A Corrigendum on
Development andvalidation of a prognostic nomogram based on DNAmethylation-driven genes for patients with ovarian cancer
by Zhou M, Hong S, Li B, Liu C, Hu M, Min J, Tang J and Hong L (2021). Front. Genet. 12:675197. doi: 10.3389/fgene.2021.675197
In the original article, there was a mistake in Table 1 as published. In Table 1, we erroneously displayed the sample number and clinical data (age, grade and stage) of GSE49997 (n = 193) dataset as the sample number and clinical data of GSE26712 (n = 185) dataset. In fact, no clinical data is provided for the GSE26712 dataset, which only provides survival data. The corrected Table 1 appears below.

TABLE 1. Clinicopathologic characteristics of ovarian cancer (OC) patients in The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) cohorts.
In the original article, there was an error. GSE26712 dataset contains 185 patients instead of 193.
A correction has been made to Dataset acquisition and pre-processing:
“The DNA methylation data of OC were downloaded from TCGA1 database. The mRNA expression profiles of normal ovarianand OC samples were downloaded from the GTEx and TCGA databases using the University of California Santa Cruz (UCSC) Xena browser (Chang et al., 2019). In addition, the microarray data of GSE9891 and GSE26712 were acquired from GEO2 to represent independent cohorts of OC. Patients without survival time or status were excluded from the study. To ensure that the established prognostic signature had better generalization, TCGA dataset was used as the training set, and GSE9891 and GSE26712 datasets were used as the validation set. Cases without a certain age, FIGO stage, and tumor grade were excluded. Finally, 358 OC patients were included in TCGA set, 273 patients in the GSE9891 set, and 185 patients in the GSE26712 set. Table 1 lists the clinical features of the patients in the training and validation sets.”
In the original article, there was a mistake in Figure 1A as published. GSE26712 dataset contains 185 patients instead of 193. The corrected Figure 1A appears below.
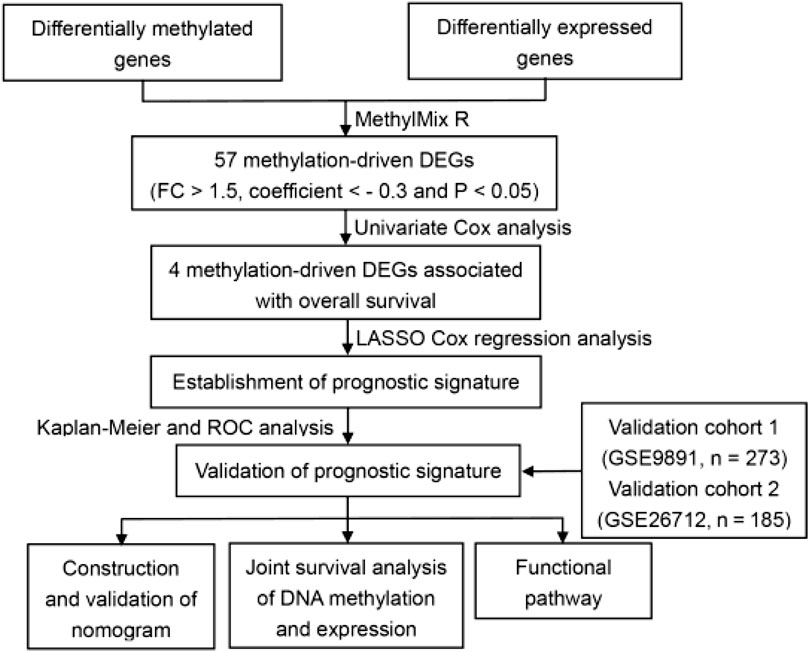
FIGURE 1. Identification of methylated related genes and flowchart of the establishment of novel prognostic signature. (A) The flowchart of the establishment of novel prognostic risk model for patients with ovarian cancer (OC). (B) The heatmap plot of 57 methylation related differentially expressed genes (DEGs) in OC. The color change from blue to red in the heatmap illustrates the trend from low to high methylation.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: ovarian cancer, methylation, CpG sites, model, overall survival, biomarkers
Citation: Zhou M, Hong S, Li B, Liu C, Hu M, Min J, Tang J and Hong L (2022) Corrigendum: Development and validation of a prognostic nomogram based on DNA methylation-driven genes for patients with ovarian cancer. Front. Genet. 13:951409. doi: 10.3389/fgene.2022.951409
Received: 24 May 2022; Accepted: 08 July 2022;
Published: 15 August 2022.
Edited and reviewed by:
Yongchun Zuo, Inner Mongolia University, ChinaCopyright © 2022 Zhou, Hong, Li, Liu, Hu, Min, Tang and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Hong, ZHJfaG9uZ2xpQHdodS5lZHUuY24=
†These authors have contributed equally to this work
 Min Zhou†
Min Zhou† Li Hong
Li Hong