Introduction
Helicobacter pylori (H. pylori) infection affects approximately half of the world’s population. Unless eradicated, it remains in the stomach throughout life. After 1983, discovery of H. pylori as persistent resident of gastric mucosa, changed the traditional thoughts and perspectives regarding (i) having sterile stomach lifelong, (ii) infectious potential links existence with various extra gastric disorders, and (iii) ongoing researches on H. pylori-induced diseases (1). Undeniably, colonization with pathogenic H. pylori results in severe gastroduodenal disorders. H. pylori is well-recognized as an established causative agent associated with a wide variety of upper gastroduodenal diseases ranged from a chronic gastritis to gastric cancer. Surprisingly, gastric colonization with H. pylori induces superficial gastritis in all infected individuals, while only a minority develops to severe symptomatic diseases (2). Notably, rationale underlying this unique distribution of diseases is driven by a sophisticated and mysterious interplay between H. pylori and its host. Broadly defined, the certain pattern of H. pylori-induced digestive disease is strongly influenced by bacterial virulence factors which draw the final outcome of infection (2, 3). It is now firmly established that biomarkers allow us for having an early diagnosis and prediction of medical conditions. Moreover, biomarkers provide opportunity to modify current available protocols to manage the infection and its associated outcomes (4). In this case, a biologic biomarker can be used for early diagnosis of certain digestive disease (e.g., gastric cancer, duodenal ulcer); and even identification of high-risk population for disease prevention. As such, biomarkers are becoming increasingly important tools in clinical settings. Thus, potential applications of biomarkers in infectious diseases such as predicting outcomes would be an interesting area of ongoing research. With this regard, the search for relevant biomarkers that diagnose/predict a clinical condition among the H. pylori-infected patients is a challenging area of research. Following several suggested H. pylori biomarkers for certain digestive diseases over the past years (5–8), still a biomarker capable of predicting definitive digestive diseases outcome is lacking for clinical settings.
Definition and Application of the Opinion
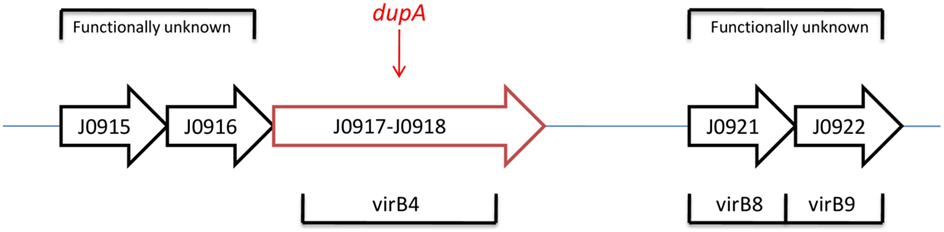
In this new opinion, under condition of identification of complete duodenal ulcer promoting (dupA) gene, our knowledge about biomarker application in H. pylori-induced diseases such as duodenal ulcer and gastric cancer will be influenced greatly. Accordingly, we propose a logical and practical opinion that detection of full length (dupA and its flanking gene) (Figure 1) H. pylori dupA gene by simple multiplex PCR assay can be used to predict digestive diseases outcome and solve the above-mentioned problems. To examine H. pylori dupA as biomarker that can serve as an indicator of the digestive disease potential, our opinion can be easily be evaluated in a cohort of population with different disease groups including gastric cancer, duodenal ulcer, and gastritis. Furthermore, primer set which can pick whole dupA gene in addition to both left and right sides of the dupA gene (Figure 1) would be preferable to investigate likely involvement of this cluster in determining final diseases outcome. To date, an actual function of dupA is still not fully understood. Moreover, flanking genes to dupA (virB4, virB8, and virB9) are also not determined. Strikingly, the opinion stands the best situation of being confirmed if dupA as part of (Type 4 secretion system) T4SS can show in vivo activity.
Discussion
Current opinion would be confirmed by a finding that dupA is forming T4SS in combination of those flanking genes. Indeed, function of flanking genes to dupA (virB4, 8, and 9) are not determined yet (Figure 1). Hence, this interesting gap promotes researchers to examine it within a cohort study. Of note, dupA gene encodes homologs of virB4 ATPase which current knowledge found it involved in DNA transfer/uptake. Involvement of T4SS in pathogenesis of H. pylori is an apparent fact. Accordingly, current evidences are indicating on potential role of dupA to form a functional T4SS. One further step would be to check those flanking genes in combination of dupA; an opinion which might be able to answer those unclear points regarding dupA after its introduction in 2005 to now (9, 10). Notably, dupA and its flanking genes are located in plasticity region (PR) of the H. pylori genome (11). It is important to point out that the vir genes exist before and after the region of the dupA locus and the surrounding six vir gene homologs (virB8, virB9, virB10, virB11, virD4, and virD2) are important in forming a novel putative T4SS (tfs3a). As a result, H. pylori strains containing full dupA are bound to be virulent due to the ability of building a complete T4SS. In a continuous cluster in H. pylori strain J99, the PR has been reported to range from jhp0914 to jhp0961 (Figure 1) (12). In H. pylori genome, PR is an area where G + C content is lower than that of the rest (35% compared with 39%), indicating on variable genes which mostly are virulence associated (11). Thus, the polymorphism pattern in this area of PR can be considered as related with different set of bacterial virulence. That would be rationally possible that sequential contribution of these genes be involved to determine final outcome of the H. pylori in colonized individuals. However, this article is first to present this possibility; an opinion can be promising if proven after those mentioned in in vivo tests.
Conclusion
This paper is an invitation for having a different look in to the biomarkers for digestive diseases. Current opinion encourage using H. pylori dupA as a predicting tool to screen certain types of digestive diseases such as duodenal ulcer and gastric cancer.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The contents of the paper are the sole responsibility of the author and do not necessarily represent the official views of any institute or organization.
References
1. Talebi Bezmin Abadi A. Therapy of Helicobacter pylori: present medley and future prospective. Biomed Res Int (2014) 2014:7. doi: 10.1155/2014/124607
2. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev (2006) 19(3):449–90. doi:10.1128/CMR.00054-05
3. Olivares D, Gisbert JP. Factors involved in the pathogenesis of Helicobacter pylori infection. Rev Esp Enferm Dig (2006) 98(5):374–86. doi:10.4321/S1130-01082006000500008
4. Chau CH, Rixe O, McLeod H, Figg WD. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res (2008) 14(19):5967–76. doi:10.1158/1078-0432.CCR-07-4535
5. Saha DR, Datta S, Chattopadhyay S, Patra R, De R, Rajendran K, et al. Indistinguishable cellular changes in gastric mucosa between Helicobacter pylori infected asymptomatic tribal and duodenal ulcer patients. World J Gastroenterol (2009) 15(9):1105–12. doi:10.3748/wjg.15.1105
6. Liu J, Liu H, Zhang T, Ren X, Nadolny C, Dong X, et al. Serum Helicobacter pylori NapA antibody as a potential biomarker for gastric cancer. Sci Rep (2014) 4:4143. doi:10.1038/srep04143
7. Talebi Bezmin Abadi A, Taghvaei T, Tabrizi FH, Mobarez AM. Biomarker in Helicobacter pylori infection: the standoff condition? Rev Soc Bras Med Trop (2013) 46(4):529–30. doi:10.1590/0037-8682-0109-0128-2013
8. Ramis IB, Vianna JS, Silva Junior LV, Von Groll A, Silva PE. cagE as a biomarker of the pathogenicity of Helicobacter pylori. Rev Soc Bras Med Trop (2013) 46(2):185–9. doi:10.1590/0037-8682-0054-2012
9. Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology (2005) 128(4):833–48. doi:10.1053/j.gastro.2005.01.009
10. Abadi ATB, Taghvaei T, Wolfram L, Kusters JG. Infection with Helicobacter pylori strains lacking dupA is associated with an increased risk of gastric ulcer and gastric cancer development. J Med Microbiol (2012) 61(1):23–30. doi:10.1099/jmm.0.027052-0
Keywords: biomarker, Helicobacter pylori, dupA, digestive system, gene cluster
Citation: Talebi Bezmin Abadi A (2014) The Helicobacter pylori dupA: a novel biomarker for digestive diseases. Front. Med. 1:13. doi: 10.3389/fmed.2014.00013
Received: 25 April 2014; Accepted: 15 May 2014;
Published online: 28 May 2014.
Edited by:
Zsolt Molnár, University of Szeged, HungaryCopyright: © 2014 Talebi Bezmin Abadi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:YW1pbi50YWxlYmlAZ21haWwuY29t
 Amin Talebi Bezmin Abadi
Amin Talebi Bezmin Abadi