- 1Department of Gynecology and Obstetrics, Key Laboratory of Obstetrics and Gynecologic and Pediatric Diseases and Birth Defects of Ministry of Education, Development and Related Diseases of Women and Children Key Laboratory of Sichuan Province, West China Second University Hospital, Sichuan University, Chengdu, Sichuan, China
- 2Clinical Trial Center, NMPA Key Laboratory for Clinical Research and Evaluation of Innovative Drugs, West China Hospital of Sichuan University, Chengdu, Sichuan, China
Ovarian clear cell carcinoma (OCCC) is a rare subtype of epithelial ovarian cancer with unique molecular characteristics, specific biological and clinical behavior, poor prognosis and high resistance to chemotherapy. Pushed by the development of genome-wide technologies, our knowledge about the molecular features of OCCC has been considerably advanced. Numerous studies are emerging as groundbreaking, and many of them are promising treatment strategies. In this article, we reviewed studies about the genomics and epigenetics of OCCC, including gene mutation, copy number variations, DNA methylation and histone modifications.
1 Introduction
Ovarian cancer is one of the most common malignancies in the female reproductive system, accounting for 2.5% of all malignancies among females (Torre et al., 2018). The prognosis of ovarian cancer is poor, and the 5-year survival rate is only 47.6%, largely driven by late-stage diagnoses (Eisenhauer, 2017). The histological and pathological type of ovarian cancer is an important factor in the prognosis of patients, as well as one of the major pieces of evidence for chemotherapy. Epithelial ovarian cancer (EOC) is the most common subtype of ovarian cancer, accounting for approximately 90% of ovarian cancers. According to the histological type, it is now widely accepted that EOC can be further subdivided into five main histological types (Lheureux et al., 2019). Among them, high-grade serous ovarian cancer (HGSOC) is the predominant subtype, accounting for 70%–80% of EOCs (Barnes et al., 2021). Other rarer subtypes include low-grade serous (<5%), endometrioid (10%), clear cell (6%), mucinous (6%) and other rare or undefined cancers (Lu and Chen, 2014).
Ovarian clear cell carcinoma (OCCC) was first named mesonephroma by Schiller in 1939 because it was thought to originate from the mesonephric gland, similar to kidney cancer (Saavedra and Sandow, 1968). Then, it was recognized as a distinct histologic subtype of ovarian cancer and termed ovarian clear cell carcinoma by the World Health Organization in 1973 (Korenaga et al., 2020). As the rare subtype of EOC, OCCC has unique histopathological characteristics and specific biological and clinical behaviors compared with other types of ovarian cancers (Kishikawa et al., 1995). OCCC is most frequently discovered in perimenopausal women and comprises approximately 5%–10% of ovarian cancers in North America and Europe (Torre et al., 2018). In many Asian countries, especially in Japan, the proportion is significantly higher at 15%–25%, probably due to genetic and environmental differences (Sung et al., 2014; Schnack et al., 2016). Most OCCC patients present with a unilateral large ovarian mass, which is more likely to be diagnosed early in the disease progression (57%–81% in stage I/II) and has a higher overall survival rate than contemporary HGSOCs (Anglesio et al., 2011). In contrast, OCCC is considered to have a drug-resistant phenotype, as the response rate to traditional paclitaxel plus carboplatin chemotherapy was lower than that of the other histologic types, ranging from 22% to 56% (Ho et al., 2004; Jin et al., 2014). Therefore, OCCC has a higher risk of relapse, and advanced OCCC (stage III/IV) has a poorer prognosis than contemporary HGSOC, with a 5-year survival rate of approximately 20% (Winter et al., 2007; del Carmen et al., 2012). Numerous studies have found that OCCC is closely associated with endometriosis and is thought to be a malignant transformation of benign ectopic endometrium on the ovary (Pearce et al., 2012; Gadducci et al., 2014; Suda et al., 2020; Shin et al., 2021). Patients with endometriosis have a ninefold increased risk of ovarian cancer (Kobayashi et al., 2007). In addition, studies have shown that tubal ligation can reduce the incidence of OCCC, which may result from preventing the formation of endometriosis lesions due to the countercurrent of menstrual blood (Kurian et al., 2005). On the other hand, the expansion of genome-wide technologies has revealed that the molecular characteristics of OCCC are distinctly different from those of other subtypes. As we known, HGSOC is recognized by near-ubiquitous TP53 (tumor protein 53) mutation and/or BRCA1/2 (breast cancer susceptibility gene 1/2) mutation, somatic and/or germline mutations in genes involved in genome-wide copy-number variation and homologous recombination repair (Kurman and Shih Ie, 2010; Cancer Genome Atlas Research, 2011; Huang et al., 2018; Barnes et al., 2021). However, existing studies have found that the pathogenesis of OCCC is similar to endometrial cancer, such as inactivation of tumor suppressor genes such as PTEN (phosphatase and tensin homologue) and ARID1A (AT-rich interactive domain 1A), activation of the PI3K/AKT/mTOR (phosphoinositide-3 kinase/protein kinase-B/mammalian) signal pathway, and high microsatellite instability (MSI) (Ueda et al., 2005; Gounaris et al., 2011; Xiao et al., 2012; Worley et al., 2015; Willis et al., 2017). With the development of next-generation sequencing, our knowledge about tumorigenesis and the mechanisms of tumor cell proliferation, immune avoidance and antitumor therapy has increased (Munoz-Galvan and Carnero, 2021). In this review, we summarized recent advances in the molecular characteristics of OCCC from the perspective of genomics and epigenomics (Figure 1).
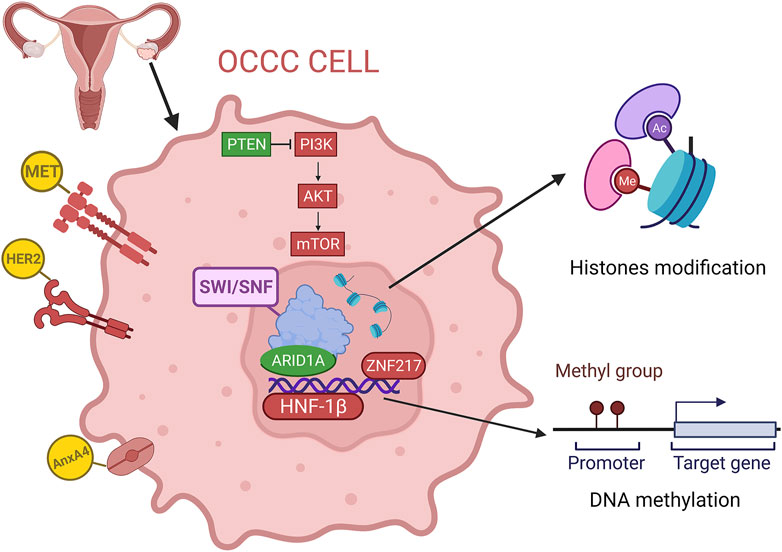
FIGURE 1. Overview of the genomics and epigenetics of OCCC, including gene mutation, copy number variations, DNA methylation and histone modification. (red for overexpression or amplification, green for deletion).
2 Genomics of ovarian clear cell carcinoma
Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are the two major approaches in genomic studies (Xiao et al., 2021). WGS or WES have been used to explore the genomic alterations involved in tumorigenesis that have been summarized in this review, such as gene mutations, copy number alterations and structural variants (Li et al., 2020; Zelli et al., 2020).
2.1 Loss of ARID1A and BAF250a expression
AT-rich interactive domain 1A (ARID1A), located on human chromosome 1p35.3, is a frequently mutated gene locus in many tumors, including OCCC, endometrial cancer, ovarian endometrioid cancer, gastric cancer, colorectal cancer and pancreatic cancer (Guan et al., 2011a; Wang et al., 2011; McConechy et al., 2012; Kadoch et al., 2013). ARID1A, encoding BRG1-associated factor 250a protein (BAF250a), is the most frequently mutated gene in OCCC, seen in 46%–57% of cases (Sato et al., 2018; Yano et al., 2019). It was reported that ARID1A mutations and loss of BAF250a expression were often evident in OCCC tumors and contiguous atypical endometriosis instead of distant endometriotic lesions (Wiegand et al., 2010). Furthermore, studies discovered that ARID1A mutations and loss of BAF250a protein accumulated in a stepwise manner during the transformation process from benign endometriosis through atypical endometriosis to OCCC, suggesting that ARID1A mutation occurred as a very early event in OCCC development (Yamamoto et al., 2012a; Xiao et al., 2012). Choi et al. classified OCCCs according to ARID1A expression status and found that ARID1A-mutated OCCCs exhibited distinct immunohistochemical features from the others, suggesting a different underlying molecular event during carcinogenesis (Choi et al., 2017). Additionally, ARID1A-mutated OCCC has shorter progression-free survival (PFS) than ARID1A-unmutated OCCC, but there was no significant difference in overall survival (OS) (Katagiri et al., 2012; Itamochi et al., 2015; Ye et al., 2016). However, Katagiri et al. (2012) revealed that among OCCC patients undergoing platinum-based chemotherapy, patients with ARID1A had shorter PFS and OS and were more resistant to chemotherapy. Moreover, Lyu et al. (2016) used siRNA to effectively silence the expression of ARID1A in the human OCCC cell line ES2 and found that cell apoptosis was decreased and its sensitivity to cisplatin chemotherapy was reduced. Both studies indicated that ARID1A loss in OCCC was a negative prognostic factor for platinum chemotherapy patients.
Mutations in ARID1A are often heterozygous non-sense or frameshift variants that are not enriched in hotspot sites (Jones et al., 2010; Wiegand et al., 2010). Studies discovered that both ARID1A alleles were affected through two mutations which were presumably biallelic or through either a mutation in one allele and loss of heterozygosity of the other allele. Thus, both homozygous and heterozygous mutations lack ARID1A protein expression (Caumanns et al., 2018a). BAF250a, encoded by ARID1A, has an N-terminus containing a LXXLL motif and a DNA binding domain consisting of 100 amino acids, which binds DNA sequences in a non-specific manner. Meanwhile, there are also three LXXLL motifs at the C-terminus, which constitute a glucocorticoid receptor binding domain that is involved in regulating the transcription and expression of several genes important in tissue repair, embryo development, cell cycle regulation, cell proliferation, aging, apoptosis and tumorigenesis (Nie et al., 2000) (Figure 2). The ARID1A protein has the ability to bind to proteins or DNA and regulate gene transcription and expression and has an important role in tissue repair, embryo development, cell cycle regulation, cell proliferation, aging, apoptosis and tumorigenesis (Nie et al., 2000; Jones et al., 2010; Nakonechny and Gilks, 2016). In addition, ARID1A protein is a key component of SWI/SNF (mating-type switching/sucrose non-fermenting), an ATP-dependent chromatin remodelling complex. This complex is necessary for the function of DNA repair proteins, such as p53, BRCA1, GADD45 (growth arrest and DNA damage gene 45) and Fanconi anemia protein, and therefore play an important role in DNA damage repair (Ribeiro-Silva et al., 2019). A subsequent study showed that the SWI/SNF complex is the most frequently mutated chromatin regulatory complex (CRC) in human cancer with a variety of mutation patterns, similar to TP53 (Kadoch et al., 2013). The SWI/SNF complex is also increasingly recognized as a tumor-inhibiting complex whose subunit mutations occur in a variety of malignancies, including non-small-cell lung cancer, bladder transitional cell carcinoma, pancreatic cancer and breast cancer (Gui et al., 2011; Zhu et al., 2015; Takao et al., 2017; Naito et al., 2019).
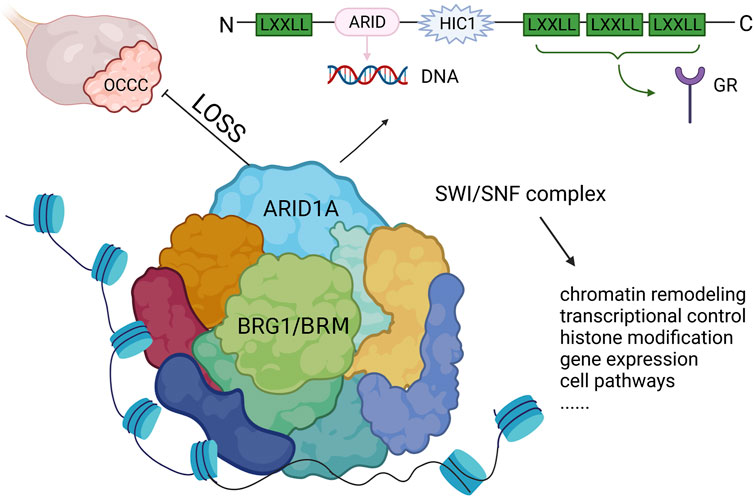
FIGURE 2. The structure of ARID1A and its role in OCCC. ARID1A mutation leads to the loss of expression and inhibits the function of SWI/SNF complex, which drives tumor malignancy. LXXLL, Leucine-x-x-leucine-leucine motif; ARID, AT-rich interactive domain; HIC1, hypermethylated in cancer 1 domain; GR, glucocorticoid receptor.
Studies have demonstrated that ARID1A mutations often coexist with PTEN or PIK3CA mutations in tumors, suggesting a synergistic role in tumorigenesis (Yamamoto et al., 2012a). Guan et al. found that loss of ARID1A alone is insufficient for tumor initiation, but additional genetic alterations were needed, such as PTEN deletion, to drive tumorigenesis (Guan et al., 2014). A recent study involving ten institutions across five countries revealed that ARID1A mutations were present almost exclusively in OCCC while TP53 mutations were in non-OCCC by examining genome-wide tumor methylation and gene expression (Cunningham et al., 2022). Another study also found that mutations in the ARID1A and TP53 genes were irrelevant in OCCC- and endometriosis-associated ovarian cancers. ARID1A mutation did not affect the binding rate of p53 protein to cyclin-dependent kinase inhibitor 1A (CDKN1A) gene and Sma and Mad-related gene (SMAD3) promoters, but both CDKN1A and SMAD3 protein expression were significantly decreased. At the same time, ARID1A could not induce the expression of CDKN1A and SMAD3 in cell lines with TP53 knockout or after RNA interference. Finally, evidence was obtained that the ARID1A/BRG1 complex interacted directly with p53, which synergistically regulated the transcription of CDKN1A and SMAD3 and negatively regulated the cell cycle. These results indicated that both ARID1A and TP53 were necessary to inhibit tumor growth, and mutations in either of them inactivated the complex and resulted in the loss of tumor suppression (Guan et al., 2011b). Zeng et al. discovered that overexpression of ARID1A in glioma cells reduced the expression of pAKT and pS6K and arrested the cell cycle in G1/S phase through the PI3K-AKT signaling pathway, leading to inhibition of cell proliferation and induction of apoptosis in glioma cells (Zeng et al., 2016). In addition, ARID1A can recruit the protein MSH2 (MutS Homolog 2) into chromatin during DNA replication, which plays an important role in mismatch repair. Loss of ARID1A is associated with mismatch repair defects, high mutant load, high PD-L1 expression, high MSI, and an increased number of tumor-infiltrating lymphocytes in many cancers (Caumanns et al., 2018a). Kurota et al. found that ARID1A mutation was associated with high PD-L1 in OCCC but not in other histologic subtypes of ovarian cancer (Kuroda et al., 2021). It was also reported that mice with ARID1A mutations were more sensitive to PD-L1 immune checkpoint blockade in ovarian and colorectal cancer models than wild-type mice, indicating that ARID1A deficiency could be coordinated with immune checkpoint blockade therapy (Shen et al., 2018). However, a small study on OCCC patients found higher expression of PD-L1 and more tumor-infiltrating lymphocytes in tumors with high MSI, but the relationship with ARID1A mutation has not been studied (Howitt et al., 2017). Nevertheless, high MSI is relatively low in OCCC (10%–14%); thus, the proportion of OCCC tumors with both high MSI and ARID1A mutations will be lower (Cai et al., 2004; Howitt et al., 2017). Therefore, exploring immunotherapy in ARID1A-mutated OCCC may be suitable for only a small number of patients. Interestingly, studies in recent years have found that ARID1A deficiency is an important factor for targeted therapies towards synthetic lethality in OCCC (Takahashi et al., 2021). HDAC2, HDAC6, BRD2 (bromodomain containing protein 2), CCNE1 (cyclin E1), and YES1 (v-YES-1 Yamaguchi sarcoma viral oncogene homolog 1) inhibitors were found to specifically cause the synthetic lethality of ARID1A mutant OCCC, providing potential options for the targeted therapy of OCCC (Miller et al., 2016; Caumanns et al., 2018a; Kawahara et al., 2021).
All the above results indicate that ARID1A is a potential tumor suppressor gene that is closely related to the tumorigenesis and chemotherapy resistance of OCCC. Therefore, new therapeutic strategies, such as ARID1A mutations, in OCCC might improve chemotherapy sensitivity, prolong survival, and even prevent malignancy from progressing from endometriosis to OCCC.
2.2 Amplifications and/or mutations of the PI3K/AKT/mTOR signaling pathway
The phosphoinositide-3 kinase/protein kinase-B/mammalian target of rapamycin (PI3K/AKT/mTOR) signaling pathway not only plays an important role in cell physiology but is also abnormally activated in a variety of human cancers, leading to cell proliferation, cell apoptosis inhibition, cell cycle acceleration, angiogenesis, and tumor metastasis (Jin et al., 2014). The PI3K/AKT/mTOR signaling pathway is mainly composed of PI3K, AKT and mTOR, which are frequently abnormal in OCCC, and any abnormality of the three can cause functional abnormalities of the PI3K/AKT/mTOR pathway (Caumanns et al., 2019). The PIK3CA gene (encoding the PI3K catalytic domain subunit) is most commonly altered in this pathway and is mutated in 30%–40% of OCCCs), whereas PTEN (antagonistic to PI3K) is not expressed in 40% of OCCCs (Hashiguchi et al., 2006; Kuo et al., 2009; Caumanns et al., 2018b). Other changes include deleterious mutations in PIK3R1 (the regulatory subunit of PI3K) and amplifications and/or mutations of the PI3K downstream signaling molecules AKT1, AKT2, and AKT3 (Friedlander et al., 2016; Itamochi et al., 2017; Murakami et al., 2017).
PI3K is a group of membrane-associated lipid kinases composed of three subunits: the p85 regulatory subunit, p55 regulatory subunit and p110 catalytic subunit (Donahue et al., 2012). PI3K is mutated or overexpressed in a variety of cancers, including ovarian, breast, prostate, gastric, colorectal, glioblastoma, endometrial and brain cancers (Elmenier et al., 2019; Kato et al., 2019). Mutations in the PIK3CA gene, which encodes the P110 α protein in PI3K, lead to overactivation of PI3K kinase activity. PIK3CA gene mutations were found in 30%–40% of OCCC patients, and PIK3CA gene mutation was also found in endometriosis lesions of these patients, indicating that the PI3K-mediated pathway played an important role in the tumorigenesis of OCCC and that PIK3CA gene mutation is one of the early events in endometriosis-related OCCC (Kuo et al., 2009; Yamamoto et al., 2011; Murakami et al., 2021). Studies have shown that PIK3CA mutation is often seen in early OCCC, and patients harboring activated PIK3CA have longer OS and better prognosis than those without PIK3CA activation, possibly due to more indolent biological properties (Abe et al., 2013; Zannoni et al., 2014). Furthermore, it was reported that coexistent ARID1A-PIK3CA mutations could promote tumorigenesis of OCCC through protumorigenic inflammatory cytokine signaling (Chandler et al., 2015). In addition, the regulatory genes PIK3R1 and PTEN were also inactivated in 40% of patients with PIK3CA activation. PIK3R1 encodes the p85 regulatory subunit in PI3K, and its mutation can lead to downregulation, which occurs in 7%–8% of OCCCs. PTEN is a coregulatory factor in PI3K activity, and its deletion coordinates with PIK3CA gene mutation in tumorigenesis. PTEN deletion has been found in OCCC, ovarian endometrioid carcinoma and endometriosis (Sato et al., 2000; Kinross et al., 2012).
AKT, also known as protein kinase B (PKB), is a 60 kDa serine/threonine protein kinase and can be divided into three subtypes (AKT1, AKT2, AKT3 or PKBα, PKBβ, PKBγ) involved in various cellular activities, such as cell survival, proliferation, invasion, apoptosis and angiogenesis (Shariati and Meric-Bernstam, 2019). As a downstream protein of PI3K, AKT is a core protein kinase in the signaling pathway that can activate and regulate numerous downstream targets. AKT is activated in many precancerous lesions and cancers, and mutations and amplifications of AKT suggest poor prognosis (LoPiccolo et al., 2007). Increased copy numbers of AKT2, located at chr19q13.2, were reported in 24% of OCCCs (Oda et al., 2018). A study found AKT2 amplification in OCCC with relatively shorter PFS by genomic analysis (Tan et al., 2011). Sasano et al. (2015) discovered that AKT was frequently activated in both early and late stages of OCCC, and the response to cyclophosphamide was more sensitive in cisplatin-resistant OCCCs whose AKT activation was also increased.
mTOR, a 289 kDa serine/threonine kinase, belongs to the PI3K-related protein kinase family and exists in the form of mTORC1 and mTORC2 complexes in vivo, which can regulate various basic cellular processes, including protein synthesis, cell growth, metabolism, senescence, regeneration and autophagy (Murugan, 2019). Numerous studies have found that mTOR is associated with a number of human diseases, such as cancers, neurodegenerative diseases and aging (Guo and Yu, 2019). Mabuchi et al. (2009) detected the expression of phosphorylated mTOR in 98 cases of primary ovarian cancer (52 cases of clear cell carcinoma and 46 cases of serous adenocarcinoma) by immunohistochemistry and showed that mTOR activation was higher in OCCC than serous ovarian cancer (86.6% vs. 50%) (Mabuchi et al., 2009). Another study also confirmed more activation of mTORC2 in OCCC than in serous ovarian cancer (Hisamatsu et al., 2013). In addition, studies have found that the level of phosphorylated mTOR in late OCCC was significantly higher than that in early OCCC (100% vs. 83.6%), indicating that the expression of phosphorylated mTOR in OCCC was significantly correlated with disease stage (Khemapech et al., 2012). However, the clinical trial of mTOR inhibitor temsirolimus has shown unsatisfactory effects in OCCC (Farley et al., 2016). Even so, another clinical trial of dual mTOR inhibitor TAK228 is still on-going (de la Rosa et al., 2022).
Activation of the PI3K/AKT/mTOR signaling pathway has become a research focus of OCCC and its chemotherapy resistance, and many signaling pathway-related inhibitors are in the stage of clinical trials. It is believed that with the deepening of studies on the regulatory mechanisms and effects of this pathway and the evolution of new inhibitors, targeted therapies through the PI3K/AKT/mTOR signaling pathway in OCCC will have far-reaching significance.
2.3 High-frequency amplification and overexpression of MET
Mesenchymal to epithelial transition factor (MET) is a proto-oncogene encoding MET kinase, which is a receptor of hepatocyte growth factor (HGF) on the cell surface (Cooper et al., 1984; Bottaro et al., 1991). MET kinase belongs to the receptor tyrosine kinase family and consists of three parts: extracellular parts (heterodimer binding α chain and ß chain by sulfide), transmembrane part (combination of the receptor signaling c-Cbl binding domain and structure domain near the membrane) and intracellular receptor tyrosine kinase domain (Eder et al., 2009). HGF binds to MET and regulates kinase activity by inducing homodimerization and phosphorylation of Y1234 and Y1235, two catalytic sites of tyrosine residues in the MET receptor. Phosphorylated carboxyl terminals containing Y1349 and Y1356 serve as docking sites for intracellular protein kinases, transmit signals downstream and activate the RAS-MAPK signaling pathway, PI3K/AKT/mTOR signaling pathway, JAK/STAT signaling pathway and other pathways to promote cell proliferation and survival (Longati et al., 1994; Furge et al., 2001; Hervieu and Kermorgant, 2018) (Figure 3). MET is expressed in epithelial cells of many organs during embryonic development and adulthood, including the liver, pancreas, prostate, kidney, muscle and bone marrow (Eder et al., 2009). However, MET is activated in tumor cells through gene amplification and overexpression, leading to a series of invasive growth processes in tumorigenesis, including cell proliferation, invasion and angiogenesis (Comoglio and Boccaccio, 2001; Boccaccio and Comoglio, 2006). Amplification of the MET gene has been found in various tumors, such as gastric, esophageal, lung and colorectal cancers (Michaels et al., 2018; Zhang et al., 2018; Seo et al., 2019; Gainor et al., 2020). The deletion of MET exon 14 skipping transcripts will lead to abnormal activation of downstream signaling pathways, which was not detected in OCCC yet. Compared with serous ovarian cancer and normal ovarian tissue, MET amplification and expression in OCCC were significantly increased (Kim et al., 2016). Yamashita et al. used real-time fluorescence quantitative PCR to analyze tumor tissues of 73 OCCC patients and found that 37.0% had MET gene amplification. The survival rate of stage I and II patients with MET gene amplification was significantly lower than that of patients without MET gene amplification. MET knockout with shRNA increased apoptosis and cell senescence, and the survival rate of OCCC cells with amplification of MET decreased (Yamashita et al., 2013). Wang et al. (2016) conducted immunohistochemistry (IHC) on the tissues of 86 OCCC patients and found that 94.2% of them had high MET expression, and the expression intensity of MET was closely related to chemotherapy resistance and poor prognosis. In addition, studies on endometriosis-related OCCC found that the incidence of MET overexpression increased with the tumorigenesis of OCCC: non-atypical precursor lesions (0%), atypical lesions (67%), relatively differentiated cancer components (92%), and poorly differentiated cancer components (100%), suggesting that accumulative MET gene copy number alterations causing MET overexpression are associated with higher tumor grade (Yamamoto et al., 2012b). These results demonstrated that MET overexpression was one of the early carcinogenesis events in MET-amplified OCCC and might promote the development of OCCC. Studies have found that dozens of MET inhibitors, such as INCB28060, PHA665752, HS10241, INC280, PF-2341066 and MK803, can inhibit ovarian cancer tumor growth and metastasis and reverse chemotherapy resistance (Zillhardt et al., 2010; Marchion et al., 2013; Moran-Jones et al., 2015; Li et al., 2016; Wang and Cheng, 2017; Han et al., 2019). The clinical trial NRG-GY001 demonstrated that the MET inhibitor cabozantinib had no clinical benefits for clear cell ovarian (Konstantinopoulos et al., 2018).
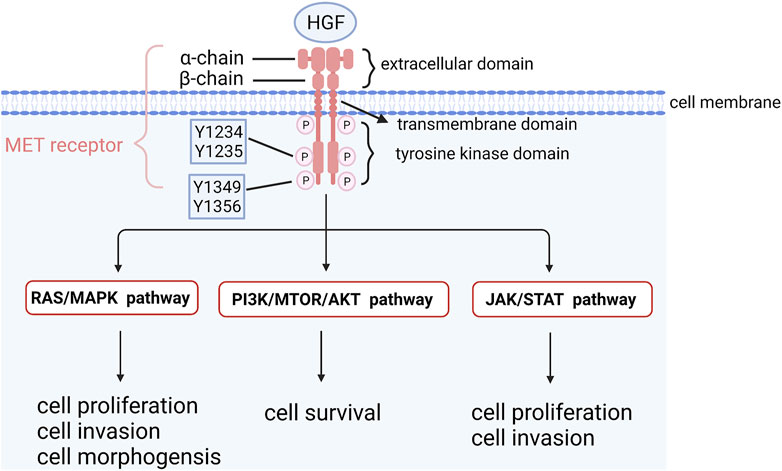
FIGURE 3. The structure of MET and its downstream signaling pathways. HGF binds to MET and regulates kinase activity by inducing homodimerization and phosphorylation of Y1234 and Y1235, two catalytic sites of tyrosine residues in MET receptor. Phosphorylated carboxyl terminal containing Y1349 and Y1356 serves as docking sites for intracellular protein kinases, transmits signals downstream and activates RAS-MAPK signaling pathway, PI3K/AKT/mTOR signaling pathway and other pathways to promote cell proliferation and survival.
In summary, the high frequency amplification and overexpression of MET in tumors and its high specificity for OCCC suggest that MET is a good candidate gene for OCCC targeted therapy. The emergence of a large number of MET-related inhibitors in recent years provides the possibility of drug screening for OCCC targeted therapy.
2.4 Amplification of HER2
Human epidermal growth factor receptor 2 (HER2) is also one of the current research focuses. Amplification of HER2 is closely related to chemotherapy resistance and poor prognosis of tumors. A study showed that HER2 positivity by ISH (in situ hybridization) was 16.7% in OCCC (Koopman et al., 2018). Tan et al. (2011) found that HER2 gene amplification and protein overexpression were observed in 14% of OCCCs, suggesting that HER2 may be a potential therapeutic target for OCCC. However, in another study, different levels of HER2 protein expression were detected by western blot in 10 OCCC cell lines, while HER2 protein expression was only detected in 5 cell lines by IHC, and HER2 gene amplification was only observed in RMG-1-cell lines with fluorescence in situ hybridization (FISH), indicating that HER2 targeted therapy might benefit only a few OCCC patients (Itamochi et al., 2007). Fujimura et al. (2002) discovered that HER2 was overexpressed in 42.9% OCCC by IHC and that three OCCC cell lines, RMG-1, HAC-II and KK, also showed positive staining for HER2, with expression levels 7.2, 6.4 and 4.5 times higher than those in normal mammary glands, respectively. Trastuzumab, as a human monoclonal antibody against HER2, significantly inhibited the growth of OCCC cell lines in vitro and RMG-1-transplanted tumor growth in vivo in a dose-dependent manner. The overall survival of trastuzumab-treated mice was also longer than that of the control group. A clinical trial on trastuzumab treating HER2-overexpressing ovarian and primary peritoneal cancers observed an overall response rate of only 7%. This might be the reason that the selected 41 patients (7 of whom were OCCC) were only tested for HER2 expression by IHC, and the gene copy number of HER2 was not evaluated (Bookman et al., 2003). Therefore, future studies on targeting HER2 for OCCC may need to evaluate both the amplification of the HER2 gene and protein overexpression level, as well as develop HER2 inhibitors with stronger targeting effects or conduct research on drug combination therapy.
2.5 Expression of HNF-1β
Hepatocyte nuclear factor-1β (HNF-1β) is a transcriptional activator that regulates gene promoters and enhancers and is expressed in the liver, digestive tract, pancreas and kidney. A study revealed that HNF-1β was the most abundantly upregulated transcription factor in OCCC, with an expression array study among OCCC and non-OCCC cell lines (Yamaguchi et al., 2010). Studies have shown that HNF-1β is an effective molecular marker for endometriosis and OCCC tumors and is overexpressed in 40% of endometriotic cysts without malignancy and in almost all cases of OCCC (Kato et al., 2006). Another study also showed that HNF1-β is overexpressed in OCCC and useful in cases of diagnostic uncertainty (82.5% sensitivity and 95.2% specificity for OCCC vs. HGSOC) (Kobel et al., 2009). However, the role of HNF-1β in OCCC and endometriosis remains uncertain. A recent study revealed the novel HNF-1β/GSK-3β/p-NF-κB pathway in OCCC, occurring in response to DNA damage (Kawahara et al., 2020). In addition, another study found that HNF-1β interacted with the NF-κB signaling pathway, increasing cell survival by altering apoptotic events, especially by upregulating Bcl-2 expression in OCCC via a mitochondria-mediated pathway (Suzuki et al., 2015). Furthermore, it was found that RNA interference could be used to reduce HNF-1β expression, which may lead to OCCC cell apoptosis (Kobayashi et al., 2009). The results suggest that HNF-1β overexpression is involved in inhibiting apoptosis in OCCC cells. However, no drugs targeting HNF-1β have been developed, and further studies on the regulatory mechanism of HNF-1β in OCCC are needed.
2.6 Overexpression of AnxA4
Annexin A4 (AnxA4), an important member of the annexin superfamily, binds to calcium ions and phospholipids simultaneously to regulate cell membrane permeability and cell membrane transport, mediating cell growth and apoptosis, and participating in tumor proliferation, invasion and metastasis, angiogenesis and chemotherapy resistance (Yao et al., 2016). Wang et al. (2017) found that AnxA4 contained a Lewis Y structure, which could modify and enhance its interaction with NF-κB p50 and promote the malignant progression of OCCC. Studies have shown that AnxA4 is overexpressed in OCCC and induces chemotherapy resistance by enhancing drug efflux (Kim et al., 2009; Kim et al., 2010). Another study confirmed that AnxA4 promoted platinum-drug resistance through ATP7A, a copper and platinum transporter (Matsuzaki et al., 2014). Furthermore, it was reported that the repeats of the calcium binding site in the AnxA4 protein could induce resistance to platinum drugs by increasing the intracellular chloride concentration (Morimoto et al., 2014). In conclusion, AnxA4 is a potential therapeutic target for OCCC. However, no drug has been proven to effectively inhibit AnxA4 expression. Therefore, the development of an effective AnxA4 inhibitor in combination with platinum therapy may be a new treatment for OCCC.
2.7 Amplification of ZNF217
The zinc finger protein 217 (ZNF217) gene, in the amplicon on chromosome 20q13.2, encodes a Krupple-like transcription factor and is a candidate oncogene expressed in 20%–30% of primary human breast cancers, with overexpression correlating with poor prognosis (Vendrell et al., 2012). ZNF217 overexpression can inhibit signaling events and promote tumor metastasis and chemotherapy resistance (Littlepage et al., 2012). ZNF217 gene amplification was reported in 31% of OCCC cases, and protein overexpression was reported in 40% of OCCC cases (Huang et al., 2014). Rahman et al. demonstrated that ZNF217 gene amplification was significantly associated with lymph node metastasis of OCCC. Compared with non-ZNF217-amplified cells, ZNF217-amplified cells showed stronger inhibition of cell migration and invasion after treatment with siRNA, suggesting that ZNF217 might be a new therapeutic target for OCCC (Rahman et al., 2012).
2.8 Other genomic alterations in ovarian clear cell carcinoma
In addition to the mutated genes listed above, some other frequently mutated genes in OCCC were also identified by WES or targeted multiple gene panel testing, including ARID1B (AT-rich interactive domain 1B), KRAS (V-Ki-ras2 Kirsten ratsarcoma viral oncogene homolog), PPP2R1A (protein phosphatase 2 regulatory subunit A, alpha, encoding serine/threonine protein phosphatase 2 scaffold subunit alpha), SMARCA4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4, encoding ATP-dependent chromatic modeller BRG1), MLL3 (mixed lineage leukemia 3), ERBB2 (V-Erb-B2 avian erythroblastic leukemia viral oncogene homolog 2, also known as HER2), TFAP2A (transcription factor AP-2 alpha), TP53 and CTNNB1 (catenin beta 1, encoding betacatenin) (Tan et al., 2011; Arildsen et al., 2017; Itamochi et al., 2017; Maru et al., 2017; Murakami et al., 2017; Kim et al., 2018; Shibuya et al., 2018). Regarding the profiles of chromosomal CNVs in OCCC, although focal CNVs at specific gene loci were less frequent in OCCC than in HGSOC, recurrent CNVs were identified at various loci (Sung et al., 2013; Okamoto et al., 2015; Uehara et al., 2015; Mabuchi et al., 2016). In addition to MET, AKT2, and ZNF217, which are listed above, it was reported that amplification of chromosome 8 (8p11.21-q11.23 and 8q22.1-q24.13) was detected in 52% of OCCCs (Uehara et al., 2015), as well as copy number loss (loss of heterozygosity or homozygous deletion) at the loci of CDKN2A/2B (Cyclin-Dependent Kinase Inhibitor 2A/2B) (9p21.3) in 17% of OCCCs (Kuo et al., 2010). Other CNVs identified include amplification of chromosome 8q (64%), 17q (46%) and 20q (54%), and deletion of 9q (21%), 13q (28%), 18q (21%), and 19p (41%) by WES (Murakami et al., 2017). Furthermore, in addition to HNF-1β, multiple microarray datasets revealed additional significantly upregulated genes in OCCC, including VCAN, HIF-1α, IL-6, p21, STAT3 and other genes related to oxidative stress and coagulation (Yamaguchi et al., 2010; Diaz et al., 2013; Matsuo et al., 2015).
3 Epigenomics of ovarian clear cell carcinoma
Epigenomics is the study of epigenetic modifications at the genome level. Epigenetic modifications act on DNA and histones in cells, including DNA methylation, DNA hydroxymethylation, posttranslational modifications of histones (acetylation, methylation, etc.) and chromatin remodelling. Epigenetic alterations in the genome have important effects on physiological processes and tumorigenesis.
3.1 Abnormal DNA methylation
DNA methylation is an epigenetic mechanism of gene regulation and plays an important role in various biological processes, such as X-chromosome inactivation, genomic imprinting, aging, and cancer (Houshdaran et al., 2010). Abnormal DNA methylation, either global DNA hypomethylation or aberrant hypermethylation of tumor suppressor genes, contributes to tumorigenesis (Gargiulo and Minucci, 2009). A variety of studies have identified that the OCCC methylation profile is distinct from the other histologic subtypes of ovarian cancer (Yamaguchi et al., 2014a; Yamaguchi et al., 2014b; Engqvist et al., 2020). Yamaguchi et al. collected genome-wide methylation and gene expression data for 14 OCCC, 32 non-OCCC and 4 non-cancerous cell lines and found inverse relationships between gene expression and methylation, including 22 hypomethylated genes and 276 hypermethylated genes, suggesting functional regulation by methylation. Furthermore, categorical and pathway analyses indicated that the hypomethylated genes were involved in the response to stress and the HNF-1β pathway and had increased expression, such as HNF-1α, HNF-1β, PAX8 (paired box gene 8) and SGK2 (serum and glucocorticoid kinase 2), while the hypermethylated genes included members of the estrogen receptor α (ERα) network genes involved in tumor development and had decreased expression (Yamaguchi et al., 2014b). Another study discovered that the low expression levels of SLIT2 in OCCC were the result of promoter hypermethylation and were associated with tumor migration (Lin et al., 2021). It was reported that MLH1 promoter hypermethylation could cause mismatch repair deficiency (MMRD) and MSI, leading to tumorigenesis in endometriosis-associated OCCC (Leskela et al., 2020). Additionally, methylation levels at the RASSF2A promoter were found to be obviously higher in endometriosis-associated OCCC than in ovarian endometriosis cyst and normal endometrial tissues (Xia et al., 2018). Senthong et al. (2014) indicated that hypomethylation of long interspersed element-1 (LINE-1) was an early molecular event participating in OCCC malignant transformation. Methylation of HIN-1 is also associated with chemotherapy resistance and poor outcomes in OCCC (Ho et al., 2015). Several other genes have also been reported to be aberrantly methylated in OCCC, including 14-3-3σ, TMS1/ASC (TMS1 (Target of Methylation induced Silencing), also known as ASC (Apoptosis Speck like protein containing a CARD)), WT1 (Wilms’ tumor 1), RASSF1A (ras association domain family 1A gene), CDH13 (cadherin 13), CACNA1A (calcium voltage-gated channel subunit alpha1 A), KCNH2 (the K (+) voltage-gated channel subfamily H member 2, also known as the human ether-a-go-go-related gene or hERG), THBS2 (thrombospondin 2), DLEC1 (deleted in lung and esophageal cancer 1), and SFRP5 (secreted frizzled-related protein 5) (Terasawa et al., 2004; Asadollahi et al., 2010; Ho et al., 2012; Cicek et al., 2013; Kelemen et al., 2013; Chen et al., 2015). In short, abnormal DNA methylation regulates specific pathways and biological functions in OCCC, influences the characteristic biology and contributes to tumorigenesis.
3.2 Modification of histones
Histones bind to DNA to form chromatin in eukaryotes, and modification of histones, including methylation, acetylation, small ubiquitin-like modifications, phosphorylation, ubiquitination and so on, can regulate gene expression by histone modification enzymes, which play an important role in tumorigenesis (Esteller, 2007; Audia and Campbell, 2016). It was reported that ARID1A suppressed histone deacetylase 6 (HDAC6) in OCCC directly, and HDAC6 nuclear expression was associated with immuno- and hypoxia tolerance and cancer stem cell phenotype in OCCC and upregulated following chemotherapy, leading to a poor prognosis (Yano et al., 2018; Yano et al., 2019). Additionally, HDAC6 inhibition selectively was found to promote apoptosis of ARID1A-inactivated cells, suggesting the use of HDAC6 inhibition in the treatment of OCCC (Altucci, 2017). Expression of HDAC 1 and 7 was also identified with a poor prognosis in OCCC (Yano et al., 2018). Approximately half of OCCC had mutations in the SWI/SNF chromatin remodelling complex subunit ARID1A, and cross-cancer analysis of the TCGA database showed that lysine-specific histone demethylase 1 (LSD1), which regulated the chromatin landscape and gene expression by demethylating proteins such as histone H3, was highly expressed in SWI/SNF-mutated tumors. Furthermore, the reversible LSD1 inhibitor SP-2577 has the ability to promote antitumor immunity and T cell infiltration in OCCC cell lines (Soldi et al., 2020). Several studies have demonstrated synthetic lethality by targeting EZH2 (enhancer of Zeste homolog 2) histone methyltransferase activity in ARID1A-mutated OCCC (Bitler et al., 2015). The results of another study suggested that the histone methyltransferase SET and MYND domain containing 2 (SMYD2) could promote OCCC viability and inhibit SMYD2-induced apoptosis in OCCC cells (Kojima et al., 2020). Wolf-Hirschhorn syndrome candidate gene-1 (WHSC1), a histone methyltransferase, has also been found to be upregulated in OCCC, leading to cell growth induction (Kojima et al., 2019). At present, there are relatively few studies on histone modification in OCCC. As the research goes further, novel molecular targeted approaches for OCCC treatment may be represented.
3.3 Chromatin remodelling complex
Chromatin remodelling complexes use the energy of ATP to move nucleosomes in the genome and play a crucial role in controlling chromatin structure and regulating gene transcription. According to their structure and function, chromatin remodelling complexes can be divided into four major groups: SWI/SNF, CHD (Chromodomain-Helicase DNA binding protein family), ISWI (Imitation SWItch family), and INO80 (INOsitol-requiring 80 family) (Clapier and Cairns, 2009). Except for ARID1A mutation leading to the inactivation of SWI/SNF, which we discussed above, it was reported that BRG1 phosphorylation inhibited the functions of the SWI/SNF complex in chromatin activation and thus derived tumor malignancy, thereby promoting the expression of various cancer-related proteins (Kimura et al., 2021). Furthermore, two other genes involved in chromatin remodelling, KMT2D (the lysine (K)-specific methyltransferase 2D gene) and SPOP (Speckle-type POZ protein), may also contribute to the tumorigenesis of OCCC (Arildsen et al., 2017). SPOP was reported to indirectly affect heterochromatin maintenance through ubiquitination of the DAXX protein in kidney cancer (Li et al., 2014). Meanwhile, the protein encoded by KMT2D is a histone methyltransferase that methylates the Lys-4 position of histone H3 and is part of a large protein complex called ASCOM (activating signal cointegrator-2 complex). ASCOM is recognized as a physiologically relevant coactivator for p53 and is involved in the p53 tumor suppression pathway, which may be significant for OCCC tumorigenesis (Lee et al., 2009). Importantly, mutations of ARID1A, SPOP and KMT2D were mutually exclusive (Arildsen et al., 2017).
4 Conclusion
As mentioned above, we summarized the gene alterations and associated clinical trials in ovarian clear cell carcinoma [Table 1, (Bookman et al., 2003; Farley et al., 2016; Miller et al., 2016; Alldredge and Eskander, 2017; Konstantinopoulos et al., 2018; de la Rosa et al., 2022)]. In summary, OCCC has distinct genomic and epigenomic profiles compared to other histologic types of EOC. With the help of clinical sequencing in OCCC, numerous types of genomic and epigenomic alterations have been identified, which may shed light on novel therapeutic options in OCCC. Although OCCC is a rare tumor in gynecological cancers, the molecular targets are similar to those of tumors in other organs, especially renal clear cell carcinoma. Thus, we hope our review will contribute to the development of precise medicine against OCCC.
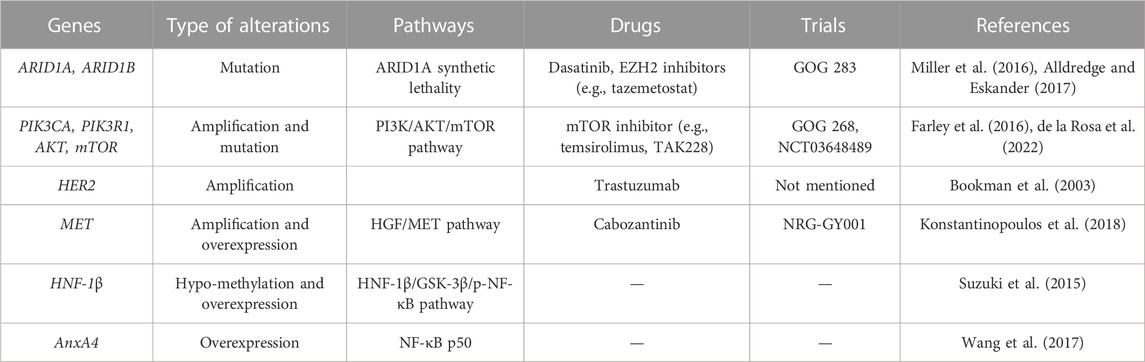
TABLE 1. Summary of the gene alterations and associated clinical trials in ovarian clear cell carcinoma.
Author contributions
Conceptualization, AT and XL; writing, review, and editing, AT and XD; supervision, XL and XZ; funding acquisition, XZ and XL All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Si Chuan Province, grant number No. 2022NSFSC1531; the National Natural Science Foundation of China, grant number No. 81821002; Sichuan Science and Technology Program, grant number 2021YJ0011; Foundation of Development and Related Diseases of Women and Children Key Laboratory of Sichuan Province, grant number No. 2022003; Foundation of Development and Related Diseases of Women and Children Key Laboratory of Sichuan Province, grant number No. 2023004. The APC was funded by West China Second University Hospital.
Acknowledgments
Figures in this review were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, A., Minaguchi, T., Ochi, H., Onuki, M., Okada, S., Matsumoto, K., et al. (2013). PIK3CA overexpression is a possible prognostic factor for favorable survival in ovarian clear cell carcinoma. Hum. Pathol. 44 (2), 199–207. doi:10.1016/j.humpath.2012.05.005
Alldredge, J. K., and Eskander, R. N. (2017). EZH2 inhibition in ARID1A mutated clear cell and endometrioid ovarian and endometrioid endometrial cancers. Gynecol. Oncol. Res. Pract. 4, 17. doi:10.1186/s40661-017-0052-y
Altucci, L. (2017). A key HDAC6 dependency of ARID1A-mutated ovarian cancer. Nat. Cell Biol. 19 (8), 889–890. doi:10.1038/ncb3588
Anglesio, M. S., Carey, M. S., Kobel, M., Mackay, H., and Huntsman, D. G.Vancouver Ovarian Clear Cell Symposium Speakers (2011). Clear cell carcinoma of the ovary: A report from the first ovarian clear cell symposium, june 24th, 2010. Gynecol. Oncol. 121 (2), 407–415. doi:10.1016/j.ygyno.2011.01.005
Arildsen, N. S., Jonsson, J. M., Bartuma, K., Ebbesson, A., Westbom-Fremer, S., Masback, A., et al. (2017). Involvement of chromatin remodeling genes and the rho GTPases RhoB and CDC42 in ovarian clear cell carcinoma. Front. Oncol. 7, 109. doi:10.3389/fonc.2017.00109
Asadollahi, R., Hyde, C. A., and Zhong, X. Y. (2010). Epigenetics of ovarian cancer: From the lab to the clinic. Gynecol. Oncol. 118 (1), 81–87. doi:10.1016/j.ygyno.2010.03.015
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8 (4), a019521. doi:10.1101/cshperspect.a019521
Barnes, B. M., Nelson, L., Tighe, A., Burghel, G. J., Lin, I. H., Desai, S., et al. (2021). Distinct transcriptional programs stratify ovarian cancer cell lines into the five major histological subtypes. Genome Med. 13 (1), 140. doi:10.1186/s13073-021-00952-5
Bitler, B. G., Fatkhutdinov, N., and Zhang, R. (2015). Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin. Ther. Targets 19 (11), 1419–1422. doi:10.1517/14728222.2015.1062879
Boccaccio, C., and Comoglio, P. M. (2006). Invasive growth: A MET-driven genetic programme for cancer and stem cells. Nat. Rev. Cancer 6 (8), 637–645. doi:10.1038/nrc1912
Bookman, M. A., Darcy, K. M., Clarke-Pearson, D., Boothby, R. A., and Horowitz, I. R. (2003). Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: A phase II trial of the gynecologic Oncology group. J. Clin. Oncol. 21 (2), 283–290. doi:10.1200/JCO.2003.10.104
Bottaro, D. P., Rubin, J. S., Faletto, D. L., Chan, A. M., Kmiecik, T. E., Vande Woude, G. F., et al. (1991). Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251 (4995), 802–804. doi:10.1126/science.1846706
Cai, K. Q., Albarracin, C., Rosen, D., Zhong, R., Zheng, W., Luthra, R., et al. (2004). Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum. Pathol. 35 (5), 552–559. doi:10.1016/j.humpath.2003.12.009
Cancer Genome Atlas Research, N. (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474 (7353), 609–615. doi:10.1038/nature10166
Caumanns, J. J., Berns, K., Wisman, G. B. A., Fehrmann, R. S. N., Tomar, T., Klip, H., et al. (2018). Integrative kinome profiling identifies mTORC1/2 inhibition as treatment strategy in ovarian clear cell carcinoma. Clin. Cancer Res. 24 (16), 3928–3940. doi:10.1158/1078-0432.CCR-17-3060
Caumanns, J. J., van Wijngaarden, A., Kol, A., Meersma, G. J., Jalving, M., Bernards, R., et al. (2019). Low-dose triple drug combination targeting the PI3K/AKT/mTOR pathway and the MAPK pathway is an effective approach in ovarian clear cell carcinoma. Cancer Lett. 461, 102–111. doi:10.1016/j.canlet.2019.07.004
Caumanns, J. J., Wisman, G. B. A., Berns, K., van der Zee, A. G. J., and de Jong, S. (2018). ARID1A mutant ovarian clear cell carcinoma: A clear target for synthetic lethal strategies. Biochim. Biophys. Acta Rev. Cancer 1870 (2), 176–184. doi:10.1016/j.bbcan.2018.07.005
Chandler, R. L., Damrauer, J. S., Raab, J. R., Schisler, J. C., Wilkerson, M. D., Didion, J. P., et al. (2015). Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 6, 6118. doi:10.1038/ncomms7118
Chen, C. A., Chiang, Y. C., Chang, M. C., Hu, Y. H., You, S. L., Cheng, Y. Y. K., et al. (2015). Gene methylation profiles as prognostic markers in ovarian clear cell and endometrioid adenocarcinomas. Am. J. Transl. Res. 7 (1), 139–152.
Choi, J. Y., Han, H. H., Kim, Y. T., Lee, J. H., Kim, B. G., Kang, S., et al. (2017). Ovarian clear cell carcinoma sub-typing by ARID1A expression. Yonsei Med. J. 58 (1), 59–66. doi:10.3349/ymj.2017.58.1.59
Cicek, M. S., Koestler, D. C., Fridley, B. L., Kalli, K. R., Armasu, S. M., Larson, M. C., et al. (2013). Epigenome-wide ovarian cancer analysis identifies a methylation profile differentiating clear-cell histology with epigenetic silencing of the HERG K+ channel. Hum. Mol. Genet. 22 (15), 3038–3047. doi:10.1093/hmg/ddt160
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi:10.1146/annurev.biochem.77.062706.153223
Comoglio, P. M., and Boccaccio, C. (2001). Scatter factors and invasive growth. Semin. Cancer Biol. 11 (2), 153–165. doi:10.1006/scbi.2000.0366
Cooper, C. S., Park, M., Blair, D. G., Tainsky, M. A., Huebner, K., Croce, C. M., et al. (1984). Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 311 (5981), 29–33. doi:10.1038/311029a0
Cunningham, J. M., Winham, S. J., Wang, C., Weiglt, B., Fu, Z., Armasu, S. M., et al. (2022). DNA methylation profiles of ovarian clear cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 31 (1), 132–141. doi:10.1158/1055-9965.EPI-21-0677
de la Rosa, C. N., Krell, J., Day, E., Clarke, A., Reddi, M., Webber, L., et al. (2022). Statistical analysis plan for the Dual mTorc Inhibition in advanCed/recurrent Epithelial ovarian, fallopian tube or primary peritoneal cancer (of clear cell, endometrioid and high-grade serous type, and carcinosarcoma) trial (DICE). Trials 23 (1), 13. doi:10.1186/s13063-021-05669-9
del Carmen, M. G., Birrer, M., and Schorge, J. O. (2012). Clear cell carcinoma of the ovary: A review of the literature. Gynecol. Oncol. 126 (3), 481–490. doi:10.1016/j.ygyno.2012.04.021
Diaz, E. S., Walts, A. E., Karlan, B. Y., and Walsh, C. S. (2013). Venous thromboembolism during primary treatment of ovarian clear cell carcinoma is associated with decreased survival. Gynecol. Oncol. 131 (3), 541–545. doi:10.1016/j.ygyno.2013.09.005
Donahue, T. R., Tran, L. M., Hill, R., Li, Y., Kovochich, A., Calvopina, J. H., et al. (2012). Integrative survival-based molecular profiling of human pancreatic cancer. Clin. Cancer Res. 18 (5), 1352–1363. doi:10.1158/1078-0432.CCR-11-1539
Eder, J. P., Vande Woude, G. F., Boerner, S. A., and LoRusso, P. M. (2009). Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin. Cancer Res. 15, 2207–2214. doi:10.1158/1078-0432.CCR-08-1306
Eisenhauer, E. A. (2017). Real-world evidence in the treatment of ovarian cancer. Ann. Oncol. 28, viii61–viii65. doi:10.1093/annonc/mdx443
Elmenier, F. M., Lasheen, D. S., and Abouzid, K. A. M. (2019). Phosphatidylinositol 3 kinase (PI3K) inhibitors as new weapon to combat cancer. Eur. J. Med. Chem. 183, 111718. doi:10.1016/j.ejmech.2019.111718
Engqvist, H., Parris, T. Z., Biermann, J., Ronnerman, E. W., Larsson, P., Sundfeldt, K., et al. (2020). Integrative genomics approach identifies molecular features associated with early-stage ovarian carcinoma histotypes. Sci. Rep. 10 (1), 7946. doi:10.1038/s41598-020-64794-8
Esteller, M. (2007). Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8 (4), 286–298. doi:10.1038/nrg2005
Farley, J. H., Brady, W. E., Fujiwara, K., Nomura, H., Yunokawa, M., Tokunaga, H., et al. (2016). A phase II evaluation of temsirolimus in combination with carboplatin and paclitaxel followed by temsirolimus consolidation as first-line therapy in the treatment of stage III-IV clear cell carcinoma of the ovary. J. Clin. Oncol. 34 (15), 5531. doi:10.1200/jco.2016.34.15_suppl.5531
Friedlander, M. L., Russell, K., Millis, S., Gatalica, Z., Bender, R., and Voss, A. (2016). Molecular profiling of clear cell ovarian cancers: Identifying potential treatment targets for clinical trials. Int. J. Gynecol. Cancer 26 (4), 648–654. doi:10.1097/IGC.0000000000000677
Fujimura, M., Katsumata, N., Tsuda, H., Uchi, N., Miyazaki, S., Hidaka, T., et al. (2002). HER2 is frequently over-expressed in ovarian clear cell adenocarcinoma: Possible novel treatment modality using recombinant monoclonal antibody against HER2, trastuzumab. Jpn. J. Cancer Res. 93 (11), 1250–1257. doi:10.1111/j.1349-7006.2002.tb01231.x
Furge, K. A., Kiewlich, D., Le, P., Vo, M. N., Faure, M., Howlett, A. R., et al. (2001). Suppression of Ras-mediated tumorigenicity and metastasis through inhibition of the Met receptor tyrosine kinase. Proc. Natl. Acad. Sci. U. S. A. 98, 10722–10727. doi:10.1073/pnas.191067898
Gadducci, A., Lanfredini, N., and Tana, R. (2014). Novel insights on the malignant transformation of endometriosis into ovarian carcinoma. Gynecol. Endocrinol. 30 (9), 612–617. doi:10.3109/09513590.2014.926325
Gainor, J. F., Stevens, S. E., Willers, H., Shih, H. A., and Heist, R. S. (2020). Intracranial activity of gefitinib and capmatinib in a patient with previously treated non-small cell lung cancer harboring a concurrent EGFR mutation and MET amplification. J. Thorac. Oncol. 15 (1), e8–e10. doi:10.1016/j.jtho.2019.07.024
Gargiulo, G., and Minucci, S. (2009). Epigenomic profiling of cancer cells. Int. J. Biochem. Cell Biol. 41 (1), 127–135. doi:10.1016/j.biocel.2008.07.021
Gounaris, I., Charnock-Jones, D. S., and Brenton, J. D. (2011). Ovarian clear cell carcinoma--bad endometriosis or bad endometrium? J. Pathol. 225 (2), 157–160. doi:10.1002/path.2970
Guan, B., Mao, T. L., Panuganti, P. K., Kuhn, E., Kurman, R. J., Maeda, D., et al. (2011). Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 35 (5), 625–632. doi:10.1097/PAS.0b013e318212782a
Guan, B., Rahmanto, Y. S., Wu, R. C., Wang, Y., Wang, Z., Wang, T. L., et al. (2014). Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J. Natl. Cancer Inst. 106 (7), dju146. doi:10.1093/jnci/dju146
Guan, B., Wang, T. L., and Shih Ie, M. (2011). ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71 (21), 6718–6727. doi:10.1158/0008-5472.CAN-11-1562
Gui, Y., Guo, G., Huang, Y., Hu, X., Tang, A., Gao, S., et al. (2011). Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43 (9), 875–878. doi:10.1038/ng.907
Guo, Z., and Yu, Q. (2019). Role of mTOR signaling in female reproduction. Front. Endocrinol. (Lausanne) 10, 692. doi:10.3389/fendo.2019.00692
Han, Y., Chen, M. K., Wang, H. L., Hsu, J. L., Li, C. W., Chu, Y. Y., et al. (2019). Synergism of PARP inhibitor fluzoparib (HS10160) and MET inhibitor HS10241 in breast and ovarian cancer cells. Am. J. Cancer Res. 9 (3), 608–618.
Hashiguchi, Y., Tsuda, H., Inoue, T., Berkowitz, R. S., and Mok, S. C. (2006). PTEN expression in clear cell adenocarcinoma of the ovary. Gynecol. Oncol. 101 (1), 71–75. doi:10.1016/j.ygyno.2005.09.047
Hervieu, A., and Kermorgant, S. (2018). The role of PI3K in met driven cancer: A recap. Front. Mol. Biosci. 5, 86. doi:10.3389/fmolb.2018.00086
Hisamatsu, T., Mabuchi, S., Matsumoto, Y., Kawano, M., Sasano, T., Takahashi, R., et al. (2013). Potential role of mTORC2 as a therapeutic target in clear cell carcinoma of the ovary. Mol. Cancer Ther. 12 (7), 1367–1377. doi:10.1158/1535-7163.MCT-12-1185
Ho, C. M., Huang, C. J., Huang, C. Y., Wu, Y. Y., Chang, S. F., and Cheng, W. F. (2012). Promoter methylation status of HIN-1 associated with outcomes of ovarian clear cell adenocarcinoma. Mol. Cancer 11, 53. doi:10.1186/1476-4598-11-53
Ho, C. M., Huang, C. J., Huang, S. H., Chang, S. F., and Cheng, W. F. (2015). Demethylation of HIN-1 reverses paclitaxel-resistance of ovarian clear cell carcinoma through the AKT-mTOR signaling pathway. BMC Cancer 15, 789. doi:10.1186/s12885-015-1744-5
Ho, C. M., Huang, Y. J., Chen, T. C., Huang, S. H., Liu, F. S., Chang Chien, C. C., et al. (2004). Pure-type clear cell carcinoma of the ovary as a distinct histological type and improved survival in patients treated with paclitaxel-platinum-based chemotherapy in pure-type advanced disease. Gynecol. Oncol. 94 (1), 197–203. doi:10.1016/j.ygyno.2004.04.004
Houshdaran, S., Hawley, S., Palmer, C., Campan, M., Olsen, M. N., Ventura, A. P., et al. (2010). DNA methylation profiles of ovarian epithelial carcinoma tumors and cell lines. PLoS One 5 (2), e9359. doi:10.1371/journal.pone.0009359
Howitt, B. E., Strickland, K. C., Sholl, L. M., Rodig, S., Ritterhouse, L. L., Chowdhury, D., et al. (2017). Clear cell ovarian cancers with microsatellite instability: A unique subset of ovarian cancers with increased tumor-infiltrating lymphocytes and PD-1/PD-L1 expression. Oncoimmunology 6 (2), e1277308. doi:10.1080/2162402X.2016.1277308
Huang, H. N., Huang, W. C., Lin, C. H., Chiang, Y. C., and Kuo, K. T. (2014). Chromosome 20q13.2 ZNF217 locus amplification correlates with decreased E-cadherin expression in ovarian clear cell carcinoma with PI3K-Akt pathway alterations. Hum. Pathol. 45 (11), 2318–2325. doi:10.1016/j.humpath.2014.07.020
Huang, K. L., Mashl, R. J., Wu, Y., Ritter, D. I., Wang, J., Oh, C., et al. (2018). Pathogenic germline variants in 10,389 adult cancers. Cell 173 (2), 355–370 e14. doi:10.1016/j.cell.2018.03.039
Itamochi, H., Kigawa, J., Kanamori, Y., Oishi, T., Bartholomeusz, C., Nahta, R., et al. (2007). Adenovirus type 5 E1A gene therapy for ovarian clear cell carcinoma: A potential treatment strategy. Mol. Cancer Ther. 6 (1), 227–235. doi:10.1158/1535-7163.MCT-05-0499
Itamochi, H., Oishi, T., Oumi, N., Takeuchi, S., Yoshihara, K., Mikami, M., et al. (2017). Whole-genome sequencing revealed novel prognostic biomarkers and promising targets for therapy of ovarian clear cell carcinoma. Br. J. Cancer 117 (5), 717–724. doi:10.1038/bjc.2017.228
Itamochi, H., Oumi, N., Oishi, T., Shoji, T., Fujiwara, H., Sugiyama, T., et al. (2015). Loss of ARID1A expression is associated with poor prognosis in patients with stage I/II clear cell carcinoma of the ovary. Int. J. Clin. Oncol. 20 (5), 967–973. doi:10.1007/s10147-015-0811-x
Jin, Y., Li, Y., and Pan, L. (2014). The target therapy of ovarian clear cell carcinoma. Onco Targets Ther. 7, 1647–1652. doi:10.2147/OTT.S49993
Jones, S., Wang, T. L., Shih, I. M., Mao, T. L., Nakayama, K., Roden, R., et al. (2010). Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330 (6001), 228–231. doi:10.1126/science.1196333
Kadoch, C., Hargreaves, D. C., Hodges, C., Elias, L., Ho, L., Ranish, J., et al. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45 (6), 592–601. doi:10.1038/ng.2628
Katagiri, A., Nakayama, K., Rahman, M. T., Rahman, M., Katagiri, H., Nakayama, N., et al. (2012). Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod. Pathol. 25 (2), 282–288. doi:10.1038/modpathol.2011.161
Kato, N., Sasou, S., and Motoyama, T. (2006). Expression of hepatocyte nuclear factor-1beta (HNF-1beta) in clear cell tumors and endometriosis of the ovary. Mod. Pathol. 19 (1), 83–89. doi:10.1038/modpathol.3800492
Kato, N., Sato, Y., Kamataki, A., Fukase, M., Uchigasaki, S., and Kurose, A. (2019). PIK3CA hotspot mutations and cyclooxygenase-2 expression in ovarian clear cell carcinomas: A close association with stromal features. Hum. Pathol. 86, 32–37. doi:10.1016/j.humpath.2018.11.013
Kawahara, N., Mizutani, A., Matsubara, S., Takeda, Y., and Kobayashi, H. (2020). GSK-3β mediates the effects of HNF-1β overexpression in ovarian clear cell carcinoma. Exp. Ther. Med. 20 (5), 122. doi:10.3892/etm.2020.9250
Kawahara, N., Yamada, Y., and Kobayashi, H. (2021). CCNE1 is a putative therapeutic target for arid1a-mutated ovarian clear cell carcinoma. Int. J. Mol. Sci. 22 (11), 5869. doi:10.3390/ijms22115869
Kelemen, L. E., Kobel, M., Chan, A., Taghaddos, S., and Dinu, I. (2013). Differentially methylated loci distinguish ovarian carcinoma histological types: Evaluation of a DNA methylation assay in FFPE tissue. Biomed. Res. Int. 2013, 815894. doi:10.1155/2013/815894
Khemapech, N., Pitchaiprasert, S., and Triratanachat, S. (2012). Prevalence and clinical significance of mammalian target of rapamycin phosphorylation (p-mTOR) and vascular endothelial growth factor (VEGF) in clear cell carcinoma of the ovary. Asian Pac J. Cancer Prev. 13 (12), 6357–6362. doi:10.7314/apjcp.2012.13.12.6357
Kim, A., Enomoto, T., Serada, S., Ueda, Y., Takahashi, T., Ripley, B., et al. (2009). Enhanced expression of Annexin A4 in clear cell carcinoma of the ovary and its association with chemoresistance to carboplatin. Int. J. Cancer 125 (10), 2316–2322. doi:10.1002/ijc.24587
Kim, A., Serada, S., Enomoto, T., and Naka, T. (2010). Targeting annexin A4 to counteract chemoresistance in clear cell carcinoma of the ovary. Expert Opin. Ther. Targets 14 (9), 963–971. doi:10.1517/14728222.2010.511180
Kim, H. J., Yoon, A., Ryu, J. Y., Cho, Y. J., Choi, J. J., Song, S. Y., et al. (2016). c-MET as a potential therapeutic target in ovarian clear cell carcinoma. Sci. Rep. 6, 38502. doi:10.1038/srep38502
Kim, S. I., Lee, J. W., Lee, M., Kim, H. S., Chung, H. H., Kim, J. W., et al. (2018). Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecol. Oncol. 148 (2), 375–382. doi:10.1016/j.ygyno.2017.12.005
Kimura, A., Arakawa, N., Kagawa, H., Kimura, Y., and Hirano, H. (2021). Phosphorylation of Ser1452 on BRG1 inhibits the function of the SWI/SNF complex in chromatin activation. J. Proteomics 247, 104319. doi:10.1016/j.jprot.2021.104319
Kinross, K. M., Montgomery, K. G., Kleinschmidt, M., Waring, P., Ivetac, I., Tikoo, A., et al. (2012). An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Invest 122 (2), 553–557. doi:10.1172/JCI59309
Kishikawa, T., Sakamoto, M., Ino, Y., Kubushiro, K., Nozawa, S., and Hirohashi, S. (1995). Two distinct patterns of peritoneal involvement shown by in vitro and in vivo ovarian cancer dissemination models. Invasion Metastasis 15 (1-2), 11–21.
Kobayashi, H., Sumimoto, K., Moniwa, N., Imai, M., Takakura, K., Kuromaki, T., et al. (2007). Risk of developing ovarian cancer among women with ovarian endometrioma: A cohort study in shizuoka, Japan. Int. J. Gynecol. Cancer 17 (1), 37–43. doi:10.1111/j.1525-1438.2006.00754.x
Kobayashi, H., Yamada, Y., Kanayama, S., Furukawa, N., Noguchi, T., Haruta, S., et al. (2009). The role of hepatocyte nuclear factor-1beta in the pathogenesis of clear cell carcinoma of the ovary. Int. J. Gynecol. Cancer 19 (3), 471–479. doi:10.1111/IGC.0b013e3181a19eca
Kobel, M., Kalloger, S. E., Carrick, J., Huntsman, D., Asad, H., Oliva, E., et al. (2009). A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am. J. Surg. Pathol. 33 (1), 14–21. doi:10.1097/PAS.0b013e3181788546
Kojima, M., Sone, K., Oda, K., Hamamoto, R., Kaneko, S., Oki, S., et al. (2020). The histone methyltransferase SMYD2 is a novel therapeutic target for the induction of apoptosis in ovarian clear cell carcinoma cells. Oncol. Lett. 20 (5), 153. doi:10.3892/ol.2020.12014
Kojima, M., Sone, K., Oda, K., Hamamoto, R., Kaneko, S., Oki, S., et al. (2019). The histone methyltransferase WHSC1 is regulated by EZH2 and is important for ovarian clear cell carcinoma cell proliferation. BMC Cancer 19 (1), 455. doi:10.1186/s12885-019-5638-9
Konstantinopoulos, P. A., Brady, W. E., Farley, J., Armstrong, A., Uyar, D. S., and Gershenson, D. M. (2018). Phase II study of single-agent cabozantinib in patients with recurrent clear cell ovarian, primary peritoneal or fallopian tube cancer (NRG-GY001). Gynecol. Oncol. 150 (1), 9–13. doi:10.1016/j.ygyno.2018.04.572
Koopman, T., van der Vegt, B., Dijkstra, M., Bart, J., Duiker, E., Wisman, G. B. A., et al. (2018). HER2 immunohistochemistry in endometrial and ovarian clear cell carcinoma: Discordance between antibodies and with in-situ hybridisation. Histopathology 73 (5), 852–863. doi:10.1111/his.13704
Korenaga, T. R., Ward, K. K., Saenz, C., McHale, M. T., and Plaxe, S. (2020). The elevated risk of ovarian clear cell carcinoma among Asian Pacific Islander women in the United States is not affected by birthplace. Gynecol. Oncol. 157 (1), 62–66. doi:10.1016/j.ygyno.2020.01.034
Kuo, K. T., Mao, T. L., Chen, X., Feng, Y., Nakayama, K., Wang, Y., et al. (2010). DNA copy numbers profiles in affinity-purified ovarian clear cell carcinoma. Clin. Cancer Res. 16 (7), 1997–2008. doi:10.1158/1078-0432.CCR-09-2105
Kuo, K. T., Mao, T. L., Jones, S., Veras, E., Ayhan, A., Wang, T. L., et al. (2009). Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am. J. Pathol. 174 (5), 1597–1601. doi:10.2353/ajpath.2009.081000
Kurian, A. W., Balise, R. R., McGuire, V., and Whittemore, A. S. (2005). Histologic types of epithelial ovarian cancer: Have they different risk factors? Gynecol. Oncol. 96 (2), 520–530. doi:10.1016/j.ygyno.2004.10.037
Kurman, R. J., and Shih Ie, M. (2010). The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 34 (3), 433–443. doi:10.1097/PAS.0b013e3181cf3d79
Kuroda, Y., Chiyoda, T., Kawaida, M., Nakamura, K., Aimono, E., Yoshimura, T., et al. (2021). ARID1A mutation/ARID1A loss is associated with a high immunogenic profile in clear cell ovarian cancer. Gynecol. Oncol. 162 (3), 679–685. doi:10.1016/j.ygyno.2021.07.005
Lee, J., Kim, D. H., Lee, S., Yang, Q. H., Lee, D. K., Lee, S. K., et al. (2009). A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl. Acad. Sci. U. S. A. 106 (21), 8513–8518. doi:10.1073/pnas.0902873106
Leskela, S., Romero, I., Cristobal, E., Perez-Mies, B., Rosa-Rosa, J. M., Gutierrez-Pecharroman, A., et al. (2020). Mismatch repair deficiency in ovarian carcinoma: Frequency, causes, and consequences. Am. J. Surg. Pathol. 44 (5), 649–656. doi:10.1097/PAS.0000000000001432
Lheureux, S., Gourley, C., Vergote, I., and Oza, A. M. (2019). Epithelial ovarian cancer. Lancet 393 (10177), 1240–1253. doi:10.1016/S0140-6736(18)32552-2
Li, E., Hu, Z., Sun, Y., Zhou, Q., Yang, B., Zhang, Z., et al. (2016). Small molecule inhibitor of c-Met (PHA665752) suppresses the growth of ovarian cancer cells and reverses cisplatin resistance. Tumour Biol. 37 (6), 7843–7852. doi:10.1007/s13277-015-4318-x
Li, G., Ci, W., Karmakar, S., Chen, K., Dhar, R., Fan, Z., et al. (2014). SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell 25 (4), 455–468. doi:10.1016/j.ccr.2014.02.007
Li, Y., Zhang, X., Gao, Y., Shang, C., Yu, B., Wang, T., et al. (2020). Development of a genomic signatures-based predictor of initial platinum-resistance in advanced high-grade serous ovarian cancer patients. Front. Oncol. 10, 625866. doi:10.3389/fonc.2020.625866
Lin, C. J., Huang, W. R., Wu, C. Z., and Tseng, R. C. (2021). Changes in SLIT2 expression are associated with the migration of human ovarian clear cell carcinoma cells. Oncol. Lett. 22 (1), 551. doi:10.3892/ol.2021.12812
Littlepage, L. E., Adler, A. S., Kouros-Mehr, H., Huang, G., Chou, J., Krig, S. R., et al. (2012). The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2 (7), 638–651. doi:10.1158/2159-8290.CD-12-0093
Longati, P., Bardelli, A., Ponzetto, C., Naldini, L., and Comoglio, P. M. (1994). Tyrosines1234-1235 are critical for activation of the tyrosine kinase encoded by the MET proto-oncogene (HGF receptor). Oncogene 9, 49–57.
LoPiccolo, J., Granville, C. A., Gills, J. J., and Dennis, P. A. (2007). Targeting Akt in cancer therapy. Anticancer Drugs 18 (8), 861–874. doi:10.1097/CAD.0b013e3280cc2c6f
Lu, Z., and Chen, J. (2014). Introduction of WHO classification of tumours of female reproductive organs, fourth edition. Zhonghua Bing Li Xue Za Zhi 43 (10), 649–650.
Lyu, C., Zhang, Y., Zhou, X., and Lang, J. (2016). ARID1A gene silencing reduces the sensitivity of ovarian clear cell carcinoma to cisplatin. Exp. Ther. Med. 12 (6), 4067–4071. doi:10.3892/etm.2016.3863
Mabuchi, S., Kawase, C., Altomare, D. A., Morishige, K., Sawada, K., Hayashi, M., et al. (2009). mTOR is a promising therapeutic target both in cisplatin-sensitive and cisplatin-resistant clear cell carcinoma of the ovary. Clin. Cancer Res. 15 (17), 5404–5413. doi:10.1158/1078-0432.CCR-09-0365
Mabuchi, S., Sugiyama, T., and Kimura, T. (2016). Clear cell carcinoma of the ovary: Molecular insights and future therapeutic perspectives. J. Gynecol. Oncol. 27 (3), e31. doi:10.3802/jgo.2016.27.e31
Marchion, D. C., Bicaku, E., Xiong, Y., Bou Zgheib, N., Al Sawah, E., Stickles, X. B., et al. (2013). A novel c-Met inhibitor, MK8033, synergizes with carboplatin plus paclitaxel to inhibit ovarian cancer cell growth. Oncol. Rep. 29 (5), 2011–2018. doi:10.3892/or.2013.2329
Maru, Y., Tanaka, N., Ohira, M., Itami, M., Hippo, Y., and Nagase, H. (2017). Identification of novel mutations in Japanese ovarian clear cell carcinoma patients using optimized targeted NGS for clinical diagnosis. Gynecol. Oncol. 144 (2), 377–383. doi:10.1016/j.ygyno.2016.11.045
Matsuo, K., Hasegawa, K., Yoshino, K., Murakami, R., Hisamatsu, T., Stone, R. L., et al. (2015). Venous thromboembolism, interleukin-6 and survival outcomes in patients with advanced ovarian clear cell carcinoma. Eur. J. Cancer 51 (14), 1978–1988. doi:10.1016/j.ejca.2015.07.012
Matsuzaki, S., Enomoto, T., Serada, S., Yoshino, K., Nagamori, S., Morimoto, A., et al. (2014). Annexin A4-conferred platinum resistance is mediated by the copper transporter ATP7A. Int. J. Cancer 134 (8), 1796–1809. doi:10.1002/ijc.28526
McConechy, M. K., Ding, J., Cheang, M. C., Wiegand, K., Senz, J., Tone, A., et al. (2012). Use of mutation profiles to refine the classification of endometrial carcinomas. J. Pathol. 228 (1), 20–30. doi:10.1002/path.4056
Michaels, P. D., Robinson, H., Nardi, V., Iafrate, A. J., Le, L., and Lennerz, J. K. (2018). MET amplification in esophageal squamous carcinoma. Int. J. Surg. Pathol. 26 (8), 731–732. doi:10.1177/1066896918764301
Miller, R. E., Brough, R., Bajrami, I., Williamson, C. T., McDade, S., Campbell, J., et al. (2016). Synthetic lethal targeting of arid1a-mutant ovarian clear cell tumors with dasatinib. Mol. Cancer Ther. 15 (7), 1472–1484. doi:10.1158/1535-7163.MCT-15-0554
Moran-Jones, K., Brown, L. M., and Samimi, G. (2015). INC280, an orally available small molecule inhibitor of c-MET, reduces migration and adhesion in ovarian cancer cell models. Sci. Rep. 5, 11749. doi:10.1038/srep11749
Morimoto, A., Serada, S., Enomoto, T., Kim, A., Matsuzaki, S., Takahashi, T., et al. (2014). Annexin A4 induces platinum resistance in a chloride-and calcium-dependent manner. Oncotarget 5 (17), 7776–7787. doi:10.18632/oncotarget.2306
Munoz-Galvan, S., and Carnero, A. (2021). Leveraging genomics, transcriptomics, and epigenomics to understand the biology and chemoresistance of ovarian cancer. Cancers (Basel) 13 (16), 4029. doi:10.3390/cancers13164029
Murakami, K., Kanto, A., Sakai, K., Miyagawa, C., Takaya, H., Nakai, H., et al. (2021). Frequent PIK3CA mutations in eutopic endometrium of patients with ovarian clear cell carcinoma. Mod. Pathol. 34, 2071–2079. doi:10.1038/s41379-021-00861-3
Murakami, R., Matsumura, N., Brown, J. B., Higasa, K., Tsutsumi, T., Kamada, M., et al. (2017). Exome sequencing landscape analysis in ovarian clear cell carcinoma shed light on key chromosomal regions and mutation gene networks. Am. J. Pathol. 187 (10), 2246–2258. doi:10.1016/j.ajpath.2017.06.012
Murugan, A. K. (2019). mTOR: Role in cancer, metastasis and drug resistance. Semin. Cancer Biol. 59, 92–111. doi:10.1016/j.semcancer.2019.07.003
Naito, T., Udagawa, H., Umemura, S., Sakai, T., Zenke, Y., Kirita, K., et al. (2019). Non-small cell lung cancer with loss of expression of the SWI/SNF complex is associated with aggressive clinicopathological features, PD-L1-positive status, and high tumor mutation burden. Lung Cancer 138, 35–42. doi:10.1016/j.lungcan.2019.10.009
Nakonechny, Q. B., and Gilks, C. B. (2016). Ovarian cancer in hereditary cancer susceptibility syndromes. Surg. Pathol. Clin. 9 (2), 189–199. doi:10.1016/j.path.2016.01.003
Nie, Z., Xue, Y., Yang, D., Zhou, S., Deroo, B. J., Archer, T. K., et al. (2000). A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell Biol. 20 (23), 8879–8888. doi:10.1128/MCB.20.23.8879-8888.2000
Oda, K., Hamanishi, J., Matsuo, K., and Hasegawa, K. (2018). Genomics to immunotherapy of ovarian clear cell carcinoma: Unique opportunities for management. Gynecol. Oncol. 151 (2), 381–389. doi:10.1016/j.ygyno.2018.09.001
Okamoto, A., Sehouli, J., Yanaihara, N., Hirata, Y., Braicu, I., Kim, B. G., et al. (2015). Somatic copy number alterations associated with Japanese or endometriosis in ovarian clear cell adenocarcinoma. PLoS One 10 (2), e0116977. doi:10.1371/journal.pone.0116977
Pearce, C. L., Templeman, C., Rossing, M. A., Lee, A., Near, A. M., Webb, P. M., et al. (2012). Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case-control studies. Lancet Oncol. 13 (4), 385–394. doi:10.1016/S1470-2045(11)70404-1
Rahman, M. T., Nakayama, K., Rahman, M., Katagiri, H., Katagiri, A., Ishibashi, T., et al. (2012). Gene amplification of ZNF217 located at chr20q13.2 is associated with lymph node metastasis in ovarian clear cell carcinoma. Anticancer Res. 32 (8), 3091–3095.
Ribeiro-Silva, C., Vermeulen, W., and Lans, H. (2019). SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair (Amst) 77, 87–95. doi:10.1016/j.dnarep.2019.03.007
Saavedra, J. A., and Sandow, J. (1968). Clear cell adenocarcinoma of the ovary (Saphir and Lackner) and so-called mesonephroma ovarii (Schiller) as combined tumors. Review of the literature and case report. Arch. Gynakol 206 (2), 131–153. doi:10.1007/BF00666920
Sasano, T., Mabuchi, S., Kuroda, H., Kawano, M., Matsumoto, Y., Takahashi, R., et al. (2015). Preclinical efficacy for AKT targeting in clear cell carcinoma of the ovary. Mol. Cancer Res. 13 (4), 795–806. doi:10.1158/1541-7786.MCR-14-0314
Sato, E., Nakayama, K., Razia, S., Nakamura, K., Ishikawa, M., Minamoto, T., et al. (2018). ARID1B as a potential therapeutic target for arid1a-mutant ovarian clear cell carcinoma. Int. J. Mol. Sci. 19 (6), 1710. doi:10.3390/ijms19061710
Sato, N., Tsunoda, H., Nishida, M., Morishita, Y., Takimoto, Y., Kubo, T., et al. (2000). Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: Possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 60 (24), 7052–7056.
Schnack, T. H., Hogdall, E., Nedergaard, L., and Hogdall, C. (2016). Demographic clinical and prognostic factors of primary ovarian adenocarcinomas of serous and clear cell histology-A comparative study. Int. J. Gynecol. Cancer 26 (1), 82–90. doi:10.1097/IGC.0000000000000585
Senthong, A., Kitkumthorn, N., Rattanatanyong, P., Khemapech, N., Triratanachart, S., and Mutirangura, A. (2014). Differences in LINE-1 methylation between endometriotic ovarian cyst and endometriosis-associated ovarian cancer. Int. J. Gynecol. Cancer 24 (1), 36–42. doi:10.1097/IGC.0000000000000021
Seo, S., Ryu, M. H., Ryoo, B. Y., Park, Y., Park, Y. S., Na, Y. S., et al. (2019). Clinical significance of MET gene amplification in metastatic or locally advanced gastric cancer treated with first-line fluoropyrimidine and platinum combination chemotherapy. Chin. J. Cancer Res. 31 (4), 620–631. doi:10.21147/j.issn.1000-9604.2019.04.06
Shariati, M., and Meric-Bernstam, F. (2019). Targeting AKT for cancer therapy. Expert Opin. Investig. Drugs 28 (11), 977–988. doi:10.1080/13543784.2019.1676726
Shen, J., Ju, Z., Zhao, W., Wang, L., Peng, Y., Ge, Z., et al. (2018). ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 24 (5), 556–562. doi:10.1038/s41591-018-0012-z
Shibuya, Y., Tokunaga, H., Saito, S., Shimokawa, K., Katsuoka, F., Bin, L., et al. (2018). Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosom. Cancer 57 (2), 51–60. doi:10.1002/gcc.22507
Shin, H. Y., Yang, W., Chay, D. B., Lee, E. J., Chung, J. Y., Kim, H. S., et al. (2021). Tetraspanin 1 promotes endometriosis leading to ovarian clear cell carcinoma. Mol. Oncol. 15 (4), 987–1004. doi:10.1002/1878-0261.12884
Soldi, R., Ghosh Halder, T., Weston, A., Thode, T., Drenner, K., Lewis, R., et al. (2020). The novel reversible LSD1 inhibitor SP-2577 promotes anti-tumor immunity in SWItch/Sucrose-NonFermentable (SWI/SNF) complex mutated ovarian cancer. PLoS One 15 (7), e0235705. doi:10.1371/journal.pone.0235705
Suda, K., Cruz Diaz, L. A., Yoshihara, K., Nakaoka, H., Yachida, N., Motoyama, T., et al. (2020). Clonal lineage from normal endometrium to ovarian clear cell carcinoma through ovarian endometriosis. Cancer Sci. 111 (8), 3000–3009. doi:10.1111/cas.14507
Sung, C. O., Choi, C. H., Ko, Y. H., Ju, H., Choi, Y. L., Kim, N., et al. (2013). Integrative analysis of copy number alteration and gene expression profiling in ovarian clear cell adenocarcinoma. Cancer Genet. 206 (5), 145–153. doi:10.1016/j.cancergen.2013.04.002
Sung, P. L., Chang, Y. H., Chao, K. C., and Chuang, C. M.Task Force on Systematic Review and Meta-analysis of Ovarian Cancer (2014). Global distribution pattern of histological subtypes of epithelial ovarian cancer: A database analysis and systematic review. Gynecol. Oncol. 133 (2), 147–154. doi:10.1016/j.ygyno.2014.02.016
Suzuki, E., Kajita, S., Takahashi, H., Matsumoto, T., Tsuruta, T., and Saegusa, M. (2015). Transcriptional upregulation of HNF-1β by NF-κB in ovarian clear cell carcinoma modulates susceptibility to apoptosis through alteration in bcl-2 expression. Lab. Invest 95 (8), 962–972. doi:10.1038/labinvest.2015.73
Takahashi, K., Takenaka, M., Okamoto, A., Bowtell, D. D. L., and Kohno, T. (2021). Treatment strategies for arid1a-deficient ovarian clear cell carcinoma. Cancers (Basel) 13 (8), 1769. doi:10.3390/cancers13081769
Takao, C., Morikawa, A., Ohkubo, H., Kito, Y., Saigo, C., Sakuratani, T., et al. (2017). Downregulation of ARID1A, a component of the SWI/SNF chromatin remodeling complex, in breast cancer. J. Cancer 8 (1), 1–8. doi:10.7150/jca.16602
Tan, D. S., Iravani, M., McCluggage, W. G., Lambros, M. B. K., Milanezi, F., Mackay, A., et al. (2011). Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clin. Cancer Res. 17 (6), 1521–1534. doi:10.1158/1078-0432.CCR-10-1688
Terasawa, K., Sagae, S., Toyota, M., Tsukada, K., Ogi, K., Satoh, A., et al. (2004). Epigenetic inactivation of TMS1/ASC in ovarian cancer. Clin. Cancer Res. 10 (6), 2000–2006. doi:10.1158/1078-0432.ccr-0932-03
Torre, L. A., Trabert, B., DeSantis, C. E., Miller, K. D., Samimi, G., Runowicz, C. D., et al. (2018). Ovarian cancer statistics. CA Cancer J. Clin. 68 (4), 284–296. doi:10.3322/caac.21456
Ueda, H., Watanabe, Y., Nakai, H., Hemmi, H., Koi, M., and Hoshiai, H. (2005). Microsatellite status and immunohistochemical features of ovarian clear-cell carcinoma. Anticancer Res. 25 (4), 2785–2788.
Uehara, Y., Oda, K., Ikeda, Y., Koso, T., Tsuji, S., Yamamoto, S., et al. (2015). Integrated copy number and expression analysis identifies profiles of whole-arm chromosomal alterations and subgroups with favorable outcome in ovarian clear cell carcinomas. PLoS One 10 (6), e0128066. doi:10.1371/journal.pone.0128066
Vendrell, J. A., Thollet, A., Nguyen, N. T., Ghayad, S. E., Vinot, S., Bieche, I., et al. (2012). ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 72 (14), 3593–3606. doi:10.1158/0008-5472.CAN-11-3095
Wang, H., Deng, L., Cai, M., Zhuang, H., Zhu, L., Hao, Y., et al. (2017). Annexin A4 fucosylation enhances its interaction with the NF-kB p50 and promotes tumor progression of ovarian clear cell carcinoma. Oncotarget 8 (64), 108093–108107. doi:10.18632/oncotarget.10226
Wang, H. M., Tan, M. Z., Zhang, S., Li, X., Gao, J., Zhang, D. Y., et al. (2016). Expressions of CD44,CD47,and c-met in ovarian clear cell carcinoma and their clinical significance. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 38 (6), 720–725. doi:10.3881/j.issn.1000-503X.2016.06.016
Wang, J., and Cheng, J. X. (2017). c-Met inhibition enhances chemosensitivity of human ovarian cancer cells. Clin. Exp. Pharmacol. Physiol. 44 (1), 79–87. doi:10.1111/1440-1681.12672
Wang, K., Kan, J., Yuen, S. T., Shi, S. T., Chu, K. M., Law, S., et al. (2011). Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43 (12), 1219–1223. doi:10.1038/ng.982
Wiegand, K. C., Shah, S. P., Al-Agha, O. M., Zhao, Y., Tse, K., Zeng, T., et al. (2010). ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363 (16), 1532–1543. doi:10.1056/NEJMoa1008433
Willis, B. C., Sloan, E. A., Atkins, K. A., Stoler, M. H., and Mills, A. M. (2017). Mismatch repair status and PD-L1 expression in clear cell carcinomas of the ovary and endometrium. Mod. Pathol. 30 (11), 1622–1632. doi:10.1038/modpathol.2017.67
Winter, W. E., Maxwell, G. L., Tian, C., Carlson, J. W., Ozols, R. F., Rose, P. G., et al. (2007). Prognostic factors for stage III epithelial ovarian cancer: A gynecologic Oncology group study. J. Clin. Oncol. 25 (24), 3621–3627. doi:10.1200/JCO.2006.10.2517
Worley, M. J., Liu, S., Hua, Y., Kwok, J. S. L., Samuel, A., Hou, L., et al. (2015). Molecular changes in endometriosis-associated ovarian clear cell carcinoma. Eur. J. Cancer 51 (13), 1831–1842. doi:10.1016/j.ejca.2015.05.011
Xia, Y., Xiong, N., and Huang, Y. (2018). Relationship between methylation status of RASSF2A gene promoter and endometriosis-associated ovarian cancer. J. Biol. Regul. Homeost. Agents 32 (1), 21–28.
Xiao, W., Awadallah, A., and Xin, W. (2012). Loss of ARID1A/BAF250a expression in ovarian endometriosis and clear cell carcinoma. Int. J. Clin. Exp. Pathol. 5 (7), 642–650.
Xiao, W., Ren, L., Chen, Z., Fang, L. T., Zhao, Y., Lack, J., et al. (2021). Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat. Biotechnol. 39 (9), 1141–1150. doi:10.1038/s41587-021-00994-5
Yamaguchi, K., Huang, Z., Matsumura, N., Mandai, M., Okamoto, T., Baba, T., et al. (2014). Epigenetic determinants of ovarian clear cell carcinoma biology. Int. J. Cancer 135 (3), 585–597. doi:10.1002/ijc.28701
Yamaguchi, K., Mandai, M., Oura, T., Matsumura, N., Hamanishi, J., Baba, T., et al. (2010). Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene 29 (12), 1741–1752. doi:10.1038/onc.2009.470
Yamaguchi, K., Matsumura, N., Mandai, M., Baba, T., Konishi, I., and Murphy, S. K. (2014). Epigenetic and genetic dispositions of ovarian carcinomas. Oncoscience 1 (9), 574–579. doi:10.18632/oncoscience.82
Yamamoto, S., Tsuda, H., Miyai, K., Takano, M., Tamai, S., and Matsubara, O. (2012). Accumulative copy number increase of MET drives tumor development and histological progression in a subset of ovarian clear-cell adenocarcinomas. Mod. Pathol. 25 (1), 122–130. doi:10.1038/modpathol.2011.143
Yamamoto, S., Tsuda, H., Takano, M., Iwaya, K., Tamai, S., and Matsubara, O. (2011). PIK3CA mutation is an early event in the development of endometriosis-associated ovarian clear cell adenocarcinoma. J. Pathol. 225 (2), 189–194. doi:10.1002/path.2940
Yamamoto, S., Tsuda, H., Takano, M., Tamai, S., and Matsubara, O. (2012). Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod. Pathol. 25 (4), 615–624. doi:10.1038/modpathol.2011.189
Yamashita, Y., Akatsuka, S., Shinjo, K., Yatabe, Y., Kobayashi, H., Seko, H., et al. (2013). Met is the most frequently amplified gene in endometriosis-associated ovarian clear cell adenocarcinoma and correlates with worsened prognosis. PLoS One 8 (3), e57724. doi:10.1371/journal.pone.0057724
Yano, M., Katoh, T., Miyazawa, M., Miyazawa, M., Ogane, N., Miwa, M., et al. (2019). Clinicopathological correlation of ARID1A status with HDAC6 and its related factors in ovarian clear cell carcinoma. Sci. Rep. 9 (1), 2397. doi:10.1038/s41598-019-38653-0
Yano, M., Yasuda, M., Sakaki, M., Nagata, K., Fujino, T., Arai, E., et al. (2018). Association of histone deacetylase expression with histology and prognosis of ovarian cancer. Oncol. Lett. 15 (3), 3524–3531. doi:10.3892/ol.2018.7726
Yao, H., Sun, C., Hu, Z., and Wang, W. (2016). The role of annexin A4 in cancer. Front. Biosci. (Landmark Ed. 21, 949–957. doi:10.2741/4432
Ye, S., Yang, J., You, Y., Cao, D., Huang, H., Wu, M., et al. (2016). Clinicopathologic significance of HNF-1β, AIRD1A, and PIK3CA expression in ovarian clear cell carcinoma: A tissue microarray study of 130 cases. Med. Baltim. 95 (9), e3003. doi:10.1097/MD.0000000000003003
Zannoni, G. F., Improta, G., and Fraggetta, F. (2014). PIK3CA in ovarian clear cell carcinoma. Hum. Pathol. 45 (12), 2514. doi:10.1016/j.humpath.2014.07.023
Zelli, V., Compagnoni, C., Cannita, K., Capelli, R., Capalbo, C., Di Vito Nolfi, M., et al. (2020). Applications of next generation sequencing to the analysis of familial breast/ovarian cancer. High. Throughput 9 (1), 1. doi:10.3390/ht9010001
Zeng, Y., Liu, Z., Yang, J., Liu, Y., Huo, L., Li, Z., et al. (2016). ARID1A is a tumour suppressor and inhibits glioma cell proliferation via the PI3K pathway. Head. Neck Oncol. 5 (1), 6.
Zhang, M., Li, G., Sun, X., Ni, S., Tan, C., Xu, M., et al. (2018). MET amplification, expression, and exon 14 mutations in colorectal adenocarcinoma. Hum. Pathol. 77, 108–115. doi:10.1016/j.humpath.2018.03.024
Zhu, B., Tian, J., Zhong, R., Tian, Y., Chen, W., Qian, J., et al. (2015). Genetic variants in the SWI/SNF complex and smoking collaborate to modify the risk of pancreatic cancer in a Chinese population. Mol. Carcinog. 54 (9), 761–768. doi:10.1002/mc.22140
Keywords: ovarian clear cell carcinoma, genomics, epigenomics, gynecological oncology, review
Citation: Tong A, Di X, Zhao X and Liang X (2023) Review the progression of ovarian clear cell carcinoma from the perspective of genomics and epigenomics. Front. Genet. 14:952379. doi: 10.3389/fgene.2023.952379
Received: 25 May 2022; Accepted: 06 February 2023;
Published: 16 February 2023.
Edited by:
Marianna Aprile, Department of Biomedical Sciences (CNR), ItalyReviewed by:
Ying-Cheng Chiang, National Taiwan University, TaiwanLuke Benjamin Hesson, University of New South Wales, Australia
Copyright © 2023 Tong, Di, Zhao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Liang, eGlhb2xpYW5nOTEwMUAxNjMuY29t
 An Tong
An Tong Xiangjie Di
Xiangjie Di Xia Zhao
Xia Zhao Xiao Liang
Xiao Liang