduction channel for sound transduction in the hair cells of the mammalian inner ear (Kurima et al., 2002; Vreugde et al., 2002; Pan et al., 2013). However, they are very broadly expressed which indicated that they might also functions in other tissues. In mammals, three other tmc genes (tmc3, tmc4, and tmc7) are also expressed in hair cells but their functions in hearing are largely unknown (Kurima et al., 2003; Kawashima et al., 2015; Scheffer et al., 2015). In C. elegans, TMC proteins are expressed in both neurons and muscle cells (Chatzigeorgiou et al., 2013; Zhang et al., 2015; Yue et al., 2018). Cetmc1 is required for the ASH nociceptive neuron-mediated alkaline and salt chemo-sensation (Chatzigeorgiou et al., 2013; Wang et al., 2016). Recent studies showed that TMC proteins in C. elegans mediate a background Na+-leak conductance in the egg-laying circuit (HSN neurons and vulval muscles) (Yue et al., 2018). In Drosophila, TMC protein was expressed on the larval class I and class II dendritic arborization neurons and bipolar dendrite neurons that acts in proprioception (Guo et al., 2016). The tmc gene and TMC-expressing multi-dendritic neurons in the fruit fly labellum are required for food texture detection (Zhang et al., 2016). Our studies showed that sweet neurons inhibit texture discrimination by signaling TMC-expressing mechanosensitive neurons when deciding where to deposit their eggs in Drosophila (Wu et al., 2019). These results indicated that TMC-expressing neurons play opposing roles in hardness discrimination in two different behavioral decisions.
We found that Nltmc3 was highly expressed in the FRO especially on the oviduct which indicated that this gene might influence reproductive physiology in the BPH. Indeed, knockdown of Nltmc3 led to reduction of female fertility and undeveloped ovaries (Figure 7). In C. elegans, adult worms lacking either Cetmc1 or Cetmc2 retained more eggs in the uterus and had significantly less progenies. A more severely defective egg-laying phenotype was observed in double mutant worms (Yue et al., 2018). Our previous studies showed that interference of β-adrenergic-like octopamine receptor (NlOA2B2) signaling pathway had a strong impact on the egg laying of the female BPH (Wu et al., 2017). OA2B2 has already been established in D. melanogaster to be important for ovulation of eggs (Lim et al., 2014; Li et al., 2015). However, we did not observe more retained eggs in the Nltmc3-silenced BPHs. These results indicate that silencing Nltmc3 gene has little impairment on the ovulation of BPHs. We observed a dramatic reduction in the number of mature eggs in the ovaries of females injected with dsRNA of Nltmc3, as compared with the dsgfp-injected females. Hence, our results indicated that NlTMC3 is required for ovary development and fecundity in N. lugens. In many insects, the amino acid/target of rapamycin (TOR) and insulin nutritional signaling pathways have vital roles in insect reproduction (Roy et al., 2018). Former studies have showed that silencing of the TOR gene in BPHs leads to unmatured eggs (Lu et al., 2016). The TOR nutritional signaling pathway and juvenile hormone (JH) regulation of vitellogenesis has been known for a long time (Zhai et al., 2015; Zhuo et al., 2017; Roy et al., 2018; Zhang et al., 2019). The possible involvement of NlTMC3 in TOR or JH signaling in N. lugens females needs further investigation.
In summary, we found that the Nltmc3 plays a critical role in female N. lugens ovary development, and Nltmc3 knockdown leads to reduction of female fertility. Further studies should be conducted to clarify how NlTMC3 influences female N. lugens reproduction.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/MT576067.
Author Contributions
S-FW conceived and designed the experiments. Y-LJ, Y-JZ, DG, C-YL, and J-YM performed the experiments. S-FW, Y-LJ, DG, C-YL, and C-FG analyzed the data. S-FW, Y-LJ, and Y-JZ wrote and revised the manuscript. All authors commented on the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Nos. 31772205 and 31830075), and the Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015B157).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/spidey/
- ^ http://gsds.cbi.pku.edu.cn/index.php
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0/
- ^ http://www.ebi.ac.uk/Tools/msa/clustalo
References
Chatzigeorgiou, M., Bang, S., Hwang, S. W., and Schafer, W. R. (2013). tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494, 95–99. doi: 10.1038/nature11845
Guo, Y., Wang, Y., Zhang, W., Meltzer, S., Zanini, D., Yu, Y., et al. (2016). Transmembrane channel-like (tmc) gene regulates Drosophila larval locomotion. Proc. Natl. Acad. Sci. U.S.A. 113, 7243–7248. doi: 10.1073/pnas.1606537113
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jia, Y., Zhao, Y., Kusakizako, T., Wang, Y., Pan, C., Zhang, Y., et al. (2019). TMC1 and TMC2 proteins are pore-forming subunits of mechanosensitive ion channels. Neuron 105, 310–321. doi: 10.1016/j.neuron.2019.10.017
Kawashima, Y., Kurima, K., Pan, B., Griffith, A. J., and Holt, J. R. (2015). Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467, 85–94. doi: 10.1007/s00424-014-1582-3
Keresztes, G., Mutai, H., and Heller, S. (2003). TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4:24. doi: 10.1186/1471-2164-4-24
Kurima, K., Peters, L. M., Yang, Y., Riazuddin, S., Ahmed, Z. M., Naz, S., et al. (2002). Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284. doi: 10.1038/ng842
Kurima, K., Yang, Y., Sorber, K., and Griffith, A. J. (2003). Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and Epidermodysplasia verruciformis. Genomics 82, 300–308. doi: 10.1016/s0888-7543(03)00154-x
Li, Y., Fink, C., El-Kholy, S., and Roeder, T. (2015). The octopamine receptor octß2R is essential for ovulation and fertilization in the fruitfly Drosophila melanogaster. Arch. Insect Biochem. Physiol. 88, 168–178. doi: 10.1002/arch.21211
Lim, J., Sabandal, P. R., Fernandez, A., Sabandal, J. M., Lee, H.-G., Evans, P., et al. (2014). The octopamine receptor Octβ2R regulates ovulation in Drosophila melanogaster. PLoS One 9:e104441. doi: 10.1371/journal.pone.0104441
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, K., Chen, X., Liu, W.-T., and Zhou, Q. (2016). TOR pathway-mediated juvenile hormone synthesis regulates nutrient-dependent female reproduction in Nilaparvata lugens (Stål). Int. J. Mol. Sci. 17:438. doi: 10.3390/ijms17040438
Min, S., Lee, S. W., Choi, B.-R., Lee, S. H., and Kwon, D. H. (2014). Insecticide resistance monitoring and correlation analysis to select appropriate insecticides against Nilaparvata lugens (Stål), a migratory pest in Korea. J. Asia Pac. Entomol. 17, 711–716. doi: 10.1016/j.aspen.2014.07.005
Pan, B., Akyuz, N., Liu, X.-P., Asai, Y., Nist-Lund, C., Kurima, K., et al. (2018). TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99, 736–753.e736.
Pan, B., Géléoc, G. S., Asai, Y., Horwitz, G. C., Kurima, K., Ishikawa, K., et al. (2013). TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515. doi: 10.1016/j.neuron.2013.06.019
Roy, S., Saha, T. T., Zou, Z., and Raikhel, A. S. (2018). Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 63, 489–511. doi: 10.1146/annurev-ento-020117-043258
Scheffer, D. I., Shen, J., Corey, D. P., and Chen, Z.-Y. (2015). Gene expression by mouse inner ear hair cells during development. J. Neurosci. 35, 6366–6380. doi: 10.1523/jneurosci.5126-14.2015
Vreugde, S., Erven, A., Kros, C. J., Marcotti, W., Fuchs, H., Kurima, K., et al. (2002). Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet. 30:257. doi: 10.1038/ng848
Wang, X., Li, G., Liu, J., Liu, J., and Xu, X. Z. S. (2016). TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron 91, 146–154. doi: 10.1016/j.neuron.2016.05.023
Wu, S. F., Ja, Y. L., Zhang, Y. J., and Yang, C. H. (2019). Sweet neurons inhibit texture discrimination by signaling TMC-expressing mechanosensitive neurons in Drosophila. eLife 8:e46165.
Wu, S. F., Jv, X. M., Li, J., Xu, G. J., Cai, X. Y., and Gao, C. F. (2017). Pharmacological characterisation and functional roles for egg-laying of a β-adrenergic-like octopamine receptor in the brown planthopper Nilaparvata lugens. Insect. Biochem. Mol. Biol. 87, 55–64. doi: 10.1016/j.ibmb.2017.06.008
Wu, S. F., Zeng, B., Zheng, C., Mu, X. C., Zhang, Y., Hu, J., et al. (2018). The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012-2016. Sci. Rep. 8:4586.
Yue, X., Zhao, J., Li, X., Fan, Y., Duan, D., Zhang, X., et al. (2018). TMC proteins modulate egg laying and membrane excitability through a background leak conductance in C. elegans. Neuron 97, 571–585.e575.
Zhai, Y., Sun, Z., Zhang, J., Kang, K., Chen, J., and Zhang, W. (2015). Activation of the TOR signalling pathway by glutamine regulates insect fecundity. Sci. Rep. 5:10694.
Zhang, J.-L., Yuan, X.-B., Chen, S.-J., Chen, H.-H., Xu, N., Xue, W.-H., et al. (2019). The histone deacetylase NlHDAC1 regulates both female and male fertility in the brown planthopper, Nilaparvata lugens. Open Biol. 8:180158. doi: 10.1098/rsob.180158
Zhang, L., Gualberto, D. G., Guo, X., Correa, P., Jee, C., and Garcia, L. R. (2015). TMC-1 attenuates C. elegans development and sexual behaviour in a chemically defined food environment. Nat. Commun. 6:6345.
Zhang, Y. V., Aikin, T. J., Li, Z., and Montell, C. (2016). The basis of food texture sensation in Drosophila. Neuron 91, 863–877. doi: 10.1016/j.neuron.2016.07.013
Keywords: TMC, reproduction, mechanoreceptors, Nilaparvata lugens, expression pattern
Citation: Jia Y-L, Zhang Y-J, Guo D, Li C-Y, Ma J-Y, Gao C-F and Wu S-F (2020) A Mechanosensory Receptor TMC Regulates Ovary Development in the Brown Planthopper Nilaparvata lugens. Front. Genet. 11:573603. doi: 10.3389/fgene.2020.573603
Received: 17 June 2020; Accepted: 02 September 2020;
Published: 26 October 2020.
Edited by:
Wei Guo, Institute of Zoology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2020 Jia, Zhang, Guo, Li, Ma, Gao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun-Fan Wu, d3VzZkBuamF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Ya-Long Jia†
Ya-Long Jia† Cong-Fen Gao
Cong-Fen Gao Shun-Fan Wu
Shun-Fan Wu