- 1Laboratório de Citogenética, Centro de Estudos Avançados da Biodiversidade, Instituto de Ciências Biológicas, Universidade Federal Do Pará, Belém, Brazil
- 2Laboratório de Estudos da Ictiofauna da Amazônia, Instituto Federal de Educação Ciência e Tecnologia Do Pará, Abaetetuba, Brazil
- 3Laboratório de Biologia Cromossômica, Estrutura e Função, Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Ponta Grossa, Brazil
Peckoltia is widely distributed genus in the Amazon and Orinoco basins and the Guiana Shield, containing 18 valid species, and distinct morphotypes still needing description in the scientific literature due to its great taxonomic complexity. This study performed a comparative chromosomal analysis of two undescribed Peckoltia species (Peckoltia sp. 3 Jarumã and Peckoltia sp. 4 Caripetuba) from the Brazilian Amazon using conventional chromosome bands methods and in situ localization of the repetitive DNA (5S and 18S rRNA and U1 snRNA genes and telomeric sequences). Both species presented 2n = 52 but differed in their karyotype formula, probably due to inversions or translocations. The nucleolus organizer regions (NORs) showed distal location on a probably homeologous submetacentric pair in both species, besides an extra signal in a subtelocentric chromosome in Peckoltia sp. 4 Caripetuba. Heterochromatin occurred in large blocks, with different distributions in the species. The mapping of the 18S and 5S rDNA, and U1 snDNA showed differences in locations and number of sites. No interstitial telomeric sites were detected using the (TTAGGG)n probes. Despite 2n conservationism in Peckoltia species, the results showed variation in karyotype formulas, chromosomal bands, and locations of repetitive sites, demonstrating great chromosomal diversity. A proposal for Peckoltia karyotype evolution was inferred in this study based on the diversity of location and number of chromosomal markers analyzed. A comparative analysis with other Peckoltia karyotypes described in the literature, their biogeography patterns, and molecular phylogeny led to the hypothesis that the derived karyotype was raised in the left bank of the Amazon River.
Introduction
The Loricariidae is one of the most specious family of catfish within the order Siluriformes, containing 1,016 valid species (Fricke et al., 2021). They are endemic to the Neotropical region, distributed throughout South America and part of Central America, and occur in a great diversity of habitats (Armbruster, 2004; Armbruster, 2008; Armbruster and Lujan, 2016). Analyzes based on morphological and molecular data support the recognition of six subfamilies: Lithogeninae, Delturinae, Hypoptopomatinae, Neoplecostominae, Loricariinae, and Hypostominae grouped in the tribes: Corymbophanini, Rhinelepini, Hypostomini, Pterygoplichthyini and Ancistrini (Armbruster, 2004; Lujan et al., 2015).
Peckoltia Miranda Ribeiro, 1912 (sensu Lujan et al., 2015), comprises 18 valid species, in addition to distinct morphotypes that still lack description in the scientific literature. They are widely distributed in the Amazon and Orinoco basins and the Guiana Shield (Armbruster et al., 2015; Lujan et al., 2015; Armbruster and Lujan, 2016). According to phylogenetic analyzes proposed for Loricariidae, Peckoltia genus receive strong support as monophyletic lineage (Lujan et al., 2015; Lujan et al., 2017; Roxo et al., 2019); however, a complex taxonomic identification procedure at a specific level related to a wide geographic distribution and morphological similarity is observed for these species (Armbruster et al., 2015; Armbruster and Lujan, 2016). For example, representatives of Peckoltia vittate (the type species of the genus) collected in the Amazon region (Xingu, Madeira and Orinoco rivers) presented polyphyletic lineages in molecular phylogeny by Lujan et al. (2015). Armbruster and Lujan (2016), analyzed morphological and molecular characters of samples collected in the Orinoco basin associated with P. vittate by Lujan et al. (2015) and described a new species, Peckoltia wernekei. These recent analyses agree that the diversity of Peckoltia species can be underestimated for the Amazon region.
Cytogenetic markers are important tools to analyze fish species possessing complex taxonomy (Bertollo et al., 2000; Centofante et al., 2003; Mariotto and Miyazawa, 2006) or to understand evolutionary features in groups with highly rearranged karyotypes (Nagamachi et al., 2010; Deon et al., 2020). Cytogenetic data are available for eight lineages of the genus Peckoltia, including valid species and unidentified morphotypes, collected at different points in the Amazon region. Despite all Peckoltia species share 2n = 52 chromosomes, variations in chromosome morphology, the number and position of NORs, distribution of the constitutive heterochromatin (CH) regions, and the presence of B chromosomes are observed among species of this genus (Table 1). Therefore, in Peckoltia species, many unique karyotypic features are observed that can be useful in recognizing distinct taxonomic units.
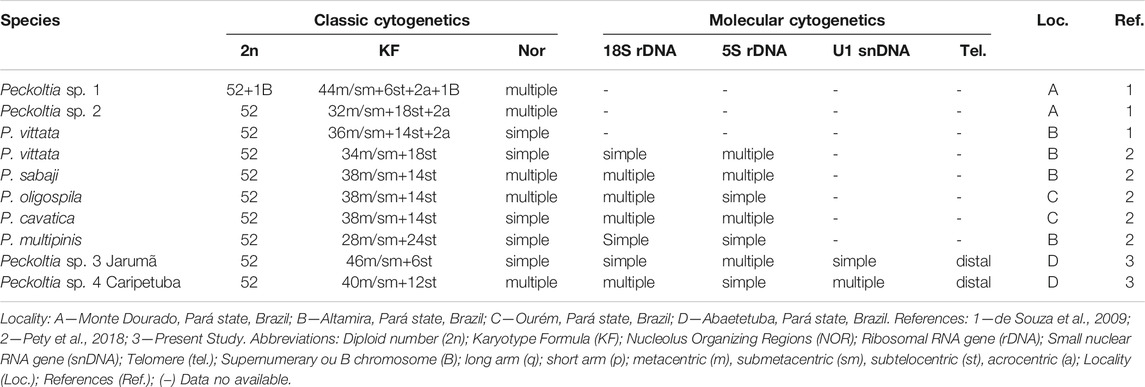
TABLE 1. Chromosomal diversity available in the literature and obtained in the present study for Peckoltia genus.
Repetitive DNAs are found in most eukaryotic genomes, representing important markers for molecular diversity analysis at the chromosomal level; they are organized in blocks (e.g., satellites and multigene families) or dispersed (e.g., transposons and retrotransposons). The contribution of the repetitive DNA for fish genome evolution has been evidenced (Vicari et al., 2010; Schemberger et al., 2019). The eukaryotic ribosomal DNA (rDNA) represents two multigene families with an organization in tandem: 45S ribosomal RNA (18S + 5.8S + 28S genes) and 5S ribosomal RNA (Long and Dawin, 1980). These genes are widely used in chromosomal studies in several organisms, including Peckoltia species (Pety et al., 2018), showing great molecular chromosomal diversity involving these sequences.
Small nuclear RNA genes (snDNA) represent another multigene family involved in the splicing and maturation process of messenger RNA encoded by the U1, U2, U4, U5 and U6 snRNA genes (Busch et al., 1982). The snDNA sequences have been used as chromosomal markers for detailed comparative chromosome analysis in several groups of organisms, including fish, reptiles and arthropods (Cabral-de-Melo et al., 2012; Almeida et al., 2017; Cavalcante et al., 2020; Dulz et al., 2020).
In this study, we describe the karyotypes of two undescribed Peckoltia species, first time sampled in the Tocantins River basin in Brazil, and compare them with cytogenetic data available in the literature. From this, we discuss the possible mechanisms of karyotypic diversification, biogeography and their evolutionary implications for this genus.
Materials and Methods
Samples
Samples of the two morphologically different but still undescribed species of Peckoltia named Peckoltia sp. 3 Jarumã and Peckoltia sp. 4 Caripetuba, after the rivers they were collected in different hydrographic points in the Tocantins River basins of northern Brazil were analyzed (Figure 1). The taxonomic identification of the sample was checked using the identification key proposed by Armbruster and Lujan (2016), Armbruster (2008). The results show that the specimens do not fit into any of the species already Figure 1described. The collection points, number of individuals, sex, and voucher of deposits in the zoological collection are shown in Table 2. The samples were obtained under a permanent field permit obtained by JCP (number 13248 from “Instituto Chico Mendes de Conservação da Biodiversidade”). The Cytogenetics Laboratory from UFPA has permit number 19/2003 from the Ministry of Environment for sample transport and permit 52/2003 to use the samples for research. The Ethics Committee (Comitê de Ética Animal da Universidade Federal do Pará) approved this research (Permit 68/2015). The specimens have been deposited in the ichthyological collection of the Museu Paraense Emílio Goeldii (MPEG) (Belém, Brazil).
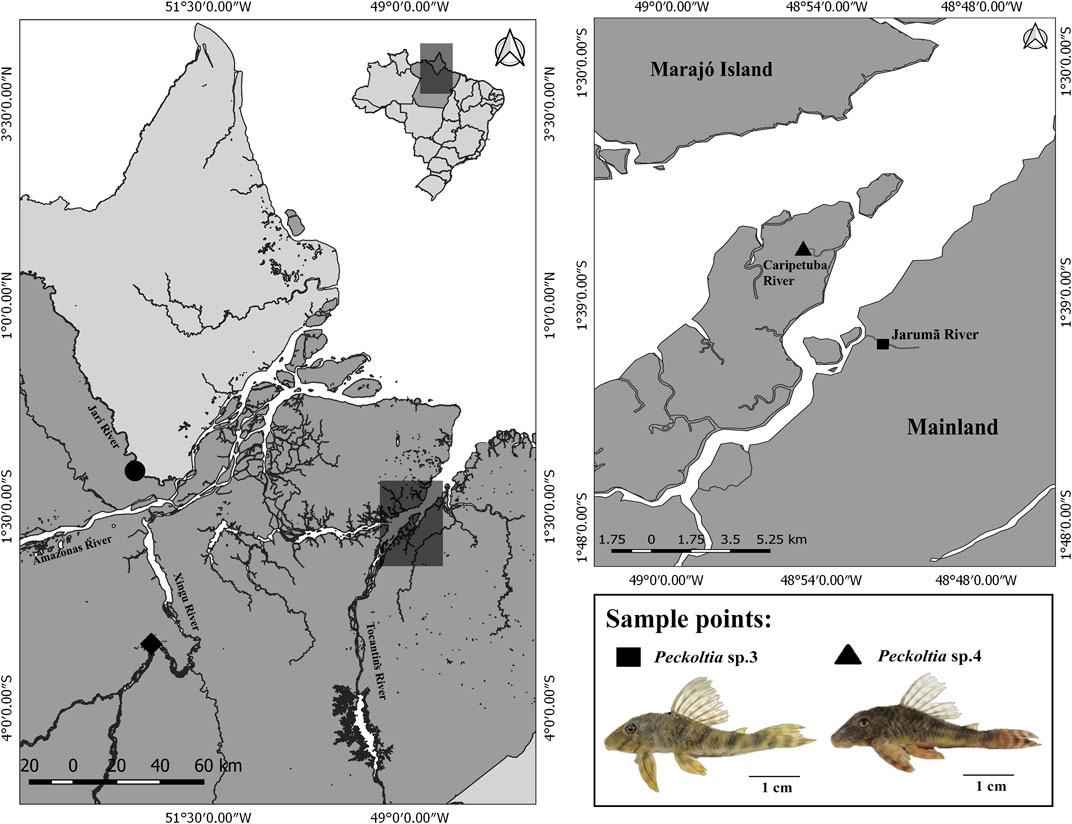
FIGURE 1. Geographical location of sampling sites for Peckoltia sp. 3 Jarumã (square) and Peckoltia sp. 4 Caripetuba (triangle) in the present study. The coordinates of the sampling sites for Peckoltia vittata (diamond), Peckoltia sp. 1, and Peckoltia sp. 2 (circle) reported by de Souza et al. (2009) are also shown. The map was made using QUANTUM-GIS (Q-GIS) v. 3.4.5. The database was obtained from Instituto Brasileiro de Geografia e Estatística—IBGE. An Peckoltia specimen is shown below. Scale bar: 1 cm. Photo by KSS.
Chromosomal Analysis
Mitotic chromosomes were obtained from kidney cells after in vivo colchicine treatment as described (Bertollo et al., 1978). The animals were anesthetized with eugenol and subsequently sacrificed for the removal of kidney cells. Metaphases were analyzed by conventional Giemsa, C-banding (Sumner, 1972) and AgNO3 staining (Howell and Black, 1980). Fluorescence in situ hybridization (FISH) was undertaken as described (Martins and Galetti, 1999) using a general telomere probe for vertebrates, 18S rDNA, 5S rDNA, and U1 snDNA probes.
Probes Labeling and in situ Localization
DNA extraction was performed using PureLink Genomic DNA Mini Kit (Invitrogen) following the manufacturer’s instructions. The probes were obtained from a PCR using genomic DNA of Peckoltia sp. 3 Jarumã and Peckoltia sp. 4 Caripetuba with primers previously described for 18S rDNA (Hatanaka and Galetti Jr, 2004), for 5S rDNA (Suarez et al., 2017) and U1 snDNA (Cabral-de Melo et al., 2012). These probes were labeled by nick-translation with biotin or digoxigenin. Telomeric probes were obtained from PCR using the set of primers F-5′(TTAGGG)5-3′ and R-5′(CCCTAA)5-3′ followed by labeling with Digoxigenin-11-dUTP (Roche Applied Science®) (Ijdo et al., 1991). Fluorescence in situ hybridization (FISH) was performed as described by Martins and Galetti (1999) using the following stringency conditions: 2.5 ng/μL of each probe, 50% formamide, 2 x SSC, 10% dextran sulfate, and hybridization at 42°C for 16 h. Fluorescent signals were detected using Streptavidin Alexa Fluor 488 (Molecular Probes, Carlsbad, CA, United States) and anti-digoxigenin rhodamine Fab fragments (Roche Applied Science, Penzberg, Germany). Chromosomes were counterstained with 0.2 μg/ml of 4′6-diamidino-2-phenylindole (DAPI) in Vectashield mounting medium (Vector, Burlingame, CA, United States).
Microscopic Analysis and Image Capture
At least 30 metaphases Giemsa-stained per individual were analyzed for confirming the diploid number, karyotypic structure, and chromosomal markers. Cytogenetic images of Giemsa-stained chromosomes were obtained using an Olympus BX41 microscope (bright field/phase) with a digital camera CCD 1300QDS and analyzed using GenASIs software version 7.2.7.34276 from ASI (Applied Spectral Imaging). FISH images were obtained using a Nikon H550S microscope and analyzed using the Nis-Elements software. Images were adjusted using Adobe Photoshop CS6 software. Chromosomal morphology was classified according to literature (Levan et al., 1964), with adaptations.
Results
Peckoltia sp. 3 Jarumã and Peckoltia sp. 4 Caripetuba species presented 2n = 52 chromosomes and karyotype formulas (KF) 46m/sm + 6st, and 40m/sm + 12st, respectively. Heteromorphic sex chromosomes were not identified in the karyotypes described here (Figures 2A,C).
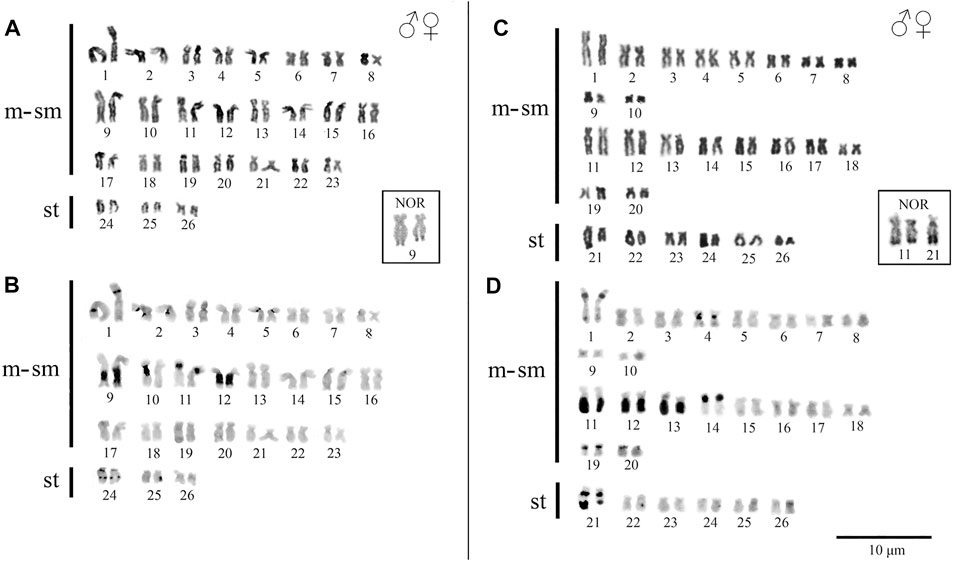
FIGURE 2. Karyotypes of Peckoltia species. In (A) conventional staining and (B) C-banding of Peckoltia sp. 3 Jarumã; in (C) conventional staining and (D) C-banding of Peckoltia sp. 4 Caripetuba. In box the Nucleolus Organizing Regions (NORs).
In Peckoltia sp. 3 Jarumã, heterochromatin occurred in the interstitial region of the short arm (p) of pairs 1p, 5p and 11p; in large blocks in the long arm (q) in pairs 9q with size heteromorphism and 12q; and in the pericentromeric and interstitial region of the 24q pair (Figure 2B). In Peckoltia sp. 4 Caripetuba, heterochromatin occurred in the interstitial region of pairs 1p, 14p, 19p and 20p; in the centromeric region of par 4; in large blocks in pairs 11q, 12q and 13q and in the pericentromeric and distal region of pair 21q, in which there is a marked heteromorphism in size between the homologues (Figure 2D). The Ag-NORs were located distal at pair 9q adjacent to the heterochromatin block in Peckoltia sp. 3 Jarumã (Figure 2 in box), and at pair 11q, coincident with heterochromatin and in one of the homologues in pair 21q in Peckoltia sp. 4 Caripetuba (Figure 2 in box).
Telomeric sequences occurred in the distal region of all chromosomes in both species, with no evidence of ITS vestiges (Figures 3A,C). The 18S rDNA, 5S rDNA, and U1 snDNA probes hybridized at the distal position of the chromosomes in the karyotypes of both species. In Peckoltia sp. 3 Jarumã, both 18S and 5S rDNA are colocalized in the 9q pair, in addition to a 5S rDNA site in one of the homologues in the 24q pair, and U1 snDNA is located in the distal region of the pair 13q (Figure 3B). In Peckoltia sp. 4 Caripetuba, 18S rDNA is located in pair 11q, in an additional site in one homologue of pair 21q; 5S rDNA is located in pair 1p; and U1 snDNA is located in the distal position of the pair 17q, with an additionnal site in one homologue of pair 21q colocalized with heterochromatin and 18S rDNA site (Figure 3D).
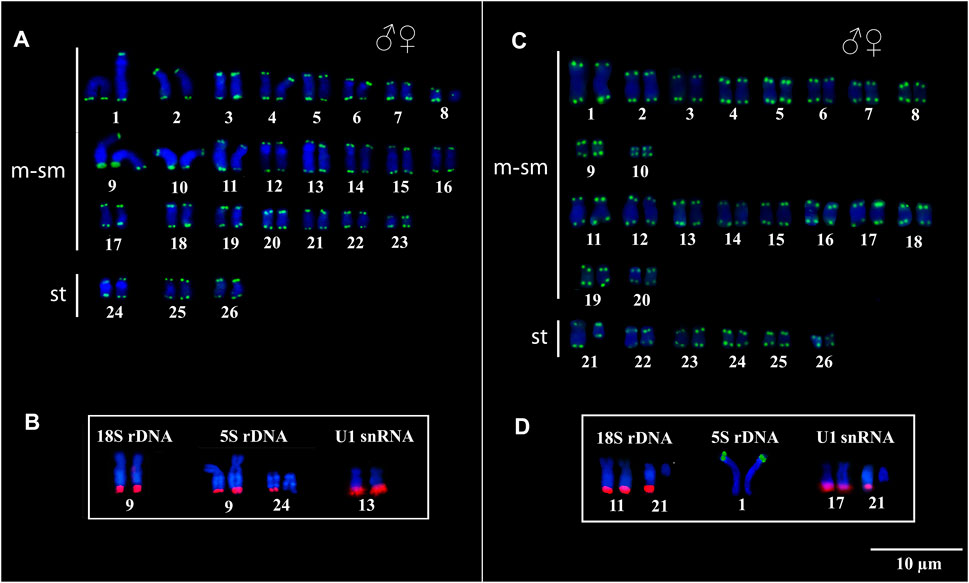
FIGURE 3. Fluorescent in situ Hybridization indicating the physical location of the Telomeric probes, 18S rDNA, 5S rDNA and U1 snDNA probes in Peckoltia species. In Peckoltia sp. 3 Jarumã: (A) Telomeric sequence (green) and (B) 18S rDNA (red), 5S rDNA (red) and U1 snDNA (red). In Peckoltia sp. 4 Caripetuba: (C) Telomeric sequence (green) and (D) 18S rDNA (red), 5S rDNA (green) and U1 snDNA (red).
Discussion
Karyotype Diversity in Peckoltia
Cytogenetic information for the species of Peckoltia is described in Table 1 and, despite the occurrence of 2n = 52 chromosomes, all species of Peckoltia show different KF. These differences can be explained by inversions or translocations, which represent important mechanisms of karyotypic diversification in Loricariidae (Kavalco et al., 2005; Mariotto et al., 2011). Alternatively, centromeric repositioning has also been proposed to cause variations in chromosome morphology with no change in diploid number (Montefalcone et al., 1999; Rocchi et al., 2012). However, the occurrence of this mechanism in Loricariidae karyotypes still needs deep investigation.
The Loricariidae has extensive chromosomal diversity, with variation in diploid number from 34 to 96 chromosomes, with a putative ancestral karyotype showing 2n = 54 chromosomes (Artoni and Bertollo, 2001). The reduction to 2n = 52 was probably due to a Robertsonian fusion in an ancient common ancestor in Ancistrini representatives with no ITS manutention (Bueno et al., 2018).
In Loricariidae, it is expected the presence of heterochromatic regions distributed in blocks on few chromosomes (Ziemniczak et al., 2012). Interestingly, in Peckoltia, extensive heterochromatic blocks are observed, some occupying a large part of the long arms of submetacentric/subtelocentric chromosomes (de Souza et al., 2009, present study). The presence of large heterochromatic blocks on morphologically similar chromosomes may suggest a shared character in Peckoltia karyotypes, as proposed previously (de Souza et al., 2009). However, it is known that heterochromatic regions are characterized by great diversity in highly repetitive DNA content (Dimitri et al., 2009) and may not reflect chromosomal homologies in Peckoltia species, as visualized among the species analyzed here by FISH with repetitive sequences. Noteworthy, the distribution of heterochromatin, and different repetitive sequences, observed in pairs 24 in Peckoltia sp. 3 Jarumã and 21 in Peckoltia sp. 4 Caripetuba, makes these chromosomes good cytotaxonomic markers, and both represent unique characteristics in each of these species.
Other plesiomorphic conditions for Loricariidae include the 18S and 5S rDNA sequences in a single pair of meta/submetacentric chromosomes (Ziemniczak et al., 2012). However, in the Ancistrini tribe, both the synteny and non-synteny between the 18S and 5S sequences are commonly observed (Mariotto et al., 2011; Ribeiro et al., 2015; Favarato et al., 2016; Prizon et al., 2016; Pety et al., 2018), showing the huge chromosome sites variation in the karyotypes of this group (Pansonato-Alves et al., 2013; Bueno et al., 2014; Pety et al., 2018). The mapping of 18S and the 5S sequences in the karyotypes here described is in agreement with that observed in other species of Peckoltia; in which extensive dispersion of these genes is observed (Pety et al., 2018) (Table 1). This dispersion of rDNA in the genomes of Loricariidae can be explained either by the association of these genes with other repetitive sequences, including transposable elements or by the evolutionary breakpoint regions close to rDNA sites promoting chromosome rearrangements (Barros et al., 2017; Glugoski et al., 2018, 2020; Deon et al., 2020). Furthermore, the heterochromatic condition involving clusters of rDNA suggests that other repetitive DNA classes, around 45S and 5S rDNA sequences, may promote their chromosomal dispersion in the Peckoltia species analyzed here, as shown for other species of Loricariidae (Glugoski et al., 2018; Deon et al., 2020).
In fish, the snRNA genes have shown great diversity of the pattern of chromosomal localization (Úbeda-Manzanaro et al., 2010; Cabral-de-Melo et al., 2012; Scacchetti et al., 2015; Yano et al., 2017). In this work, the U1 snDNA sequence was mapped for the first time in Loricariidae species, showing location in a pair of submetacentric chromosomes in both species, in addition to an extra site in one of the homologues of pair 21 of Peckoltia sp. 4 Caripetuba (Figure 3D). The snDNA sequences are considerably more stable at the chromosomal level when compared to rDNA (Cabral-de-Melo et al., 2012). However, we observed that among the karyotypes of Peckoltia analyzed here, the U1 snDNA probes show variation in the number of chromosomal sites similar to that observed for the rDNA (Table 1). These data indicate that these gene families can be equally dynamic in the genomes of species of Peckoltia. Several chromosomal sites of rDNA and snDNA sequences are observed in different groups of fish, such as species of the Loricariidae, Cichlidae and Anostomidae families; the emergence of new chromosomal sites is related to the association of these sequences with active mobile elements in these organisms (Kapitonov and Jurka, 2006; Cabral-de-Melo et al., 2012; Dulz et al., 2020). Future analyzes of rDNA and snDNA nucleotide sequences will be essential to verify the possible involvement of transposable elements in the movement of these sequences in the genomes of Peckoltia species.
Biogeography Hypothesis in Peckoltia
The putative ancestral karyotype for Loricariidae has 2n = 54, a single NOR and gene synteny for 5S and 18S rDNA sequences (Ziemniczak et al., 2012). The tribes belonging to the Hypostominae subfamily share a common ancestor (Armbruster, 2004; Lujan et al., 2015) that possibly had a 2n = 52 chromosomes (Bueno et al., 2018). Thus, variations in the 2n, multiple NOR and synteny break between 5S and 18S would represent derived characteristics that can be apomorphic or homoplasic. Analyses involving 18S and 5S rDNA and U1 snDNA show the importance of these sequences as markers of karyotype diversification in the Peckoltia genus.
Phylogenetic analyzes support the monophyly of the Peckoltia genus (Lujan et al., 2015, 2017; Roxo et al., 2019). A phylogeny for the Peckoltia clade proposed by Lujan et al. (2017), based on the concatenated sequences of two mitochondrial genes (16S, Cyt b) and three nuclear genes (RAG1, RAG2, MyH6), has two well-defined branches, which are sister groups and two branches with non-defined relationships. One of the defined branches presents P. vittata (single NOR, synteny of the 5S with 18S) (Pety et al., 2017), P. compta, P. braueri and P. lineola (karyotypes not described); and the other branch with P. sabaji (multiple NORs, non-synteny of the 5S with 18S) (Pety et al., 2018), P. furcata and P. relictum (karyotypes not described). Noteworthy, most of the specimens from the branch with P. vittata are on the right bank of the lower Amazon River, and those from the branch with P. sabaji are on the left bank. The karyotype of Peckoltia sp. 4 Caripetuba would be more similar to that of Peckoltia sp. 1 and 2 previously described (de Souza et al., 2009), collected on the left bank of the Amazon River and with karyotypic characteristics derived from the ancestral karyotype proposed for Loricariidae. Thus, it is possible that these derived features, such as multiple rDNA and NORs sites, have arisen in this region (Figure 4).
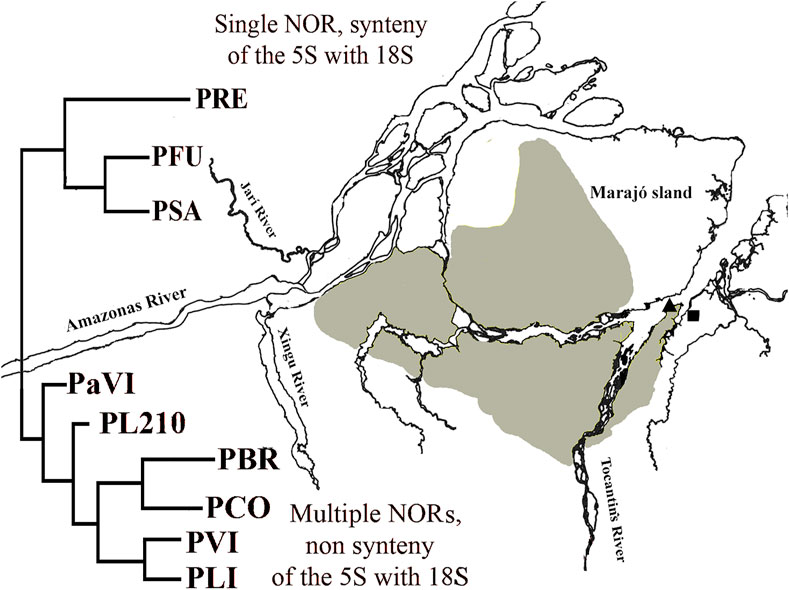
FIGURE 4. Map of the mouth of Amazon River with a simplified version of Peckoltia phylogeny (in red) by Lujan et al. (2017). PRE: Peckoltia relictum; PFU: Peckoltia furcatum; PSA: Peckoltia sabaji; PaVI: Peckoltia aff. vittata; PL210: Peckoltia n. sp. L210; PBR: Peckoltia braueri; PCO: Peckoltia compta; PVI: Peckoltia vittata; PLI: Peckoltia lineola. Geographical location of sampling sites for Peckoltia sp. 3 Jarumã (square) and Peckoltia sp. 4 Caripetuba (triangle) in the present study. Yellow: the distribution of the Plio-Pleistocene/Pleistocene Post-Barreiras sediments (Rossetti and Valeriano, 2007).
In the present work, the karyotype of Peckoltia sp. 4 Caripetuba has several derived characteristics, being found on the right bank of the Amazon River. This fact can be explained if we consider the paleogeography of the region. The continental portion of Abaetetuba (Rio Jarumã) comprises the Barreiras Formation (Miocene). In turn, the Post-Barreiras Formation (Plio-Pleistocene) filled the paleovale of the Tocantins River, diverting this river (which originally crossed Marajó Island to its northern portion) and thus splitting Marajó Island from the mainland, originating Rio Pará and the islands in front of the city of Abaetetuba-PA (Figure 4), where the Caripetuba River is found (Rossetti and Valeriano, 2007). Therefore, the rivers where the karyotype of Peckoltia sp. 4 Caripetuba is found are in islands considerably newer than the one where Peckoltia sp. 3 Jarumã is found. These islands are covered by the Post-Barreiras Formation, being connected to the western portion of Marajó Island (Tatumi et al., 2007), not related with the continental region where the Jarumã River is located. The Marajó Island, in turn, is a communication corridor, connecting biodiversity on the left bank of the Amazon River with biodiversity on the right bank (Fernandes et al., 1995). This fact may explain why the most recent karyotype (Peckoltia sp. 4 Caripetuba) is located in this region. It will be important to test by molecular markers whether the Peckoltia-associated morphotypes analyzed in this study and by de Souza et al. (2009), which have derived karyotypes, belong to the P. sabaji branch.
This study describes the karyotypes of two undescribed species of Peckoltia and compares them with available chromosomal data for the genus. The maintenance of the same 2n in the species of Peckoltia may suggest that this genus has conserved karyotypes; however, the variations observed in the KF, NOR, heterochromatin and 18S and 5S rDNA sequences between the karyotypes of the species in this study compared to those previously described suggest great interspecific diversity in the genus. Furthermore, the differential localization of the U1 snDNA sequence among the karyotypes described here corroborates the involvement of repetitive sequences in the diversification of the genomes of these species. Therefore, due to the considerable cytogenetic diversity, species-specific characters were observed, showing great potential for identifying distinct taxonomic lineages in Peckoltia, in addition to demonstrating that the karyotypic variation in this genus is much greater than conventional staining suggests. Future analyzes considering the geographic distribution of Peckoltia species with primitive versus derived karyotypic characteristics compared to molecular phylogenies may provide relevant information about its evolutionary history.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Comitê de Ética Animal da Universidade Federal do Pará.
Author Contributions
KS: Conceptualization; Data Curation; Formal analysis; Investigation; Methodology; Visualization; Writing original draft; Writing review and editing. AS: Investigation; Methodology; Resources; Visualization; Writing review and editing. AP: Investigation; Methodology; Visualization; Writing review and editing. RN: Investigation; Visualization; Writing review and editing. MV: Methodology; Resources; Visualization; Writing review and editing. JP: Data Curation; Formal analysis; Funding acquisition; Resources; Visualization; Writing review and editing. CN: Data Curation; Formal analysis; Funding acquisition; Project administration; Resources; Supervision; Visualization; Writing review and editing.
Funding
The Pro-Reitoria de Pesquisa e Pos-graduação da Universidade Federal do Para supported by paying the open access publication fees. The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support on project coordinated by CYN (Edital Pró-Amazônia Proc 047/2012); the FAPESPA for financial support (Edital Vale–Proc 2010/110447) and Banco Nacional de Desenvolvimento Econômico e Social–BNDES (2.318.697.0001) on a project coordinated by JCP. CYN (305880/2017-9), JCP (305876/2017-1) and MRV (305142/2019-4) are grateful to CNPq for Productivity Grants. This study is part of the Master dissertation of KS in Genetic and Molecular Biology who is recipient of CAPES Master Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to members of the team of the cytogenetics laboratory UFPA for the fieldwork and chromosomal preparations. To MSc. Jorge Rissino, to MSc. Shirley Nascimento and Maria da Conceição for assistance in laboratory work. Sample collections was authorized by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) and Secretaria de Estado de Meio Ambiente do Pará (SEMA-PA) under permit 020/2005 (Registration: 207419).
References
Almeida, B. R. R., Milhomem-Paixão, S. S. R., Noronha, R. C. R., Nagamachi, C. Y., Costa, M. J. R. D., Pardal, P. P. O., et al. (2017). Karyotype Diversity and Chromosomal Organization of Repetitive DNA in Tityus Obscurus (Scorpiones, Buthidae). BMC Genet. 18 (1), 35–11. doi:10.1186/s12863-017-0494-6
Armbruster, J. W., and Lujan, N. K. (2016). A New Species of Peckoltia from the Upper Orinoco (Siluriformes, Loricariidae). ZooKeys 569, 105–121. doi:10.3897/zookeys.569.6630
Armbruster, J. W., Werneke, D. C., and Tan, M. (2015). Three New Species of Saddled Loricariid Catfishes, and a Review of Hemiancistrus, Peckoltia, and Allied Genera (Siluriformes). Zookeys 480, 97–123. doi:10.3897/zookeys.480.6540
Armbruster, J. W. (2004). Phylogenetic Relationships of the Suckermouth Armoured Catfishes (Loricariidae) with Emphasis on the Hypostominae and the Ancistrinaefishes Loricariidae with Emphasis on the Hypostominae and the Ancistrinae. Zoolog. J. Linn. Soc. 141 (1), 1–80. doi:10.1111/j.1096-3642.2004.00109.x
Armbruster, J. W. (2008). The Genus Peckoltia with the Description of Two New Species and a Reanalysis of the Phylogeny of the Genera of the Hypostominae (Siluriformes: Loricariidae). Zootaxa 1822 (1), 1–76. doi:10.11646/zootaxa.1822.1.1
Artoni, R. F., and Bertollo, L. A. (2001). Trends in the Karyotype Evolution of Loricariidae Fish (Siluriformes). Hereditas 134, 201–210. doi:10.1111/j.1601-5223.2001.00201.x
Barros, A. V., Wolski, M. A. V., Nogaroto, V., Almeida, M. C., Moreira-Filho, O., and Vicari, M. R. (2017). Fragile Sites, Dysfunctional Telomere and Chromosome Fusions: what Is 5S rDNA Role. Gene 608, 20–27. doi:10.1016/j.gene.2017.01.013
Bertollo, L. A. C., Born, G. G., Dergam, J. A., Fenocchio, A. S., and Moreira-Filho, O. (2000). A Biodiversity Approach in the Neotropical Fish, Hoplias malabaricus. Karyotypic Survey, Geographic Distribution of Cytotypes and Cytotaxonomic Considerations. Chromosome Res. 8 (7), 603–613. doi:10.1023/a:1009233907558
Bertollo, L. A. C., Takahashi, C. S., and Moreira-Filho, O. (1978). Cytotaxonomic Considerations on Hoplias Lacerdae (Pisces Erythrinidae). Brazil. J. Genet. 1, 103–120.
Bueno, V., Venere, P. C., Thums Konerat, J., Zawadzki, C. H., Vicari, M. R., and Margarido, V. P. (2014). Physical Mapping of the 5S and 18S rDNA in Ten Species of Hypostomus Lacépède 1803 (Siluriformes: Loricariidae): Evolutionary Tendencies in the Genus. ScientificWorldJournal 2014, 943825. doi:10.1155/2014/943825
Bueno, V., Konerat, J. T., Zawadzki, C. H., Venere, P. C., Blanco, D. R., and Margarido, V. P. (2018). Divergent Chromosome Evolution in Hypostominae Tribes (Siluriformes: Loricariidae): Correlation of Chromosomal Data with Morphological and Molecular Phylogenies. Zebrafish 15 (5), 492–503. doi:10.1089/zeb.2018.1612
Busch, H., Reddy, R., Rothblum, L., and Choi, Y. C. (1982). SnRNAs, SnRNPs, and RNA Processing. Annu. Rev. Biochem. 51 (1), 617–654. doi:10.1146/annurev.bi.51.070182.003153
Cabral-de-Mello, D. C., Valente, G. T., Nakajima, R. T., and Martins, C. (2012). Genomic Organization and Comparative Chromosome Mapping of the U1 snRNA Gene in Cichlid Fish, with an Emphasis in Oreochromis niloticus. Chromosome Res. 20 (2), 279–292. doi:10.1007/s10577-011-9271-y
Cavalcante, M. G., Nagamachi, C. Y., Pieczarka, J. C., and Noronha, R. C. R. (2020). Evolutionary Insights in Amazonian Turtles (Testudines, Podocnemididae): Co-location of 5S rDNA and U2 snRNA and Wide Distribution of Tc1/Mariner. Biol. open 9 (4). bio049817, doi:10.1242/bio.049817
Centofante, L., Bertollo, L. A. C., Justi, A. J., and Moreira-Filho, O. (2003). Correlation of Chromosomal and Morphologic Characters in Two Astyanax Species. Ichthyol. Exploration Freshwaters 14 (4), 361–368.
De Souza, A., Nagamachi, C., Milhomem, S., Feldberg, E., and Pieczarka, J. (2009). Cytogenetic Analysis in Catfish Species of the Genus Peckoltia Miranda Ribeiro, 1912 (Teleostei: Siluriformes: Loricariidae). Comp. Cytogenet. 3, 103–109. doi:10.3897/compcytogen.v3i2.17
Deon, G. A., Glugoski, L., Vicari, M. R., Nogaroto, V., Sassi, F. d. M. C., Cioffi, M. d. B., et al. (2020). Highly Rearranged Karyotypes and Multiple Sex Chromosome Systems in Armored Catfishes from the Genus Harttia (Teleostei, Siluriformes). Genes 11, 1366. doi:10.3390/genes11111366
Dimitri, P., Caizzi, R., Giordano, E., Carmela Accardo, M., Lattanzi, G., and Biamonti, G. (2009). Constitutive Heterochromatin: a Surprising Variety of Expressed Sequences. Chromosoma 118 (4), 419–435. doi:10.1007/s00412-009-0211-y
Dulz, T. A., Azambuja, M., Nascimento, V. D., Lorscheider, C. A., Noleto, R. B., Moreira-Filho, O., et al. (2020). Karyotypic Diversification in Two Megaleporinus Species (Characiformes, Anostomidae) Inferred from In Situ Localization of Repetitive DNA Sequences. Zebrafish 17 (5), 333–341. doi:10.1089/zeb.2020.1918
Favarato, R. M., da Silva, M., de Oliveira, R. R., Artoni, R. F., Feldberg, E., and Matoso, D. A. (2016). Cytogenetic Diversity and the Evolutionary Dynamics of rDNA Genes and Telomeric Sequences in the Ancistrus Genus (Loricariidae: Ancistrini). Zebrafish 13 (2), 103–111. doi:10.1089/zeb.2015.1140
Fernandes, M. E. B., Cardoso Da Silva, J. M., and De S. E Silva Junior, J. (1995). The Monkeys of the Islands of the Amazon Estuary, Brazil: a Biogeographic analysisA Biogeographic Analysis. Mammalia 59 (2), 213–221. doi:10.1515/mamm.1995.59.2.213
Fricke, R., Eschmeyer, W. N., and Van der Laan, R. (2021). Eschmeyer's Catalog of Fishes. Genera, Species, References. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Electronic version accessed in March 25, 2021).
Glugoski, L., Deon, G. A., Schott, S., Vicari, M. R., Nogaroto, V., and Moreira-Filho, O. (2020). Comparative Cytogenetic Analyses in Ancistrus Species (Siluriformes: Loricariidae). Neotropical Ichthyology 18, e200013. doi:10.1590/1982-0224-2020-0013
Glugoski, L., Giuliano-Caetano, L., Moreira-Filho, O., Vicari, M. R., and Nogaroto, V. (2018). Co-located hAT Transposable Element and 5S rDNA in an Interstitial Telomeric Sequence Suggest the Formation of Robertsonian Fusion in Armored Catfish. Gene 650, 49–54. doi:10.1016/j.gene.2018.01.099
Hatanaka, T., and Galetti, P. M. (2004). Mapping of the 18S and 5S Ribosomal RNA Genes in the Fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122 (3), 239–244. doi:10.1007/s10709-004-2039-y
Howell, W. M., and Black, D. A. (1980). Controlled Silver-Staining of Nucleolus Organizer Regions with a Protective Colloidal Developer: a 1-step Method. Experientia 36 (8), 1014–1015. doi:10.1007/BF01953855
Ijdo, J. W., Wells, R. A., Baldini, A., and Reeders, S. T. (1991). Improved Telomere Detection Using a Telomere Repeat Probe (TTAGGG)ngenerated by PCR. Nucl. Acids Res. 19 (17), 4780. doi:10.1093/nar/19.17.4780
Kapitonov, V. V., and Jurka, J. (2006). Self-synthesizing DNA Transposons in Eukaryotes. Proc. Natl. Acad. Sci. 103 (12), 4540–4545. doi:10.1073/pnas.0600833103
Kavalco, K. F., Pazza, R., Bertollo, L. A. C., and Moreira-Filho, O. (2005). Karyotypic Diversity and Evolution of Loricariidae (Pisces, Siluriformes). Heredity 94 (2), 180–186. doi:10.1038/sj.hdy.6800595
Levan, A., Fredga, K., and Sandberg, A. A. (1964). Nomenclature for Centromeric Position on Chromosomes. Hereditas 52 (2), 201–220. doi:10.1111/j.1601-5223.1964.tb01953.x
Long, E. O., and Dawid, I. B. (1980). Repeated Genes in Eukaryotes. Annu. Rev. Biochem. 49 (1), 727–764. doi:10.1146/annurev.bi.49.070180.003455
Lujan, N. K., Armbruster, J. W., Lovejoy, N. R., and López-Fernández, H. (2015). Multilocus Molecular Phylogeny of the Suckermouth Armored Catfishes (Siluriformes: Loricariidae) with a Focus on Subfamily Hypostominae. Mol. Phylogenet. Evol. 82, 269–288. doi:10.1016/j.ympev.2014.08.020
Lujan, N. K., Cramer, C. A., Covain, R., Fisch-Muller, S., and López-Fernández, H. (2017). Multilocus Molecular Phylogeny of the Ornamental wood-eating Catfishes (Siluriformes, Loricariidae, Panaqolus and Panaque) Reveals Undescribed Diversity and Parapatric Clades. Mol. Phylogenet. Evol. 109, 321–336. doi:10.1016/j.ympev.2016.12.040
Mariotto, S., Centofante, L., Vicari, M., Artoni, R., and Moreira Filho, O. (2011). Chromosomal Diversification in Ribosomal DNA Sites in Ancistrus Kner, 1854 (Loricariidae, Ancistrini) from Three Hydrographic Basins of Mato Grosso, Brazil. Comp. Cytogenet. 5 (4), 289–300. doi:10.3897/CompCytogen.v5i4.1757
Mariotto, S., and Miyazawa, C. S. (2006). Ancistrus Cf. Dubius (Siluriformes, Ancistrinae), a Complex of Species. 1. Chromosomic Characterization of Four Populations and Occurence of Sexual Chromosomes of Type XX/XY, in the Pantanal basin of Mato Grosso, Brazil. Caryologia 59 (4), 299–304. doi:10.1080/00087114.2006.10797929
Martins, C., and Galetti Jr, P. M. (1999). Chromosomal Localization of 5S rDNA Genes in Leporinus Fish (Anostomidae, Characiformes). Chromosome Res. 7 (5), 363–367. doi:10.1023/a:1009216030316
Montefalcone, G., Tempesta, S., Rocchi, M., and Archidiacono, N. (1999). Centromere Repositioning. Genome Res. 9 (12), 1184–1188. doi:10.1101/gr.9.12.1184
Nagamachi, C. Y., Pieczarka, J. C., Milhomem, S. S., O'Brien, P. C., de Souza, A. C., and Ferguson-Smith, M. A. (2010). Multiple Rearrangements in Cryptic Species of Electric Knifefish, Gymnotus carapo (Gymnotidae, Gymnotiformes) Revealed by Chromosome Painting. BMC Genet. 11, 28. doi:10.1186/1471-2156-11-28
Pansonato-Alves, J. C., Serrano, É. A., Utsunomia, R., Scacchetti, P. C., Oliveira, C., and Foresti, F. (2013). Mapping Five Repetitive DNA Classes in Sympatric Species of Hypostomus (Teleostei: Siluriformes: Loricariidae): Analysis of Chromosomal Variability. Rev. Fish. Biol. Fish. 23 (4), 477–489. doi:10.1007/s11160-013-9303-0
Pety, A. M., Cardoso, A. L., Nagamachi, C. Y., Pieczarka, J. C., de Sousa, L. M., and Noronha, R. C. R. (2018). In Situ localization of Ribosomal Sites in Peckoltia and Ancistomus (Loricariidae: Hypostominae) from the Amazon Basin. Zebrafish 15 (3), 263–269. doi:10.1089/zeb.2017.1523
Prizon, A. C., Borin-Carvalho, L. A., Bruschi, D. P., Otávio Ribeiro, M., Magrinelli Barbosa, L., de Brito Ferreira, G. E., et al. (2016). Cytogenetic Data on Ancistrus Sp. (Siluriformes, Loricariidae) of the Paraguay River basin (MS) Sheds Light on Intrageneric Karyotype Diversification. Ccg 10 (4), 625–636. doi:10.3897/CompCytogen.v10i4.8532
Ribeiro, M. O., Noleto, R. B., Lorscheider, C. A., Porto, F. E., Prizon, A. C., Zawadzki, C. H., et al. (2015). Cytogenetic Description of Ancistrus Abilhoai (Siluriformes: Loricariidae) from Iguaçu River basin, Southern Brazil. Genet. Mol. Res. 14 (2), 4051–4057. doi:10.4238/2015.april.27.20
Rocchi, M., Archidiacono, N., Schempp, W., Capozzi, O., and Stanyon, R. (2012). Centromere Repositioning in Mammals. Heredity 108 (1), 59–67. doi:10.1038/hdy.2011.101
Rossetti, D. F., and Valeriano, M. M. (2007). Evolution of the Lowest Amazon basin Modeled from the Integration of Geological and SRTM Topographic Data. Catena 70 (2), 253–265. doi:10.1016/j.catena.2006.08.009
Roxo, F. F., Ochoa, L. E., Sabaj, M. H., Lujan, N. K., Covain, R., Silva, G. S. C., et al. (2019). Phylogenomic Reappraisal of the Neotropical Catfish Family Loricariidae (Teleostei: Siluriformes) Using Ultraconserved Elements. Mol. Phylogenet. Evol. 135, 148–165. doi:10.1016/j.ympev.2019.02.017
Scacchetti, P. C., Utsunomia, R., Pansonato-Alves, J. C., da Costa Silva, G. J., Vicari, M. R., Artoni, R. F., et al. (2015). Repetitive DNA Sequences and Evolution of ZZ/ZW Sex Chromosomes in Characidium (Teleostei: Characiformes). PLoSOne 10 (9), e0137231. doi:10.1371/journal.pone.0137231
Schemberger, M. O., Nascimento, V. D., Coan, R., Ramos, É., Nogaroto, V., Ziemniczak, K., et al. (2019). DNA Transposon Invasion and Microsatellite Accumulation Guide W Chromosome Differentiation in a Neotropical Fish Genome. Chromosoma 128 (4), 547–560. doi:10.1007/s00412-019-00721-9
Suárez, P., Pinto Barroso, I. C. G., Silva, D. d. S., Milhomem, S. S. R., Cabral-de-Mello, D. C., Martins, C., et al. (2017). Highest Diploid Number Among Gymnotiformes: First Cytogenetic Insights into Rhabdolichops (Sternopygidae). Zebrafish 14 (3), 272–279. doi:10.1089/zeb.2016.1405
Sumner, A. T. (1972). A Simple Technique for Demonstrating Centromeric Heterochromatin. Exp. Cel Res. 75, 304–306. doi:10.1016/0014-4827(72)90558-7
Tatumi, S. H., da Silva, L. P., Pires, E. L., Rossetti, D. F., Góes, A. M., and Munita, C. S. (2007). Datação de Sedimentos Pós-Barreiras no Norte Do Brasil: implicações paleogeográficas. Revista Brasileira de Geociências 38 (3), 514–524. doi:10.25249/0375-7536.2008383514524
Úbeda-Manzanaro, M., Merlo, M. A., Palazón, J. L., Cross, I., Sarasquete, C., and Rebordinos, L. (2010). Chromosomal Mapping of the Major and Minor Ribosomal Genes, (GATA)n and U2 snRNA Gene by Double-Colour FISH in Species of the Batrachoididae Family. Genetica 138 (7), 787–794. doi:10.1007/s10709-010-9460-1
Vicari, M. R., Nogaroto, V., Noleto, R. B., Cestari, M. M., Cioffi, M. B., Almeida, M. C., et al. (2010). Satellite DNA and Chromosomes in Neotropical Fishes: Methods, Applications and Perspectives. J. Fish Biol. 76 (5), 1094–1116. doi:10.1111/j.1095-8649.2010.02564.x
Yano, C. F., Bertollo, L. A. C., Rebordinos, L., Merlo, M. A., Liehr, T., Portela-Bens, S., et al. (2017). Evolutionary Dynamics of rDNAs and U2 Small Nuclear DNAs in Triportheus (Characiformes, Triportheidae): High Variability and Particular Syntenic Organization. Zebrafish 14 (2), 146–154. doi:10.1089/zeb.2016.1351
Keywords: neotropical fish, snRNA, rDNA, biodiversity, amazon
Citation: Santos da Silva K, de Souza ACP, Pety AM, Noronha RCR, Vicari MR, Pieczarka JC and Nagamachi CY (2021) Comparative Cytogenetics Analysis Among Peckoltia Species (Siluriformes, Loricariidae): Insights on Karyotype Evolution and Biogeography in the Amazon Region. Front. Genet. 12:779464. doi: 10.3389/fgene.2021.779464
Received: 18 September 2021; Accepted: 18 October 2021;
Published: 28 October 2021.
Edited by:
Tony Silveira, Federal University of Rio Grande, BrazilReviewed by:
Vladimir Pavan Margarido, Universidade Estadual do Oeste do Paraná, BrazilAnalía Del Valle Garnero, Federal University of Pampa, Brazil
Lucia Caetano, State University of Londrina, Brazil
Copyright © 2021 Santos da Silva, de Souza, Pety, Noronha, Vicari, Pieczarka and Nagamachi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cleusa Yoshiko Nagamachi, Y2xldXNhbmFnYW1hY2hpQGdtYWlsLmNvbQ==, Y2xldXNhQHVmcGEuYnI=
†These authors have contributed equally to this work
 Kevin Santos da Silva1
Kevin Santos da Silva1 Marcelo Ricardo Vicari
Marcelo Ricardo Vicari Julio Cesar Pieczarka
Julio Cesar Pieczarka Cleusa Yoshiko Nagamachi
Cleusa Yoshiko Nagamachi