- 1Louisiana Animal Disease Diagnostic Laboratory and Department of Pathobiological Sciences, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA, United States
- 2Maxwell H. Gluck Equine Research Center, University of Kentucky, Lexington, KY, United States
- 3Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois Urbana-Champaign, Urbana, IL, United States
- 4Equine Health Studies Program, Department of Veterinary Clinical Sciences, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA, United States
- 5Department of Infectious Diseases and Public Health, Jockey Club College of Veterinary Medicine, City University of Hong Kong, Kowloon, Hong Kong SAR, China
Equine arteritis virus (EAV) is the causative agent of equine viral arteritis (EVA), a respiratory, systemic, and reproductive disease of equids. Following natural infection, up to 70% of the infected stallions can remain persistently infected over 1 year (long-term persistent infection [LTPI]) and shed EAV in their semen. Thus, the LTP-infected stallions play a pivotal role in maintaining and perpetuating EAV in the equine population. Previous studies identified equine C-X-C motif chemokine ligand 16 (CXCL16) as a critical host cell factor determining LTPI in the stallion’s reproductive tract. Two alleles (CXCL16S and CXCL16r) were identified in the equine population and correlated with the susceptibility or resistance of a CD3+ T cell subpopulation in peripheral blood to in vitro EAV infection, respectively. Interestingly, CXCL16S has been linked to the establishment of LTPI in stallions, and thus, genotyping stallions based on CXCL16S/r would allow identification of those at the highest risk of establishing LTPI. Thus, we developed a TaqMan® allelic discrimination qPCR assay for the genotyping of the equine CXCL16 gene based on the identification of a single nucleotide polymorphism in position 1,073 based on NCBI gene ID: 100061442 (or position 527 based on Ensembl: ENSECAG00000018406.2) located in exon 2. One hundred and sixty horses from four breeds were screened for the CD3+ T cell susceptibility phenotype to EAV infection by flow cytometry and subsequently sequenced to determine CXCL16 allelic composition. Genotyping by Sanger sequencing determined that all horses with the resistant CD3+ T cell phenotype were homozygous for CXCL16r while horses with the susceptible CD3+ T cell phenotype carried at least one CXCL16S allele or homozygous for CXCL16S. In addition, genotypification with the TaqMan® allelic discrimination qPCR assay showed perfect agreement with Sanger sequencing and flow cytometric analysis. In conclusion, the new TaqMan® allelic discrimination genotyping qPCR assay can be used to screen prepubertal colts for the presence of the CXCL16 genotype. It is highly recommended that colts that carry the susceptible genotype (CXCL16 S/S or CXCL16S/r) are vaccinated against EAV after 6 months of age to prevent the establishment of LTPI carriers following possible natural infection with EAV.
Introduction
Equine arteritis virus (EAV) is an enveloped, positive-sense, single-stranded RNA virus that belongs to the family Arteriviridae, subfamily Equarterivirinae, and genus Alphaarterivirus in the order Nidovirales (Cavanagh, 1997; Kuhn et al., 2016; Brinton et al., 2021). EAV is the causative agent of equine viral arteritis (EVA) in horses, a respiratory and reproductive disease with worldwide distribution. It causes significant economic losses to the horse industry due to abortion storms, neonatal mortality, respiratory disease, and the establishment of long-term persistent infection (LTPI) in stallions. EAV infection can be asymptomatic or associated with a wide range of clinical signs, including dependent edema, conjunctivitis, periorbital or supraorbital edema, respiratory distress, urticaria, and leukopenia (Timoney and McCollum, 1993; McCollum et al., 1995; MacLachlan et al., 1996; Balasuriya et al., 1999; Balasuriya et al., 2007; Go et al., 2012b; Vairo et al., 2012; Balasuriya et al., 2013; Balasuriya, 2014; Campos et al., 2014; Balasuriya et al., 2016; Carossino et al., 2017a). Additionally, EAV infection of pregnant mares can result in abortion or birth of congenitally infected foals that develop a rapidly progressive and ultimately fatal bronchointerstitial pneumonia or pneumoenteric syndrome (Vaala et al., 1992; Del Piero, 2000). Transmission of EAV can occur through both the respiratory and the venereal routes. Following initial exposure, EAV can establish a persistent infection in the reproductive tract of stallions, resulting in continuous shedding of infectious virus in semen (Balasuriya et al., 2013; Balasuriya et al., 2016; Carossino et al., 2017a). About 10–70% of naturally infected stallions with EAV can become persistently infected carriers. The duration of EAV carrier state in stallions can be divided into short-term (i.e., a few weeks, or a few months and up to 1 year) or long term (i.e., continues to shed virus for many years or even the remainder of the animal’s lifetime) (Go et al., 2011; Balasuriya et al., 2013; Campos et al., 2014; Balasuriya and Carossino, 2017; Carossino et al., 2017b). The ampullae, an accessory sex gland present in the stallion reproductive tract, has recently been recognized as the primary site of EAV persistence in long-term persistently infected carrier stallions (Carossino et al., 2017a; Carossino et al., 2019). Carrier stallions do not exhibit clinical signs of disease or impairment of fertility, which guarantees the maintenance and perpetuation of EAV in the equine population between breeding seasons.
Previous studies in our group have demonstrated that EAV can infect a distinct subpopulation of peripheral CD3+ T lymphocytes from certain horses in vitro, while CD3+ T lymphocytes from other horses remain resistant to infection. A subsequent genome-wide association study (GWAS) revealed that the ability of EAV to infect CD3+ T lymphocytes in vitro (susceptibility phenotype) is strongly associated with a haplotype in the region of equine chromosome 11 (ECA11) (Go et al., 2011; Go et al., 2012a). Further genomic and proteomic analysis demonstrated that the equine C-X-C motif chemokine ligand 16 (CXCL16) gene, located in this region within ECA11, acts as a cellular receptor for EAV (Sarkar et al., 2016b). More importantly, it has been demonstrated that the ability of EAV to infect CD3+ T lymphocytes and establish long-term carrier status in stallions is strongly associated with four single nucleotide polymorphisms (SNPs) in the exon 2 of the CXCL16 gene (A1073→T1073; G1099→C1099; T1102→A/G1102; G1108→A1108; based on the annotation for the NCBI equine reference CXCL16 [gene ID: 100061442] sequence or A527→T527; G553→C553; T556→A/G556; G562→A562; based on the annotation for the Ensembl equine reference CXCL16 [ENSECAG00000018406.2] sequence), coding for the susceptibility allele, namely CXCL16S (Sarkar et al., 2016a). It has been demonstrated that CXCL16S acts as a cellular receptor for EAV while CXCL16R, coded by the resistance allele CXCL16r, does not. Concurrently, stallions that are either homozygous or heterozygous for the susceptibility allele (CXCL16 S/S, CXCL16S/r) can become EAV LTPI carriers, while the stallions that are homozygous for the resistant allele (CXCL16r/r) do not become carriers or they will be only for a limited period [i.e., short-term carriers or shedders (<1 year)] (Carossino et al., 2017a). The presence of the susceptible genotype (CXCL16S) differs between breeds, as does EAV seroprevalence (Go et al., 2011; Sarkar et al., 2016a; Socha et al., 2020). A high percentage of Standardbred, Saddlebred, Wielkopolska breed, Polish coldblood, and Silesian breed horses have been shown to carry the susceptible genotype. In contrast, the CXCL16r homozygous genotype is most often present in Quarter Horses, Thoroughbreds, Arabian horses, Hucul horses, and Malopolska horses. CXCL16 is, therefore, a critical host factor in the establishment and maintenance of EAV LTPI in the stallion and serves as a “hub” gene likely driving a specific transcriptional network for the establishment of LTPI in the ampullae (Carossino et al., 2019).
Determination of the CXCL16 genotype in stallions is critical to identify those at greatest risk of becoming LTPI EAV carriers and assist with targeted vaccination practices to prevent the occurrence of the carrier state. In this study, we developed a TaqMan® allelic discrimination qPCR assay for rapid and precise genotyping of the CXCL16 gene. This allelic discrimination qPCR assay was validated on 160 horses previously sequenced for the CXCL16 gene, showing perfect agreement with conventional Sanger sequencing and flow cytometric analysis of CD3+ T lymphocyte’s phenotype.
Materials and Methods
Ethics Approval and Consent to Participate
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky, Lexington, KY and Louisiana State University, Baton Rouge, LA approved protocol numbers 2011-0888 and 18-122, respectively. The explicit owner informed consent for the inclusion of animals in this study was not stated.
Animals
A total of 160 horses from four different breeds, American Saddlebred (SB), Quarter Horse (QH), Standardbred (STB), and Thoroughbred (TB), were randomly selected for this study (n = 40 of each breed) (Sarkar et al., 2016a). These horses were randomly selected from farms in central Kentucky.
Virus and Antibodies
The virulent Bucyrus strain of EAV (VBS; ATCC, Manassas, VA, Cat# VR-796) was used at a multiplicity of infection (MOI) of two for in vitro infection of PBMCs. The equine CD3 specific monoclonal antibody (MAb), UC F6G, kindly provided by Jeff Stott, University of California, Davis, was used as the primary antibody. The R-phycoerythrin (R-PE)-conjugated F (ab')2 fragment of goat anti-mouse IgG1 (SouthernBiotech, Birmingham, AL, Cat# 1,072-09, RRID: AB_2794434) was used as the secondary antibody. Alexa Fluor 488-labeled MAb against the non-structural protein 1 (nsp1; MAb 12A4) was used to detect EAV antigen (Wagner et al., 2003; Go et al., 2010).
Blood Collection, Isolation of PBMCs, and Flow Cytometry
Flow cytometric analysis of EAV infected equine peripheral blood mononuclear cells (PBMCs) was performed as previously described (Go et al., 2010; Go et al., 2011). Briefly, blood was collected from each horse using Vacutainer tubes containing 15% EDTA solution (Kendall Healthcare, Mansfield, MA). Next, PBMCs were isolated from the buffy-coat fraction by centrifugation using Ficoll-Plaque Plus (Amersham Biosciences, Piscataway, NJ) and infected with EAV VBS strain for 36 h. The susceptible or resistant phenotype of each animal was defined by dual-color flow cytometric analysis of in vitro EAV-infected CD3+ T lymphocytes. Flow cytometry data were collected using a BD FACS Calibur™ flow cytometer with approximately 100,000 events and analyzed with BD CellQuest Pro (RRID: SCR_014489) and FlowJo (RRID: SCR_008520) version 10 software.
DNA Extraction
Genomic DNA (gDNA) was obtained from 2×107 PBMCs of each horse by using the Gentra Puregene Blood kit (Qiagen, Valencia, CA, Cat #158389), following the manufacturer’s instructions. The gDNA concentration was assessed using a NanoDrop 1000 Spectrophotometer (Thermo Scientific NanoDrop 1000 Spectrophotometer, RRID: SCR_020560) at an absorbance ratio of optical density at 260/280 nm by following the manufacturer’s instructions and stored at −20°C until used.
Equine CXCL16 Gene Sequencing
A polymerase chain reaction (PCR) was performed to amplify a 280 bp sequence encompassing the four SNPs in the CXCL16 gene as previously described (Sarkar et al., 2016a) (Figure 1). The 18 µl mixture contained 100 ng of gDNA, 0.15 mM of forward primer (5′-CGGTGGGTTGGAGGCTAA-3′), 0.15 mM of reverse primer (5′- GACCAGAGAGGGTCCCAGA-3′) and 10 µl of AmpliTaq Gold™ Fast PCR Master Mix (Applied Biosystems, Waltham, MA, Cat# 4390939). Amplification consisted of the following steps: initial denaturation at 95°C for 10 min and 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 30 s; and a final extension at 72°C for 10 min. Amplicons were then shipped overnight for sequencing to Eurofins Genomics (Louisville, KY, United States). DNA sequences were analyzed using Chromas Lite (Technelysium Pty Ltd., South Brisbane, Australia).
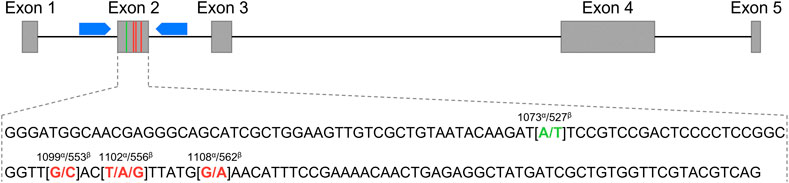
FIGURE 1. Schematic representation of the Equus caballus C-X-C motif chemokine ligand 16 gene (CXCL16) organization. The grey boxes represent exons 1 to 5, and the black connecting lines represent introns. The blue pentagons correspond to the primers used for exon 2 sequencing. The exon 2 sequence is depicted below. The SNP at position 1073α/527β used for the genotyping qPCR assay is shown in green, and the three other SNPs at positions 1099α/553β, 1102α/556β and 1108α/562β are marked in red. α: annotation based on the NCBI CXCL16 gene ID: 100061442; β: annotation based on Ensembl CXCL16 gene: ENSECAG00000018406.2.
Equine CXCL16 Genotyping Assay
A Custom TaqMan® SNP Genotyping Assay (ID: AHX1KFO; Applied Biosystems, Waltham, MA, Cat# 4332077) was designed to amplify a product encompassing the SNP at position 1,073 (A/T) of the equine CXCL16 gene based on NCBI gene ID: 100061442 (or position 527 based on Ensembl: ENSECAG00000018406.2, respectively). The two fluorogenic MGB probes were designed with VIC and FAM dyes to allow genotyping targeting, respectively, Adenine (CXCL16r) or Thymine (CXCL16S) at this position. Real-time PCR was performed on a 25 µl reaction mixture containing 12.5 µl of TaqMan® GTXpress™ Master Mix (Applied Biosystems, Cat# 4401892), 1.25 µl of TaqMan® genotyping assay mix (20×) containing primers and probes, 6.25 µl of DNase-free water, and 5 µl of gDNA diluted at 1 ng/μl in nuclease-free water. Thermal cycling and endpoint analyses were performed on a 7,500 Fast Real-Time PCR System (Life Technologies) using the following conditions: initial DNA polymerase activation step at 95°C for 20 s and 40 cycles of denaturation at 95°C for 3 s and annealing of primers/extension at 60°C for 30 s. The pre-read and post-read were taken at 25°C, according to the TaqMan® GTXpress™ Master Mix protocol.
Results
Determination of CD3+ T Lymphocyte Phenotypes by Flow Cytometry
Following in vitro infection of PBMCs with EAV virulent Bucyrus strain (VBS), a small subpopulation of the CD3+ T lymphocytes susceptible to EAV infection (3.9 ± 0.4% to 7.2 ± 0.6%) was identified in 104 horses using flow cytometry (Figure 2; upper right quadrant). Consequently, these horses were classified as having a CD3+ T-cell “susceptible” phenotype. In contrast, no CD3+ T lymphocytes susceptible to EAV infection were identified in 56 horses, and these were classified as having CD3+ T-cell “resistant” phenotype (Table 1).
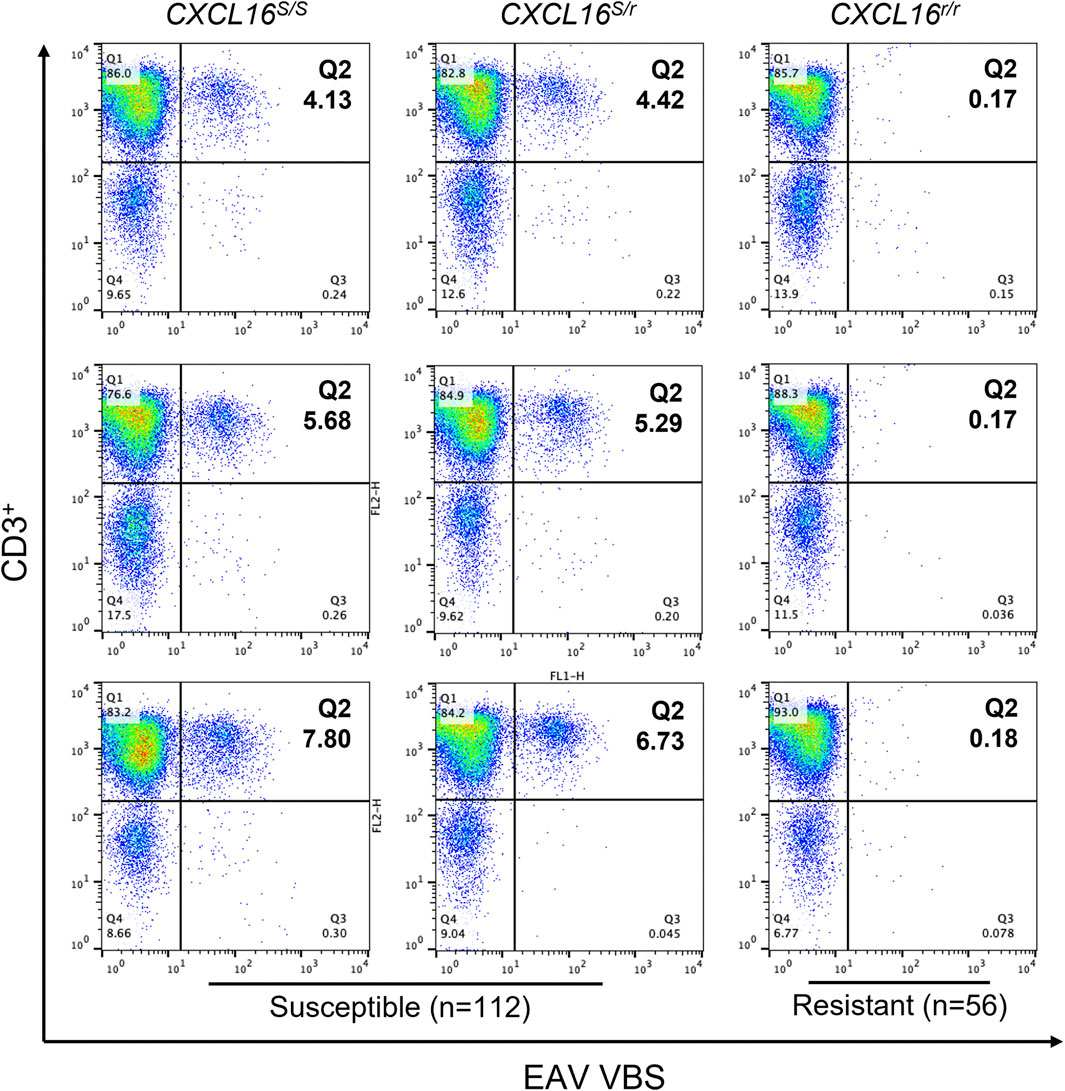
FIGURE 2. Susceptibility of CD3+ T lymphocytes to EAV VBS infection in vitro. Equine PBMCs were infected with EAV VBS at an MOI of two and harvested after 36 h in culture. PBMCs were stained with anti-CD3 and anti-nsp1 antibodies, and a dual-color immunofluorescence flow cytometry analysis was performed. Three representative plots corresponding to horses with each of the three CXCL16 genotypes are shown. Plots were generated using FlowJo version 10 software. The upper right quadrant (Q2) shows the sub-population of CD3+ T lymphocytes infected with EAV VBS (CD3+ T-cell “susceptible” subpopulation). The numbers in Q2 represent percentages.
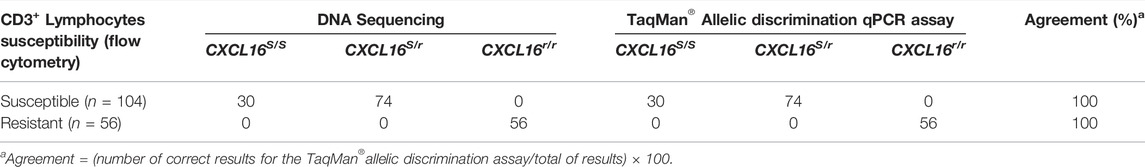
TABLE 1. Determination of CXCL16 genotypes by DNA sequencing and TaqMan® allelic discrimination qPCR assay and phenotyping via flow cytometric analysis of EAV-infected PBMCs (n = 160).
CXCL16 Genotype Determination by Sequencing
A 280 bp sequence encompassing the four SNPs in the exon 2 of the CXCL16 was obtained by Sanger sequencing (Figure 1) for the 160 horses. Thirty horses were homozygous for the CXCL16S allele, 74 were heterozygous (CXCL16S/r), and 56 horses were homozygous for the CXCL16r allele (Table 1). Sequencing data showed that all horses with the susceptible CD3+ T lymphocyte phenotype had the CXCL16 S/S (n = 30) or the CXCL16S/r (n = 74) genotype. In contrast, all the horses with the resistant CD3+ T lymphocyte phenotype had the CXCL16r/r genotype (n = 56).
Performance of an Allelic Discrimination TaqMan® qPCR Assay for Rapid CXCL16 Genotyping
The CXCL16 genotype of the horses was then analyzed using the novel TaqMan® allelic discrimination qPCR assay targeting the A/T SNP at position 1,073 of the equine CXCL16 gene based on the NCBI gene ID: 100061442 (or position 527 based on Ensembl: ENSECAG00000018406.2, respectively), and results are plotted in Figure 3. Three distinct clusters were graphically obtained, corresponding to horses with the CXCL16 S/S (n = 30), CXCL16S/r (n = 74), and CXCL16r/r (n = 56) genotypes. The agreement between the TaqMan® allelic discrimination qPCR assay and Sanger sequencing (gold standard) was 100% (Table 1).
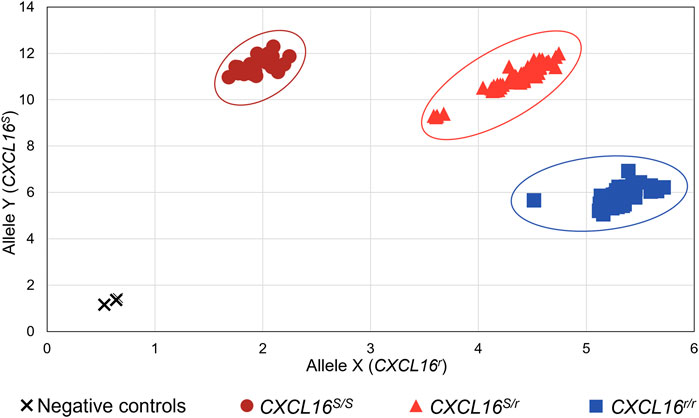
FIGURE 3. Equine CXCL16 allelic discrimination assay output and allele determination. The TaqMan® allelic discrimination qPCR assay was used for genotyping CXCL16S and CXCL16r alleles of 160 horses. The X-axis represents the fluorescence intensity for the CXCL16r allele-specific probe labeled with VIC. The Y-axis represents the fluorescence intensity for the CXCL16S allele-specific probe labeled with FAM. Dark red circles: homozygous CXCL16 S/S (n = 30); light red triangles: heterozygous CXCL16S/r (n = 74); blue squares, homozygous CXCL16r/r (n = 56); black cross; negative controls (no DNA). The negatives controls, without gDNA, were located in the bottom left of the graph and were distinct from the positive samples.
Distribution of Genotypes by Breed
Genotype distribution by breed is shown in Figure 4. In concordance with our previous studies, Standardbred (92%) and Saddlebred (83%) horses most frequently carried the CXCL16S allele, followed by Quarter Horses (55%) and, lastly, Thoroughbreds, in which the CXCL16S allele was detected in approximately one-third (34%) of the population screened.
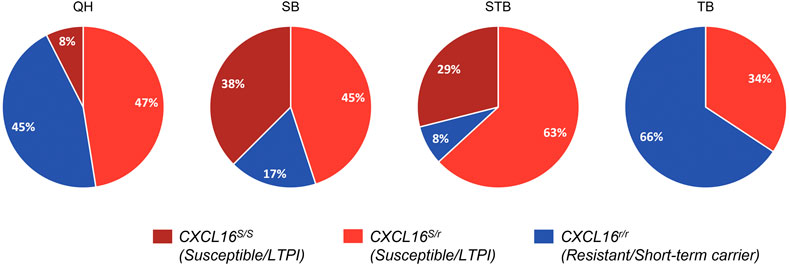
FIGURE 4. CXCL16 genotype distribution among diverse horse breeds. Distribution of the three different CXCL16 genotypes among different horse breeds included in the study (n = 40 for each breed). Dark red: CXCL16 S/S; light red: CXCL16S/r; blue: CXCL16r/r. LTPI: Long-term persistent infection.
Discussion
Single nucleotide polymorphisms (SNPs) are the simplest and most common source of genetic polymorphisms that can cause individual differences in susceptibility to various diseases. Over three million SNPs have been identified in the equine genome (Doan et al., 2012), corresponding to one SNP per 800 bp. Several genomic studies conducted in our laboratory have shown that four SNPs located in the exon 2 of the equine CXCL16 gene are linked to the establishment of EAV LTPI in the stallion (Sarkar et al., 2016a; Carossino et al., 2017b; Carossino et al., 2019). Indeed, the protein coded by CXCL16S was demonstrated to act as a cellular receptor for EAV, while the protein coded by CXCL16R does not (Sarkar et al., 2016a; Sarkar et al., 2016b). Therefore, there is a critical need for genotyping the equine CXCL16 gene to rationalize targeted vaccination of stallions with the greatest risk of becoming EAV LTPI carriers and possibly assist in breeding selection. SNP detection using real-time PCR-based assays has been extensively reported (Ehmer et al., 2008; Bowers et al., 2010; Manga and Dvořák, 2010; Kamau et al., 2012; Fedick et al., 2013; Salehi et al., 2015; Heissl et al., 2017). In this study, we have successfully developed a TaqMan® allelic discrimination qPCR assay for genotyping of the equine CXCL16 gene.
The four SNPs in exon 2 of CXCL16 correlate with the presence of a subpopulation of CD3+ T lymphocytes in peripheral blood, which is susceptible to EAV infection in vitro. Previously, it has been shown that carrier stallions homozygous or heterozygous for the CXCL16S allele carry this specific CD3+ T lymphocyte subpopulation susceptible to EAV infection in vitro (Go et al., 2012a). However, identifying stallions based on this phenotype requires in vitro EAV infection of cultured PBMCs followed by dual-color flow cytometric analysis, making this technique time-consuming and technically challenging. In addition, this technique does not differentiate horses that are homozygous from those that are heterozygous for the CXCL16S allele (Go et al., 2011). Conventional Sanger sequencing is considered as the gold standard, with high sensitivity for genotypification; however, this methodology is also time-consuming, and occasionally results are affected by inadequate samples, yielding low-quality reads (Sarkar et al., 2016a). In contrast, TaqMan® qPCR assays provide a high-throughput, rapid and straightforward method for genotyping analysis. When comparing the time required to obtain the result, the TaqMan® allelic discrimination qPCR assay is the most effective method, with results obtained in less than a day (Figure 5). Notably, the 100% agreement obtained between Sanger sequencing and TaqMan® assay demonstrates the power and reliability of the latter. This method is a powerful, cost-effective, and rapid screening tool for the genotypification of the CXCL16 gene. Furthermore, this process is similar to any other real-time PCR assay and can be performed in any molecular laboratory equipped with a real-time thermocycler. In this study, the assay was performed on a 96-well format, but it can easily be converted to a 384-well format for the high-throughput screening of horses.
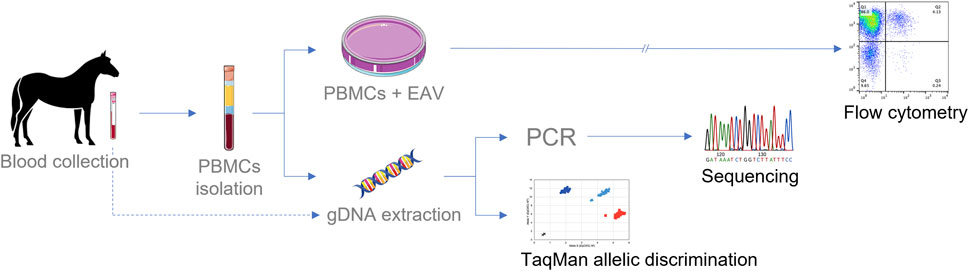
FIGURE 5. Schematic diagram for testing EAV susceptibility phenotype in CD3+ T lymphocytes and genotypification of CXCL16. Culturing PBMCs in the presence of EAV followed by flow cytometric analysis is the most extended method for phenotype determination (>36 h). Sanger sequencing of the CXCL16 gene requires an average of two to three working days for complete analysis. The TaqMan® allelic discrimination qPCR assay is the technique that provides the fastest results, with results in less than 24 h gDNA can be extracted from PBMCs or directly from whole blood.
To conclude, the TaqMan® allelic discrimination qPCR assay described in this study provides a new method for medium to high-throughput genotyping of the equine CXCL16 alleles with perfect agreement compared to Sanger sequencing allowing accurate identification of stallions at the most significant risk of becoming EAV carriers. Thus, it will assist with targeted vaccination practices with particular emphasis on stallions carrying the CXCL16S allele to prevent the occurrence of the carrier state. In addition, this test also opens avenues for selective breeding, which is critical for equine breeding enterprises worldwide and from a disease control perspective.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky, Lexington, KY (protocol number 2011-0888) and Louisiana State University, Baton Rouge, LA (protocol number 18–122). Written informed consent for participation was not obtained from the owners because this study was conducted as part of standard clinical practice and as such does not require informed consent.
Author Contributions
UB conceived the idea, contributed to study design, supervised the project, obtained extramural funding, and edited the manuscript. YYG initially identified and established the relationship between in vitro CD3+ T cell susceptibility to EAV infection and long-term persistent infection in the carrier stallion. CT, MC, and KG designed the allelic discrimination PCR. CT, YYG, UB, EB, JE, FA, and MK collected the samples and performed the experiments and sequencing. CT, YYG, EB, and JE analyzed the data. CT wrote the manuscript with input from MC, UB, and YYG.
Funding
This work is supported by the NIH-USDA NIFA R01 Research Grant Program Dual Purpose with Dual Benefit: Research in Biomedicine and Agriculture Using Agriculturally Important Domestic Animal Species grant number 2019-67016-29102 (award number AWD-47990-1) from the USDA National Institute of Food and Agriculture to UBRB. YYG is supported by City University of Hong Kong Intramural funding (APRC Project number 9610530).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank horse owners who kindly provided samples for the study. We also thank Edward Squires at the Maxwell H. Gluck Equine Research Center for coordinating the collection of blood samples from various farms. Finally, the authors are grateful to the University of Kentucky’s Maine Chance Farm staff and the Equine Health Studies Program (EHSP) staff at the School of Veterinary Medicine, Louisiana State University for their excellent care of the horses and collection of blood samples.
References
Balasuriya, U. B., and Carossino, M. (2017). Reproductive Effects of Arteriviruses: Equine Arteritis Virus and Porcine Reproductive and Respiratory Syndrome Virus Infections. Curr. Opin. Virol. 27, 57–70. doi:10.1016/j.coviro.2017.11.005
Balasuriya, U. B. R., Carossino, M., and Timoney, P. J. (2016). Equine Viral Arteritis: A Respiratory and Reproductive Disease of Significant Economic Importance to the Equine Industry. Equine Vet. Educ. 30, 497–512. doi:10.1111/eve.12672
Balasuriya, U. B. R. (2014). Equine Viral Arteritis. Vet. Clin. North America: Equine Pract. 30, 543–560. doi:10.1016/j.cveq.2014.08.011
Balasuriya, U. B. R., Go, Y. Y., and MacLachlan, N. J. (2013). Equine Arteritis Virus. Vet. Microbiol. 167, 93–122. doi:10.1016/j.vetmic.2013.06.015
Balasuriya, U. B. R., Snijder, E. J., Heidner, H. W., Zhang, J., Zevenhoven-Dobbe, J. C., Boone, J. D., et al. (2007). Development and Characterization of an Infectious cDNA Clone of the Virulent Bucyrus Strain of Equine Arteritis Virus. J. Gen. Virol. 88, 918–924. doi:10.1099/vir.0.82415-0
Balasuriya, U. B. R., Snijder, E. J., van Dinten, L. C., Heidner, H. W., Wilson, W. D., Hedges, J. F., et al. (1999). Equine Arteritis Virus Derived from an Infectious cDNA Clone Is Attenuated and Genetically Stable in Infected Stallions. Virology 260, 201–208. doi:10.1006/viro.1999.9817
Bowers, J. R., Engelthaler, D. M., Ginther, J. L., Pearson, T., Peacock, S. J., Tuanyok, A., et al. (2010). BurkDiff: A Real-Time PCR Allelic Discrimination Assay for Burkholderia Pseudomallei and B. Mallei. PLoS ONE 5, e15413. doi:10.1371/journal.pone.0015413
Brinton, M. A., Gulyaeva, A. A., Balasuriya, U. B. R., Dunowska, M., Faaberg, K. S., Goldberg, T., et al. (2021). ICTV Virus Taxonomy Profile: Arteriviridae 2021. J. Gen. Virol. 102. doi:10.1099/jgv.0.001632
Campos, J. R., Breheny, P., Araujo, R. R., Troedsson, M. H. T., Squires, E. L., Timoney, P. J., et al. (2014). Semen Quality of Stallions Challenged with the Kentucky 84 Strain of Equine Arteritis Virus. Theriogenology 82, 1068–1079. doi:10.1016/j.theriogenology.2014.07.004
Carossino, M., Dini, P., Kalbfleisch, T. S., Loynachan, A. T., Canisso, I. F., Cook, R. F., et al. (2019). Equine Arteritis Virus Long-Term Persistence Is Orchestrated by CD8+ T Lymphocyte Transcription Factors, Inhibitory Receptors, and the CXCL16/CXCR6 axis. Plos Pathog. 15, e1007950. doi:10.1371/journal.ppat.1007950
Carossino, M., Loynachan, A. T., Canisso, I. F., Cook, R. F., Campos, J. R., Nam, B., et al. (2017a). Equine Arteritis Virus Has Specific Tropism for Stromal Cells and CD8 + T and CD21 + B Lymphocytes but Not for Glandular Epithelium at the Primary Site of Persistent Infection in the Stallion Reproductive Tract. J. Virol. 91. doi:10.1128/JVI.00418-17
Carossino, M., Wagner, B., Loynachan, A. T., Cook, R. F., Canisso, I. F., Chelvarajan, L., et al. (2017b). Equine Arteritis Virus Elicits a Mucosal Antibody Response in the Reproductive Tract of Persistently Infected Stallions. Clin. Vaccin. Immunol 24. doi:10.1128/CVI.00215-17
Cavanagh, D. (1997). Nidovirales: a New Order Comprising Coronaviridae and Arteriviridae. Arch. Virol. 142, 629–633.
Doan, R., Cohen, N. D., Sawyer, J., Ghaffari, N., Johnson, C. D., and Dindot, S. V. (2012). Whole-Genome Sequencing and Genetic Variant Analysis of a Quarter Horse Mare. BMC Genomics 13, 78. doi:10.1186/1471-2164-13-78
Ehmer, U., Lankisch, T. O., Erichsen, T. J., Kalthoff, S., Freiberg, N., Wehmeier, M., et al. (2008). Rapid Allelic Discrimination by TaqMan PCR for the Detection of the Gilbert's Syndrome Marker UGT1A1*28. J. Mol. Diagn. 10, 549–552. doi:10.2353/jmoldx.2008.080036
Fedick, A., Su, J., Jalas, C., Northrop, L., Devkota, B., Ekstein, J., et al. (2013). High-Throughput Carrier Screening Using TaqMan Allelic Discrimination. PLoS ONE 8, e59722. doi:10.1371/journal.pone.0059722
Go, Y. Y., Bailey, E., Cook, D. G., Coleman, S. J., MacLeod, J. N., Chen, K.-C., et al. (2011). Genome-Wide Association Study Among Four Horse Breeds Identifies a Common Haplotype Associated with In Vitro CD3 + T Cell Susceptibility/Resistance to Equine Arteritis Virus Infection. J. Virol. 85, 13174–13184. doi:10.1128/JVI.06068-11
Go, Y. Y., Bailey, E., Timoney, P. J., Shuck, K. M., and Balasuriya, U. B. R. (2012a). Evidence that In Vitro Susceptibility of CD3 + T Lymphocytes to Equine Arteritis Virus Infection Reflects Genetic Predisposition of Naturally Infected Stallions to Become Carriers of the Virus. J. Virol. 86, 12407–12410. doi:10.1128/JVI.01698-12
Go, Y. Y., Cook, R. F., Fulgêncio, J. Q., Campos, J. R., Henney, P., Timoney, P. J., et al. (2012b). Assessment of Correlation between In Vitro CD3+ T Cell Susceptibility to EAV Infection and Clinical Outcome Following Experimental Infection. Vet. Microbiol. 157, 220–225. doi:10.1016/j.vetmic.2011.11.031
Go, Y. Y., Zhang, J., Timoney, P. J., Cook, R. F., Horohov, D. W., and Balasuriya, U. B. R. (2010). Complex Interactions between the Major and Minor Envelope Proteins of Equine Arteritis Virus Determine its Tropism for Equine CD3 + T Lymphocytes and CD14 + Monocytes. J. Virol. 84, 4898–4911. doi:10.1128/JVI.02743-09
Heissl, A., Arbeithuber, B., and Tiemann-Boege, I. (2017). High-Throughput Genotyping with TaqMan Allelic Discrimination and Allele-specific Genotyping Assays. Methods Mol. Biol. 1492, 29–57. doi:10.1007/978-1-4939-6442-0_3
Kamau, E., Alemayehu, S., Feghali, K. C., Tolbert, L. S., Ogutu, B., and Ockenhouse, C. F. (2012). Development of a TaqMan Allelic Discrimination Assay for Detection of Single Nucleotides Polymorphisms Associated with Anti-malarial Drug Resistance. Malar. J. 11, 23. doi:10.1186/1475-2875-11-23
Kuhn, J. H., Lauck, M., Bailey, A. L., Shchetinin, A. M., Vishnevskaya, T. V., Bào, Y., et al. (2016). Reorganization and Expansion of the Nidoviral Family Arteriviridae. Arch. Virol. 161, 755–768. doi:10.1007/s00705-015-2672-z
MacLachlan, N. J., Balasuriya, U. B. R., Rossitto, P. V., Hullinger, P. A., Patton, J. F., and Wilson, W. D. (1996). Fatal Experimental Equine Arteritis Virus Infection of a Pregnant Mare: Immunohistochemical Staining of Viral Antigens. J. Vet. Diagn. Invest. 8, 367–374. doi:10.1177/104063879600800316
Manga, I., and Dvořák, J. (2010). TaqMan Allelic Discrimination Assay for A1 and A2 Alleles of the Bovine CSN2 Gene. Czech J. Anim. Sci. 55, 307–312. doi:10.17221/89/2009-CJAS
McCollum, W. H., Timoney, P. J., and Tengelsen, L. A. (1995). Clinical, Virological and Serological Responses of Donkeys to Intranasal Inoculation with the KY-84 Strain of Equine Arteritis Virus. J. Comp. Pathol. 112, 207–211. doi:10.1016/s0021-9975(05)80062-3
Salehi, M., Mojaver, M., Mokarian, F., and Kazemi, M. (2015). Specific TaqMan Allelic Discrimination Assay for Rs1477196 and Rs9939609 Single Nucleotide Polymorphisms of FTO Gene Demonstrated that There Is No Association between These SNPs and Risk of Breast Cancer in Iranian Women. Adv. Biomed. Res. 4, 136. doi:10.4103/2277-9175.161532
Sarkar, S., Bailey, E., Go, Y. Y., Cook, R. F., Kalbfleisch, T., Eberth, J., et al. (2016a). Allelic Variation in CXCL16 Determines CD3+ T Lymphocyte Susceptibility to Equine Arteritis Virus Infection and Establishment of Long-Term Carrier State in the Stallion. Plos Genet. 12, e1006467. doi:10.1371/journal.pgen.1006467
Sarkar, S., Chelvarajan, L., Go, Y. Y., Cook, F., Artiushin, S., Mondal, S., et al. (2016b). Equine Arteritis Virus Uses Equine CXCL16 as an Entry Receptor. J. Virol. 90, 3366–3384. doi:10.1128/JVI.02455-15
Socha, W., Larska, M., and Rola, J. (2020). Molecular Investigation of Allelic Variants of EqCXCL16 Gene in Equine Arteritis Virus Infected Stallions of Selected Horse Breeds in Poland. Infect. Genet. Evol. 85, 104455. doi:10.1016/j.meegid.2020.104455
Timoney, P. J., and McCollum, W. H. (1993). Equine Viral Arteritis. Vet. Clin. North America: Equine Pract. 9, 295–309. doi:10.1016/s0749-0739(17)30397-8
Vaala, W. E., Hamir, A. N., Dubovi, E. J., Timoney, P. J., and Ruiz, B. (1992). Fatal, Congenitally Acquired Infection with Equine Arteritis Virus in a Neonatal Thoroughbred. Equine Vet. J. 24, 155–158. doi:10.1111/j.2042-3306.1992.tb02803.x
Vairo, S., Vandekerckhove, A., Steukers, L., Glorieux, S., Van den Broeck, W., and Nauwynck, H. (2012). Clinical and Virological Outcome of an Infection with the Belgian Equine Arteritis Virus Strain 08P178. Vet. Microbiol. 157, 333–344. doi:10.1016/j.vetmic.2012.01.014
Wagner, H. M., Balasuriya, U. B. R., and James MacLachlan, N. (2003). The Serologic Response of Horses to Equine Arteritis Virus as Determined by Competitive Enzyme-Linked Immunosorbent Assays (C-ELISAs) to Structural and Non-structural Viral Proteins. Comp. Immunol. Microbiol. Infect. Dis. 26, 251–260. doi:10.1016/S0147-9571(02)00054-1
Keywords: equine arteritis virus, EAV, equine viral arteritis, EVA, C-X-C motif chemokine ligand 16, CXCL16, allelic discrimination, genotyping
Citation: Thieulent CJ, Carossino M, Balasuriya UBR, Graves K, Bailey E, Eberth J, Canisso IF, Andrews FM, Keowen ML and Go YY (2022) Development of a TaqMan® Allelic Discrimination qPCR Assay for Rapid Detection of Equine CXCL16 Allelic Variants Associated With the Establishment of Long-Term Equine Arteritis Virus Carrier State in Stallions. Front. Genet. 13:871875. doi: 10.3389/fgene.2022.871875
Received: 08 February 2022; Accepted: 23 March 2022;
Published: 13 April 2022.
Edited by:
Falko Steinbach, University of Surrey, United KingdomReviewed by:
Tara G. McDaneld, U. S. Meat Animal Research Center, United StatesRussell S. Fraser, University of Prince Edward Island, Canada
Copyright © 2022 Thieulent, Carossino, Balasuriya, Graves, Bailey, Eberth, Canisso, Andrews, Keowen and Go. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Young Go, eXVueWdvQGNpdHl1LmVkdS5oaw==
 Come J. Thieulent
Come J. Thieulent Mariano Carossino1
Mariano Carossino1 Igor F. Canisso
Igor F. Canisso Frank M. Andrews
Frank M. Andrews Yun Young Go
Yun Young Go