- 1Department of Vegetable Science, College of Horticulture and Forestry, Punjab Agricultural University, Ludhiana, India
- 2School of Agricultural Biotechnology, College of Agriculture, Punjab Agricultural University, Ludhiana, India
Cucumber is an important vegetable crop that provides an accessible draft genome, which has significantly expedited research in various fields of molecular genetics. Cucumber breeders have been employing various methodologies to improve the yield and quality of the crop. These methodologies comprise enhancement of disease resistance, use of gynoecious sex types and their association with parthenocarpy, alterations in plant architecture, and enhancement of genetic variability. The genetics of sex expression are a complex trait in cucumbers but are very significant for the genetic improvement of cucumber crop. This review comprises an explanation of the current status of gene(s) involvement and its expression studies, the inheritance of genes, molecular markers, and genetic engineering associated with sex determination, as well as a discussion of the role of ethylene in sex expression and sex-determining genes of the ACS family. There is no doubt that gynoecy is an important trait among all sex forms of cucumber for heterosis breeding, but if it is associated with parthenocarpy, fruit yield can be enhanced to a greater extent under favorable conditions. However, little information is available with regard to parthenocarpy in gynoecious-type cucumber. This review sheds light on the genetics and molecular mapping of sex expression and could be beneficial especially to cucumber breeders and other scientists working on crop improvement via traditional and molecular assistant approaches.
1 Introduction
Cucumber (Cucumis sativus L.) belongs to the Cucurbitaceae family or vine family; the wild relative C. sativus var. hardwickii is believed to be its ancestor in India, and its highly diverse wild and cultivated forms are found in the Indian sub-continent (Sebastian et al., 2010). The Cucumis genus comprises 66 species, out of which cucumber alone possesses seven pairs of chromosomes with 2n = 14 and a genome size of 367 Mb; the rest have chromosomes as a multiple of 12 (2n = 24) (Weng, 2020).
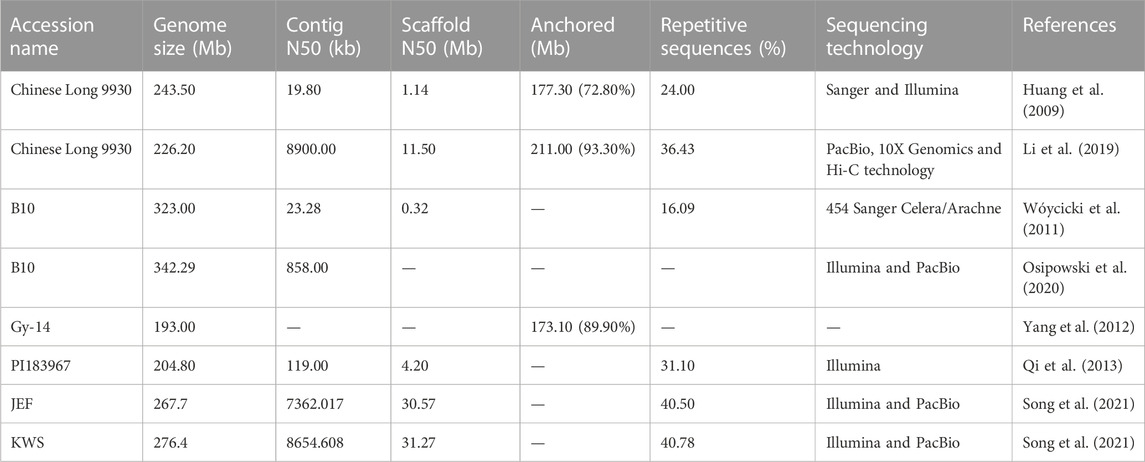
Cucumber was the first among the various horticultural crops to have a fully accessible draft genome (Huang et al., 2009; Wóycicki et al., 2011; Yang et al., 2012; Qi et al., 2013; Osipowski et al., 2020; Song et al., 2021), which significantly expedited research in crop improvement, breeding processes, and genome editing. The relatively smaller genomic size and its short life span (varying from 4 to 5 months) offer advantages for experiments pertaining to genetic studies in cucumber. The past decades have provided substantial progress in developing bioinformatics resources and facilitating the mapping and cloning of genes and quantitative trait loci (QTLs) of horticultural important traits of cucumber (Weng, 2020; Ma et al., 2022). To date, genomes have been sequenced for six accessions of cucumber, namely, Chinese Long 9930, Gy-14, B10, PI183967, JEF, and KWS (Table 1). The Chinese variety “Chinese Long 9930” (GenBank: GCA_000004075.2) was sequenced by Huang et al. (2009) (updated by Li et al., 2019). The American variety “Gy14” (http://wenglab.horticulture.wisc.edu/) was sequenced by the USDA-ARS (United States Department of Agriculture-Agricultural Research Unit) Vegetable Crops Research Unit, Wisconsin (Cavagnaro et al., 2010). The European Borszczagowski line “B10” (GenBank: GCA_000224045.1) was sequenced by the Polish consortium of cucumber genome sequencing (Wóycicki et al., 2011), and it was updated by Osipowski et al. (2020) with PacBio reads (GenBank: LKUO00000000.2). About 94% of the cucumber genome is covered by B10 and is considered the most comprehensive draft version to date (Osipowski et al., 2020). Recently, Turek et al. (2023) gave more insight into the genome of B10v3 accession by comparing it with the genome of cucumber lines Gy-14 and 9930. Recent advances in genome sequencing have provided stimulating opportunities to accelerate research in cucumber breeding.
2 Role of genes in sex determination
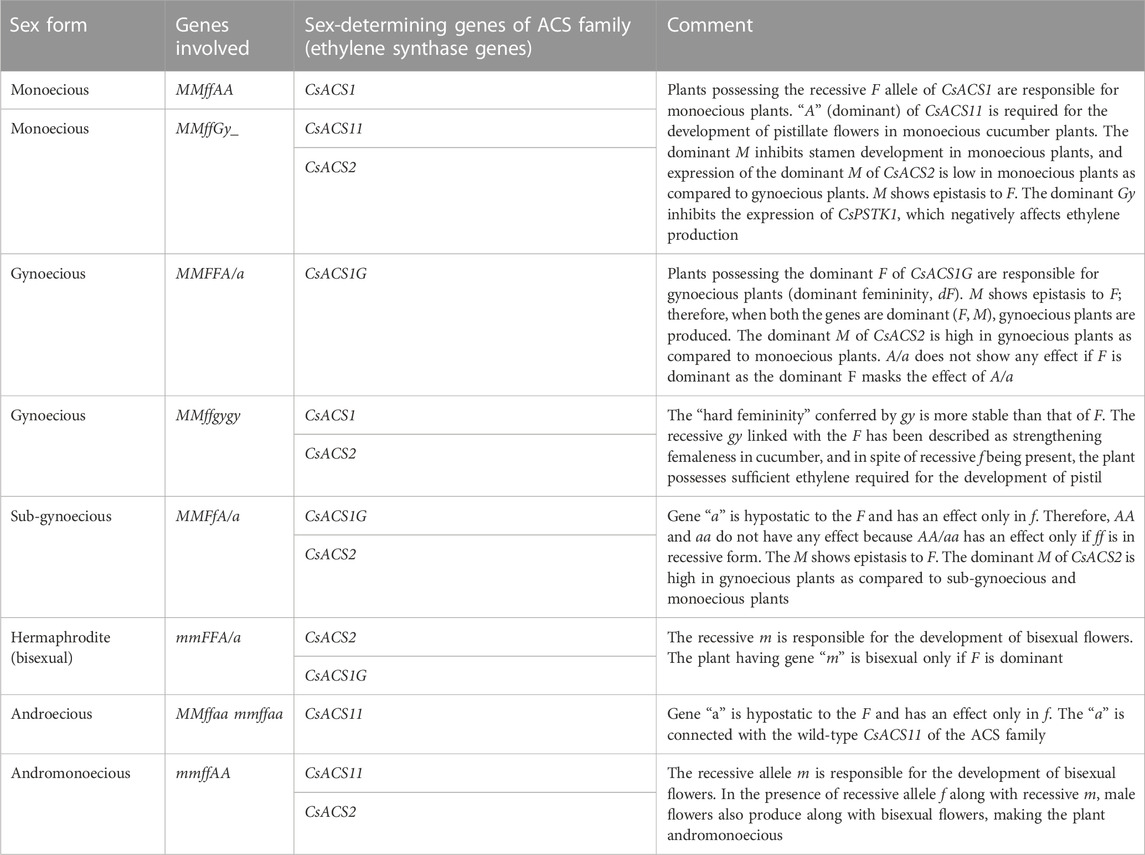
Cucumber plants can have three types of flowers: staminate/male, pistillate/female, and hermaphrodite/bisexual/perfect (Figure 1). Early studies described the role of three genes for sex determination in cucumber: F (femaleness), m (andromonoecy), and a (androecy), belonging to the aminocyclopropane-1-carboxylic acid synthase (ACS) gene family (CsACS1G, CsACS2, CsACS11, and CsACO2, corresponding to F, m, a, and a-1, respectively) (Kubicki, 1969; Pierce and Wehner, 1990; Trebitsh et al., 1997; Mibus and Tatlioglu, 2004; Li et al., 2009). Thus, the different sex expressions in a cucumber plant may be represented as monoecious (MMffAA), gynoecious (MMFFA/a), sub-gynoecious (MMFfA/a), andromonoecious (mmffAA), hermaphroditic (mmFFA/a), and androecious (MMffaa/mmffaa) (Li et al., 2020) (Table 2). The dominant F allele is responsible for the gynoecious plant, whereas the recessive F allele results in monoecious plants. Linkage of CsACS1 with the F locus was first reported by Trebitsh et al. (1987), who found that the monoecious (ff) cucumber plant possessed a single copy of CsACS1; however, an additional copy of CsACS1 (known as CsACS1G) was present in the gynoecious (FF) plant, which co-segregates with the F locus (Table 2). Moreover, the inhibition of CsACS1G leading to a monoecious nature confirmed the role of the F locus in sex determination (Mibus and Tatlioglu, 2004). Li et al. (2020) reported three genes in gynoecious plants: CsACS1, CsACS1G, and CsMYB. The length of the promoter sequence of CsACS1G is longer compared to the promoter sequence of CsACS1 (Yamasaki et al., 2000). Miao et al. (2011) developed a genetic map comprising microsatellite markers, which located F on chromosome 6. The recessive allele m is responsible for the development of bisexual flowers, and a pleiotropic gene action for the m allele has been observed as m alleles also resulted in a spherical fruit shape along with a bisexual flower (Li et al., 2009) (Figure 1). Yamasaki et al. (2001) described the epistatic nature of M with respect to F and that the plants with the mmff genotype are andromonoecious, mmFF has bisexual flowers, and MMFF has female flowers, whereas MMff plants are monoecious. The M locus is also known to be the duplicate of CsACS2 (Li et al., 2009; Table 2), and the level of CsACS2 may be regulated via ethylene produced by CsACS1 (Kamachi et al., 2000). CsACS2 is highly expressed in gynoecious plants as compared to monoecious plants (Yamasaki et al., 2003), and it accumulates in a few flowers of monoecious lines located at specific nodes (Wang et al., 2012). Gene “a” is responsible for androecy and is hypostatic to the F and has an effect in plants containing only “ff”. The “a” gene is connected with wild-type CsACS11 of the ACS family (Boualem et al., 2009). Plants with the genetic constitution of ffaa are entirely male. The gene was cloned from the rare androecious cucumber variety “EREZ” (Boualem et al., 2015). The recessive “gy” locus is due to the mutation in the WIP (CmWIP1–CsWIP1) gene (Martin et al., 2009; Boualem et al., 2015; Chen et al., 2016), and it resulted in “hard femininity,” which is more stable compared to the “F” gene. The “gy” locus is linked with the alleged serine/threonine kinase gene CsPSTK1, which is involved in ethylene biosynthesis/signaling and sex determination. In the presence of recessive gy, CsPSTK1 encourages ethylene signaling cascade, and regardless of the occurrence of the recessive “f” allele, the plant possesses a sufficient amount of ethylene required for pistil promotion. Such an amount of ethylene also induces the M gene, which inhibits stamen development. Thus, a cucumber plant with the ffMMGy genotype is monoecious, and the genotype ffMMgygy is gynoecious (Pawełkowicz et al., 2012). The CsWIP1 and F genes are located on chromosomes 4 and 6, respectively. The In-F (intensive female) gene was found to result in a high ratio of female flowers in monoecious plants (lacking F). Furthermore, a plant with both F and In-F genes failed to produce male flowers upon treatment with gibberellic acid (Kubicki, 1969). Kubicki (1974) used artificial mutagenesis to identify the hermaphrodite (h) allele, which produces bisexual flowers with normal ovaries resembling female flowers in their shape and pollination ability. tr (trimonoecious) controls the formation of all types of flowers, i.e., male, female, and bisexual; however, the bisexual flowers have superior ovaries (hypogynous), while female flowers are epigynous (inferior ovaries) (Kubicki, 1969). Cucumber sex determination is also regulated by other genes/QTLs and the application of growth regulators.
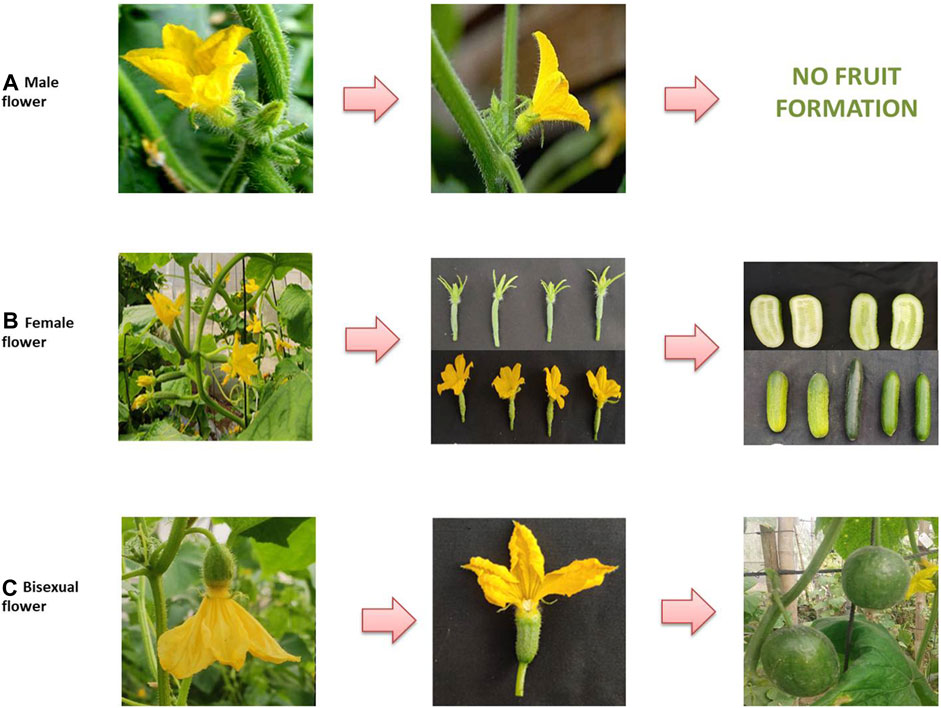
FIGURE 1. Types of flower sexes and their resulting fruit morphology in cucumber: (A) male flowers with the presence of stamens and thin pedicels resulting in no fruit formation; (B) female flower with the presence of pistils, ovaries, and thick pedicels resulting in cylindrical fruits; (C) bisexual flowers with the presence of both male and female organs and intermediate pedicels resulting in round fruits.
3 Inheritance of sex determination
The type and strength of sex expression are important traits to consider in the commercial production of cucumber as the disparities in this parameter can influence the date of harvest and crop yield.
3.1 Gynoecy
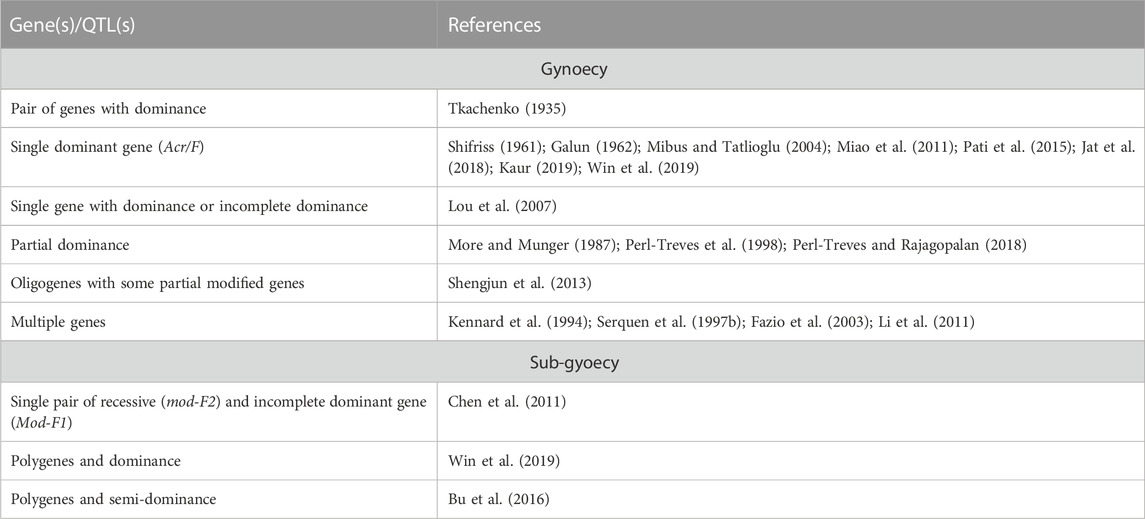
Gynoecy is an important genetic mechanism that is exploited for hybrid development. Due to their high yield, earliness, and more female flowers, gynoecy-based cucumber hybrids are achieving popularity among vegetable growers. The phenotypic recognition of the gynoecious trait at the early stages of breeding lines is challenging as the trait is under the influence of environmental factors. Thus, the identification of phenotypically pure gynoecious lines becomes difficult. Therefore, the inheritance of gynoecy needs to be efficiently studied for employing breeding procedures for transferring gynoecious genes into desirable cucumber genotypes. The inheritance of the gynoecious trait reported by different scientists is given in Table 3.
Tkachenko (1935) was the first to study the inheritance of gynoecious sex expression in the Japanese variety and reported that “femaleness” was regulated by a pair of genes and this character was dominant to the male character. Galun (1962) and Shifriss (1961) confirmed the monogenic inheritance by discovering the genetic factor Acr (later renamed as the F locus) responsible for speeding up the number of female flowers in gynoecious lines. The number of female flowers is directly related to the dominant allele (F) in a dosage-dependent manner (Shifriss, 1961; Shifriss et al., 1964). The cross of gynoecious × monoecious plants has been studied by various researchers to comprehend the inheritance of gynoecious sex expression, who observed that the gynoecious sex expression is ruled by various factors: the single dominant gene (Jat et al., 2018; Kaur, 2019; Mibus and Tatlioglu, 2004; Miao et al., 2011; Pati et al., 2015; Win et al., 2015), single gene with dominance or incomplete dominance (Lou et al., 2007), partial dominance (More and Munger, 1987; Perl-Treves et al., 1998; Perl-Treves and Rajagopalan, 2018), and oligogenes with modifications in some background genes (Shengjum et al., 2013). In contrast, Li et al. (2011) reported it to be regulated by multiple genes having quantitative inheritance, and they further added the significant influence of seasons on female flowering in diverse germplasms. The polygenic inheritance pattern was also reported by Fazio et al. (2003), Kennard et al. (1994), and Serquen et al. (1997a). Mibus and Tatlioglu (2004) used monoecious (MMff) and gynoecious (MMFF) cucumber lines for the molecular characterization of F/f and observed that the F/f locus is linked with CsACS1G, concluding that the dominant nature of the F allele impacts the amplification of CsACS1G in MMFF and MMFf.
3.2 Sub-gynoecy
The inheritance of the sub-gynoecious trait in cucumber was studied by different scientists, and their results are given in Table 3. Chen et al. (2011) used C. sativus L. var sativus cv. 97-17 and S-2-98 as parents to investigate the inheritance of sub-gynoecy in cucumber plants and observed that both the sub-gynoecious inbred lines were regulated by a single pair of recessive genes and also a single pair of incompletely dominant genes, which could enhance the intensity of femaleness. Moreover, they inherited independently with F and M and were labelled as mod-F2 and Mod-F1, respectively. Bu et al. (2016) indicated that sub-gynoecy is (semi-) dominant to the monoecy in the F1 generation. However, in the BC1 and F2 populations, continuous variation was observed that indicated the polygenic nature of the sub-gynoecious trait. Similarly, Win et al. (2019) reported the polygenic and dominant nature of the sub-gynoecious trait.
4 Molecular markers-assisted breeding for sex expression
4.1 Gynoecy
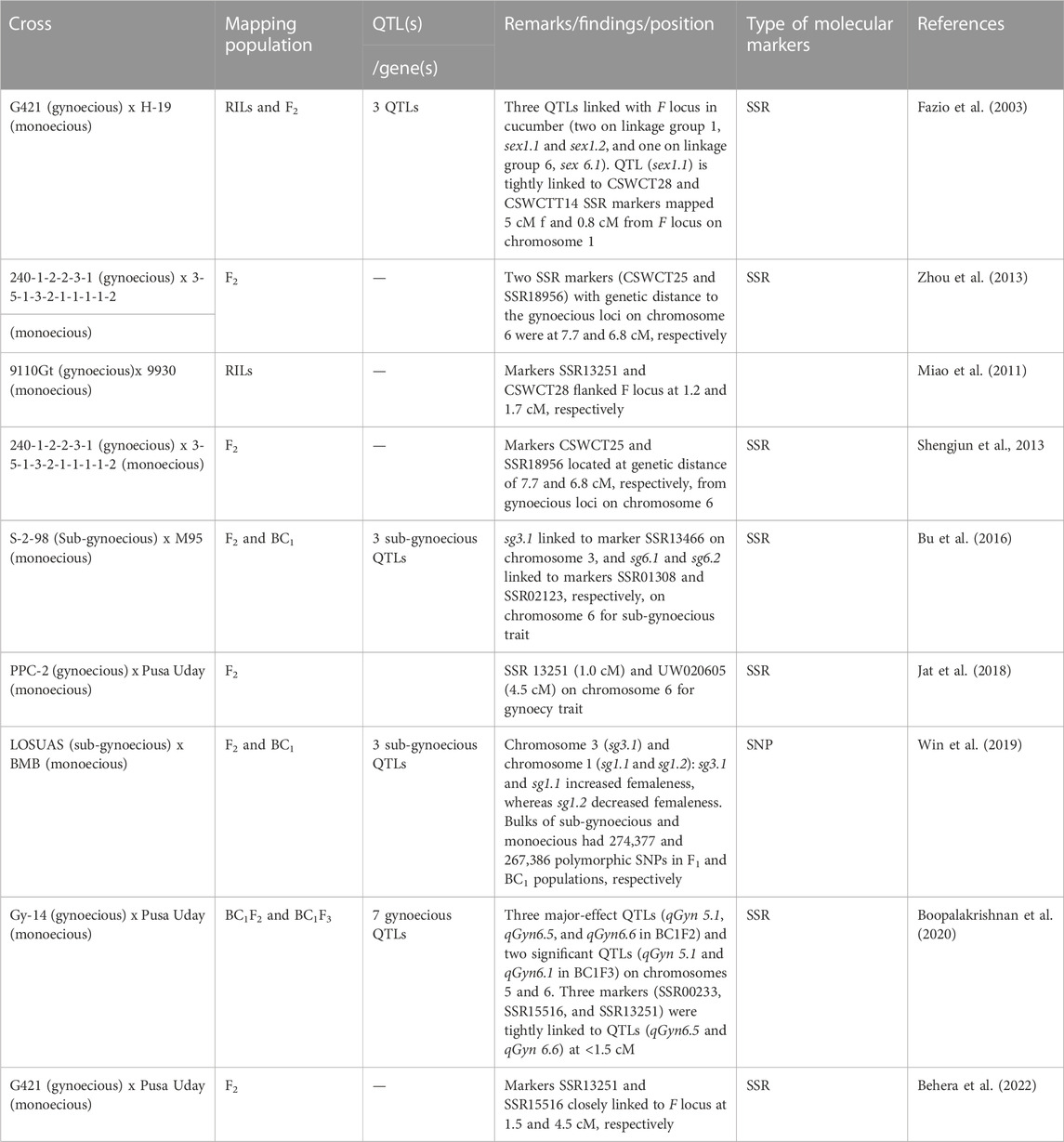
Cucumber’s short genome size makes it ideal for researchers to use molecular breeding techniques to develop cultivars (Table 2). The mapping of gynoecious trait in cucumber and the identification of markers linked to the gynoecious locus are important in marker-assisted backcross breeding for transferring the gynoecious trait to horticulturally desired cucumber genotypes to speed up the cucumber breeding program. Cucumber has a small genetic base and little polymorphism (Kennard et al., 1994). Even though 30 low-resolution linkage maps using RAPDs or AFLPs have been assembled, their dominating nature renders their use unsuitable in MAS (Yuan et al., 2008). However, while such molecular markers could help in identifying gynoecious lines, their dominant traits could not tell the difference between homozygous and heterozygous gynoecious lines. Because heterozygous gynoecious lines may have a less stable gynoecious phenotype than the homozygous lines, an updated molecular marker is still needed to overcome this obstacle. The adoption of DNA markers, particularly SSRs for genes/QTLs that favor mapping, was prompted by whole-genome sequencing of cucumber (Huang et al., 2009). The different molecular markers reported by various scientists for the gynoecious trait in cucumber are given in Table 4. Some polymorphic SSR markers connected to the cucumber gynoecious locus have been described (Zhou et al., 2013; Zhang et al., 2015; Zhu et al., 2016) (Table 5); however, their usefulness is still limited due to the greater genetic distances of the markers from the F locus. Fazio et al. (2003) discovered three QTLs linked with the F locus in cucumber (two on LG 1, sex1.1 and sex1.2, and one on LG 6, sex6.1) and found that the marker CSWCT28 is tightly linked to sex1.1 (LOD = 13.0), which is mapped 5 cM from the F locus on a distal end of linkage group 1. The F locus-linked primer pair P2S/P3A was reported to cover the whole precise genomic sequence of the gene CsACS1G (Knopf and Trebitsh, 2006). This primer pair spanned the recombination event between the CsACS1 and CsBCAT genes, amplifying the upstream of the CsACS1G gene (Wu et al., 2012). The breakpoint of the 30.2-kb duplicated region that designates the F locus is marked by the markers Primer F and Primer R (Zhang et al., 2015). Yuan et al. (2008) also identified three QTLs for female flower ratio, out of which two were found on chromosome 2 (sex2.1 and sex2.2) and one on chromosome 6 (sex6.1). Guo et al. (2010) sequenced a high number of ESTs from cucumber flower buds of two sex types and discovered differentially expressed genes and potential SSR and SNP markers in these two sex-type flowers. These EST sequences are a useful resource for future functional genomics studies, marker creation, and cucumber breeding. Bommesh et al. (2019) worked on the development and maintenance of gynoecious inbred lines from various cucumber hybrids and made selections in the F2 generation and carried them to the F4 population, which were validated phenotypically and genotypically through SSR markers. They also identified two polymorphic markers, SSR-02021 and SSR-18718, for the trait.
Miao et al. (2011) generated a linkage map with 148 recombinant inbred lines (RILs) and microsatellite markers derived from a cross between the two inbred lines 9110Gt and 9930. It was known that chromosome 6 harbors the gene of gynoecious sex expression (F). Furthermore, two markers flanking the F locus, CSWCT28 and SSR13251, were identified at locations 1.2 and 1.7 cM away from the F locus, respectively. They also revealed three significant QTLs for the node at which the first female flower appears in the RILs derived from monoecious (ff) x gynoecious (FF) cucumber lines. Chen et al. (2011) discovered two sub-gynoecious loci, mod-F2 and Mod-F1, which were regulated by a single pair of recessive genes and incompletely dominant genes, respectively. Zhang et al. (2015) reported a copy number variation (CNV) containing four genes that define the F locus and produce gynoecious cucumber plants with exclusively female flowers and fruit at practically every node. A recent 30.2-kb duplication at a meiotically unstable site resulted in the CNV, which was likely caused by micro-homology-mediated break-induced replication. Jat et al. (2018) investigated molecular mapping of the gynoecious (F) locus and discovered markers SSR13251 and UW020605, which were tightly linked to the F locus in cucumber, with stretch distances of 1.0 and 4.5 cM, respectively. Boopalakrishnan et al. (2020) detected seven gynoecious QTLs on chromosomes 5 and 6 in the BC1F2 population of cucumber, of which the three major-effect QTLs were qGyn 5.1, qGyn 6.5, and qGyn 6.6, and the markers SSR00233, SSR15516, and SSR13251 flanked the QTLs qGyn6.5 and qGyn6.6. In this way, understanding the genes that control floral traits could aid vegetable breeders in developing new, more stable, and climate-resilient gynoecious lines to make hybrid seed production easier.
4.2 Sub-gynoecy
A sub-gynoecy nature demonstrates a high level of female character, which might be useful as a substitute for a gynoecious nature under regular or limited development settings. The different molecular markers reported by various scientists for the sub-gynoecious trait in cucumber are given in Table 4. Bu et al. (2016) developed F2 and BC1 populations from S-2-98 (sub-gynoecious) and M95 (monoecious) to explore sub-gynoecy quantitatively and discovered three QTLs (sg3.1, sg6.1, and sg6.2) on chromosomes 3 and 6 with tightly linked markers SSR13466, SSR01308, and SSR02123, respectively, for the sub-gynoecious trait. The peak marker for sex6.1 given by Fazio et al. (2003) was located at a genetic distance of 63.8 cM from marker SSR02123 and was tightly linked to sg6.2 on chromosome 6. Another major-effect QTL (sg3.1) comprising a 799 kb genomic region accounted for 54.6% of phenotypic variation. Win et al. (2019) utilized sub-gynoecious “LOSUAS” and monoecious “BMB” and developed the F2 and BC1 populations to identify the quantitative nature of sub-gynoecy in cucumbers. They identified one major-effect QTL on chromosome 3 (sg3.1) and two minor QTLs on the short and long arms of chromosome 1 (sg1.1 and sg1.2). The major- and minor-effect QTLs were able to increase and decrease the degree of femaleness, respectively. A gene responsible for encoding GA20-oxidase was designated as candidate gene for the sgy3.1 locus.
Phenotypically, the sub-gynoecious nature, which is independent of the F gene but bears similarity to the one that is heterozygous at the F locus (Ff), usually initiates with male flowers development at the first few nodes (1–10) followed by continuous female flowers at later nodes. When QTL mapping for the percentage of female flowers was tested using populations derived from gynoecious (FF) × monoecious (ff) crosses, the major-effect QTL was found in accordance with the F locus (Fazio et al., 2003; Yuan et al., 2008). These observations suggested that multiple genetic factors are responsible for controlling the percentage of female flowers, although F and sgy3.1 played a pivotal role in gynoecious and sub-gynoecious plants, respectively.
5 Gene expression studies and transcriptome analysis
The plant transcriptome provides essential information about the gene expression level related to plant growth, cellular differentiation, and morphogenesis. The expression profiles of the genes vary temporally and spatially, which reflects the important role of the transcriptome in plant development. Various techniques used to study gene expression include transcriptome profiling, suppression subtractive hybridization (SSH), microarray analysis, cDNA-amplified fragment length polymorphism, and reverse transcription polymerase chain reaction (Wu et al., 2010). In relation to sex differentiation in cucumber, Wu et al. (2010) identified transcription factors including rf2 (translation releasing factor 2), PnLHY2, EREBP-9 (ethylene-responsive transcription factor), MYB, IF-2 (translation initiation factor), BHLH, BTF3, WAKY, DREB3, and TGA2, which were differentially expressed in gynoecious mutants and monoecious inbred lines. The spatial expression of EREBP-9 was observed specifically in the ovule at the development of the first phase. EREBP-9 transcription factors have a DNA-binding domain that binds to APETALA2 (Riechmann et al., 1998).
Various studies on transcriptome analysis have identified differentially expressed genes in gynoecious and monoecious cucumber lines. Transcriptome sequencing and comparative analysis of expressed sequence tags and mRNA sequences in gynoecious and hermaphrodite cucumber plants identified two zinc-finger transcription factors belonging to the CmWIP1 family that were highly expressed in gynoecious and hermaphrodite plants (Guo et al., 2010). CmWIP1 expression causes carpel abortion, which results in the development of unisexual male flowers (Martin et al., 2009). Moreover, a gene belonging to the BZR1-BES1 family (brassinosteroid signaling pathway gene family) was highly expressed in hermaphrodite flowers (Guo et al., 2010). Shoot tips of gynoecious cucumbers treated with gibberellic acid exhibited high expression of genes SAUR32, WRKY41, CYP450, MYB21, NAC2, NAC72, SAUR21, 1AA13, CKX1, and ETR1, as identified through transcriptome and qRT-PCR analysis (Zhang et al., 2017; Zhang et al., 2019).
Pan et al. (2018) performed the transcriptome analysis of apical shoots of near-isogenic lines of cucumber, which included gynoecious, monoecious, and hermaphrodite, and revealed that B-class floral homeotic genes CsPI and CsAP3 were highly expressed in monoecious lines but suppressed in gynoecious lines. Meanwhile, the C-class floral homeotic gene CsAG2 was highly expressed in gynoecious lines. They also reported the gene HECATE showing 45 times more expression in gynoecious lines as compared to monoecious and hermaphrodite lines. The gene is known to be involved in auxin-mediated gynoecy in Arabidopsis (Gremski et al., 2007). Other genes that were found to be highly expressed in gynoecious lines included CsIAA29, ETR1, and gene encoding histidine phosphotransfer protein. Pawelkowicz et al. (2019) identified the genes expressing in gynoecious and monoecious cucumbers and grouped them into various categories including transcription factors (APETALA, SEPALLATA, AGAMOUS, SEEDSTICK, HAT5, WUS, KNOX STM SHOOT MERISTEMLESS, and PISTILLATA), genes involved in processes of hormones (ETR, ERS, EIN, MTO1, CKX7, GAMYB1, ACS1G, and BRI1), metal ions (genes encoding metal oxides), cell wall synthesis, cytochromes (BCS1, BAS1, CYP75B1, CYP71B2, CYP716A1, and CYP78A10), sugar and lipid metabolism, and ubiquitination processes.
Sex determination in cucumber is also influenced by light as exposure of cucumber seedlings to red and blue light in the ratio 2:1 resulted in the development of female flowers (Song et al., 2018). The transcriptome analysis revealed that the genes involved in hormone signaling pathways constituted the majority of differentially expressed genes in seedlings treated with a higher proportion of red light. The development of female flowers in response to blue light was found to be regulated by the upregulation of genes involved in auxin and abscisic acid signal transduction pathways, photosynthesis, starch, and sucrose metabolism (Zhou et al., 2018). Wang et al. (2018) performed a transcriptome analysis of the monoecious cucumber cultivar “C09-123,” grown under conditions of low (26°C/12°C) and high (26°C/24°C) temperatures, to study their effect on sex differentiation and observed that male flower development was induced by high night temperatures governed by the genes AP3, PI, SEP1, and GASA (gibberellin-regulated family protein).
6 Genetic engineering for sex expression
Genetic engineering involves the cloning of a gene of interest and its delivery to the required plant to achieve a specific phenotype with stable expression. The technique has been successfully employed in cucumber to achieve desired mutants. In cucumbers, the genetic transformation efficiency ranges from 1% to about 23% (Feng et al., 2023). The foremost study of CRISPR/Cas9 gene editing in cucumber was carried out to develop transgenic-free cucumber plants with resistance against viruses (Chandrasekaran et al., 2016). However, little has been explored with regard to sex expression as the trait is highly complex and regulated by environmental factors. Recently, the optimization of CRISPR/Cas9 system has been achieved by employing a stronger CsU6 promoter and a GFP tag in the technique, which makes it possible to select between transformed and transgene-free mutants in the progeny (Feng et al., 2019). Transgene-free cucumber plants with editing in CsWIP1, CsVFB1, CsMLO8, and CsGAD1 have also been developed. The first study to report knocking off CsWIP1 to increase femaleness in cucumbers has laid the pathway to achieve a gynoecious character by using CRISPR/Cas9-mediated mutagenesis (Hu et al., 2017). After changing CmWIP1 (carpel development inhibitor) to CsWIP1, a significant increase in the formation of gynoecious inbred lines in cucumber was noticed, which might be used for hybrid seed production. Zhang et al. (2021) successfully introduced the complete genomic region of ACS1G into monoecious plants to transform them into gynoecious lines. These studies have helped to establish protocols for genetic engineering targeted at gynoecious sex expression in cucumber.
7 Conclusion
Cucumber was the foremost among the various horticultural crops to have a fully accessible draft genome in three varieties (Chinese Long 9930, Gy14, and B10) due to its small genome size (367 Mb) and short life span (varying from 4 to 5 months), which offered substantial progress in developing bioinformatics resources, facilitating the mapping and cloning of genes, and discovering QTLs of important horticultural traits in cucumber. The inheritance of the gynoecious sex expression has been studied by various researchers, who have reported that the gynoecious sex expression is governed by various factors: the single dominant gene, a single gene with dominance or incomplete dominance, partial dominance, oligogenes with some background genes modified, and polygenes. There is no doubt that gynoecious sex expression is genetically controlled, but environmental conditions such as longer days and higher temperatures induce male flower formation, whereas female flower development is aided by shorter days and lower temperatures. The interaction of genes and the environment has immediate consequences, such as the instability of gynoecious sex expression in cucumber. The mapping of gynoecy in cucumber and identification of markers linked to the gynoecious locus is important in marker-assisted backcross breeding for transferring the gynoecious trait to horticulturally desirable cucumber genotypes in order to speed up the cucumber breeding program. However, some SSR markers connected to the cucumber gynoecious locus have been developed; nonetheless, their usefulness is still limited due to the greater genetic distances from the markers to the F locus. Different scientists have reported the QTLs linked with the F locus on different chromosomes (1, 3, 5, 6), which creates confusion. Therefore, there is a need to identify the QTL(s) linked with the genes controlling the gynoecious trait. Moreover, genetic engineering can be further used to knock out the genes responsible for male flower induction in the monoecious lines and can help in the development of stable gynoecious lines.
Author contributions
RD wrote majority of the review article. HK wrote the section on transcriptomics, prepare figure and prepare the review according to journal’s format. PM edited the manuscript. E added few tables and some text in the review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Department of Science and Technology under the Science and Engineering Research Board-POWER grant (SPG/2021/000999).
Acknowledgments
The authors thank the director of the School of Agricultural Biotechnology for providing the infrastructural facilities to carry out this research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Behera, T. K., Boopalakrishnan, G., Jat, G. S., Munshi, A. D., Choudhary, H., Ravindran, A., et al. (2022). Deriving stable tropical gynoecious inbred lines of slicing cucumber from American pickling cucumber using MABB. Hortic. Environ. Biotechnol. 63, 273–274. doi:10.1007/s13580-021-00392-5
Bommesh, J. C., Pitchaimuthu, M., Ravishankar, K. V., and Varalakshmi, B. (2019). Development and maintenance of tropical gynoecious inbred lines in cucumber (Cucumis sativus) and validation by DNA markers. Agric. Res. 9, 161–168. doi:10.1007/s40003-019-00423-9
Boopalakrishnan, G., Jayavel, S., Behera, T. K., Munshi, A. D., Choudhary, H., Singh, A., et al. (2020). Mapping and linkage analysis of epistatic QTLs for gynoecious trait (F) in cucumber (Cucumis sativus L). J. Hortic. Sci. Biotechnol. doi:10.1080/14620316.2020.1855084
Boualem, A., Troadec, C., Camps, C., Lemhemdi, A., Morin, H., Sari, M. A., et al. (2015). A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Sci 350, 688–691. doi:10.1126/science.aac8370
Boualem, A., Troadec, C., Kovalski, I., Sari, M. A., Perl-Treves, R., and Bendahmane, A. (2009). A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS ONE 4 (7), 6144. doi:10.1371/journal.pone.0006144
Bu, F., Chen, H., Shi, Q., Zhou, Q., Gao, D., Zhang, Z., et al. (2016). A major quantitative trait locus conferring subgynoecy in cucumber. Theor. Appl. Genet. 129, 97–104. doi:10.1007/s00122-015-2612-z
Cavagnaro, P. F., Senalik, D. A., Yang, L., Simon, P. W., Harkins, T. T., Kodira, C. D., et al. (2010). Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L). BMC Genomics 11, 569. doi:10.1186/1471-2164-11-569
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17 (7), 1140–1153. doi:10.1111/mpp.12375
Chen, H., Sun, J., Li, S., Cui, Q., Zhang, H., Xin, F., et al. (2016). An ACC oxidase gene essential for cucumber carpel development. Mol. Plant. 9, 1315–1327. doi:10.1016/j.molp.2016.06.018
Chen, H., Tian, Y., Lu, X., and Liu, X. (2011). The inheritance of two novel subgynoecious genes in cucumber (Cucumis sativus L). Sci. Hortic. 127, 464–467. doi:10.1016/j.scienta.2010.11.004
Fazio, G., Staub, J. E., and Stevens, M. R. (2003). Genetic mapping and QTL analysis of horticultural traits in cucumber (Cucumis sativus L) using recombinant inbred lines. Theor. Appl. Genet. 107, 864–874. doi:10.1007/s00122-003-1277-1
Feng, J., Wang, N., Li, Y., Wang, H., Zhang, W., Wang, H., et al. (2023). Recent progress in genetic transformation and gene editing technology in cucurbit crops. Agron 13 (3), 755. doi:10.3390/agronomy13030755
Feng, S., Zhang, J., Mu, Z., Wang, Y., Wen, C., Wu, T., et al. (2019). Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. doi:10.1007/s00122-019-03484-0
Galun, E. (1962). Study of the inheritance of sex expression in the cucumber. The interaction of major genes with modifying genetic and non-genetic factors. Genetica 32, 134–163. doi:10.1007/BF01816091
Gremski, K., Ditta, G., and Yanofsy, M. F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Develop 134, 3593–3601. doi:10.1242/dev.011510
Guo, S., Zheng, Y., Joung, J. G., Liu, S., Zhang, Z., Crasta, O. R., et al. (2010). Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genom 11, 384. doi:10.1186/1471-2164-11-384
Hu, B., Li, D., Liu, X., Qi, J., Gao, D., Zhao, S., et al. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 10, 1575–1578. doi:10.1016/j.molp.2017.09.005
Huang, S., Li, R. Q., Zhang, Z. H., Li, L., Gu, X. F., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi:10.1038/ng.475
Jat, G. S., Munshi, A. D., Behera, T. K., Choudhary, H., Dash, P., Ravindran, A., et al. (2018). Genetics and molecular mapping of gynoecious (F) locus in cucumber (Cucumis sativus L). J. Hort. Sci. Biotechnol. 94, 24–32. doi:10.1080/14620316.2018.1449671
Kamachi, S., Mizusawa, H., Mazuura, S., and Sakai, S. (2000). Expression of two 1-aminocyclopropane-1-carboxylate synthase genes, CS-ACS1 and CS-ASC2, correlated with sex phenotypes in cucumber plants (Cucumis sativus L). Plant Biotechnol. 17, 69–74. doi:10.5511/PLANTBIOTECHNOLOGY.17.69
Kaur, T. (2019). Inheritance of gynoecium, yield and quality traits of cucumber (Cucumis sativus L). M.Sc. thesis, Punjab Agricultural University: Ludhiana, India.
Kennard, W. C., Poelter, K., and amd Staub, J. E. (1994). Linkages among RFLP, RAPD, isozyme, disease resistance, and morphological markers in narrow and wide crosses of cucumber. Theor. Appl. Genet. 819, 42–48. doi:10.1007/BF00226980
Knopf, R. R., and Trebitsh, T. (2006). The female-specific CS-ACS1G gene of cucumber.A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 47, 1217–1228. doi:10.1093/pcp/pcj092
Kubicki, B. (1969). Investigation of sex determination in cucumber (Cucumis sativusL). Genet. Pol. 10, 123–143.
Kubicki, B. (1974). New sex types in cucumber and their uses in breeding work. Proc. XIX Intl Hort. Congr., 475–485.
Li, J., Qin, Z., Zhou, X., Xin, M., and Wu, T. (2011). Genetic analysis of gynoecious in cucumber (Cucumis sativus L). Sci. Agric. Sin. 44, 3169–3176.
Li, Q., Li, H., Huang, W., Xu, Y., Zhou, S., Wang, S., et al. (2019). A chromosome-scale genome assembly of cucumber (Cucumis sativus L). GigaSci 8. doi:10.1093/gigascience/giz072
Li, Z., Han, Y., Niu, H., Wang, Y., Jiang, B., and Yiqun, W. (2020). Gynoecy instability in cucumber (Cucumis sativus L) is due to unequal crossover at the copy number variation-dependent Femaleness (F) locus. Hortic. Res. 7, 32. doi:10.1038/s41438-020-0251-2
Li, Z., Huang, S., Liu, S., Pan, J., Zhang, Z., Tao, Q., et al. (2009). Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genet 182, 1381–1385. doi:10.1534/genetics.109.104737
Li, Z., Wang, S., Tao, Q., Pan, J., Si, L., Gong, Z., et al. (2012). A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L). J. Exp. Bot. 63, 4475–4484. doi:10.1093/jxb/ers123
Lou, F. Q., Chen, F. J., Chen, Z. L., and Wolokau, N. J. (2007). Identification of an AFLP marker linked to a locus controlling gynoecy in cucumber and its conversion into SCAR marker, useful for plant breeding. Acta Hort. 763, 75–82. doi:10.17660/Acta.Hortic.2007.763.10
Ma, L., Wang, Q., Zheng, Y., Guo, J., Yuan, S., Fu, A., et al. (2022). Cucurbitaceae genome evolution, gene function, and molecular breeding. Hortic. Res. 9. uhab057. doi:10.1093/hr/uhab057
Martin, A., Troadec, C., Boualem, A., Rajab, M., Fernandez, R., Morin, H., et al. (2009). A transposon-induced epigenetic change leads to sex determination in melon. Nature 461, 1135–1138. doi:10.1038/nature08498
Miao, H., Zhang, S., Wang, X., Zhang, Z., Li, M., Mu, S., et al. (2011). A linkage map of cultivated cucumber (Cucumis sativus L) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182, 167–176. doi:10.1007/s10681-011-0410-5
Mibus, H., and Tatlioglu, C. T. (2004). Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L). Theor. Appl. Genet. 109, 1669–1676. doi:10.1007/s00122-004-1793-7
More, T. A., and Munger, H. A. (1987). Effect of temperature and photoperiod on gynoecious sex expression and stability in cucumber. Veg. Sci. 14, 42–50.
Osipowski, P., Pawełkowicz, M., Wojcieszek, M., Skarzynska, A., Pryszcz, L., and Plader, W. (2020). A high-quality cucumber genome assembly enhances computational comparative genomics. Mol. Genet. Genomics 295, 177–193. doi:10.1007/s00438-019-01614-3
Pan, J., Wang, G., Wen, H., Du, H., Lian, H., He, H., et al. (2018). Differential gene expression caused by the F and M loci provides insight into ethylene-mediated female flower differentiation in cucumber. Front. Plant Sci. doi:10.3389/fpls.2018.01091
Pati, K., DasMunshi, A., and Behera, T. K. (2015). Inheritance of gynoecism in cucumber (Cucumis sativus L) using genotype gbs-1 as gynoecious parent. Genetika 47 (1), 349–356. doi:10.2298/GENSR1501349P
Pawełkowicz, M., Osipowski, P., Wojcieszek, M., Woycicki, R., Witkowicz, J., Hincha, D., et al. (2012). Identification and characterization of genes connected with flower morphogenesis in cucumber. Biotechnol 93, 123–134. doi:10.5114/bta.2012.46577
Pawelkowicz, M., Pryszcz, L., Skarzynska, A., Woycicki, R. K., Posyniak, K., Rymuszka, J., et al. (2019). Comparative transcriptome analysis reveals new molecular pathways for cucumber genes related to sex determination. Plant Reprod. 32, 193–216. doi:10.1007/s00497-019-00362-z
Perl-Treves, R., Kahana, A., Rosenman, N., Xiang, Y., and Silberstein, L. (1998). Expression of multiple AGAMOUS-Like genes in male and female flowers of cucumber (Cucumis sativus L). Plant Cell Physiol. 39 (7), 701–710. doi:10.1093/oxfordjournals.pcp.a029424
Perl-Treves, R., and Rajagopalan, P. A. (2018). “Close yet separate: Patterns of male and female floral development in monecious species,” in Annual plant reviews volume 20: Flowering and its manipulation. Editor C. Ainsworth (Wiley Online Library), 117–146.
Pierce, L. K., and Wehner, T. C. (1990). Review of genes and linkage groups in cucumber. Hort. Sci. 25, 605–615. doi:10.21273/HORTSCI.25.6.605
Qi, J., Liu, X., Shen, D., Miao, H., Xie, B., Li, X., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45 (12), 1510–1515. doi:10.1038/ng.2801
Riechmann, J. L., and Meyerowitz, E. M. (1998) The AP2/EREBP family of plant transcription factors. J. Biol. Chem. 379, 633–646. doi:10.1515/bchm.1998.379.6.633Robinson
Sebastian, P., Schaefer, H., Telford, I. R. H., and Renner, S. (2010). Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. 107, 14269–14273. doi:10.1073/pnas.1005338107
Serquen, F. C., Bacher, J., and Staub, J. E. (1997a). Genetic analysis of yield components in cucumber (Cucumis sativus L) at low plant density. J. Am. Soc. Hort. Sci. 122, 522–528. doi:10.21273/JASHS.122.4.522
Serquen, F. C., Bacher, J., and Staub, J. E. (1997b). Mapping and QTL analysis of horticultural traits in a narrow cross in cucumber (Cucumis sativus L) using random-amplified polymorphic DNA markers. Mol. Breed. 3, 257–268. doi:10.1023/A:1009689002015
Shengjun, Z., Peng, Z., Yuquing, Z., Xinjuan, C., and Liping, C. (2013). Identification of SSR markers linked to gynoecious loci in cucumber (Cucumis sativus L). J. Zhejiang Univer. 39, 291–298. doi:10.3785/j.issn.1008-9209.2012.11.141
Shifriss, O., George, W. L., and Quinones, J. A. (1964). Gynodioecism in cucumbers. Genet 49, 285–291.
Shifriss, O. (1961). Sex control in cucumbers. J. Hered. 52, 5–12. doi:10.1093/oxfordjournals.jhered.a107021
Song, J., Zhang, Y., Song, S., Su, W., Chen, R., Sun, G., et al. (2018). Comparative RNA-Seq analysis on the regulation of cucumber sex differentiation under different ratios of blue and red light. Bot. Stud. 59, 21. doi:10.1186/s40529-018-0237-7
Song, K., Shin, Y., Jung, M., Subramaniyam, S., Lee, K. P., Oh, E. A., et al. (2021). Chromosome-scale genome assemblies of two Korean cucumber inbred lines. Front. Genet. 12. doi:10.3389/fgene.2021.733188
Tkachenko, N. N. (1935). Preliminary results of a genetic investigation of the cucumber,Cucumis sativusL. Bull. Appl. Plant Breed. 9, 311–356.
Trebitsh, T., Rudich, J., and Riov, J. (1987). Auxin, biosynthesis of ethylene and sex expression in cucumber (Cucumis sativus). Plant Growth Regul. 5, 105–113. doi:10.1007/BF00024738
Trebitsh, T., Staub, J. E., and O’Neill, S. D. (1997). Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the Female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 113, 987–995. doi:10.1104/pp.113.3.987
Turek, S., Plader, W., Hoshi, Y., Skarzynska, A., and Pawełkowicz, M. (2023). Insight into the organization of the B10v3 cucumber genome by integration of biological and bioinformatic data. Int. J. Mol. Sci. 24, 4011. doi:10.3390/ijms24044011
Wang, C., Xin, M., Zhou, X., Liu, W., Liu, D., and Qin, Z. (2018). Transcriptome profiling reveals candidate genes associated with sex differentiation induced by night temperature in cucumber. Sci. Hort. 232, 162–169. doi:10.1016/j.scienta.2017.12.018
Wang, Y. H., Behera, T. K., and Kole, C. (2012). Genetics, genomics and breeding of cucurbits. New York: CRC Press. doi:10.1201/b11436
Weng, Y. (2020). Cucumis sativus chromosome evolution, domestication, and genetic diversity. Plant Breed. Rev. 44, 79–111. doi:10.1002/9781119717003.ch4
Win, K. T., Zhang, C., Song, K., Lee, J. H., and Lee, S. (2015). Development and characterization of a co-dominant molecular marker via sequence analysis of a genomic region containing the Female (F) locus in cucumber (CucumissativusL.). Mol. Breed. 35, 229. doi:10.1007/s11032-015-0424-0
Win, K. T., Zhang, C., Silva, R. R., Lee, J. H., Kim, Y. C., and Lee, S. (2019). Identification of quantitative trait loci governing subgynoecy in cucumber. Theor. Appl. Genet. 132, 1505–1521. doi:10.1007/s00122-019-03295-3
Wóycicki, R., Witkowicz, J., Gawroński, P., Dąbrowska, J., Lomsadze, A., Pawełkowicz, M., et al. (2011). Thegenome sequence of the north-European cucumber (Cucumis sativus L) unravels evolutionary adaptation mechanisms in plants. PLoS ONE 6 (7), 22728. doi:10.1371/journal.pone.0022728
Wu, T., Qin, Z., Feng, Z., Zhou, X., Xin, M., and Du, Y. (2012). Functional analysis of the promoter of a female specific cucumber CsACS1G gene. Plant Mol. Biol. Rep. 30, 235–241. doi:10.1007/s11105-011-0318-1
Wu, T., Qin, Z., Zhou, W., Feng, Z., and Du, Y. (2010). Transcriptome profile analysis of floral sex determination in cucumber. J. Plant Physiol. 167, 905–913. doi:10.1016/j.jplph.2010.02.004
Yamasaki, S., Fujii, N., Matsuura, S., Mizusawa, H., and Takahashi, H. (2001). The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 42 (6), 608–619. doi:10.1093/pcp/pce076
Yamasaki, S., Fujii, N., and Takahashi, H. (2003). Photoperiodic regulation ofCS-ACS2, CS-ACS4 and CS-ERS gene expression contributes to the femaleness of cucumber flowers through diurnal ethylene production under short-day conditions. Plant Cell Environ. 26, 537–546. doi:10.1046/j.1365-3040.2003.00984.x
Yamasaki, S., Fujii, N., and Takahashi, H. (2000). The ethylene-regulated expression of CS-ETR2 and CS-ERS genes in cucumber plants and their possible involvement with sex expression in flowers. PlantCell Physiol. 41 (5), 608–616. doi:10.1093/pcp/41.5.608
Yang, L., Koo, D. H., Li, Y., Zhang, X., Luan, F., Havey, M. J., et al. (2012). Chromosome rearrangements during domestication of cucumber as revealed by high- density genetic mapping and draft genome assembly. Plant J. 71, 895–906. doi:10.1111/j.1365-313X.2012.05017.x
Yuan, X., Pan, J., Cai, R., Guan, Y., Liu, L., Zhang, W., et al. (2008). Genetic mapping and QTL analysis of fruit and flower related traits in cucumber (Cucumis sativus L) using recombinant inbred lines. Euphytica 164, 473–491. doi:10.1007/s10681-008-9722-5
Zhang, H., Li, S., Yang, L., Cai, G., Chen, H., Gao, D., et al. (2021). Gain-of-function of the 1-aminocyclopropane-1-carboxylate synthase gene ACS1G induces female flower development in cucumber gynoecy. Plant Cell 33 (2), 306–321. doi:10.1093/plcell/koaa018
Zhang, K. J., Song, H., Bo, K. L., Li, J., Ma, Z., Lou, Q. F., et al. (2015). QTL mapping of multi-locule-number trait in Xishuangbanna cucumber. Sci. Agri. Sin. 48, 3211–3220. doi:10.3864/j.issn.0578-1752.2015.16.011
Zhang, S., Miao, H., Song, Z., Liu, P., Wang, Y., Wehner, T. C., et al. (2017). Molecular mapping and candidate gene analysis for fruit epidermal structure in cucumber. Plant Breed. doi:10.1111/pbr.12510
Zhang, Y., Zhao, G., Li, Y., Mo, N., Zhang, J., and Liang, Y. (2019). Transcriptome analysis implies that GA regulates sex expression via ethylene-dependent and ethylene-independent pathways in cucumber (Cucumis sativus L). Front. Plant Sci. doi:10.3389/fpls.2017.00010
Zhou, S., Zhang, P., Zhu, Y., Chen, X., and Chen, L. (2013). Identification of SSR marker linked to gynoecious loci in cucumber (Cucumis sativus L). J. Zhejiang Univ. Agric. Life Sci. 39, 291–298. doi:10.3785/J.ISSN.1008-9209.2012.11.141
Zhou, Y., Ahammed, G. J., Wang, Q., Wu, C., Wan, C., and Yang, Y. (2018). Transcriptomic insights into the blue light-induced female floral sex expression in cucumber (Cucumis sativus L). Sci. Rep. 8, 14261. doi:10.1038/s41598-018-32632-7
Zhu, W. Y., Huang, L., Chen, L., Yang, J. T., Wu, J. N., Qu, M. L., et al. (2016). A high-density genetic linkage map for cucumber (Cucumis sativus L): Based on specific length amplified fragment (SLAF) sequencing and QTL analysis of fruit traits in cucumber. Front. Plant Sci. 19, 437. doi:10.3389/Ffpls.2016.00437
Keywords: gene expression, genes, gynoecy, inheritance, molecular markers
Citation: Dhall RK, Kaur H, Manchanda P and Eshanee (2023) Genetics and marker-assisted breeding for sex expression in cucumber. Front. Genet. 14:1180083. doi: 10.3389/fgene.2023.1180083
Received: 05 March 2023; Accepted: 18 May 2023;
Published: 01 June 2023.
Edited by:
Joanna Jankowicz-Cieslak, International Atomic Energy Agency, AustriaReviewed by:
Kailiang Bo, Chinese Academy of Agricultural Sciences, ChinaMagdalena Ewa Pawełkowicz, Warsaw University of Life Sciences, Poland
Copyright © 2023 Dhall, Kaur, Manchanda and Eshanee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: R. K. Dhall, cmFqaW5kZXJrdW1hckBwYXUuZWR1,
†ORCID: R. K. Dhall, orcid.org/0000-0003-3055-9780
 R. K. Dhall
R. K. Dhall Harleen Kaur
Harleen Kaur Pooja Manchanda
Pooja Manchanda Eshanee
Eshanee