Abstract
Context:
There is a great need in clinical research with imaging to collect, to store, to organize, and to process large amount of varied data according to legal requirements and research obligations. In practice, many laboratories or clinical research centers working in imaging domain have to manage innumerous images and their associated data without having sufficient information technology skills and resources to develop and to maintain a robust software solution. Since conventional infrastructure and data storage systems for medical image such as “Picture Archiving and Communication System” may not be compatible with research needs, we propose a solution: ArchiMed, a complete storage and visualization solution developed for clinical research.
Material and methods:
ArchiMed is a service-oriented server application written in Java EE™, which is integrated into local clinical environments (imaging devices, post-processing workstations, others devices, etc.) and allows to safely collect data from other collaborative centers. It ensures all kinds of imaging data storage with a “study-centered” approach, quality control, and interfacing with mainstream image analysis research tools.
Results:
With more than 10 millions of archived files for about 4TB stored with 116 studies, ArchiMed, in function for 5 years at CIC-IT1 of Nancy-France, is used every day by about 60 persons, among whom are engineers, researchers, clinicians, and clinical trial project managers.
Introduction
One main challenge of clinical research with imaging is the need to collect, to store, to organize, and to process large amount of various data according to legal requirements and research obligations.
In practice, most of labs and Contract Research Organizations (CRO) manage many studies or protocols at the same time with several classes of contributors, including radiologists, researchers, physicians, and project managers who perform different kinds of data analysis. These contributors have to manage innumerous images and their associated data without having sufficient IT skills and resources to develop and to maintain a robust software solution.
The first approach to solve the important problem is to use conventional clinical data storage systems for medical image such as “Picture Archiving and Communication System” (PACS) (Choplin et al., 1992; van de Wetering et al., 2006). Those systems, available in most of clinical centers are designed to allow to store and transfer only Digital Imaging and Communications in Medicine (DICOM)2 images.
The first observation that can be made is that those clinical data storage system offer “patient centered” data structure. Such structure does not match specific research needs in terms of data usage, large-scale processing, and connection with mainstream image analysis research tools. It does not allow fine-tune rights and restriction management for each user according to different studies, either.
DICOM is the most used medical image format but has some limitation for research. It is required in research to store and process various other formats like image in MR raw data,3 physiological signals (Odille et al., 2008), analysis results, or segmentations.
Moreover, taking into account the diversity of research contributors and the big data quantity in clinical research context, every proposed system must be user friendly and respond quickly. More than just a question of appearance, the user experience of graphical user interface is the key point for clinicians or researchers who want to search data efficiently, import a large number of images, or load a dataset into the software that they work with every day.
Thus, we need a solution that is able to store all kinds of files (DICOM, raw data, etc.) with metadata, allows “batch processing” for large-scale studies and has to be fully interoperable within a research environment. This means:
Easy and safe data transfer from/to local clinical environments;
Easy and safe data import from the outside (e.g., multicenter trials);
Easy access to the data from mainstream image analysis research tools.
Considering legal requirements of clinical research, in order to comply with the local law, which the data hosting institutes have to follow (e.g., MR-001 CNIL4 reference methodology),5 such a system has to ensure data confidentiality using study based access restriction and built-in de-identification (Kushida et al., 2012; Tucker et al., 2016). Take the French law as an example, according to article R. 1123-61—decree of August 29, 2008 (French Public Health Code),6 clinical centers should ensure the long-term conservation of data for at least 15 years after the end of study. This means that not only the storage system must guarantee file and database integrity (Tucker et al., 2016) but also it must offer a quality insurance process to check data validity before integration.
Others emerging research picture archiving system solutions like Shanoir7 or CATI8 are specialized in neuroimaging data management. These solutions are not really adapted for multi organs and many other file formats storage.
For all the aforementioned reasons, it is understood that all these sensitive data must be stored inside a safe, centralized, and isolated system, effectively excluding short-term data support like CD/DVD/USB-keys and non-secured shared location such as network drives, external hard drives, or common public cloud storages.
In response to all these needs and requirement, we introduce ArchiMed, a complete, centralized, and modular storage and visualization solution developed for clinical research. Designed with a “study-centered” approach, which better fits the research workflow and organization needs, the server application developed in Java™ EE is fully integrated into the local clinical environment (imaging devices, post-processing workstations, others devices, etc.), and is able to safely collect data from collaborative centers and ensures all kinds of data storage, quality control, and interfacing with mainstream image analysis research tools.
Materials and Methods
General Description and Software Architecture
ArchiMed is based on a three-tier architecture.9 It has been designed to be a service oriented application to integrate environments with multiple clients/users (Figure 1).
Figure 1
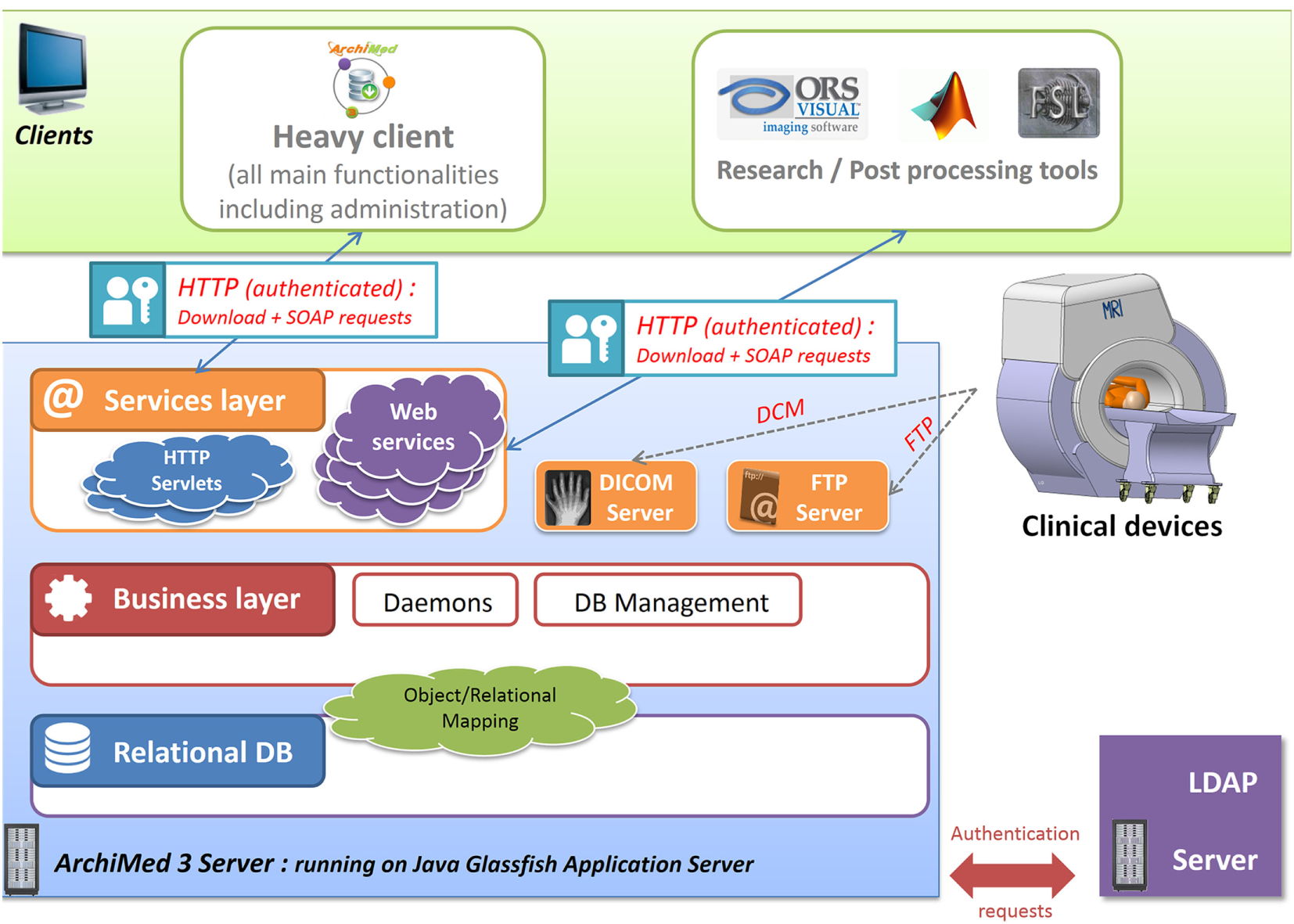
ArchiMed three-tier architecture: describe server 3 layers architecture (data, logic/business, and services) and interactions with client applications and devices.
The server side of the application is implemented in Java-EE™10 (Goncalves, 2009) to be deployable on any operating system with Java™. It is hosted on a local network and running on an open source Glassfish application server.11
Data layer and underlying database is currently deployed on a MySQL™ Relational Database Management System (RDBMS) but is also compatible with any other database management system (e.g., Oracle, Microsoft SQL Server, etc.); thanks to Object Relational Mapping.12 This high-level abstraction technique creating virtual object database can be used from within the programing language independently of the host RDBMS. Business logic layer is one component of the server part that manages how data can be created, displayed, stored, and changed. Some background tasks (daemons) are dedicated to database cleaning, new image importation, and users action queue management.
The web container layer or service layer, only accessible as authenticated user, provides HTTP SOAP web services13 for database querying and HTTP Servlet14 to perform file download/upload. FTP15 (Postel and Reynolds, 1985) and DICOM services are also available to transfer files from/to clinical devices (scanners, PACS) or post-processing workstations.
All these services are reachable via any third-party application to access and exploit data locally and also by a built-in Java™ heavy client to administrate, to browse, to visualize, and to manage data from ArchiMed. Multi-OS and secured; this client is always up to date using a version check and auto-update mechanism.
Regarding authentication requirements, ArchiMed can be linked to any existing LDAP16 (Koutsonikola and Vakali, 2004; Zeilenga, 2006)-based user directory system (like active directory) and consequently users can use their own operating system login credentials.
Multiprocessing, Access Speed and Throughputs
Knowing the big amount of data to manage (up to 20,000 images for a standard fMRI17 exam or up to 2 GB of files for a heart exam with raw data and physiological records, etc.), it is critically important to optimize network stream and parallel access to data.
Every consultation request to ArchiMed server is stateless and treated over HTTP protocol as an independent transaction that is unrelated to any previous request and is executed in a separated process. The downloading rate of data stream is predominantly limited by local area network throughput; server load is estimated as insignificant compare to network flow.
To avoid inconsistency in case of parallel contradictory operations, user actions (data import, transfer, delete, and modification) and every “Create, Update, and Delete” query that involves data change are managed in queues executed asynchronously in transactional18 context.
Data Structure
Database and inherent file system are designed with a “study-centered” approach, which better fits the research workflow and organization needs.
ArchiMed data tree is organized around four major node types inherited from DICOM standard: study, exam, series, and file (Figure 2).
Study node: regroups exams for a study/protocol. Associated information of this node type is
- ○
Study code: a unique ID of the study
- ○
Study description
- ○
Stated investigators and authorized users
- ○
…
- ○
Exam node: groups all data linked to one case of the study. Each case corresponds to an inclusion in the protocol. Associated information of this node type is
- ○
Exam code: a unique ID of the case corresponding to the subject identification number inside the protocol
- ○
Exam description
- ○
Exam date, time
- ○
Last access date
- ○
…
- ○
File type/modality node: groups all data with the same file format (file type). In most case, there is a file type for each modality (scanner-type) like DCM_MR (MRI DICOMs), DCM_CT (CT DICOMS), and AEC (physiological signal customized file format).
Series node: may regroup file of a specific acquisition/sequence. Associated information of this node type is
- ○
Series number
- ○
Series description (acquisition sequence name)
- ○
…
- ○
File node: single file/image node. As the leaf of the data tree, this node is the representation of the data file that physically presents in the file system. Associated information of this node type is
- ○
File URL: the address of the file
- ○
Insertion date
- ○
Specific metadata: many other metadata depending on the file type (acquisition parameters, voxel size, matrix dimensions, sampling frequency, etc.)
- ○
Figure 2

ArchiMed data tree nodes: describe node tree organization and underlying data structure.
All these metadata and node information extracted from file headers are inserted into the database during the insertion process using customized rules for each different file type.
It is possible to add a new customer recognized format or file type into our system by programmatically defining which metadata need to be extracted from the file and how to read them. Then database dynamically adapt its structure to integrate this new type.
Some meta information are generics and some others are common to all types (Exam code, location URL, dates, etc.), while some others are specific to a certain file type (e.g., echo time and repetition time for MR image data, rescale slope, and rescale intercept for CT image data, customized comment for result or physiological data file, etc.) (Figure 3).
Figure 3
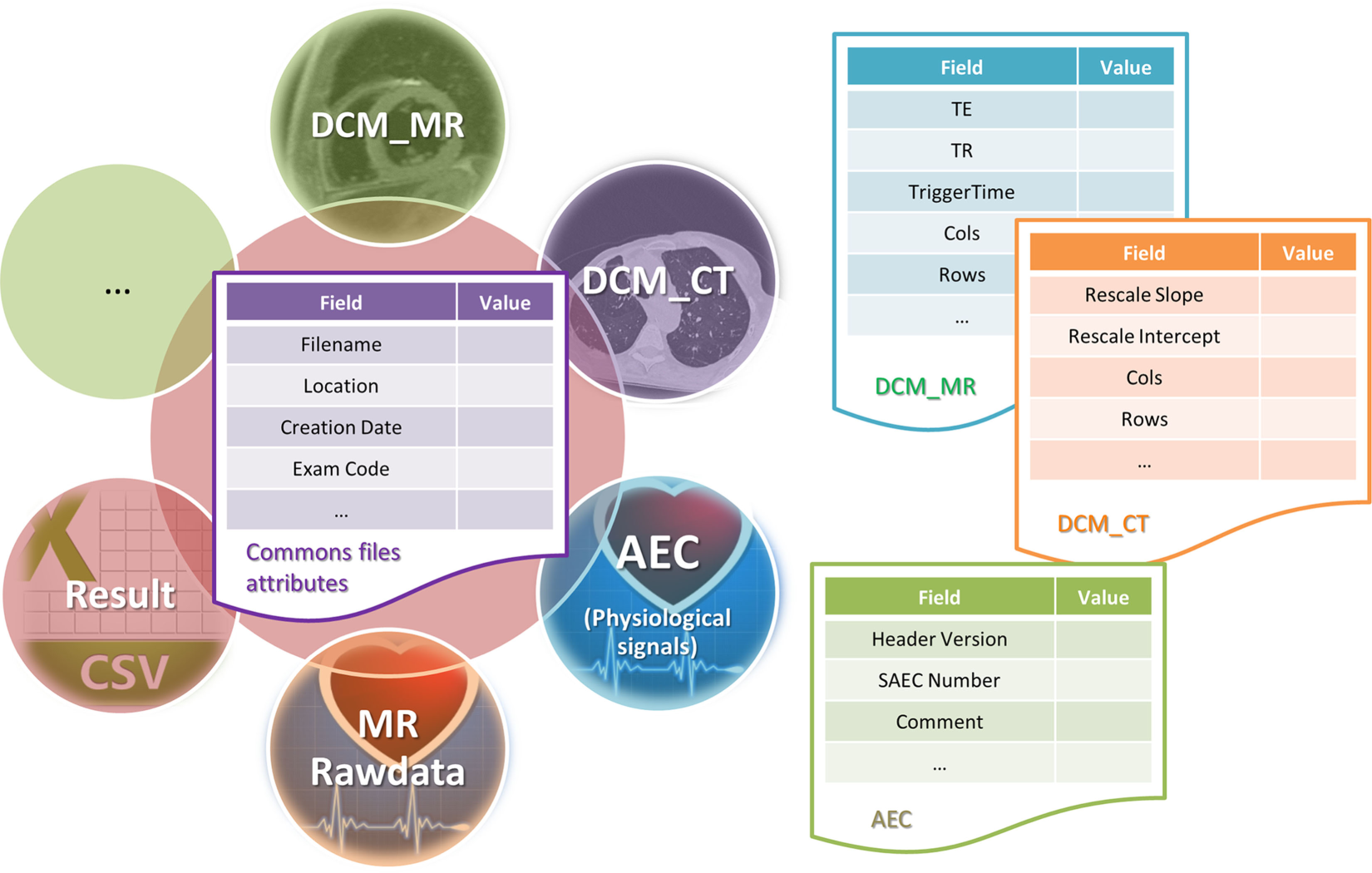
Files and metadata: describe how metadata are stored as database entities according to the file type.
Derived data generated by specific local tools such as reconstruction and post-processing software can be inserted inside ArchiMed as “Result” files and linked to the initials data by sharing the same exam node and series node (relationship between data are intrinsically defined inside the database schema). Every result file type can embed other analysis file (CSV, segmentations file, MESH, etc.).
Integrity and Quality Control
Above all, it is important to underline that for security reasons, in order to keep full local control of data. ArchiMed has been design to be hosted on a local network and not accessible from outside (via internet network). This considerably limits the risk of intrusion and subsequent data loss or damage.
To avoid unfortunate deletion, move, or file corruption, ArchiMed does not allow direct access to file systems. Data are only accessible via authenticated HTTP requests19 (Fielding et al., 1999), which greatly limits direct access to physical files in order to reduce human factor error.
A “recycle bin” temporary storage retains data deleted by users for several days before permanently erasing them from the file system and ArchiMed storage eases built-in standard backup and archiving system.
As previously stated, ArchiMed can support usual user and group management
viaa standard LDAP connection. It is, therefore, possible to integrate it into an environment with existing right management system (such as Microsoft Windows Active Directory
20or any other LDAP based user’s directory). This way, as a member of a group, a user will be able to perform different categories of actions, such as visualization, adding, removing, and downloading (view). It allows a flexible and sufficiently sensitive right management and data access for every nodes (study, exam, series, file). By default, there are four levels of rights with different access privileges for each of them
Limited user: view only,
Standard user: view and import (data insertion),
Power user: view, import, update, and delete (into recycle bin),
Administrator: view, import, update, delete, permanently delete, and configuration tasks.
Finally, keeping in mind that only human experts can definitely validate or reject acquired data before inserting into the database, we separated the data integration into two steps
In the first step, data are automatically transferred in a temporary archive. At this point, users, who are in charge of data transfer, must check data validity by checking file information and going through all the images.
In the second step, data are “transferred” to the final storage, assigned to a specific study. and eventually de-identified.
Although more time is needed than a single-pass automatic import, we believe that these two steps are necessary to prevent image insertion error.
Confidentiality
In addition to action user right explained above, administrator can grant access to a study to users. Thereby, only clinicians, researchers, engineer, and project managers identified in the protocol of the study can access to the data of this study. At this level, there is no consideration of group; access grant to a study is allowed individually.
Data files from different study are physically separated in different cache directories and totally inaccessible without authentication.
ArchiMed offers a built-in de-identification21 (Kushida et al., 2012) functionality that automatically replaces or erases identifying fields (name, date of birth, acquisition center, etc.) from data headers before or after import.
In accordance to legal requirements of clinical research and because research centers are not supposed to keep any other trace of patients/volunteers across the studies, exam code unique ID is the only available information about the case inclusion inside ArchiMed database, multicenter data storage.
In multicenter clinical trial context, the main source or error in collected data are due to bad de-identification (removed fields, missing data) or bad electronic support quality (corrupted CD, wrong data, etc.). Since ArchiMed server is hosted on a local network not accessible from outside, it cannot replace non-secure CD transfers and safely collect data from other collaborative centers. Therefore, we developed Eureka, a secure transfer tools coupled with ArchiMed, which allows external centers to send de-identified data through internet into ArchiMed (Figure 4).
Figure 4
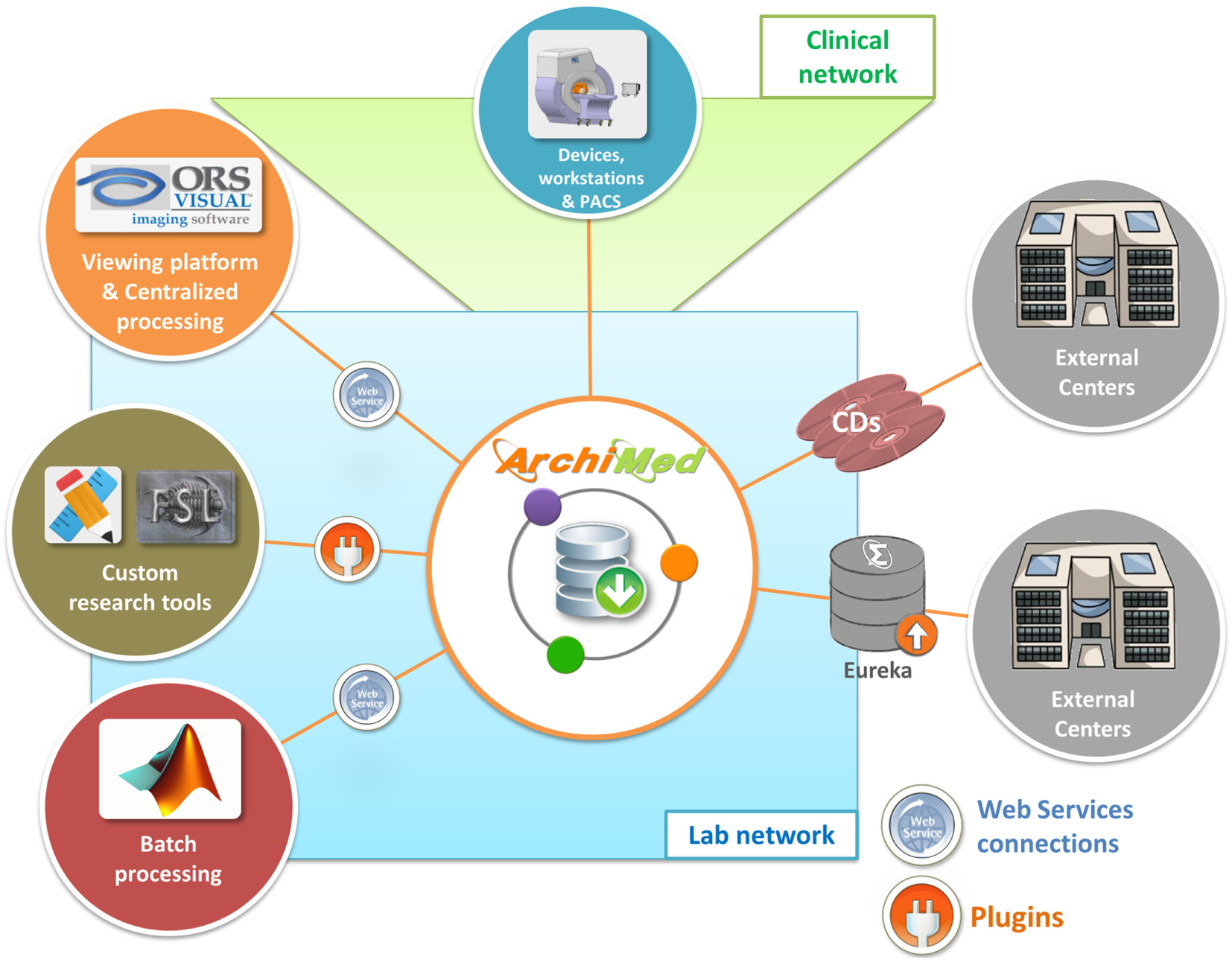
Research environment: show how ArchiMed interacts with research environment (devices, software, and external centers).
The secondary goal of this auxiliary application is to standardize and to keep control on data de-identification and transfer while allowing centers’ operators to visualize and check image before sending.
Interoperability
To connect ArchiMed to the clinical environment, a DICOM transfer protocol is implemented (Figure 4). Behaving like a DICOM node, it can receive/send images from/to clinical devices (e.g., MR, CT, etc.), PACS, or workstation. Specific files like MRI raw data are sent via FTP.
Web services and Servlets technologies make ArchiMed interoperable on HTTP. This means that every application can query the database and upload/download files.
Moreover Plugin Development Toolkit (PDK) provides Java developers with the tools necessary to create plugins that extend the functionalities of ArchiMed client application. Developed plugins can be global or associated to a specific node (exams, series, and files).
Results
Current version of ArchiMed is in function for 5 years at CIC-IT of Nancy-France and used every day by about 60 people, among whom are researchers, clinicians, and clinical trial project managers for local or multicenter studies.
Studies: 116 (including 7 multicenter studies)
Stored exams: ~10,000
Stored files: ~10,000,000
Disk size: ~4 TB
Database size: ~7 GB
Figure 5 shows data import activity and amount of data for all studies.
Figure 5
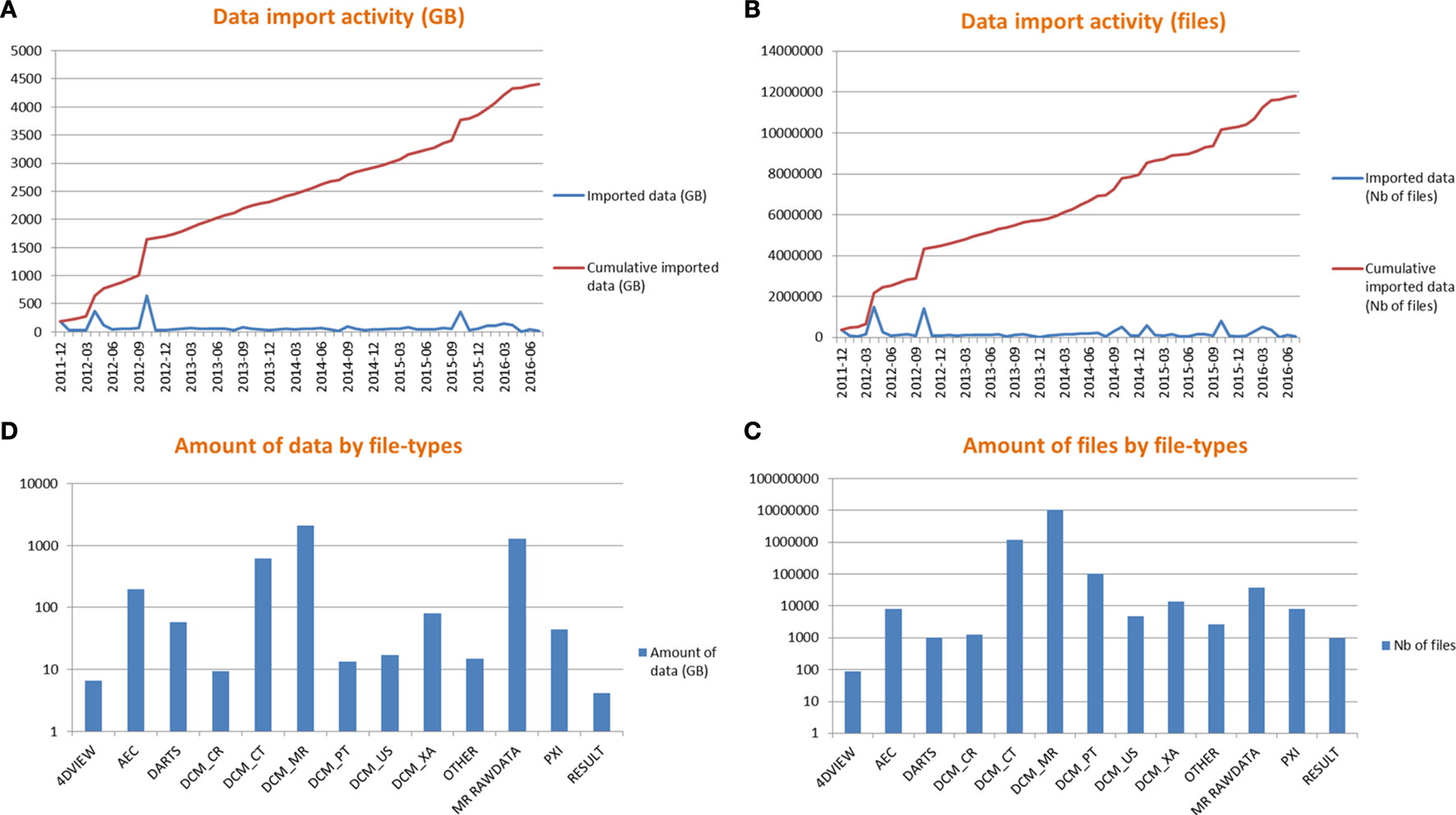
Data import activity (A,B) and amount of data by file types (C,D).
Studies Examples
“MRI Methodology” CIC-IT Protocol (Local Study Code: 2008-0003)
Started in 2008 (before the deployment of the current ArchiMed version) and still active, this research protocol, designed to improve MRI technology and sequences, represents the most important activity using ArchiMed.
Stored exams: 1,246
MR Dicom files: ~3,000,000 (~300 GB)
AEC (physiological signals) files: ~ 4,000 (~100 GB)
MR raw data: ~14,000 (~600 GB)
On daily practice, images and associated files acquired from clinical MRIs are directly sent into ArchiMed temporary storage through DICOM protocol or FTP connections, before being validated, identified (if necessary), and transferred to the corresponding study cache by project manager.
THRACE (Local Study Code: 2009-007)
Started in 2009, this multicenter protocol has been designed to assess effectiveness of endovascular mechanical thrombectomy for acute ischemic stroke (Bracard et al., 2016). CIC-IT of Nancy-France is responsible of collecting, archiving, and analyzing of all images from all the 26 participating centers.
Centers: 26
Stored exams: 1,402
MR Dicom files: ~300,000 (~100 GB)
CT Dicom files: ~400,000 (~200 GB)
XA Dicom files (angiography): ~10,000 (~80 GB)
DICOM images were sent to CIC-IT via CD in earlier days, which is the source of many practical problems (especially concerning de-identification, missing data, or corrupted files) with a significant impact in term of time and resources.
The THRACE project experience has motivated the creation of Eureka for transferring files from externals centers.
User Interface
Installed on more than 50 computers, the Java client application is the most used way to access ArchiMed. It has been designed to be easy to use with a comprehensive user interface similar to those commonly used in clinical imaging software applications (Figure 6).
Figure 6

ArchiMed client GUI: connection menu (A), main options menu (B), images and data browser interface (C).
Plugins and External Software Connections
Interoperable HTTP service interface is already used by different external applications, developed either in C++, Java™ or Matlab™ environments such as “ORS Visual ArchiMed Loader” (loading DICOM from ArchiMed to “ORS Visual™22” viewing and processing platform) or “Matlab ArchiMed Connector” (loading Dicom or other file into Matlab™23 by querying ArchiMed) (Figure 7).
Figure 7
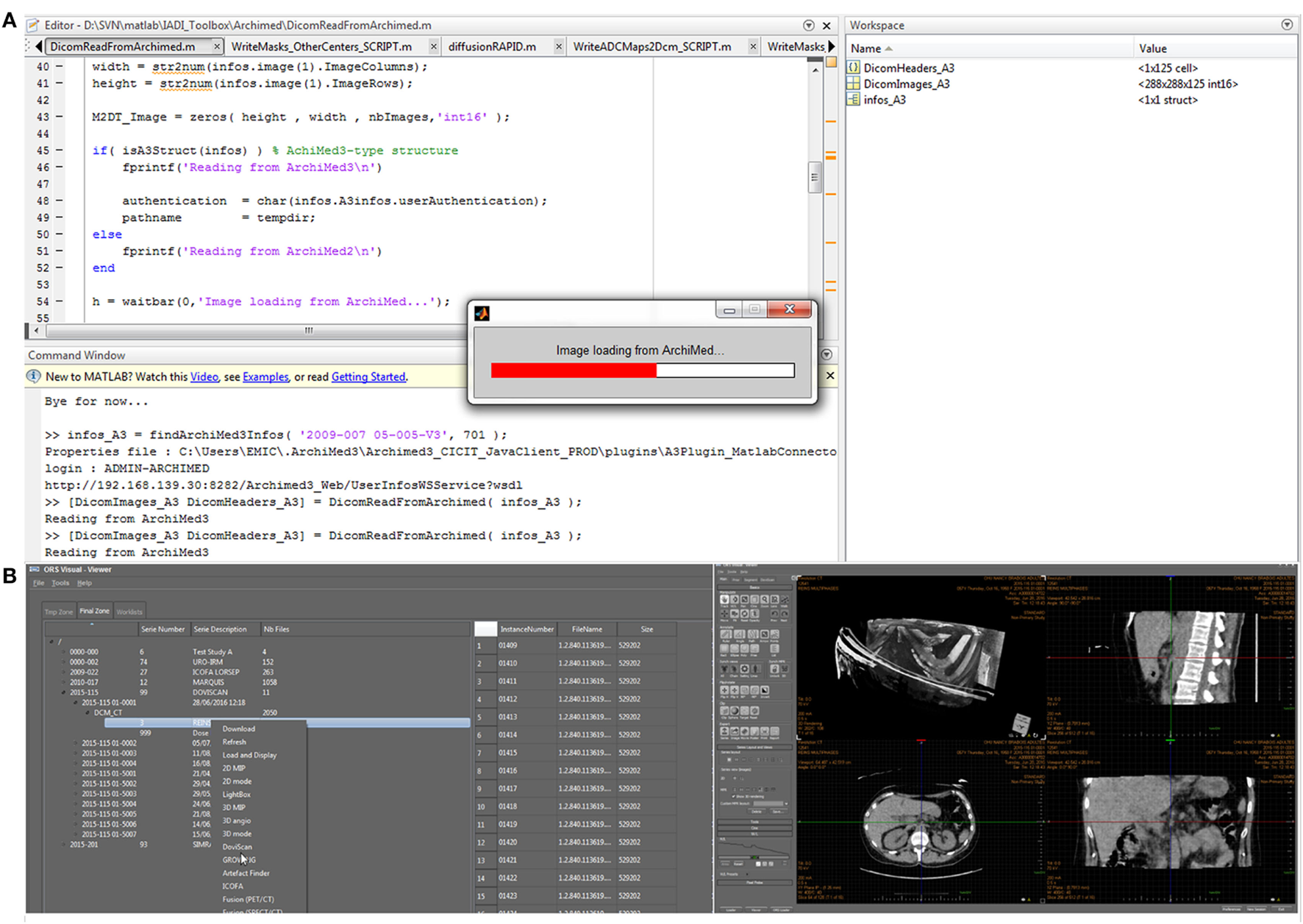
ArchiMed interoperability over HTTP. Loading image data from ArchiMed into Matlab (A) or a ORS Visual 3D dicom viewer (B).
To extend ArchiMed client application functionalities, more than 20 plugins have already been developed for specific processing, case report form, statistical analysis, external database filling, and connection with other software programs, etc. (Figure 8).
Figure 8
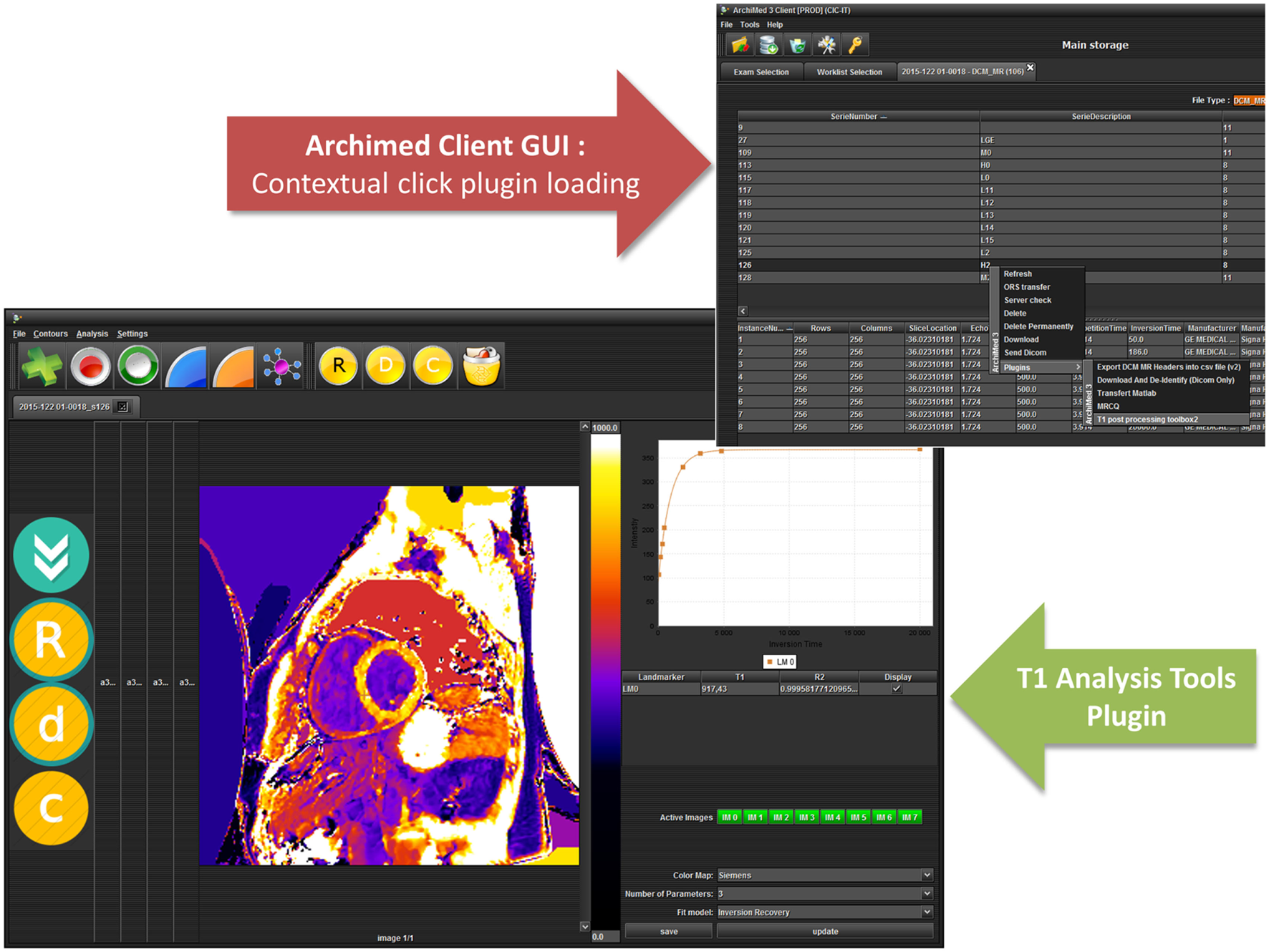
ArchiMed integrated plugins—T1 mapping tools example.
See Table 1 for examples of plugins that have been already built.
Table 1
| Plugin | Description |
|---|---|
| Dcm to Nifti converter | Convert a DICOM Series to Nifti format file |
| Matlab connector plugin | Connect with Matlab in two ways: push from ArchiMed client to Matlab or pull directly from server using web services |
| XXXX plugin | Exam electronic case report form and data analysis for XXXX protocol |
| PC analysis | Phase contrast—pulse wave velocity MR image analysis |
| FSL_FA | Compute ADC and FA maps from DICOM Series using FSL (brain imaging analysis) |
| DICOM export | Export images or movies from stored DICOM |
| MR quality control | Extract quality control parameters from a specific normalized protocol |
| DCM Series splitter | Split DICOM series according to specified criterions |
| App Launcher | Launch defined externals applications |
| XXXX pre-screening | Data check and reviewing plugin for XXXX protocol |
| Colormap display | Display DICOM images using different colormaps (useful for mapping) |
| Download and de-identify | Download DICOM files/series/exams with de-identifying custom tags |
Some existing ArchiMed plugins.
Discussion and Conclusion
ArchiMed is a complete storage and visualization solution respecting legal requirements and research obligations. It is in function for 5 years at CIC-IT of Nancy-France and is a handy research data management system for our 60 staff, among whom are researchers, clinicians, and clinical trial project managers for local or multicenter studies.
Initially, based on the internal needs of our lab to safely store and easily access imaging data, it has met all our expectations and is used in more than 100 clinical protocols at CIC-IT of Nancy-France, France since 2011.
Collaborations
It turns out that the functional organization of clinical imaging data covered by ArchiMed is not only our own needs. Many labs and research centers specialized in imaging face the same issues and are experiencing difficulties in finding a solution dedicated for research and meeting legal requirements for data preservation and confidentiality. This is why ArchiMed has won great attention from our partners in France. The very first external deployment of our ArchiMed system was with CIC-IT of Tours24 under their request. Since 2015, ArchiMed has been under function for their local clinical protocols. More installations are currently being considered.
In the context of the French research infrastructure in imaging (France Life Imaging, FLI), we will interconnect all research data storage systems in France. The FLI-IAM25 workgroup will use already existing data storage and information processing facilities (including CATI,26 Shanoir,27 and ArchiMed) and will increase their capacities for the FLI infrastructure.
Current Limitations and Improvement
About Metadata and Traceability
So far, metadata extraction and de-identification rules only depend on file types and are not related to a specific study. It can be problematic especially when we consider that some metadata can be relevant in a specific study context but unnecessary in another one (e.g., MR diffusion tensor directions will be an important factor for brain study while heart’s beat parameter will be more relevant in cardiovascular study). For these reasons, we are thinking about building in a study profile which defines, by study, what header information will be extracted and available as metadata in the database and what header information will be deleted or de-identified.
Moreover, current version of our software only logs errors and information about data insertion/modification/delete (at exam node level). For recording user activities and tracking workflow better, it should be interesting to keep reports of data consultation/download by user and client application destination (processing tool, viewing platform, etc.).
Cloud
ArchiMed has been created to be a local solution, only accessible from a local area network by authorized and identified users. However, in the age of Cloud computing (Rimal et al., 2009), it will be proper to offer a secured and fully online version of ArchiMed. We are seriously considering upgrading it as a cloud service based on a Software as a service (SaaS)28 (Levinson, 2007) model. Current service oriented architecture renders ArchiMed technically compatible with cloud infrastructure, but the impact of external storage in terms of data security, confidentiality, and legal obligations have to be taken into account.
In conclusion, ArchiMed is a well-adapted research PACS. It is working for more than 5 years at CIC-IT of Nancy-France and perfectly matches with clinical research needs in terms of workflow, organization, legal requirement, and usability.
Glossary
CNIL—Commission Nationale de l’Informatique et des Libertés (National Commission on Informatics and Liberty) is an independent French administrative regulatory body whose mission is to ensure that data privacy law is applied to the collection, storage, and use of personal data.
DICOM—Digital Imaging and Communications in Medicine is a standard for handling, storing, printing, and transmitting information in medical imaging. It includes a file format definition and a network communications protocol.
FTP—The File Transfer Protocol is a standard network protocol used to transfer computer files between a client and server on a computer network.
HTTP—The Hypertext Transfer Protocol is an application protocol for distributed, collaborative, hypermedia information systems.
Java™—Java is a general-purpose computer programing language that is concurrent, class-based, object-oriented, and specifically designed to have as few implementation dependencies as possible.
Java-EE™—Java Platform, Enterprise Edition is a widely used enterprise computing platform developed under the Java Community Process.
LDAP—The Lightweight Directory Access Protocol is an open, vendor-neutral, industry standard application protocol for accessing and maintaining distributed directory information services over an Internet Protocol (IP) network.
MRI—Magnetic resonance imaging is a medical imaging technique used in radiology to image the anatomy and the physiological processes of the body.
PACS—Picture Archiving and Communication System is a medical imaging technology which provides economical storage and convenient access to images from multiple modalities (source machine types).
SOAP—Simple Object Access Protocol is a protocol specification for exchanging structured information in the implementation of web services in computer networks.
Statements
Author contributions
All authors listed have made substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
We acknowledge all members of CIC-IT of Nancy-France to taking part in the different tests and discussions regarding software improvements and debug.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1.^CIC-IT: Clinical investigation center for Technology and Innovation.
2.^«DICOM Homepage», http://medical.nema.org/.
3.^Image raw data: image signal before any treatment or process. In MRI context, raw data contain “Fourier transform” of the MR image measured, before any reconstruction or filtering.
4.^Commission nationale de l’informatique et des libertés (CNIL), https://www.cnil.fr/.
5.^Méthodologie de référence MR-001 pour les traitements de données personnelles opérées dans le cadre des recherches biomédicales, https://www.cnil.fr/sites/default/files/atoms/files/mr-001.pdf.
6.^Article R1123-61 https://www.legifrance.gouv.fr/affichCodeArticle.do?cidTexte=LEGITEXT000006072665&idArticle=LEGIARTI000006908442&dateTexte=&categorieLien=cid.
7.^Shanoir (Sharing NeurOImaging Resources), http://www.shanoir.org/.
8.^CATI Neuroimaging, http://cati-neuroimaging.com.
9.^3-tier architecture is a client–server typically composed of a presentation tier, a domain logic tier, and a data storage tier. This is the most used architecture for service oriented applications.
10.^Java EE at a glance, http://www.oracle.com/technetwork/java/javaee/overview/index.html.
11.^Glassfish Application Server, https://glassfish.java.net/.
12.^Mapping Objects to Relational Databases: O/R Mapping In Detail, http://www.agiledata.org/essays/mappingObjects.html.
13.^Web Services Architecture, https://www.w3.org/TR/ws-arch/.
14.^Java Servlet Technology Overview, http://www.oracle.com/technetwork/java/javaee/servlet/index.html.
15.^File Transfer Protocol, https://tools.ietf.org/html/rfc959.
16.^Lightweight Directory Access Protocol (LDAP): the Protocol, https://tools.ietf.org/html/rfc4511.
17.^fMRI: functional magnetic resonance imaging or functional MRI (fMRI) is a functional neuroimaging procedure using MRI technology that measures brain activity by detecting changes associated with blood flow.
18.^Transactional processing is designed to maintain a database or file system’s integrity ensuring that interdependent operations on the system are either all completed successfully or all canceled successfully.
19.^Hypertext Transfer Protocol, https://www.rfc-editor.org/info/rfc2616.
20.^Microsoft Active Directory, https://technet.microsoft.com/en-us/library/cc977985.aspx.
21.^De-identification (DICOM in general). https://wiki.nci.nih.gov/display/Imaging/De-identification+(DICOM+in+general).
22.^ORS – Radiology Software, PACS, DICOM Viewer and Medical Imaging, http://www.theobjects.com/en/.
23.^MATLAB™ – MathWorks, http://mathworks.com/products/matlab/.
24.^Clinical Investigation Center of Tours (France), http://cic-it-tours.fr/.
25.^France Life Imaging – Information Analysis and Management (IAM) Node, https://project.inria.fr/fli/en/.
26.^CATI Neuroimaging, http://cati-neuroimaging.com.
27.^Shanoir (Sharing NeurOImaging Resources), http://www.shanoir.org/.
28.^SaaS (Software as a Service), https://en.wikipedia.org/wiki/Software_as_a_service.
References
1
BracardS.DucrocqX.MasJ. L.SoudantM.OppenheimC.MoulinT.et al (2016). Mechanical thrombectomy after intravenous alteplase versus alteplase alone after stroke (THRACE): a randomised controlled trial. Lancet Neurol.15, 1138–47.10.1016/S1474-4422(16)30177-6
2
ChoplinR. H.BoehmeJ. M.MaynardC. D. (1992). Picture archiving and communication systems: an overview. Radiographics12, 127–129.10.1148/radiographics.12.1.1734458
3
FieldingR.GettysJ.MogulJ.FrystykH.MasinterL.LeachP.et al (1999). Hypertext Transfer Protocol – HTTP/1.1. Available at: https://www.rfc-editor.org/info/rfc2616
4
GoncalvesA. (2009). Beginning Java EE 6 Platform with GlassFish 3: From Novice to Professional, 1st Edn. APress.
5
KoutsonikolaV.VakaliA. (2004). LDAP: framework, practices, and trends. IEEE Internet Comput.8, 66–72.10.1109/MIC.2004.44
6
KushidaC. A.NicholsD. A.JadrnicekR.MillerR.WalshJ. K.GriffinK. (2012). Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med. Care50(Suppl.), S82–S101.10.1097/MLR.0b013e3182585355
7
LevinsonM. (2007). Software As a Service (SaaS) Definition and Solutions. CIO. Available at: http://www.cio.com/article/2439006/web-services/software-as-a-service–saas–definition-and-solutions.html
8
OdilleF.VuissozP.-A.MarieP.-Y.FelblingerJ. (2008). Generalized reconstruction by inversion of coupled systems (GRICS) applied to free-breathing MRI. Magn. Reson. Med.60, 146–157.10.1002/mrm.21623
9
PostelJ.ReynoldsJ. (1985). File Transfer Protocol. Available at: https://tools.ietf.org/html/rfc959
10
RimalB. P.ChoiE.LumbI. (2009). “A taxonomy and survey of cloud computing systems,” in Fifth International Joint Conference on INC, IMS and IDC, 2009. NCM ’09 (Seoul: IEEE), 44–51.
11
TuckerK.BransonJ.DilleenM.HollisS.LoughlinP.NixonM. J.et al (2016). Protecting patient privacy when sharing patient-level data from clinical trials. BMC Med. Res. Methodol.16(Suppl. 1):77.10.1186/s12874-016-0169-4
12
van de WeteringR.BatenburgR.VersendaalJ.LedermanR.FirthL. (2006). A balanced evaluation perspective: picture archiving and communication system impacts on hospital workflow. J. Digit. Imaging19, 10–17.10.1007/s10278-006-0628-2
13
ZeilengaK. (2006). Lightweight Directory Access Protocol (LDAP): Technical Specification Road Map. Available at: https://tools.ietf.org/html/rfc4510.html
Summary
Keywords
clinical research, infrastructure, data storage system, imaging, database, web services, centralized resources, data sharing
Citation
Micard E, Husson D, CIC-IT Team and Felblinger J (2016) ArchiMed: A Data Management System for Clinical Research in Imaging. Front. ICT 3:31. doi: 10.3389/fict.2016.00031
Received
31 August 2016
Accepted
05 December 2016
Published
20 December 2016
Volume
3 - 2016
Edited by
Michel Dojat, INSERM, France
Reviewed by
Alex Pappachen James, Nazarbayev University, Kazakhstan; Avan Suinesiaputra, University of Auckland, New Zealand
Updates
Copyright
© 2016 Micard, Husson, CIC-IT Team and Felblinger.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emilien Micard, e.micard@chru-nancy.fr
Specialty section: This article was submitted to Computer Image Analysis, a section of the journal Frontiers in ICT
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.