- 1Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, Neuchâtel, Switzerland
- 2Faculty of Biology and Medicine, University of Lausanne, Service de Pneumologie, Centre Hospitalier Universitaire Vaudois (CHUV), Lausanne, Switzerland
Fungal infections are estimated to be the main cause of death for more than 1.5 million people worldwide annually. However, fungal pathogenicity has been largely neglected. This is notably the case for pulmonary fungal infections, which are difficult to diagnose and to treat. We are currently facing a global emergence of antifungal resistance, which decreases the chances of survival for affected patients. New therapeutic approaches are therefore needed to face these life-threatening fungal infections. In this review, we will provide a general overview on respiratory fungal infections, with a focus on fungi of the genus Aspergillus. Next, the immunological and microbiological mechanisms of fungal pathogenesis will be discussed. The role of the respiratory mycobiota and its interactions with the bacterial microbiota on lung fungal infections will be presented from an ecological perspective. Finally, we will focus on existing and future innovative approaches for the treatment of respiratory fungal infections.
Respiratory Fungal Infections
Fungal pathogens are estimated to lead to more than 1.5 million deaths every year worldwide, with a global burden exceeding one billion (1). Despite this, the issue of fungal pathogenicity has been largely neglected (2, 3). Over the past two decades, the prevalence of invasive fungal diseases has increased considerably (1). This has also been acknowledged in the case of healthcare-associated invasive fungal infection (4, 5), for which a call to action was recently issued by the scientific community (6). Moreover, the increased prevalence of invasive fungal diseases correlates with an increasing number of vulnerable at-risk patients, which include among others, immunosuppressed individuals due to transplants, AIDS, cancer, corticosteroid therapies or autoimmune diseases, or patients undergoing major surgery (1, 7).
The most prevalent human fungal pathogens are the airborne opportunists Aspergillus spp., Cryptococcus spp., and Pneumocystis spp., as well as the human-associated commensal and polymorphic fungal species Candida albicans (7, 8). These fungi are responsible for more than 90% of all reported fungal disease-related deaths (9). The latest estimates of the annual burden of fungal diseases amount to more than 14 million cases for all diseases within the pulmonary aspergillosis spectrum, over 200,000 cases of cryptococcal meningitis, 500,000 cases of Pneumocystis jirovecii pneumonia, 700,000 cases of invasive candidiasis, and over 10 million cases of asthma with fungal sensitisation (1, 9, 10).
In the current review, we will first look at the full spectrum of diseases caused by fungi of the genus Aspergillus, with a particular focus on the pathogenesis and the underlying immunological mechanisms. Then, lung ecology, and more specifically the interaction of the respiratory mycobiota (fungal composition) with the bacterial microbiota and the virome will be discussed in the context of fungal infection. Finally, we will discuss the current therapeutic approaches, as well as future perspectives in therapeutic strategies for the fight against pulmonary aspergillosis.
Pulmonary Aspergillosis: A Wide Spectrum of Diseases
Aspergillus spp. are the most frequently isolated filamentous fungi in humans and animals (11–13). These primarily saprotrophic fungi are widespread in the environment and can be found in soil and decaying biomass, especially in compost piles, where they participate in the degradation of organic matter (14–16). Their wide environmental distribution can be explained by the competitiveness and adaptability of the Aspergillus genus (17). Indeed, Aspergillus spp. are able to use multiple organic substrates and adapt to a broad range of environmental conditions (12). These fungi show a remarkable phenotypic plasticity in their ecology and stress-responses, which are believed to be at the basis of the success of Aspergillus spp. as opportunistic pathogens.
Fungi of the genus Aspergillus are associated with a large variety of clinical manifestations ranging from allergic reactions to life-threatening invasive infections. Such infections are generally caused by Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, Aspergillus nidulans, and Aspergillus terreus, with Aspergillus fumigatus being responsible for 90% of the reported cases (12). Respiratory infections due to Aspergillus spp. are caused by inhalation of airborne conidia, i.e., asexual spores (14). These fungi produce large quantities of small airborne conidia with a size of 2–5 μm in diameter (15, 18), whose concentration can range from 1 to 100 per m3 in air, but can reach up to 108 per m3 in some environments (13). A human inhales approximately 100–1,000 conidia per day, which can reach the lung alveoli due to their small size (15). In immunocompetent individuals, inhaled conidia are usually efficiently cleared either by mucociliary movement or through phagocytosis by macrophages (Figure 1) (18). However, depending on the immunological status of the host, Aspergillus spp. can lead to a variety of pathologies (12).
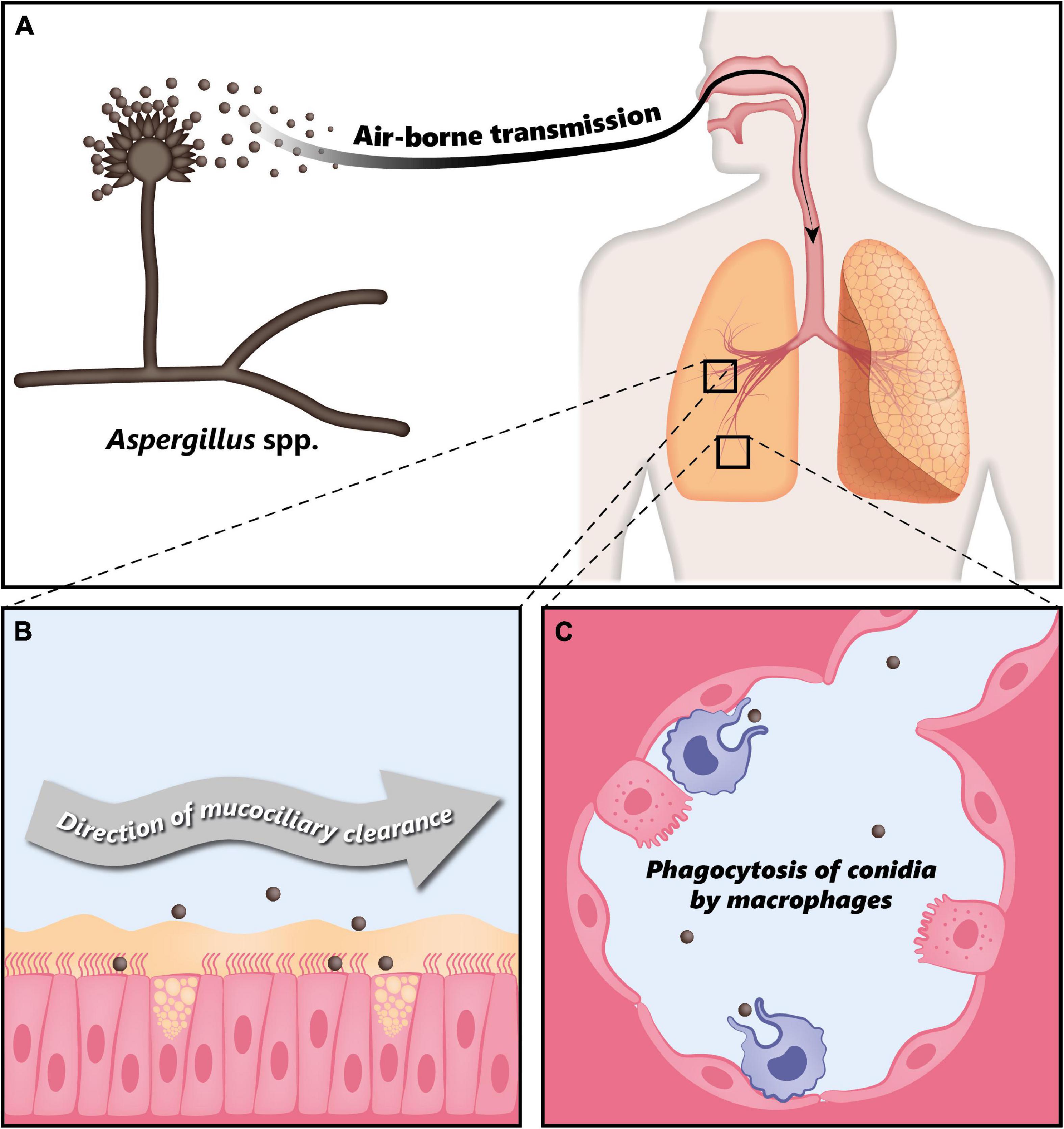
Figure 1. Clearance of Aspergillus conidia in the immunocompetent host. (A) Aspergillus spp. conidia are transmitted through air and every individual inhales thousands of conidia every day. (B) In the immunocompetent host, most of the inhaled conidia are trapped by the mucus layer secreted by the tracheal and bronchial epithelium, and are efficiently eliminated through mucociliary clearance. (C) Due to their small size, conidia can eventually reach the alveoli, where they are phagocytosed by alveolar macrophages. Modified from Palmieri (170).
Pulmonary aspergillosis is classified into three different groups with distinct clinical manifestations (11, 13, 19). The disease spectrum of pulmonary aspergillosis spans from hypersensitivity responses (asthma or allergic bronchopulmonary aspergillosis—ABPA), to colonization (i.e., presence of the fungus without any clinical, radiological or laboratory indications of active fungal disease), to infection (chronic or invasive aspergillosis). Figure 2 presents a diagram showing the disease spectrum of pulmonary aspergillosis depending on the host status.
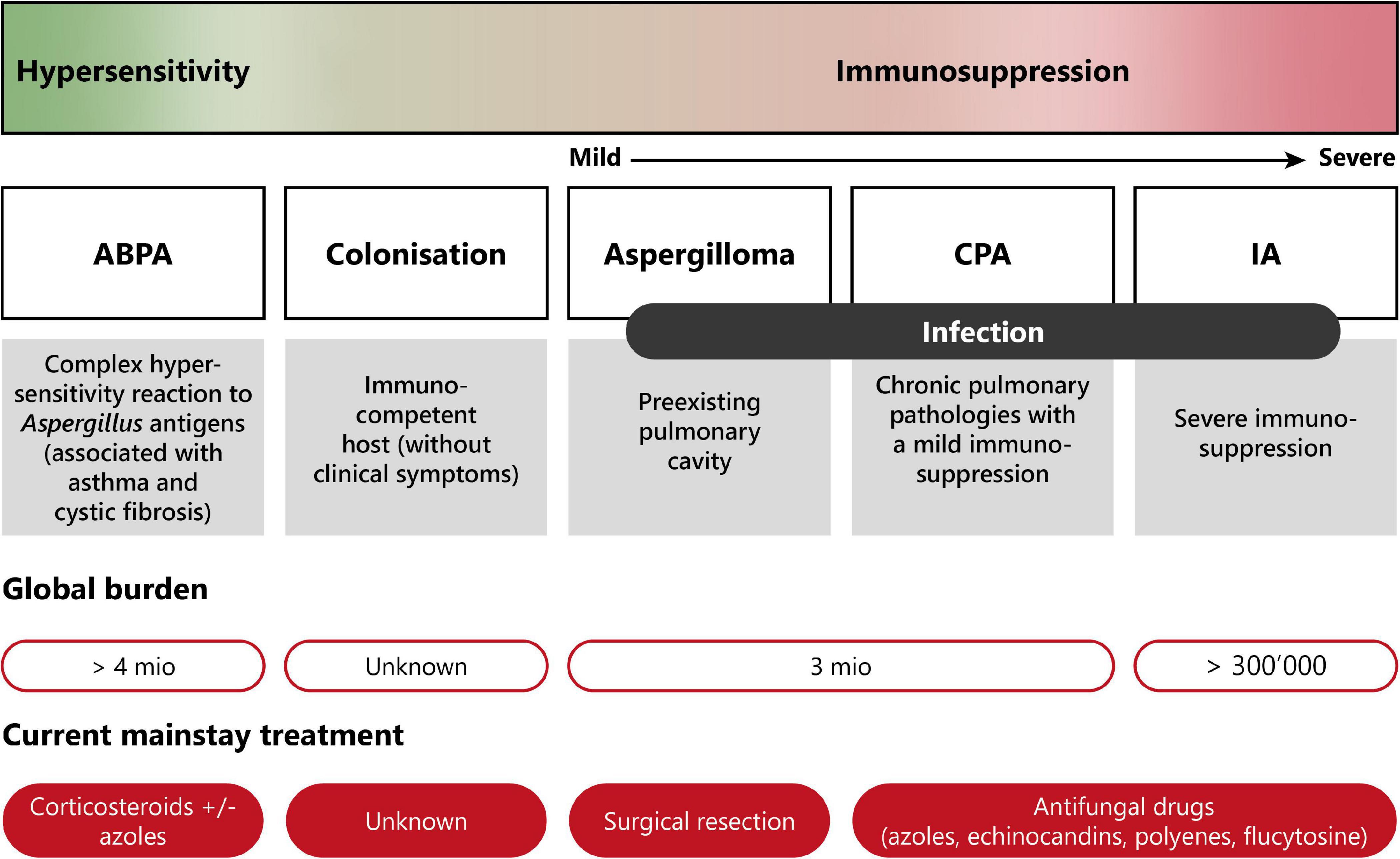
Figure 2. Disease spectrum of pulmonary aspergillosis. This diagram summarizes the diverse pathologies caused by Aspergillus spp., due to improper elimination of conidia by mucociliary clearance or macrophage phagocytosis following inhalation. Depending on the immune status of the host, pulmonary aspergillosis can range from allergic reaction (hypersensitivity) to life-threatening invasive infection (severe immunosuppression). Moreover, current burden and treatment information are indicated. ABPA, allergic bronchopulmonary aspergillosis; CPA, chronic pulmonary aspergillosis; IA, invasive aspergillosis. Modified from Palmieri (170).
Hypersensitivity Responses
Although other fungi can cause allergic bronchopulmonary mycoses (ABPM), the vast majority of hypersensitivity responses are associated to Aspergillus spp. These can range from fungal asthma to allergic bronchopulmonary aspergillosis (ABPA), the latter being a complex type I, III, and IV hypersensitivity response observed notably in patients with cystic fibrosis (CF) or chronic asthma (13). Hypersensitivity to Aspergillus is characterized by high levels of Aspergillus-specific IgE (19). ABPA affects close to 5 million patients worldwide (1, 10).
Colonization
In immunocompetent hosts, Aspergillus spp. may colonize the lungs without any clinical manifestations (10). In a study from Soubani and colleagues where Aspergillus spp. were isolated from sputum samples of 66 elderly hospitalized patients, 92% cases were determined to be Aspergillus colonization and only 4.5% fulfilled the criteria of invasive aspergillosis (20). Appropriate diagnostics and close monitoring should be considered in order to discriminate simple colonization from invasive infection (10, 11, 19). In immunocompromised patients, prior fungal colonization in the lower respiratory airways is considered an important risk factor for the development of invasive aspergillosis (10).
Chronic Pulmonary Aspergillosis
Aspergillus spp. can also cause a chronic, non-invasive form of infection called chronic pulmonary aspergillosis (CPA). One form of CPA is aspergilloma, which is characterized by the proliferation of the fungus inside a pre-existing cavity, leading to the development of a fungus ball (21). Aspergilloma typically occurs in immunocompromised patients previously suffering from lung pathologies such as tuberculosis, lung abscess, cysts, or tumors (21). Another form is chronic cavitary pulmonary aspergillosis, also called chronic necrotizing aspergillosis or complex aspergilloma. This is an inflammatory form of the infection characterized by the production of serum IgG antibodies directed to Aspergillus, elevated acute-phase inflammation markers, and the absence of pulmonary or vascular invasion. CPA usually occurs in immunocompetent or mildly immunosuppressed patients. CPA, including aspergilloma, is estimated to affect more than 3 million people worldwide (1).
Invasive Pulmonary Aspergillosis
On the other side of the disease spectrum, invasive pulmonary aspergillosis (IPA) is the most severe and life-threatening form of Aspergillus infection occurring in immunosuppressed patients. IPA affects more than 300,000 patients annually and its mortality rate ranges from 30 to 80% (1, 10). IPA is characterized by the invasion of the lung tissue by Aspergillus hyphae, which can be followed by angioinvasion and dissemination to other organs in patients with prolonged neutropenia (22). Other at-risk patients include individuals who underwent hematopoietic stem cell transplantation, solid-organ transplantation, prolonged corticosteroid therapy, or those who have AIDS (11, 13, 19). Aspergillus spp. are the most common opportunistic fungal pathogens causing invasive pulmonary aspergillosis in lung transplant recipients, with an incidence of 40.5 cases per 1,000 patients annually, despite the use of prophylactic antifungal treatments (23). Invasive aspergillosis most commonly occurs within 1 year after transplantation, with the majority of the cases reported within the first 6 months (23, 24).
Fungal Infections in Chronic Respiratory Diseases
In healthy individuals, innate immune responses and specifically macrophages and ciliated bronchial epithelial cells contribute to the efficient clearance of fungal conidia. In patients with chronic respiratory diseases, such as chronic obstructive pulmonary disease (COPD), asthma, and CF, these clearance mechanisms are impaired predisposing to fungal colonization and infection. In the following paragraphs, we will summarize the role of fungal pathogens in specific pulmonary diseases, focusing especially on Aspergillus spp.
Chronic Obstructive Pulmonary Disease
Chronic obstructive pulmonary disease patients are often affected by exacerbations due to bacterial infections. The use of long-term inhaled corticosteroids and courses of oral steroids to treat exacerbations can predispose to fungal colonization and infection, as these treatments lead to impaired host immunity (25–27). In addition, in COPD, environmental fungal sensitization has been associated with frequent exacerbations (28). Bafadhel and colleagues reported that positive cultures for filamentous fungi are common in COPD, however, this finding was not related to exacerbations (26). Moreover, A. fumigatus sensitization was associated with poor lung function and, interestingly, patients with a positive A. fumigatus culture were on higher inhaled corticosteroid doses and had higher total and percentage sputum neutrophil counts (26). Recently, Tiew and colleagues evaluated the airway mycobiome in COPD patients in a multicenter study and observed that COPD patients with very frequent exacerbations (≥3 per year) had an increased number of fungal interactions (29), which is suggestive of a more complex mycobiome. Using unsupervised hierarchical clustering of the COPD mycobiome, the authors reported two distinct patient clusters with variable clinical outcomes: the first cluster was characterized by Saccharomyces and increased symptoms, whereas the second cluster was characterized by Aspergillus, Curvularia, and Penicillium and demonstrated poorer clinical outcomes with increased exacerbations and higher mortality (29).
Innate immune cells and macrophages in particular play an important role in the first line of pulmonary host defense. In COPD patients, it has been demonstrated that alveolar macrophages exhibit reduced phagocytic capacity (30, 31). Monocyte-derived macrophages from both smokers and COPD patients were shown to be defective in their phagocytic and pro-inflammatory cytokine responses following A. fumigatus exposure (32). This impairment in macrophage function may consequently contribute to fungal germination, dissemination and infection, and lung damage in COPD patients. There is an increasing interest in understanding the direct influence of fungal colonization and infection on COPD pathogenesis and exacerbations. However, future research should continue to consider the indirect effects of bacterial composition alterations on fungal community composition in the lung through inter-kingdom interactions, and the potential consequences for COPD patients.
All diseases within the aspergillosis spectrum can be found in COPD patients although their prevalence differ. Tiew and colleagues have recently reviewed this topic in depth (33).
Asthma
Genetic and environmental factors drive asthma development, progression and risk for exacerbation. Changes in fungal community composition in the gut are associated with susceptibility to develop asthma in humans (34, 35), however, causality has not been established yet. Analysis of the airway mycobiome in asthma patients (fungal-sensitized and non-fungal sensitized) and healthy controls indicated that both the sputum and bronchoalveolar lavage (BAL) mycobiome was dominated by three species: A. fumigatus, C. albicans, and Mycosphaerella tassiana, irrespective of health status (36). Interestingly, other fungi such as Aspergillus tubingensis, a member of the A. niger species complex, was also prominent in the BAL fluid. Alterations in the balance of fungi detected in the lung were found to be associated with several disease markers, including asthma status and duration, and inflammatory biomarkers.
Fungi can play an important role in asthma development as fungal colonization and sensitization often take place in early life. Moreover, fungi are predominant triggers of asthma exacerbations. Environmental presence of fungi, such as A. alternata, in house dust has been associated with active asthma symptoms (37). In addition, a meta-analysis of seven studies revealed that indoor presence of Cladosporium, Alternaria, Aspergillus, and Penicillium was associated with enhanced asthma exacerbations in both children and adults (38). Descriptive and mechanistic studies have started to reveal the influence of alterations in the gut bacterial and fungal composition on asthma development (discussed in section “Gut-Lung Axis”). However, the influence of a change in the composition of the pulmonary bacterial community due to asthma, on the susceptibility to develop a respiratory fungal infection, and subsequent asthma exacerbation, is currently unexplored.
Cystic Fibrosis
Cystic fibrosis is a rare autosomal recessive disorder that causes severe damage to the lungs, digestive system and other organs. The microenvironment in the lungs of CF patients, which is characterized by depletion of the airway surface liquid layer leading to impaired mucociliary clearance, is ideal for microbial colonization (39). Bacterial pathogens, and most commonly Pseudomonas aeruginosa, are known contributors to disease progression and exacerbations (40). Although fungi are often isolated from the lower airways of CF patients, the clinical impact of their presence and especially for the development of non-allergic fungal disease is poorly understood (41, 42). The most frequently detected fungi in the airways of CF patients are Aspergillus spp., notably A. fumigatus, and Candida spp. (43). The prevalence of Aspergillus colonization was highest in adolescents and young adults (44). Risk factors for Aspergillus colonization in people with CF include age and the use of inhaled corticosteroids and antibiotics (44). Moreover, aggressive use of antibiotics have been suggested to contribute to the increase in fungal colonization (45). This suggests that inter-kingdom interactions may be important in containing fungal colonization in the respiratory tract.
Interestingly, A. fumigatus colonization in people with CF is often preceded by P. aeruginosa infection (46). In patients infected with both A. fumigatus and P. aeruginosa, more severe clinical outcomes have been observed when compared to those infected with P aeruginosa alone (47). P. aeruginosa can have both antifungal and growth stimulating effects on A. fumigatus resulting in: (1) inhibition, (2) reciprocal antagonism, and (3) cooperation (48), which we will briefly describe here.
The main mechanisms by which P. aeruginosa can inhibit A. fumigatus’ growth is via the release of phenazines, including pyocyanin, phenazine-1-carboxamide, 1-hydroxyphenazine, and phenazine-1-carboxylic acid. Phenazines are small diffusible quorum-sensing molecules, which easily penetrate A. fumigatus conidia, and are considered a strong virulence factor of P. aeruginosa (49–51). The quorum-sensing system allows bacteria to assess cell density and to regulate physiological activities accordingly, which consequently modulate the pathogenicity of the microorganisms (48, 52). Moreover, nutrient availability and competition is also involved in this inhibitory process. As an example, P. aeruginosa and A. fumigatus can both compete for the utilization of iron as a central nutrient for their survival (53). Although P. aeruginosa has a variety of fungicidal mechanism, alterations in these fungicidal capacities have been observed in clinical isolates from CF patients (52, 54), indicating that there can be shifts between inhibition and cooperation. Several cooperative interactions have been described which stimulate growth and potentially contribute to disease progression. Phenazines can stimulate fungal growth via increasing iron bioavailability (50). Moreover, pyochelin, a siderophore, can be used by A. fumigatus as a ferrochelator. Interestingly, dimethyl sulfide, a volatile organic compound released by P. aeruginosa, can communicate with A. fumigatus and create a positive growth environment resulting in stimulation of fungal growth (48, 55).
The Climax-Attack model (CAM) is a theory which has been proposed for CF a few years ago grounded on basic ecological principles (56). In this theory it is postulated that there are two major functional communities in CF pulmonary disease. The attack community consists of transient viral and microbial populations that induce strong innate immune responses. Consequent alterations in the immune response create a microenvironment that facilitates a climax chronic community having a reduced growth rate and being inherently resistant to antibiotic therapy (56, 57). Soret and colleagues provided important information on the involvement of the mycobiome in the CAM model in CF pulmonary exacerbations. They inferred an inter-kingdom network by plotting bacterial genera significantly correlated with at least one fungal genus and vice versa. Network analysis revealed three main clusters organized around Aspergillus, Candida and Scedosporium genera (57). The positively correlated OTUs predicted interactions of these three fungal genera with bacteria belonging to Capnocytophaga, Parvimonas, Streptococcus, or Veillonella. In addition, these interactions were assessed using in vitro co-cultures between A. fumigatus and Streptococcus mitis or Streptococcus oralis and confirmed that both S. mitis and S. oralis enhanced A. fumigatus growth. Such translational studies in which principles from ecology are used to understand disease can potentially form the basis for the future development of therapeutic strategies to combat exacerbations.
SARS-CoV-2 Co-infection
The current SARS-CoV-2 coronavirus pandemic created the perfect arena for the establishment of opportunistic fungal co-infections. The use of high dose systemic glucocorticoids, which are widely used as an anti-inflammatory medication for COVID-19 (58–60), together with epithelial cell damage in the lung following SARS-CoV-2 infection, expose patients to opportunistic fungal infections, such as COVID-19 associated invasive pulmonary aspergillosis (CAPA) (61). To date, over 100 cases of CAPA have been reported (61). Moreover, invasive mucormycosis, also known as “black fungus,” has been largely reported in convalescent COVID-19, particularly in India and other Asian countries, in patients with uncontrolled diabetes mellitus or immunosuppression (62). The main reason for the increase in invasive mucormycosis cases has been pointed out to be the elevated iron levels in the serum of convalescent COVID-19 patients (62). Complementary to dexamethasone, Tocilizumab, an IL-6 receptor antagonist, has been widely used to treat COVID-19 in critically ill patients (63–65). IL-6 plays a critical role not only in the cytokine storm in severe COVID-19 (63, 65), but also in the innate immunity against fungal pathogens such as A. fumigatus (66). Accordingly, IL-6 inhibition has been associated with an increase in secondary infections in COVID-19 patients (67). A recent case report described an invasive Aspergillus infection in a COVID-19 patient following treatment with tocilizumab (65). Moreover, IL-6 inhibiting drugs may predispose COVID-19 patients to invasive mucormycosis (62).
Mechanisms of Fungal Infections: From Innate Immune Response to pH Modulation
Innate Immunity Against Aspergillus spp.
Despite constant exposure to A. fumigatus conidia, most people do not develop fungal disease. This suggests an efficient clearance of the conidia by the innate immune system in immunocompetent individuals before the adaptive immune system is activated (18). Upon inhalation, most resting conidia arriving in the respiratory tract are deposited against the airway fluid, due to turbulent airflow caused by the branching pattern of the respiratory tract (18). The trapped conidia are then removed by the ciliary action of the respiratory epithelium, which is the first line of defense in the lung (18, 68).
Host Recognition of Fungal Pathogen-Associated Molecular Patterns
Due to their small size, some of the inhaled conidia can reach the respiratory alveoli. After 4–5 h, resting conidia become swollen, and if not cleared, germinate and form hyphae within 12–15 h after arrival into the lungs (18). The maturation of conidia triggers a morphological change leading to the loss of the thin hydrophobic RodA protein layer, thus exposing the immunogenic components of the inner cell wall (69–71). These cell wall pathogen-associated molecular patterns (PAMPs) include polysaccharides such as β-D-glucan, mannan, chitin, and galactomannan, all of which are recognized by different pattern recognition receptors (PRRs) (15, 72). A. fumigatus conidia and hyphae can be recognized by the host via both soluble (pentraxins, complement proteins, and pulmonary collectins) and cell-associated microbial PRRs [Toll-like receptors (TLRs) and C-type Lectin receptors (CLRs)] (73, 74).
Pentraxins, such as pentraxin-3 (PTX3), are secreted by various cells, including neutrophils, dendritic cells, mononuclear phagocytes, and pulmonary epithelial cells (75). They bind to galactomannan on A. fumigatus conidia and facilitate recognition by phagocytes such as alveolar macrophages (76, 77). In addition, pulmonary collectins include lung surfactant proteins A and D and serve as opsonins. They bind to A. fumigatus conidial carbohydrate structures in a calcium-dependent manner. Surfactant proteins A and D have also been shown to promote the agglutination of conidia and their binding to neutrophils and alveolar macrophages, and improve the phagocytosis and killing of conidia by neutrophils (78).
Toll-like receptors recognition of pathogens triggers a signaling cascade leading to the activation of transcriptional factors such as NF-κB, which controls the expression of pro- and anti-inflammatory cytokines and chemokines (79). The universal adaptor molecule MyD88 has been shown to play a significant role in the signaling of TLRs, which induce the production of various inflammatory cytokines and reactive oxygen species (79). TLR2 and TLR4 have been implicated in the recognition of A. fumigatus conidia and hyphae (80). However, available data concerning their roles in A. fumigatus-associated immunity are conflicting. Indeed, the A. fumigatus-associated PAMPs for TLR2 and TLR4 remain undetermined. TLR9 has also been shown to play a role in innate immunity against A. fumigatus by recognizing fungal unmethylated CpG DNA (81). Dectin-1 is a CLR primary receptor that recognizes fungal β-glucan and that is essential for the mediation of the proinflammatory response (80), and is widely expressed on innate immune cells including macrophages, dendritic cells, and neutrophils (82–84). Dectin-1 can also induce the expression of the anti-inflammatory cytokine IL-10, indicating its dual role in modulating the inflammatory response (85). Dectin-2, another type of CLR, has recently been shown to be implicated in the innate immune response against A. fumigatus. Macrophages and dendritic cells express Dectin-2 and recognizes α-mannan in the fungal cell wall’s outer layer. Accordingly, in response to A. fumigatus infection, alveolar macrophages upregulate Dectin-2. Moreover, Dectin-2 was shown to mediate an NF-κB-dependent proinflammatory response against swollen conidia (80). Finally, DC-SIGN is expressed at the surface of dendritic cells and some macrophages, and binds to Aspergillus conidia via the recognition of fungal galactomannan (18). Genetic polymorphism in the above-mentioned PRRs, as well as in cytokines, chemokines, and immune receptors genes, has been associated with an increased susceptibility to pulmonary aspergillosis (86–88). For instance, the Dectin-1 variant Y238X has been shown to impair the production of several cytokines such as IFN-γ and IL-10 by human peripheral mononuclear cells, leading to an increased susceptibility to invasive aspergillosis in patients receiving hematopoietic stem cell transplantation (HSCT) (89). Furthermore, the Asp299Gly polymorphism in TLR-4 is highly associated with chronic cavitary pulmonary aspergillosis (86).
Cellular Immune Responses
Clearance of Aspergillus Conidia by Innate Immune Cells and Epithelial Cells
Alveolar macrophages (AMs), neutrophils and epithelial cells constitute the first line of defense against inhaled A. fumigatus conidia (18). Alveolar macrophages phagocytose and kill conidia either via oxidative mechanisms through the generation of reactive oxygen species (ROS), or by non-oxidative mechanisms through phagosomal acidification (80). Corticosteroids have been shown to impair the capacity of AMs to kill conidia (18). Neutrophils were initially thought to kill hyphae exclusively, however, they have also been essential in killing germinating conidia. Neutrophils bind and phagocytose swollen conidia to trigger respiratory burst and degranulation. While the size of the hyphae prevents phagocytosis, direct contact with neutrophils can induce oxidative and non-oxidative mechanisms to damage the hyphae (18). Moreover, respiratory epithelial cells, i.e., bronchial and alveolar epithelial cells, as well as endothelial cells, have been shown to participate actively in the innate immune response against Aspergillus fumigatus strains Af293 and CEA10 by phagocytosing and killing conidia in vitro (18, 90–94). Furthermore, human peripheral blood monocytes have also been shown to internalize Aspergillus conidia and inhibit their germination and hyphal growth (95). Interestingly, both classical (CD14+CD16–) and non-classical monocytes (CD14+CD16+) were found to successfully internalize conidia (96). However, only classical monocytes were able to inhibit Aspergillus hyphal development (96). Lastly, dendritic cells (DCs) also have well-documented roles in the defense against A. fumigatus. Immature DCs (iDCs) have been shown to phagocytose opsonized and non-opsonized conidia and hyphae, both of which are recognized through PRRs such as Dectin-1, among others. TNF-α, IL-6, IL-12, IL-1α, and IL-1β are the central proinflammatory cytokines produced by iDCs upon recognition of A. fumigatus conidia and hyphae (84, 97, 98).
Innate Lymphoid Cells and Innate-Like T Cells
Innate Lymphoid Cells (ILCs), γδ T cells, and mucosal associated invariant T cells (MAIT cells) have also been reported to have important roles in the innate immune response against Aspergillus spp. ILCs, and particularly type 3 ILCs (ILC3s), are commonly found in barrier epithelial surfaces, where they contribute to the maintenance of mucosal homeostasis, elimination of pathogens, regulation of the inflammatory response, as well as tissue remodeling (99). γδ T cells are innate-like T cells displaying features of both the innate and adaptive immune system. They are mainly involved in the immune surveillance and defense against pathogens in various peripheral tissues, including the lung (100). γδ T cells have been reported to produce IL-17A following challenge with A. fumigatus conidia (101). Moreover, mice lacking γδ T cells were more susceptible to A. fumigatus infection (102). Together with ILC3s, γδ T cells were shown to produce IL-22, which is critical for an efficient clearance of A. fumigatus (101, 103). Furthermore, deficiency in IL-22 production resulted in an impaired production of cytokines and chemokines and an impaired clearance of A. fumigatus in the murine lung (101, 103). MAIT cells are a subset of CD8+ unconventional T cells which are abundant in mucosal surfaces such as lung (100), and constitute up to 10% of the total T cells present in the peripheral blood (104). Upon activation, MAIT cells release proinflammatory cytokines such as IFNγ, TNFα, IL-17, and IL-22, and are able to kill pathogens through the production and release of cytotoxic compounds such as perforin and granzymes (100, 104). MAIT cells have been shown to be activated by T cell receptor (TCR)-dependent mechanisms through direct contact with antigen presenting cells such as dendritic cells in the case of bacteria and yeasts (105, 106), and TCR-independent mechanisms via cytokines in the case of viruses (107, 108). However, the activation of MAIT cells by filamentous fungal pathogens such A. fumigatus is not yet well-understood (104). Jahreis and colleagues showed that a fast TCR-dependent response was elicited by MAIT cells against several Aspergillus species, including A. fumigatus, A. terreus, and A. flavus (104). This response is characterized by the upregulation of activation markers, such as the CD69 antigen, and the release of cytotoxic compounds such as granzyme A and perforin (104). Finally, Natural Killer (NK) cells have also been shown to have a role in the innate immune defense against A. fumigatus (18).
Adaptive Immunity
Following a fungal encounter and the initial activation of the innate immune system, adaptive immunity is rapidly organized to clear the pathogen efficiently. Indeed, three different CD4+ T-helper cell lineages have been shown to play crucial roles in pulmonary aspergillosis: Th1, Th2, and Th17 (109–111). Th1 cells response is associated with protective immunity through the secretion of the pro-inflammatory cytokines TNFα and IFNγ, which promote antifungal activity of macrophages and neutrophils at the site of infection (110, 112, 113). Interestingly, Th1 cells have been shown to induce a fungus-specific Th1 immunity to an epitope of the A fumigatus cell wall glucanase Crf1. This antigen can be presented by DCs through three common major histocompatibility complex (MHC) class II alleles, which induces memory Th1 cells that are cross-reactive to C. albicans (114).
Alternatively, Th2 cells response is rather associated with non-protective immunity through the activation of M2 macrophages and decrease of Th1 response (110, 113). Th2 responses are predominant in patients suffering from ABPA, and are characterized by a decrease in IFNγ and an increase in IL-4 and IL-10 production, which in turn promotes humoral responses, through IgE production, and allergy (112, 113). Interestingly, fungal PAMPs have been shown to act as adjuvants enhancing T cell responses (115–117). This is notably the case of chitin, which is present in the fungal cell wall, in allergic disorders such as asthma (118–122). Dubey and colleagues showed that mice pre-treated with chitin before being challenged with A. fumigatus extract had high IgE levels (123).
The role of Th17 cell response is less clear. On the one hand, Zelante and colleagues showed that production of IL-17 and IL-23 by Th17 is not protective in a murine A. fumigatus infection model and inhibits Th1 cells development and antifungal activity of neutrophils in vitro (124). This was confirmed by blocking IL-17 and IL-23 production which showed increased clearance of A. fumigatus. This protective effect of IL-17 and IL-23 has been confirmed in an acute aspergillosis murine model, where mice were sensitized with A. fumigatus (125). The IL-17 and IL-23 -producing cells were identified by the authors as eosinophils (125). On the other hand, Jolink and collaborators showed that IL-17 provides a protective immunity by decreasing lung fungal burden in a murine infection model (126). Moreover, a cross-reactive Th17 response to C. albicans and A. fumigatus has also been described during acute ABPA in humans (127). This strong cross-reactive response is suggested to be rather induced by C. albicans-specific Th17 cells upon encounter with A. fumigatus, than by naïve T cells (127).
Lastly, regulatory T (Treg) cells have also been shown to have a protective effect in the immune response in aspergillosis. Treg cells have been shown to regulate the inflammatory response caused by a strong Th1 response in the early phase of A. fumigatus infection, as well as in the case of allergic reaction due to Th2 responses (113, 128, 129).
There is evidence of oligoclonal expansion of T cells upon exposure to A. fumigatus. For instance, stimulation with the allergen Asp f 1 induced an oligoclonal expansion of antigen-specific T cells directed to this antigen in ABPA and non-ABPA patients (130). Furthermore, the p41 epitope of the A. fumigatus extracellular cell wall glucanase Crf1 has also been shown to induce an antigen-specific oligoclonal T cell response in HSCT patients (114).
Virulence Factors and Immune Evasion in Aspergillus fumigatus
Aspergillus fumigatus has evolved several virulence factors to escape innate immune responses. These virulence factors include the rodlet layer, DHN-melanin, ROS detoxifying enzymes, and toxins. Resting conidia are surrounded by a rodlet layer composed of hydrophobic RodA proteins (18, 80, 131, 132). This protein coat masks cell wall β-1,3-glucans, and thus prevents the detection of conidia by the innate immune response (69–71). Moreover, the rodlet layer was found to participate in the adherence of conidia to the pulmonary epithelium (131). DHN-melanin is the major melanin pigment that gives a gray-green color to A. fumigatus (133). It protects the integrity of the genome in conidia from ultraviolet light, as well as ROS (131, 134). It also masks fungal PAMPs, similar to the rodlet layer, and permits the fungus to evade phagocytosis by interfering with the acidification of the phagolysosome notably by interfering with intracellular Ca2+ signaling (135, 136). Moreover, A. fumigatus possesses a number of different enzymes for detoxifying ROS produced by host phagocytic cells such as macrophages. These enzymes include catalases, superoxide dismutase, glutathione transferases, fatty acid oxygenases, and efflux pumps, all of them either detoxifying H2O2 or superoxides, or expulsing ROS extracellularly (131, 134). Finally, A. fumigatus secretes several toxins considered as secondary metabolites, that further enable it to evade the immune response. These toxins are virulence factors crucial for A. fumigatus pathogenesis. For instance, gliotoxin has been shown to have several immunosuppressive effects, including inhibition of phagocytosis and neutrophil-derived ROS production, as well as proapoptotic activities (137, 138). Moreover, gliotoxin decreases ciliary movement and angiogenesis (131, 134, 139). All these immunosuppressive effects are thought to result from inhibition of the NF-κB signaling pathway (140), which as mentioned above is a key mediator of inflammatory responses (141).
pH Modulation as a Strategy to Colonize the Host Tissues
Environmental pH is an extremely important factor influencing not only fungal growth and development, but also fungal physiology (142, 143). Indeed, pH modulation has been shown to affect fungal enzyme activity (144), and to be a crucial element controlling fungal pathogenicity. Fungal infections are often accompanied by a shift in pH in the surrounding host tissue (142), through the secretion of either acids or alkali (143). The specific roles of acidification and alkalinization in fungal pathogenesis discussed in this review are summarized in Figure 3.
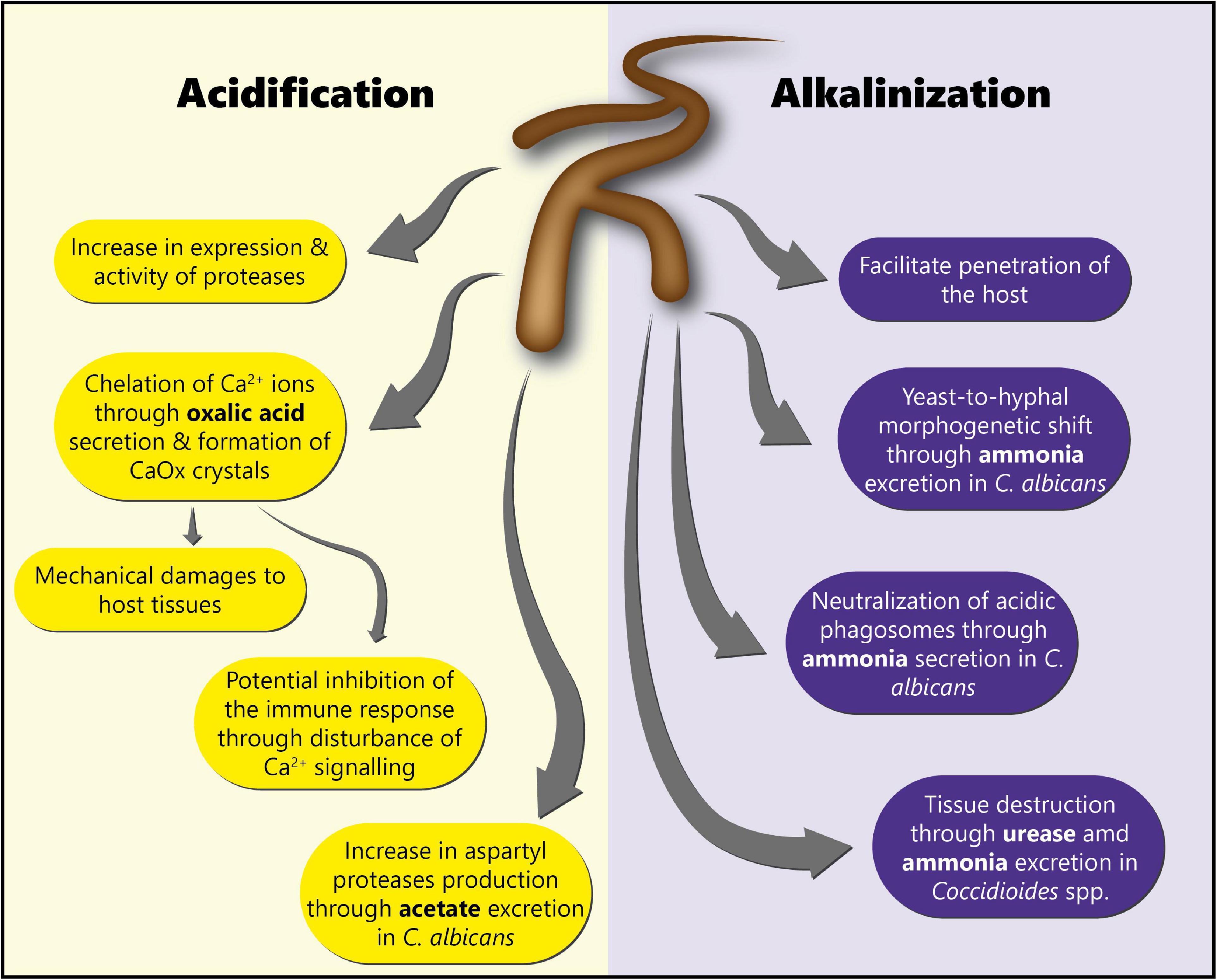
Figure 3. Roles of acidification and alkalinization mechanisms in fungal pathogenesis. This scheme summarizes the different roles of acidification and alkalinization mechanisms in fungal pathogenesis for the colonization of the host tissues.
Interestingly, fungi, including Aspergillus spp. and Candida spp. are known to produce low molecular weight organic acids (LMWOAs) such as oxalate, citrate, malate, formate, acetate, and succinate, which contribute to pH modulation (145, 146). Pathogenic fungi acidify their environment in order to enhance the activity of enzymes, as well as to damage the host tissues (143, 147). For instance, in the case of C. albicans, acidification through acetate excretion has been shown to allow the production of aspartyl proteases (143, 148–150), which are major virulence factors in this pathogen (151).
Oxalic acid is a known pathogenicity factor for the phytopathogenic fungi Sclerotinia sclerotiorum and Botrytis cinerea (152, 153). This acid is secreted in the host tissues and accumulates in the form of oxalate, leading to a pH decrease. Additionally, as oxalate is a strong chelator of divalent metallic cations, it can sequester calcium ions, with multiple possible structural and physiological consequences for the host (146). In the case of plant pathogens, the formation of calcium oxalate (CaOx) crystals in the middle lamella weakens the cell wall structure and facilitates infection. Moreover, oxalate can inhibit plant defenses and induce programmed cell death, which is also beneficial for necrotrophic pathogens (17, 146, 153).
The significance of a similar production of oxalic acid or other LMWOAs by Aspergillus spp. to infect the human lung is an active topic of research. Indeed, although many aspects of the ecology of Aspergillus spp. have been investigated in relation to their pathogenicity, one aspect that has been largely ignored is its ability to lower the pH of its environment. This may be necessary for their capacity to colonize or cause infection, via the secretion of oxalic acid or other low molecular weight organic acids. Several studies have reported the presence of CaOx crystals in the case of pulmonary aspergillosis (154–164), and the detection of CaOx crystals has been proposed as an easy tool for differential diagnosis (162). In most of the reported cases, oxalate deposition was associated with A. niger infection, but some reports also include infection caused by A. flavus or A. fumigatus (159). Oxalic acid and oxalate crystals are thought to cause host tissue damage (including in pulmonary blood vessels), as well as tissue injury via iron-dependent generation of free radicals (157, 163). A mechanical role of CaOx crystals was recently reported by Yi et al. (164) in a case of pulmonary angioinvasive aspergillosis in a Burkitt’s lymphoma patient with severe neutropenia, with pathophysiological examinations showing the presence of CaOx crystals around and within the walls of blood vessel. Aside from mechanical damage to the host tissues, the formation of CaOx could also have a dramatic effect on cell physiology. Indeed, calcium is an extremely important secondary messenger in many cell types, including those of the immune system (165). During immune stimulation, Ca2+ mobilization from extracellular medium or cellular compartments is essential to increase intracellular Ca2+ concentration (166), and thus Ca2+ chelation has been shown to inhibit the immune response in vitro (167, 168). All this suggests a potential role of oxalic acid also in the inhibition of the immune response. Despite these converging indications, a potential link between oxalic acid production and pathogenicity for Aspergillus spp. in animals has only been recently proposed (169, 170). Indeed, we demonstrated that oxalic acid production by A. niger led to a strong pH decrease, as well as calcium ion sequestration and precipitation in the form of CaOx crystals in differentiated 3D bronchial epithelial tissues. Moreover, we showed that the addition of the soil oxalotrophic bacterium Cupriavidus oxalaticus inhibited the growth of A. niger, and reverted pH values and free calcium concentrations back to physiological levels. Furthermore, CaOx crystals were no longer observed, suggesting the consumption of oxalic acid by C. oxalaticus (169, 170). However, the exact contribution of oxalic acid in the pathogenesis of A. niger still needs to be determined. Conversely, oxalic acid production by Candida spp. has never been confirmed experimentally, although these fungi are listed as oxalate-producers in the Human Metabolome Database (HMDB0002329).
Fungal pathogens are also known to manipulate the pH of their environment through alkalinization. This is notably the case of C. albicans. Alkalinization has been shown to facilitate the invasion of the host tissues, and the evasion of the immune system through neutralization of acidic macrophages phagosomes (150). Alkalinization of the host environment occurs through the excretion and accumulation of ammonia (NH3), which is then converted into ammonium ions (NH4+) by the urease (143, 150). Moreover, a lack of carbon is required for ammonia-mediated alkalinization to occur (142). Furthermore, C. albicans has been shown to auto-induce its switch to the hyphal growth form through the release of ammonia (171). Coccidioides spp. has been reported to excrete urease and ammonia to destroy the host tissue (172, 173). St Leger and colleagues have shown that A. fumigatus produced small amounts of ammonia in minimal medium, leading to a pH increase and allowing the production of active proteases (174). However, to the best of our knowledge, the role of environmental alkalinization in the pathogenesis of A. fumigatus has never been investigated.
Lung Ecology
Lung Homeostasis
Contrary to the gut, whose microbiota has been extensively studied, the lungs were considered sterile for a long time (175). However, the lung is now known to harbor a diverse microbiota composed of bacteria, fungi and viruses (176, 177). The bacterial composition in the lung has been studied in depth over the past years, although many studies have remained descriptive and in-depth mechanistic analyses are scarce. In the healthy lung, the microbiota is dominated by the genera Prevotella, Streptococcus, and Veillonella (178, 179). In patients with an acute or chronic respiratory disease the composition differs (180–183). Recent data provide clear evidence for the role of the airway microbiota (184), for instance, in the modulation of the host immune response or mucus production (185).
Whereas, the influence of bacterial community composition on lung homeostasis and disease has been a very active area of investigation, our knowledge on the involvement of fungi in these processes is still limited. Using fungal ribosomal RNA gene sequencing of BAL and sputum samples, it has been demonstrated that the mycobiota of the healthy lung mainly consists of environmental agents Davidiellaceae, Cladosporium, Aspergillus, Eurotium, Penicillium, and Candida (181). Epithelial cells of inner and outer body surfaces (e.g., intestine, skin, and lung) are the first physical barrier which interacts with commensals and are also important producers of antimicrobial peptides and immune mediators that regulate immune homeostasis and host defense (186, 187). Fungi colonize all barrier surfaces, and investigations into the influence of fungi on host immunity during homeostasis and disease are more advanced in the intestine and the skin. The influence of fungal seeding and colonization on immune maturation in the lung remains to be investigated, and we can learn from the findings in the intestine and skin and translate those to the lung in the future (188–190).
Gut-Lung Axis
In-depth analysis of the microbiome has revealed profound differences in composition between various body compartments (191, 192). The importance of inter-organ communication and the role of the microbiome herein has been increasingly recognized (193). Respiratory diseases have not only been associated with microbial dysbiosis in the lung, but also in the gut (194). The gut microbiota composition has been shown to influence the immune responses of distant organs, including lungs, through the systemic dissemination of metabolites such as short chain fatty acids (SCFAs), or through the direct seeding of bacteria from the gut to the lungs by gastro-oesophageal reflux and microaspiration. This crosstalk between the gut and the lung compartment is called the gut-lung axis (195). The metabolites or bacteria can consequently have a stimulatory effect on the local immune cells (194–196), and ultimately impair or contribute to the development or progression of respiratory disease.
Recent studies in which fungal composition in the gut was altered using antifungal drugs, provide evidence for an immunoprotective role of the gut mycobiota (197, 198). For example, prolonged oral administration of antifungal drugs led to an exacerbation of allergic airway inflammation in an experimental mouse model. Antifungal treatment led to alterations in several fungal species, with an increase in Aspergillus, Wallemia and Epicoccum spp., and an observed decrease in Candida spp. The enhanced allergic airway inflammatory response was recapitulated when orally supplementing mice with a mixture of the three enriched fungi. This indicates that disruption of commensal fungi can influence both local and distal immune responses. Whether oral antifungal drugs also affect lung mycobiome composition is currently unexplored.
Disruption of bacterial communities using antibiotics can induce fungal dysbiosis and vice versa, suggesting an important role for inter-kingdom interactions (197). Indeed, the bacterial microbiome is likely to have an influence on the composition of the fungal microbiome, either directly through bacterial-fungal interactions or indirectly through its impact on host immunity, thus making the lung environment more permissive or restrictive to fungal growth (196). Interestingly, using a gnotobiotic approach, van Tilburg and colleagues demonstrated that the presence of gut bacteria, but not fungi, in early life could reduce allergic airway inflammation in a respiratory OVA sensitization and challenge model. These outcomes suggested that homeostatic control of allergic airway inflammation is dependent on bacterial presence, and that intestinal colonization with fungi can skew this inflammatory response (199).
Although a link between the lung bacterial microbiota composition and the disease outcome in patients with invasive pulmonary aspergillosis has been recently demonstrated (200), many knowledge gaps still exist regarding the link between the pulmonary microbiota and Aspergillus spp. infections. For instance, alteration of the air-blood barrier, and in particular enhanced access to the extracellular matrix, is a known risk factor for fungal infection (201), especially in the case of Aspergillus (202). Therefore, investigating the role of the lung microbiota and specific bacterial community compositions on the strengthening of the air-blood barrier is of clinical importance. The integration of principles from ecological theory will be key to elucidate the bacterial—fungal interactions of the gut and pulmonary microbiota and their human host. This will contribute to identifying microbial groups from within the airway microbiota, or their metabolites, for the development of therapeutic tools to control Aspergillus (179, 196).
Current Therapeutic Strategies
The prompt diagnosis and treatment of invasive fungal pulmonary infections such as pulmonary aspergillosis is crucial to prevent associated complications and fatal outcomes. As the clinical presentations and radiological changes are non-specific, biopsy and histopathological analysis remains the gold standard for securing the diagnosis, but is frequently contraindicated in clinically marginal patients. Moreover, colonization is difficult to discriminate from a true invasive infection, and fungal blood cultures are insensitive (10, 203, 204). Therefore, serum and BAL biomarkers such as galactomannan and β-D-glucan, or PCR to detect fungi are being increasingly employed to establish the diagnosis (205).
Current available treatments for pulmonary aspergillosis are limited. They include the use of antifungal drugs—such as azoles, echinocandins, polyenes, and flucytosine, as primary treatment in the case of CPA or IPA, surgical resection in the case of patients suffering from aspergilloma and presenting associated complications such as severe hemoptysis, and corticosteroids in the case of ABPA, with or without the administration of antifungal drugs (206). The standard of care for fungal pulmonary infection is similar across different underlying lung diseases. Treatment would rather depend on the clinical manifestations: for example, no treatment in the case of colonization, corticosteroids and/or antifungal and/or newer biologics in case of allergic sensitisation/ABPA, and antifungal agents for invasive infection. The frequency by which these clinical manifestations are observed would depend on the underlying lung disease. For example, invasive infection is seen almost exclusively in immunocompromised patients, ABPA in patients with asthma or CF, whereas colonization can be observed in any lung disease associated with structural damage (COPD, CF, and non-CF bronchiectasis).
Resistance to antifungal drugs has increased dramatically in the last decades. While antibiotic resistance has been widely recognized in bacteria, antifungal resistance in opportunistic fungal pathogens has not yet received sufficient consideration. The reasons for the rise in antifungal resistance are multiple. Indeed, there is a limited arsenal of active antifungal compounds available on the market that are being used in both agriculture and human health, thus fostering the emergence and rapid spread of cross-resistance in human opportunistic fungal pathogens (2).
This is particularly well highlighted in the case of A. fumigatus resistance to azoles. Azoles are frontline antifungal compounds used in crop protection and in human and animal health (207, 208). They have a fungistatic effect on yeasts such as Candida albicans, while acting as fungicides against filamentous molds such as A. fumigatus. Their fungicidal effect against A. fumigatus is linked to defects in cell wall remodeling, resulting in loss of cell wall integrity and death (209). However, despite the fungicidal effect of triazoles in filamentous fungi, their application in agriculture at sub-inhibitory concentrations has led to the emergence and rapid spread of resistance among natural populations of A. fumigatus in soil (2, 210). This resistance to azoles may compromise the success of treatment in human patients suffering from A. fumigatus infections (211). Azole fungicides such as propiconazole, difenoconazole, or tebuconazole have a very similar structure when compared to those used in clinical practice, and their use is correlated with the increased emergence of clinical azole-resistant A. fumigatus strains (208). The emergence of this resistance led to the hypothesis that the extensive use of azole fungicides in agriculture selected for azole resistant A. fumigatus in the environment (212). This is supported by several studies reporting the presence of azole-resistant A. fumigatus strains in patients never treated with azoles (16, 213). This accounts for two thirds of patients suffering from azole-resistant aspergillosis. Resistance has been attributed to specific mutations in the tandem repeat (TR) of the cyp51A gene promoter region, which is involved in the biosynthesis of ergosterol in fungi (16, 214). Clinical azole-resistant A. fumigatus strains have also shown cross-resistance to azoles commonly used in agriculture (16). Thus, considering an integrated disease management approach through the One Health initiative that brings together scientists, medical doctors, veterinarians, and plant pathologists, is needed to reduce our reliance on chemical control alone and to stop the spread of resistance among opportunistic pathogens (2).
In case of pulmonary aspergillosis refractory to azoles, echinocandins, such as caspofungin or micafungin, or polyenes, such as amphotericin B, are used as second-line antifungal treatments (211, 215). Echinocandins inhibit the synthesis of the fungal cell wall component β-(1,3)-D-glucan by targeting the β-(1,3)-glucan synthase (216). Little is known on the resistance mechanism to echinocandins in A. fumigatus, due to its limited use in the treatment of Aspergillosis (216). Echinocandin resistance in A. fumigatus has been attributed to mutations in FKS genes encoding the β-(1,3)-glucan synthase (216). The same has been reported for C. albicans (217). Amphotericin B (AmB) has been shown to bind to ergosterol in the fungal cell membrane and form pores, thus disrupting the cell membrane integrity (218, 219). AmB has also been reported to induce endogenous production of ROS leading to oxidative stress and fungal death (219). Although resistance to AmB is rare, A. terreus has been shown to be intrinsically resistant (218). Moreover, AmB resistance has also been reported in A. fumigatus (220) and A. flavus (221). AmB resistance in A. terreus may be due to high endogenous production of catalase (218). Finally, flucytosine (5-FC) is a synthetic antifungal compound which, when it is taken up in fungal cells, is first converted into 5-fluorouracil (5-FU). 5-FU is then converted into metabolites which inhibits DNA and RNA synthesis (222). 5-FC is rarely administered as a monotherapy to treat fungal infections such as aspergillosis, but rather in combination with AmB (222, 223).
Advances in Therapeutic Strategies and Future Perspectives
Current approaches used for the treatment of pulmonary aspergillosis focus on attacking the pathogen directly via the use of antifungal compounds. New antifungal drugs have been recently developed against Aspergillus spp. infection. Newly developed drugs, as well as drugs currently in development, are listed on the Aspergillosis website.1 Several of these have shown promising results and are being tested in phase 3 clinical trials (224). Olorofim (F901318, ClinicalTrials.gov Identifier: NCT05101187), from a new class of antifungal drugs called orotomides, is highly active against Aspergillus spp. It targets the dihydroorotate dehydrogenase (DHODH) which is involved in the pyrimidine biosynthesis pathway. Olorofim has been shown to inhibit conidia germination, as well as polarized hyphal growth (225). Biafungin, or Rezafungin (CD101, ClinicalTrials.gov Identifier: NCT04368559), is a novel echinocandin targeting the 1,3-β-D-glucan synthase. It shows high in vitro and in vivo activity against Aspergillus spp. (224, 226). Ibrexafungerp (SCY-078, MK-3118, ClinicalTrials.gov Identifier: NCT03059992) is a triterpenoid antifungal also inhibiting the biosynthesis of β-(1,3)-D-glucan (227). Ibrexafungerp has been shown to be effective against aspergillosis in an in vivo murine model (228). Finally, another interesting compound is ASP2397 (VL-2397), which is a novel natural antifungal compound currently in phase 2 clinical trial (ClinicalTrials.gov Identifier: NCT03327727). ASP2397 is a cyclic hexapeptide siderophore with a similar structure than ferrichrome, which has a high affinity for iron (224, 229). ASP2397 showed high antifungal activity against A. fumigatus, A. terreus and A. flavus. It was shown to inhibit conidia germination as well as hyphal growth. Moreover, ASP2397 has been reported to have a higher efficacy than posaconazole in vivo in an IPA mouse model (229). However, new therapeutic approaches are needed in order to slow down the pace of antifungal resistance emergence in fungal pathogens.
In order to cause disease, a pathogen needs a susceptible host, as well as suitable environmental conditions for its growth. These three factors, i.e., pathogen, host and environment, constitute the so-called disease triangle, which dictates the occurrence of a disease caused by a particular pathogen in a susceptible host in a particular environmental setting (152). The disease triangle concept has already been used in plant disease management for decades (230). However, one aspect that has been completely overlooked in this original disease triangle is the presence of the host microbiota and its role in disease development. A recent paper from Bernardo-Cravo et al. (231) highlighted the necessity to include the host microbiome as a fourth factor influencing the onset of a disease, as it plays a very important role in host immunity. Therefore, the disease triangle should become a disease pyramid (Figure 4) and one should consider all four factors, i.e., the host, the host microbiome, the environment, and the pathogen, when thinking about disease management.
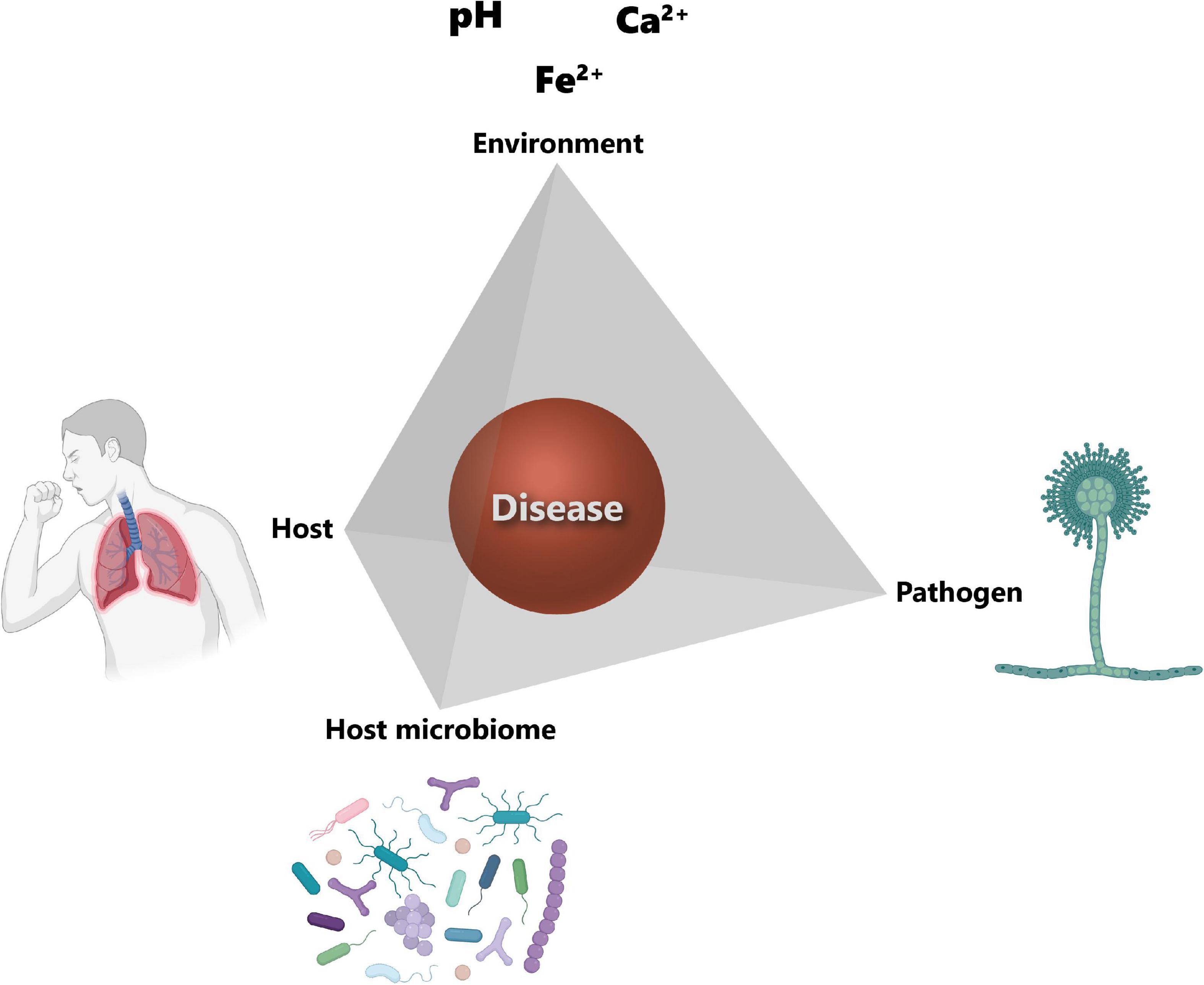
Figure 4. Disease pyramid. The onset of a disease depends on the interplay between the host, the pathogen, the environment, and the host microbiome. Host susceptibility mainly depends on genetic factors and immune status. The main factor for the successful colonization of the host tissues by the pathogen is its virulence. The environment corner refers to the host lung microenvironmental parameters and include among others pH, calcium (Ca2+) and iron (Fe2+) concentrations. Host microbiota community structure can also influence the establishment of the pathogen. Icons have been created using Biorender.com. Modified from Palmieri (170).
We recently proposed to focus on the “environment” corner of the disease pyramid to control the growth of fungal pathogens such as Aspergillus spp. through a process we named “environmental interference” (169, 170). We demonstrated that the soil oxalotrophic bacterium Cupriavidus oxalaticus is able to control the growth of the fungal pathogen A. niger. This is achieved by degrading the oxalic acid produced by A. niger, preventing acidification of the local environment favorable to infection. Therefore, C. oxalaticus inhibits the growth of the pathogenic fungus A. niger by manipulating the pH and restoring it back to a more neutral, physiological value. This environmental interference principle could be potentially extended to other important environmental factors for fungal pathogenesis, such as iron. Indeed, iron is an essential nutrient that is generally limiting, and thus its acquisition is crucial for pathogen’s virulence (232). Ghio et al. (157) reported ferric iron (Fe3+) complexation at the surface of CaOx crystals associated with A. niger infection, resulting in lung tissue injury via the generation of oxidants. However, fungal, as well as bacterial pathogens, are well-known to acquire iron through the secretion of siderophores (233, 234). A. fumigatus is genetically equipped for efficient iron acquisition, encoding four different siderophores: fusarinine C (FsC), triacetylfusarinine C (TAFC), ferricrocin (FC), and hydroxyferricrocin (HFC) (235, 236). Experimental data showed that the initial phase of lung infection with A. fumigatus is accompanied by upregulation of iron acquisition genes (237). Loss in the ability to produce siderophores, and thus to acquire iron, has been shown to be detrimental for A. fumigatus in vivo (238). Moreover, interfering with the acquisition of iron by the use of chelators inhibited the growth of A. fumigatus in a murine cornea infection model (239). Therefore, developing a bacterial biocontrol strategy based on the interference of iron acquisition by Aspergillus would provide a further option to exploit the principle of interfering with the environment. This could be achieved either by direct competition between Aspergillus spp. and the biocontrol bacteria, where these latter would produce a siderophore with a higher affinity for iron than the fungal siderophore, allowing them to acquire iron better than the fungus, or by bacterial “cheating” through stealing the siderophores produced by Aspergillus spp. Indeed, non-siderophore producing bacteria are known to steal other species’ siderophores through the use of a matching receptor (240). On the other hand, host cells internalize iron through a global process called nutritional immunity, aimed at controlling infection (241, 242). However, unlike for the gut microbiota (243), the interplay between the regular members of the airway microbiota and nutritional immunity is still largely undetermined. Indeed, while essential metals, such as iron or zinc, have been found to be altered in several respiratory diseases, the exact causes and mechanistic consequences of this metal dysregulation on the immune system and respiratory microbiota still need to be further explored (242). Interestingly, gallium, a group IIIA metal, has been proposed as an antifungal agent. Gallium is used in several medical applications ranging from cancer to calcium disorders and bone metabolism (244). Moreover, gallium nitrate III [Ga(NO3)3] has been widely used as an antibacterial agent against bacterial pathogens such as Klebsiella pneumoniae, Staphylococcus aureus or Pseudomonas aeruginosa (244). Furthermore, gallium nitrate IV has been tested in a phase 2 clinical trial for intravenous administration as an anti-infective agent against P. aeruginosa infection in CF patients (ClinicalTrials.gov Identifier: NCT02354859). Gallium is known to disrupt the iron homeostasis in bacteria and cancer cells (244). Its antifungal inhibitory effect has recently been demonstrated by Bastos and colleagues against azole-resistant A. fumigatus and multidrug-resistant Candida spp. (244). Finally, it is worth investigating whether gallium could be used to treat bacterial-fungal polymicrobial infections, which are frequent in CF patients (245, 246).
The role of the microbiota as a keystone factor influencing the onset and development of a disease provides a powerful incentive for the use of microorganisms to prevent and treat illness. These microorganisms, also referred to as live biotherapeutic products (LBPs), are defined by the FDA as “a biological product that contains live organisms, such as bacteria; is applicable to the prevention, treatment, or cure of a disease or condition of human beings; and is not a vaccine” (247). They are not intended to reach the systemic circulation, but rather exert their action through interaction with resident members of the microbiota and/or by modulating complex host-microbiota interactions. This implies a multifactorial mode of action, which is in strong contrast to the reductionist approach traditionally used in medical research (247).
The large majority of LBPs correspond to single species products. There are examples in which single species LBPs appear to confer protection against an invading pathogen, a phenomenon known as colonization resistance [i.e., Lactobacillus murinus protection against Streptococcus pneumoniae lung colonization (248); Clostridium scindens providing colonization resistance against Clostridium difficile (249)]. However, the use of single-species LBPs likely fails to capture the complexity of the multifactorial and multi-species role of the microbiota in health homeostasis. A similar concept to colonization resistance is widely used in soil ecology, namely that of disease-suppressive and conducive soils (250). This concept links the composition of the soil microbiota with the natural protection against plant fungal infections. Mendes et al. (251) showed that a specific assemblage of rhizospheric bacteria lead to disease-suppression and confer protection against the root fungal pathogen Rhizoctonia solani in sugar beet seedlings. In the same way, a specific microbiota composition could lead to more pathogen-suppressive communities, or conversely, dysbiosis of the microbiota could lead to pathogen-permissive communities. Translating soil ecological concepts into human medicine could be of great benefit, as it would provide key insights into the complex interspecies interaction dynamics within the human microbiome and between itself and the host (252, 253). Such lung microbiota-based biocontrol strategy could be applied against aspergillosis, as lung microbiota has been suggested to prevent the establishment of Aspergillus spp. in the lungs (196).
Additionally, LBPs have been shown to restore the epithelial barrier and modulate the immune response. These effects have been mostly studied in the case of administration of probiotics in the gastrointestinal tract (254). However, their potential beneficial effect on the airway epithelial barrier restoration, as well as in the immune homeostasis have been suggested by Martens et al. (254). This is notably the case of the bacterial strains Lactobacillus plantarum MB452, Lactobacillus rhamnosus GG and Streptococcus thermophiles ATCC 19258, among others (254). Recently, the use of nasal probiotic strains such as Lacticaseibacillus casei AMBR2 and Lactococcus lactis W136 has shown promising results. Indeed, Lacticaseibacillus casei AMBR2 showed a beneficial effect on the epithelial barrier function and modulation of the immune response in in vitro Calu-3 differentiated tissue co-cultured with donor-derived nasal microbiota and macrophage-like cells (255, 256). Cho and colleagues reported that Lactococcus lactis W136 suppressed the growth of patient-derived strains of the bacterial pathogen Pseudomonas aeruginosa in in vitro co-cultures (257).
Although large efforts have been made to enhance our understanding of respiratory fungal infections, the treatment of fungal pathogens poses a series of unique challenges. Fungal infections are difficult to diagnose in a timely manner, antifungal drugs and treatments are limited, and given the emergence of resistance to antifungal drugs there is a reduced effectiveness of these treatments. Moreover, severe infections often occur in subjects with significant comorbidities, including chronic respiratory diseases and a state of immunosuppression. It is thus imperative to develop novel treatment strategies for fungal infections. In order to do so, we have identified several key unanswered questions (Box 1). In view of the variability of fungal infection outcomes, and in particular those with Aspergillus spp., there is an urgent need to understand the conditions that make the respiratory tract permissive to conidial germination in susceptible individuals, and in particular, to determine whether the composition of the airway microbiota plays a role in this regard.
BOX 1. Key unanswered questions.
° What comprises a healthy lung mycobiome?
° What is the role of the mycobiome component of the respiratory microbiota in the colonization resistance to fungal pathogens?
° What is the role of commensal fungi in microbiota community stability?
° What is the involvement of commensal fungi in the maintenance of lung barrier homeostasis/function?
° How do commensal fungi contribute to pulmonary immune maturation in early life and what are the consequences of early life fungal dysbiosis for respiratory disease development?
° Which mechanisms underlie the bacterial -induced predisposition to invasive pulmonary aspergillosis and other respiratory fungal infections?
° How does nutrient availability contribute to lung microbial community composition and consequently influence fungal infection?
Future research in this field should focus on moving toward the complete characterization of the lung microbial ecosystem by shotgun metagenomic sequencing and gene expression analysis (258). Moreover, technical advances are required to enhance our ability to culture fungi from respiratory tract samples in the setting of prophylactic anti-fungal treatment, and in particular move toward building a collection of lung commensal fungal strains which can be implemented in mechanistic studies.
Author Contributions
FP and NU conceptualized and drafted the manuscript. AK, EB, PJ, and CG provided valuable discussion and input. FP, AK, EB, PJ, CG, and NU revised the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the Swiss National Science Foundation through the BRIDGE Discovery program under the grant agreement 194701 (PJ and AK), the Novartis Foundation through the FreeNovation program (PJ), the Gebert Rüf Stiftung under the grant agreement GRS-064/18 (PJ), the “Fondation Professeur Placide Nicod” (EB), and the U.S. Department of Energy, Office of Science, Biological and Environmental Research Division, under award number LANLF59T (PJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel). (2017) 3:57. doi: 10.3390/jof3040057
2. Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. (2018) 360:739–42. doi: 10.1126/science.aap7999
3. Rodrigues ML, Nosanchuk JD. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis. (2020) 14:e0007964. doi: 10.1371/journal.pntd.0007964
4. Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. (2007) 45:321–46. doi: 10.1080/13693780701218689
5. Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am. (2016) 30:1023–52.
6. Lionakis MS, Hohl TM. Call to action: how to tackle emerging nosocomial fungal infections. Cell Host Microbe. (2020) 27:859–62. doi: 10.1016/j.chom.2020.04.011
7. Pfaller MA, Diekema DJ. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J Clin Microbiol. (2004) 42:4419–31. doi: 10.1128/JCM.42.10.4419-4431.2004
8. Brunke S, Mogavero S, Kasper L, Hube B. Virulence factors in fungal pathogens of man. Curr Opin Microbiol. (2016) 32:89–95. doi: 10.1016/j.mib.2016.05.010
9. Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. (2012) 4:165rv13. doi: 10.1126/scitranslmed.3004404
10. Gago S, Denning DW, Bowyer P. Pathophysiological aspects of Aspergillus colonization in disease. Med Mycol. (2019) 57(Suppl. 2):S219–27. doi: 10.1093/mmy/myy076
11. Kousha M, Tadi R, Soubani AO. Pulmonary aspergillosis: a clinical review. Eur Respir Rev. (2011) 20:156–74.
12. Paulussen C, Hallsworth JE, Álvarez-Pérez S, Nierman WC, Hamill PG, Blain D, et al. Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microbial Biotechnol. (2017) 10:296–322. doi: 10.1111/1751-7915.12367
13. Latgé JP, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev. (2019) 33:e00140–18. doi: 10.1128/cmr.00140-18
14. Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus—what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. (2013) 9:e1003743. doi: 10.1371/journal.ppat.1003743
15. Van De Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. (2017) 15:661–74. doi: 10.1038/nrmicro.2017.90
16. Chowdhary A, Meis JF. Emergence of azole resistant Aspergillus fumigatus and one health: time to implement environmental stewardship. Environ Microbiol. (2018) 20:1299–301. doi: 10.1111/1462-2920.14055
17. de Oliveira Ceita G, Macêdo JNA, Santos TB, Alemanno L, da Silva Gesteira A, Micheli F, et al. Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora perniciosa. Plant Sci. (2007) 173:106–17. doi: 10.1016/j.plantsci.2007.04.006
18. Park SJ, Mehrad B. Innate immunity to Aspergillus species. Clin Microbiol Rev. (2009) 22:535–51. doi: 10.1128/CMR.00014-09
19. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax. (2015) 70:270–7. doi: 10.1136/thoraxjnl-2014-206291
20. Soubani AO, Khanchandani G, Ahmed HP. Clinical significance of lower respiratory tract Aspergillus culture in elderly hospitalized patients. Eur J Clin Microbiol Infect Dis. (2004) 23:491–4. doi: 10.1007/s10096-004-1137-1
21. Ofori A, Steinmetz AR, Akaasi J, Asafu Adjaye Frimpong GA, Norman BR, Obeng-Baah J, et al. Pulmonary aspergilloma: an evasive disease. Int J Mycobacteriol. (2016) 5:235–9. doi: 10.1016/j.ijmyco.2016.03.002
22. Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev. (2009) 22:447–65. doi: 10.1128/CMR.00055-08
23. Patel TS, Eschenauer GA, Stuckey LJ, Carver PL. Antifungal prophylaxis in lung transplant recipients. Transplantation. (2016) 100:1815–26. doi: 10.1097/TP.0000000000001050
24. Herrera S, Davoudi S, Farooq A, Tikkanen J, Foroutan F, Kumar D, et al. Late onset invasive pulmonary aspergillosis in lung transplant recipients in the setting of a targeted prophylaxis/preemptive antifungal therapy strategy. Transplantation. (2020) 104:2575–81. doi: 10.1097/TP.0000000000003187
25. Palmer LB, Greenberg HE, Schiff MJ. Corticosteroid treatment as a risk factor for invasive aspergillosis in patients with lung disease. Thorax. (1991) 46:15–20. doi: 10.1136/thx.46.1.15
26. Bafadhel M, McKenna S, Agbetile J, Fairs A, Desai D, Mistry V, et al. Aspergillus fumigatus during stable state and exacerbations of COPD. Eur Respir J. (2014) 43:64–71. doi: 10.1183/09031936.00162912
27. Bulpa P, Duplaquet F, Dimopoulos G, Vogelaers D, Blot S. Invasive pulmonary aspergillosis in chronic obstructive pulmonary disease exacerbations. Semin Respir Crit Care Med. (2020) 41:851–61. doi: 10.1055/s-0040-1702210
28. Tiew PY, Ko FWS, Pang SL, Matta SA, Sio YY, Poh ME, et al. Environmental fungal sensitisation associates with poorer clinical outcomes in COPD. Eur Respir J. (2020) 56:2000418. doi: 10.1183/13993003.00418-2020
29. Tiew PY, Dicker AJ, Keir HR, Poh ME, Pang SL, Mac Aogain M, et al. A high-risk airway mycobiome is associated with frequent exacerbation and mortality in COPD. Eur Respir J. (2021) 57:2002050. doi: 10.1183/13993003.02050-2020
30. Hodge S, Hodge G, Scicchitano R, Reynolds PN, Holmes M. Alveolar macrophages from subjects with chronic obstructive pulmonary disease are deficient in their ability to phagocytose apoptotic airway epithelial cells. Immunol Cell Biol. (2003) 81:289–96. doi: 10.1046/j.1440-1711.2003.t01-1-01170.x
31. Taylor AE, Finney-Hayward TK, Quint JK, Thomas CM, Tudhope SJ, Wedzicha JA, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J. (2010) 35:1039–47. doi: 10.1183/09031936.00036709
32. Wrench C, Belchamber KBR, Bercusson A, Shah A, Barnes PJ, Armstrong-James D, et al. Reduced clearance of fungal spores by chronic obstructive pulmonary disease GM-CSF- and M-CSF-derived macrophages. Am J Respir Cell Mol Biol. (2018) 58:271–3. doi: 10.1165/rcmb.2017-0351LE
33. Tiew PY, Mac Aogáin M, Ter SK, Aliberti S, Chalmers JD, Chotirmall SH. Respiratory mycoses in COPD and bronchiectasis. Mycopathologia. (2021) 186:623–38. doi: 10.1007/s11046-021-00539-z
34. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
35. Arrieta MC, Arevalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142:424–34.e410. doi: 10.1016/j.jaci.2017.08.041
36. Rick EM, Woolnough KF, Seear PJ, Fairs A, Satchwell J, Richardson M, et al. The airway fungal microbiome in asthma. Clin Exp Allergy. (2020) 50:1325–41. doi: 10.1111/cea.13722
37. Salo PM, Arbes SJ Jr, Sever M, Jaramillo R, Cohn RD, London SJ, et al. Exposure to Alternaria alternata in US homes is associated with asthma symptoms. J Allergy Clin Immunol. (2006) 118:892–8. doi: 10.1016/j.jaci.2006.07.037
38. Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol. (2015) 135:110–22. doi: 10.1016/j.jaci.2014.07.002
39. Lynch SV, Bruce KD. The cystic fibrosis airway microbiome. Cold Spring Harb Perspect Med. (2013) 3:a009738. doi: 10.1101/cshperspect.a009738
40. Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. (2015) 5:10241. doi: 10.1038/srep10241
41. Muller FM, Seidler M. Characteristics of pathogenic fungi and antifungal therapy in cystic fibrosis. Expert Rev Anti Infect Ther. (2010) 8:957–64. doi: 10.1586/eri.10.72
42. Tracy MC, Moss RB. The myriad challenges of respiratory fungal infection in cystic fibrosis. Pediatr Pulmonol. (2018) 53:S75–85. doi: 10.1002/ppul.24126
43. Kim SH, Clark ST, Surendra A, Copeland JK, Wang PW, Ammar R, et al. Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor Nrg1 as a mechanism of pathogen adaptation. PLoS Pathog. (2015) 11:e1005308. doi: 10.1371/journal.ppat.1005308
44. Duesberg U, Wosniok J, Naehrlich L, Eschenhagen P, Schwarz C. Risk factors for respiratory Aspergillus fumigatus in German cystic fibrosis patients and impact on lung function. Sci Rep. (2020) 10:18999. doi: 10.1038/s41598-020-75886-w
45. Leclair LW, Hogan DA. Mixed bacterial-fungal infections in the CF respiratory tract. Med Mycol. (2010) 48(Suppl. 1):S125–32. doi: 10.3109/13693786.2010.521522
46. Kraemer R, Deloséa N, Ballinari P, Gallati S, Crameri R. Effect of allergic bronchopulmonary aspergillosis on lung function in children with cystic fibrosis. Am J Respir Crit Care Med. (2006) 174:1211–20. doi: 10.1164/rccm.200603-423OC
47. Amin R, Dupuis A, Aaron SD, Ratjen F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest. (2010) 137:171–6. doi: 10.1378/chest.09-1103
48. Keown K, Reid A, Moore JE, Taggart CC, Downey DG. Coinfection with Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis. Eur Respir Rev. (2020) 29:200011. doi: 10.1183/16000617.0011-2020
49. Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. (2006) 61:1308–21. doi: 10.1111/j.1365-2958.2006.05306.x
50. Briard B, Bomme P, Lechner BE, Mislin GL, Lair V, Prevost MC, et al. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep. (2015) 5:8220. doi: 10.1038/srep08220
51. Briard B, Mislin GLA, Latge JP, Beauvais A. Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J Fungi (Basel). (2019) 5:48. doi: 10.3390/jof5020048
52. Sass G, Nazik H, Penner J, Shah H, Ansari SR, Clemons KV, et al. Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med Mycol. (2019) 57(Suppl. 2):S228–32. doi: 10.1093/mmy/myy087
53. Sass G, Ansari SR, Dietl AM, Déziel E, Haas H, Stevens DA. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS One. (2019) 14:e0216085. doi: 10.1371/journal.pone.0216085
54. Kerr J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J Infect. (1994) 28:305–10. doi: 10.1016/s0163-4453(94)91943-7
55. Briard B, Heddergott C, Latge JP. Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus. mBio. (2016) 7:e00219. doi: 10.1128/mBio.00219-16
56. Conrad D, Haynes M, Salamon P, Rainey PB, Youle M, Rohwer F. Cystic fibrosis therapy: a community ecology perspective. Am J Respir Cell Mol Biol. (2013) 48:150–6. doi: 10.1165/rcmb.2012-0059PS
57. Bozkurt C, Karaoz E, Adakli Aksoy B, Aydogdu S, Fisgin T. The use of allogeneic mesenchymal stem cells in childhood steroid-resistant acute graft-versus-host disease: a retrospective study of a single-center experience. Turk J Haematol. (2019) 36:186–92. doi: 10.4274/tjh.galenos.2019.2019.0090
58. Ahmed MH, Hassan A. Dexamethasone for the treatment of coronavirus disease (COVID-19): a review. SN Compr Clin Med. (2020) 2:1–10. doi: 10.1007/s42399-020-00610-8
59. Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. (2021) 384:693–704. doi: 10.1056/NEJMoa2021436
60. Romanou V, Koukaki E, Chantziara V, Stamou P, Kote A, Vasileiadis I, et al. Dexamethasone in the treatment of COVID-19: primus inter pares? J Pers Med. (2021) 11:556. doi: 10.3390/jpm11060556
61. Arastehfar A, Carvalho A, Houbraken J, Lombardi L, Garcia-Rubio R, Jenks JD, et al. Aspergillus fumigatus and aspergillosis: from basics to clinics. Stud Mycol. (2021) 100:100115. doi: 10.1016/j.simyco.2021.100115
62. Pushparaj K, Kuchi Bhotla H, Arumugam VA, Pappusamy M, Easwaran M, Liu WC, et al. Mucormycosis (black fungus) ensuing COVID-19 and comorbidity meets – magnifying global pandemic grieve and catastrophe begins. Sci Total Environ. (2022) 805:150355. doi: 10.1016/j.scitotenv.2021.150355
63. Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. (2020) 55:105954. doi: 10.1016/j.ijantimicag.2020.105954
64. Deana C, Vetrugno L, Bassi F, De Monte A. Tocilizumab administration in COVID-19 patients: water on the fire or gasoline? Med Mycol Case Rep. (2021) 31:32–4. doi: 10.1016/j.mmcr.2021.01.002
65. Witting C, Quaggin-Smith J, Mylvaganam R, Peigh G, Angarone M, Flaherty JD. Invasive pulmonary aspergillosis after treatment with tocilizumab in a patient with COVID-19 ARDS: a case report. Diagn Microbiol Infect Dis. (2021) 99:115272. doi: 10.1016/j.diagmicrobio.2020.115272
66. Cenci E, Mencacci A, Casagrande A, Mosci P, Bistoni F, Romani L. Impaired antifungal effector activity but not inflammatory cell recruitment in interleukin-6-deficient mice with invasive pulmonary aspergillosis. J Infect Dis. (2001) 184:610–7. doi: 10.1086/322793
67. Kimmig LM, Wu D, Gold M, Pettit NN, Pitrak D, Mueller J, et al. IL-6 inhibition in critically Ill COVID-19 patients is associated with increased secondary infections. Front Med (Lausanne). (2020) 7:583897. doi: 10.3389/fmed.2020.583897
68. Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. (2015) 16:27–35. doi: 10.1038/ni.3045
69. Paris S, Debeaupuis JP, Crameri R, Carey M, Charlès F, Prévost MC, et al. Conidial hydrophobins of Aspergillus fumigatus. Appl Environ Microbiol. (2003) 69:1581–8. doi: 10.1128/aem.69.3.1581-1588.2003
70. Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, et al. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. (2005) 1:e30. doi: 10.1371/journal.ppat.0010030
71. Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. (2009) 460:1117–21. doi: 10.1038/nature08264
72. Cunha C, Carvalho A, Esposito A, Bistoni F, Romani L. DAMP signaling in fungal infections and diseases. Front Immunol. (2012) 3:286. doi: 10.3389/fimmu.2012.00286
73. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol. (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
74. Parente R, Doni A, Bottazzi B, Garlanda C, Inforzato A. The complement system in Aspergillus fumigatus infections and its crosstalk with pentraxins. FEBS Lett. (2020) 594:2480–501. doi: 10.1002/1873-3468.13744
75. Kang Y, Yu Y, Lu L. The role of pentraxin 3 in aspergillosis: reality and prospects. Mycobiology. (2020) 48:1–8. doi: 10.1080/12298093.2020.1722576
76. Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. (2002) 420:182–6. doi: 10.1038/nature01195
77. Balhara J, Koussih L, Zhang J, Gounni AS. Pentraxin 3: an immuno-regulator in the lungs. Front Immunol. (2013) 4:127. doi: 10.3389/fimmu.2013.00127
78. Madan T, Eggleton P, Kishore U, Strong P, Aggrawal SS, Sarma PU, et al. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. (1997) 65:3171–9. doi: 10.1128/iai.65.8.3171-3179.1997
79. Jannuzzi GP, de Almeida JRF, Paulo LNM, de Almeida SR, Ferreira KS. Intracellular PRRs activation in targeting the immune response against fungal infections. Front Cell Infect Microbiol. (2020) 10:591970. doi: 10.3389/fcimb.2020.591970
80. Margalit A, Kavanagh K. The innate immune response to Aspergillus fumigatus at the alveolar surface. FEMS Microbiol Rev. (2015) 39:670–87. doi: 10.1093/femsre/fuv018
81. Ramirez-Ortiz ZG, Specht CA, Wang JP, Lee CK, Bartholomeu DC, Gazzinelli RT, et al. Toll-like receptor 9-dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA. Infect Immun. (2008) 76:2123–9. doi: 10.1128/iai.00047-08
82. Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, et al. Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med. (2002) 196:407–12. doi: 10.1084/jem.20020470
83. Taylor PR, Brown GD, Reid DM, Willment JA, Martinez-Pomares L, Gordon S, et al. The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. (2002) 169:3876–82. doi: 10.4049/jimmunol.169.7.3876
84. Mezger M, Kneitz S, Wozniok I, Kurzai O, Einsele H, Loeffler J. Proinflammatory response of immature human dendritic cells is mediated by dectin-1 after exposure to Aspergillus fumigatus germ tubes. J Infect Dis. (2008) 197:924–31. doi: 10.1086/528694
85. Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. (2005) 1:e42. doi: 10.1371/journal.ppat.0010042
86. Carvalho A, Pasqualotto AC, Pitzurra L, Romani L, Denning DW, Rodrigues F. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis. (2008) 197:618–21. doi: 10.1086/526500
87. de Boer MG, Jolink H, Halkes CJ, van der Heiden PL, Kremer D, Falkenburg JH, et al. Influence of polymorphisms in innate immunity genes on susceptibility to invasive aspergillosis after stem cell transplantation. PLoS One. (2011) 6:e18403. doi: 10.1371/journal.pone.0018403
88. Lupiañez CB, Canet LM, Carvalho A, Alcazar-Fuoli L, Springer J, Lackner M, et al. Polymorphisms in host immunity-modulating genes and risk of invasive aspergillosis: results from the AspBIOmics consortium. Infect Immun. (2015) 84:643–57. doi: 10.1128/iai.01359-15
89. Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, et al. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood. (2010) 116:5394–402. doi: 10.1182/blood-2010-04-279307
90. Paris S, Boisvieux-Ulrich E, Crestani B, Houcine O, Taramelli D, Lombardi L, et al. Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infect Immun. (1997) 65:1510–4. doi: 10.1128/iai.65.4.1510-1514.1997
91. Bertuzzi M, Hayes GE, Icheoku UJ, van Rhijn N, Denning DW, Osherov N, et al. Anti-Aspergillus activities of the respiratory epithelium in health and disease. J Fungi (Basel). (2018) 4:8. doi: 10.3390/jof4010008
92. Clark HR, Powell AB, Simmons KA, Ayubi T, Kale SD, Mitchell AP. Endocytic markers associated with the internalization and processing of Aspergillus fumigatus conidia by BEAS-2B cells. mSphere. (2019) 4:e00663–18. doi: 10.1128/mSphere.00663-18
93. Bigot J, Guillot L, Guitard J, Ruffin M, Corvol H, Balloy V, et al. Bronchial epithelial cells on the front line to fight lung infection-causing Aspergillus fumigatus. Front Immunol. (2020) 11:1041. doi: 10.3389/fimmu.2020.01041
94. Keizer EM, Wösten HAB, de Cock H. EphA2-dependent internalization of A. fumigatus conidia in A549 lung cells is modulated by DHN-melanin. Front Microbiol. (2020) 11:534118. doi: 10.3389/fmicb.2020.534118
95. Schiefermeier-Mach N, Haller T, Geley S, Perkhofer S. Migrating lung monocytes internalize and inhibit growth of Aspergillus fumigatus conidia. Pathogens. (2020) 9:983. doi: 10.3390/pathogens9120983
96. Serbina NV, Cherny M, Shi C, Bleau SA, Collins NH, Young JW, et al. Distinct responses of human monocyte subsets to Aspergillus fumigatus conidia. J Immunol. (2009) 183:2678–87. doi: 10.4049/jimmunol.0803398
97. Bozza S, Gaziano R, Spreca A, Bacci A, Montagnoli C, di Francesco P, et al. Dendritic cells transport conidia and hyphae of Aspergillus fumigatus from the airways to the draining lymph nodes and initiate disparate Th responses to the fungus. J Immunol. (2002) 168:1362–71. doi: 10.4049/jimmunol.168.3.1362
98. Morton CO, Varga JJ, Hornbach A, Mezger M, Sennefelder H, Kneitz S, et al. The temporal dynamics of differential gene expression in Aspergillus fumigatus interacting with human immature dendritic cells in vitro. PLoS One. (2011) 6:e16016. doi: 10.1371/journal.pone.0016016
99. Yang D, Guo X, Huang T, Liu C. The role of group 3 innate lymphoid cells in lung infection and immunity. Front Cell Infect Microbiol. (2021) 11:586471. doi: 10.3389/fcimb.2021.586471
100. Dunne MR, Wagener J, Loeffler J, Doherty DG, Rogers TR. Unconventional T cells – new players in antifungal immunity. Clin Immunol. (2021) 227:108734. doi: 10.1016/j.clim.2021.108734
101. Reeder KM, Mackel JJ, Godwin MS, Dunaway CW, Blackburn JP, Patel RP, et al. Role of common gamma-chain cytokines in lung interleukin-22 regulation after acute exposure to Aspergillus fumigatus. Infect Immun. (2018) 86:e00157–18. doi: 10.1128/IAI.00157-18
102. Romani L, Fallarino F, De Luca A, Montagnoli C, D’Angelo C, Zelante T, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. (2008) 451:211–5. doi: 10.1038/nature06471
103. Gessner MA, Werner JL, Lilly LM, Nelson MP, Metz AE, Dunaway CW, et al. Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus. Infect Immun. (2012) 80:410–7. doi: 10.1128/IAI.05939-11
104. Jahreis S, Bottcher S, Hartung S, Rachow T, Rummler S, Dietl AM, et al. Human MAIT cells are rapidly activated by Aspergillus spp. in an APC-dependent manner. Eur J Immunol. (2018) 48:1698–706. doi: 10.1002/eji.201747312
105. Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
106. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
107. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci USA. (2016) 113:10133–8. doi: 10.1073/pnas.1610750113
108. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun. (2016) 7:11653. doi: 10.1038/ncomms11653
109. Cramer RA, Rivera A, Hohl TM. Immune responses against Aspergillus fumigatus: what have we learned? Curr Opin Infect Dis. (2011) 24:315–22. doi: 10.1097/QCO.0b013e328348b159
110. Sales-Campos H, Tonani L, Cardoso CR, Kress MR. The immune interplay between the host and the pathogen in Aspergillus fumigatus lung infection. Biomed Res Int. (2013) 2013:693023. doi: 10.1155/2013/693023
111. Dewi IMW, van de Veerdonk FL, Gresnigt MS. The multifaceted role of T-helper responses in host defense against Aspergillus fumigatus. J Fungi (Basel). (2017) 3:55. doi: 10.3390/jof3040055
112. Bellocchio S, Bozza S, Montagnoli C, Perruccio K, Gaziano R, Pitzurra L, et al. Immunity to Aspergillus fumigatus: the basis for immunotherapy and vaccination. Med Mycol. (2005) 43(Suppl. 1):S181–8. doi: 10.1080/14789940500051417
113. Arias M, Santiago L, Vidal-Garcia M, Redrado S, Lanuza P, Comas L, et al. Preparations for invasion: modulation of host lung immunity during pulmonary aspergillosis by gliotoxin and other fungal secondary metabolites. Front Immunol. (2018) 9:2549. doi: 10.3389/fimmu.2018.02549
114. Stuehler C, Khanna N, Bozza S, Zelante T, Moretti S, Kruhm M, et al. Cross-protective TH1 immunity against Aspergillus fumigatus and Candida albicans. Blood. (2011) 117:5881–91. doi: 10.1182/blood-2010-12-325084
115. Chaudhary N, Staab JF, Marr KA. Healthy human T-cell responses to Aspergillus fumigatus antigens. PLoS One. (2010) 5:e9036. doi: 10.1371/journal.pone.0009036
116. Levitz SM, Huang H, Ostroff GR, Specht CA. Exploiting fungal cell wall components in vaccines. Semin Immunopathol. (2015) 37:199–207. doi: 10.1007/s00281-014-0460-6
117. Bartemes KR, Kita H. Innate and adaptive immune responses to fungi in the airway. J Allergy Clin Immunol. (2018) 142:353–63. doi: 10.1016/j.jaci.2018.06.015
118. Zhu Z, Zheng T, Homer RJ, Kim Y-K, Chen NY, Cohn L, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. (2004) 304:1678–82. doi: 10.1126/science.1095336
119. Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. (2007) 357:2016–27. doi: 10.1056/NEJMoa073600
120. Reese TA, Liang H-E, Tager AM, Luster AD, Van Rooijen N, Voehringer D, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. (2007) 447:92–6. doi: 10.1038/nature05746
121. Kobayashi T, Iijima K, Radhakrishnan S, Mehta V, Vassallo R, Lawrence CB, et al. Asthma-related environmental fungus, alternaria, activates dendritic cells and produces potent Th2 adjuvant activity. J Immunol. (2009) 182:2502. doi: 10.4049/jimmunol.0802773
122. Arae K, Morita H, Unno H, Motomura K, Toyama S, Okada N, et al. Chitin promotes antigen-specific Th2 cell-mediated murine asthma through induction of IL-33-mediated IL-1β production by DCs. Sci Rep. (2018) 8:11721. doi: 10.1038/s41598-018-30259-2
123. Dubey LK, Moeller JB, Schlosser A, Sorensen GL, Holmskov U. Chitin enhances serum IgE in Aspergillus fumigatus induced allergy in mice. Immunobiology. (2015) 220:714–21. doi: 10.1016/j.imbio.2015.01.002
124. Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. (2007) 37:2695–706. doi: 10.1002/eji.200737409
125. Guerra ES, Lee CK, Specht CA, Yadav B, Huang H, Akalin A, et al. Central role of IL-23 and IL-17 producing eosinophils as immunomodulatory effector cells in acute pulmonary aspergillosis and allergic asthma. PLoS Pathog. (2017) 13:e1006175. doi: 10.1371/journal.ppat.1006175
126. Jolink H, de Boer R, Hombrink P, Jonkers RE, van Dissel JT, Falkenburg JH, et al. Pulmonary immune responses against Aspergillus fumigatus are characterized by high frequencies of IL-17 producing T-cells. J Infect. (2017) 74:81–8. doi: 10.1016/j.jinf.2016.10.010
127. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell. (2019) 176:1340–55.e1315. doi: 10.1016/j.cell.2019.01.041
128. Montagnoli C, Fallarino F, Gaziano R, Bozza S, Bellocchio S, Zelante T, et al. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J Immunol. (2006) 176:1712–23. doi: 10.4049/jimmunol.176.3.1712
129. Murdock BJ, Shreiner AB, McDonald RA, Osterholzer JJ, White ES, Toews GB, et al. Coevolution of TH1, TH2, and TH17 responses during repeated pulmonary exposure to Aspergillus fumigatus conidia. Infect Immun. (2011) 79:125–35. doi: 10.1128/IAI.00508-10
130. Chauhan B, Hutcheson PS, Slavin RG, Bellone CJ. T-cell receptor bias in patients with allergic bronchopulmonary aspergillosis. Hum Immunol. (2002) 63:286–94. doi: 10.1016/s0198-8859(02)00361-0
131. Chotirmall SH, Mirkovic B, Lavelle GM, McElvaney NG. Immunoevasive Aspergillus virulence factors. Mycopathologia. (2014) 178:363–70. doi: 10.1007/s11046-014-9768-y
132. Singh S, Kanaujia R, Rudramurthy M. Immunopathogenesis of aspergillosis. In: M Razzaghi-Abyaneh editor. The Genus Aspergillus – Pathogenicity, Mycotoxin Production and Industrial Applications [Working Title]. London: IntechOpen. (2021).
133. Valiante V, Baldin C, Hortschansky P, Jain R, Thywißen A, Straßburger M, et al. The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. Mol Microbiol. (2016) 102:321–35. doi: 10.1111/mmi.13462
134. Abad A, Fernández-Molina JV, Bikandi J, Ramírez A, Margareto J, Sendino J, et al. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. (2010) 27:155–82. doi: 10.1016/j.riam.2010.10.003
135. Kyrmizi I, Ferreira H, Carvalho A, Figueroa JAL, Zarmpas P, Cunha C, et al. Calcium sequestration by fungal melanin inhibits calcium-calmodulin signalling to prevent LC3-associated phagocytosis. Nat Microbiol. (2018) 3:791–803. doi: 10.1038/s41564-018-0167-x
136. Ferling I, Dunn JD, Ferling A, Soldati T, Hillmann F. Conidial melanin of the human-pathogenic fungus Aspergillus fumigatus disrupts cell autonomous defenses in amoebae. mBio. (2020) 11:e00862–20. doi: 10.1128/mBio.00862-20
137. Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, Galvez EM, et al. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot Cell. (2007) 6:1562–9. doi: 10.1128/ec.00141-07
138. de Castro PA, Colabardini AC, Moraes M, Horta MAC, Knowles SL, Raja HA, et al. Regulation of gliotoxin biosynthesis and protection in Aspergillus species. PLoS Genet. (2022) 18:e1009965. doi: 10.1371/journal.pgen.1009965
139. Amitani R, Taylor G, Elezis EN, Llewellyn-Jones C, Mitchell J, Kuze F, et al. Purification and characterization of factors produced by Aspergillus fumigatus which affect human ciliated respiratory epithelium. Infect Immun. (1995) 63:3266–71. doi: 10.1128/iai.63.9.3266-3271.1995
140. Fitzpatrick LR, Wang J, Le T. In vitro and in vivo effects of gliotoxin, a fungal metabolite: efficacy against dextran sodium sulfate-induced colitis in rats. Dig Dis Sci. (2000) 45:2327–36. doi: 10.1023/a:1005630723111
141. Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. (2017) 2:17023. doi: 10.1038/sigtrans.2017.23
142. Fernandes TR, Segorbe D, Prusky D, Di Pietro A. How alkalinization drives fungal pathogenicity. PLoS Pathog. (2017) 13:e1006621. doi: 10.1371/journal.ppat.1006621
143. Vylkova S. Environmental pH modulation by pathogenic fungi as a strategy to conquer the host. PLoS Pathog. (2017) 13:e1006149. doi: 10.1371/journal.ppat.1006149
144. Alkan N, Espeso EA, Prusky D. Virulence regulation of phytopathogenic fungi by pH. Antioxid Redox Signal. (2013) 19:1012–25. doi: 10.1089/ars.2012.5062
145. Dutton MV, Evans CS. Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can J Microbiol. (1996) 42:881–95. doi: 10.1139/m96-114
146. Palmieri F, Estoppey A, House GL, Lohberger A, Bindschedler S, Chain PSG, et al. Oxalic Acid, A Molecule at the Crossroads of Bacterial-Fungal Interactions. GM Gadd, S Sariaslani, editors. Cambridge, MA: Academic Press (2019). p. 49–77.
147. Prusky D, Yakoby N. Pathogenic fungi: leading or led by ambient pH? Mol Plant Pathol. (2003) 4:509–16. doi: 10.1046/j.1364-3703.2003.00196.x
148. Samaranayake LP, Hughes A, Weetman DA, MacFarlane TW. Growth and acid production of Candida species in human saliva supplemented with glucose. J Oral Pathol. (1986) 15:251–4. doi: 10.1111/j.1600-0714.1986.tb00617.x
149. Tsuboi R, Matsuda K, Ko IJ, Ogawa H. Correlation between culture medium pH, extracellular proteinase activity, and cell growth of Candida albicans in insoluble stratum corneum-supplemented media. Arch Dermatol Res. (1989) 281:342–5. doi: 10.1007/BF00412979
150. Mba IE, Nweze EI. Mechanism of Candida pathogenesis: revisiting the vital drivers. Eur J Clin Microbiol Infect Dis. (2020) 39:1797–819. doi: 10.1007/s10096-020-03912-w
151. Singh DK, Németh T, Papp A, Tóth R, Lukácsi S, Heidingsfeld O, et al. Functional characterization of secreted aspartyl proteases in Candida parapsilosis. mSphere. (2019) 4:1–16. doi: 10.1128/mSphere.00484-19
152. Moore D, Robson G, Trinci T. 21st Century Guidebook to Fungi. Cambridge: Cambridge University Press (2011).
153. van Kan JAL, Shaw MW, Grant-Downton RT. Botrytis species: relentless necrotrophic thugs or endophytes gone rogue? Mol Plant Pathol. (2014) 15:957–61. doi: 10.1111/mpp.12148
154. Kurrein F, Path FRC, Green GH, Rowles SL. Localized deposition of calcium oxalate around a pulmonary Aspergillus niger fungus ball. Am J Clin Pathol. (1975) 64:556–63. doi: 10.1093/ajcp/64.4.556
155. Kauffman CA, Wilson KH, Schwartz DB. Necrotizing pulmonary aspergillosis with oxalosis: nekrotisierende pulmonale aspergillose mit oxalose. Mycoses. (1984) 27:535–8. doi: 10.1111/j.1439-0507.1984.tb01984.x
156. Lee SH, Barnes WG, Schaetzel WP. Pulmonary aspergillosis and the importance of oxalate crystal recognition in cytology specimens. Arch Pathol Lab Med. (1986) 110:1176–9.
157. Ghio AJ, Peterseim DS, Roggli VL, Piantadosi CA. Pulmonary oxalate deposition associated with Aspergillus niger infection. An oxidant hypothesis of toxicity. Am Rev Respir Dis. (1992) 145:1499–502. doi: 10.1164/ajrccm/145.6.1499
158. Muntz FHA. Oxalate-producing pulmonary aspergillosis in an alpaca. Vet Pathol. (1999) 36:631–2. doi: 10.1354/vp.36-6-631
159. Pabuççuoǧlu U. Aspects of oxalosis associated with aspergillosis in pathology specimens. Pathol Res Pract. (2005) 201:363–8. doi: 10.1016/j.prp.2005.03.005
160. Kuwabara H, Shibayama Y. Pulmonary aspergilloma with prominent oxalate deposition. Indian J Pathol Microbiol. (2012) 55:589–589. doi: 10.4103/0377-4929.107838
161. Oda M, Saraya T, Wakayama M, Shibuya K, Ogawa Y, Inui T, et al. Calcium oxalate crystal deposition in a patient with aspergilloma due to Aspergillus niger. J Thorac Dis. (2013) 5:E174–8. doi: 10.3978/j.issn.2072-1439.2013.08.45
162. Maeno T, Sasaki M, Shibue Y, Mimura K, Oka H. Calcium oxalate in the sputum may aid in the diagnosis of pulmonary aspergillosis: a report of two cases. Med Mycol Case Rep. (2015) 8:32–6. doi: 10.1016/j.mmcr.2015.01.003
163. Payne CL, Dark MJ, Conway JA, Farina LL. A retrospective study of the prevalence of calcium oxalate crystals in veterinary Aspergillus cases. J Vet Diagn Invest. (2017) 29:51–8. doi: 10.1177/1040638716672254
164. Yi Y, Cho SY, Lee DG, Jung JI, Park YJ, Lee KY. Invasive pulmonary aspergillosis due to Aspergillus awamori: role of calcium oxalate crystal precipitation mimicking mucormycosis. Mycopathologia. (2020) 185:409–11. doi: 10.1007/s11046-019-00405-z
166. Grinstein S, Klip A. Calcium homeostasis and the activation of calcium channels in cells of the immune system. Bull N Y Acad Med. (1989) 65:69–79.
167. Diamantstein T, Odenwald MV. Control of the immune response in vitro by calcium ions. I. The antagonistic action of calcium ions on cell proliferation and on cell differentiation. Immunology. (1974) 27:531–41.
168. Diamantstein T, Ulmer A. The control of immune response in vitro by Ca2+. II. The Ca2+ dependent period during mitogenic stimulation. Immunology. (1975) 28:121–5.
169. Palmieri F, Palmieri I, Noormamode N, Estoppey A, Ishak MO, Kelliher JM, et al. Biocontrol of Aspergillus niger in 3D-lung cell tissues by oxalotrophic bacteria. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.08.20.259929
170. Palmieri F. Bacterial Oxalotrophy as an Alternative Biocontrol Approach for the Fight Against Pulmonary Aspergillosis. Ph.D. thesis. Neuchâtel: Université de Neuchâtel (2021).
171. Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio. (2011) 2:e00055–11. doi: 10.1128/mBio.00055-11
172. Mirbod-Donovan F, Schaller R, Hung CY, Xue J, Reichard U, Cole GT. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect Immun. (2006) 74:504–15. doi: 10.1128/IAI.74.1.504-515.2006
173. Wise HZ, Hung CY, Whiston E, Taylor JW, Cole GT. Extracellular ammonia at sites of pulmonary infection with Coccidioides posadasii contributes to severity of the respiratory disease. Microb Pathog. (2013) 59-60:19–28. doi: 10.1016/j.micpath.2013.04.003
174. St Leger RJ, Nelson JO, Screen SE. The entomopathogenic fungus Metarhizium anisopliae alters ambient pH, allowing extracellular protease production and activity. Microbiology (Reading). (1999) 145(Pt 10):2691–9. doi: 10.1099/00221287-145-10-2691
175. Chang D, Dela Cruz CS, Sharma L. Challenges in understanding lung microbiome: it is not like the gut microbiome. Respirology. (2020) 25:244–5. doi: 10.1111/resp.13759
176. Mitchell AB, Oliver BGG, Glanville AR. Translational aspects of the human respiratory virome. Am J Respir Crit Care Med. (2016) 194:1458–64. doi: 10.1164/rccm.201606-1278CI
177. Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. (2020) 10:9. doi: 10.3389/fcimb.2020.00009
178. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. (2011) 184:957–63. doi: 10.1164/rccm.201104-0655OC
179. Foesel BU, Pfeiffer S, Raj ACD, Etschmann SK, Schloter M. Applying ecological theories in research: lessons learned from microbial ecology and evolution? ERS Monogr. (2019) 2019:50–66. doi: 10.1183/2312508X.10015718
180. Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. (2012) 186:536–45. doi: 10.1164/rccm.201204-0693OC
181. van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis. (2013) 13:69. doi: 10.1186/1471-2334-13-69
182. Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. (2014) 384:691–702. doi: 10.1016/S0140-6736(14)61136-3
183. Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med. (2015) 191:932–42. doi: 10.1164/rccm.201409-1583OC
184. Budden KF, Shukla SD, Rehman SF, Bowerman KL, Keely S, Hugenholtz P, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. (2019) 7:907–20. doi: 10.1016/S2213-2600(18)30510-1
185. Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, et al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. (2018) 9:1168. doi: 10.3389/fphys.2018.01168
186. Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. (2018) 39:677–96. doi: 10.1016/j.it.2018.04.002
187. Li XV, Leonardi I, Iliev ID. Gut mycobiota in immunity and inflammatory disease. Immunity. (2019) 50:1365–79. doi: 10.1016/j.immuni.2019.05.023
188. Deray G, le Hoang P, Cacoub P, Assogba U, Grippon P, Baumelou A. Oral contraceptive interaction with cyclosporin. Lancet. (1987) 1:158–9. doi: 10.1016/s0140-6736(87)91988-x
189. Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. (2017) 17:635–46. doi: 10.1038/nri.2017.55
190. Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. (2019) 16:331–45. doi: 10.1038/s41575-019-0121-2
191. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
192. Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. (2014) 14:827–35. doi: 10.1038/nri3769
193. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
194. Ubags NDJ, Marsland BJ. Mechanistic insight into the function of the microbiome in lung diseases. Eur Respir J. (2017) 50:1602467. doi: 10.1183/13993003.02467-2016
195. Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc. (2015) 12(Suppl. 2):S150–6. doi: 10.1513/AnnalsATS.201503-133AW
196. Kolwijck E, van de Veerdonk FL. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur J Immunol. (2014) 44:3156–65. doi: 10.1002/eji.201344404
197. Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. (2016) 19:865–73. doi: 10.1016/j.chom.2016.05.003
198. Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, et al. Response to fungal dysbiosis by gut-resident CX3CR1(+) mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe. (2018) 24: 847–56.e844. doi: 10.1016/j.chom.2018.11.003
199. van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin JB, et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun. (2020) 11:2577. doi: 10.1038/s41467-020-16431-1
200. Hérivaux A, Willis JR, Mercier T, Lagrou K, Gonçalves SM, Gonçales RA, et al. Lung microbiota predict invasive pulmonary aspergillosis and its outcome in immunocompromised patients. Thorax. (2021) 77:283–91. doi: 10.1136/thoraxjnl-2020-216179
201. Kottom TJ, Köhler JR, Thomas CF Jr, Fink GR, Limper AH. Lung epithelial cells and extracellular matrix components induce expression of Pneumocystis carinii STE20, a gene complementing the mating and pseudohyphal growth defects of STE20 mutant yeast. Infect Immun. (2003) 71:6463–71. doi: 10.1128/iai.71.11.6463-6471.2003
202. Kunst H, Wickremasinghe M, Wells A, Wilson R. Nontuberculous mycobacterial disease and Aspergillus-related lung disease in bronchiectasis. Eur Respir J. (2006) 28:352–7. doi: 10.1183/09031936.06.00139005
203. Schelenz S, Barnes RA, Barton RC, Cleverley JR, Lucas SB, Kibbler CC, et al. British society for medical mycology best practice recommendations for the diagnosis of serious fungal diseases. Lancet Infect Dis. (2015) 15:461–74. doi: 10.1016/s1473-3099(15)70006-x
204. Lass-Florl C. How to make a fast diagnosis in invasive aspergillosis. Med Mycol. (2019) 57(Suppl. 2):S155–60. doi: 10.1093/mmy/myy103
205. Haydour Q, Hage CA, Carmona EM, Epelbaum O, Evans SE, Gabe LM, et al. Diagnosis of fungal infections. A systematic review and meta-analysis supporting American thoracic society practice guideline. Ann Am Thorac Soc. (2019) 16:1179–88. doi: 10.1513/AnnalsATS.201811-766OC
206. Dhooria S, Sehgal IS, Muthu V, Agarwal R. Treatment of allergic bronchopulmonary aspergillosis: from evidence to practice. Future Microbiol. (2020) 15:365–76. doi: 10.2217/fmb-2019-0276
207. Kleinkauf N, Verweij PE, Arendrup MC, Donnelly PJ, Cuenca-Estrella M, Fraaije B, et al. Risk Assessment on the Impact of Environmental Usage Of Triazoles on the Development and Spread of Resistance to Medical Triazoles in Aspergillus Species. Stockholm: ECDC (2013).
208. Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc Lond B Biol Sci. (2016) 371:20150460. doi: 10.1098/rstb.2015.0460
209. Geißel B, Loiko V, Klugherz I, Zhu Z, Wagener N, Kurzai O, et al. Azole-induced cell wall carbohydrate patches kill Aspergillus fumigatus. Nat Commun. (2018) 9:3098. doi: 10.1038/s41467-018-05497-7
210. Garcia-Rubio R, Gonzalez-Jimenez I, Lucio J, Mellado E. Characterization of Aspergillus fumigatus cross-resistance between clinical and DMI azole drugs. Appl Environ Microbiol. (2020) 87:e02539–20. doi: 10.1128/AEM.02539-20
211. Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, et al. Treatment of aspergillosis: clinical practice guidelines of the infectious diseases society of America. Clin Infect Dis. (2008) 46:327–60. doi: 10.1086/525258
212. Verweij PE, Zhang J, Debets AJM, Meis JF, van de Veerdonk FL, Schoustra SE, et al. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: a dilemma for clinical management. Lancet Infect Dis. (2016) 16:e251–60. doi: 10.1016/s1473-3099(16)30138-4
213. Arastehfar A, Carvalho A, van de Veerdonk FL, Jenks JD, Koehler P, Krause R, et al. COVID-19 associated pulmonary aspergillosis (CAPA)-from immunology to treatment. J Fungi (Basel). (2020) 6:91. doi: 10.3390/jof6020091
214. Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. (2001) 39:2431–8. doi: 10.1128/jcm.39.7.2431-2438.2001
215. Flückiger U, Marchetti O, Bille J, Eggimann P, Zimmerli S, Imhof A, et al. Treatment options of invasive fungal infections in adults. Swiss Med Wkly. (2006) 136:447–63.
216. Satish S, Perlin DS. Echinocandin resistance in Aspergillus fumigatus has broad implications for membrane lipid perturbations that influence drug-target interactions. Microbiol Insights. (2019) 12:1–4. doi: 10.1177/1178636119897034
217. Perlin DS. Echinocandin resistance in Candida. Clin Infect Dis. (2015) 61(Suppl. 6):S612–7. doi: 10.1093/cid/civ791
218. Blum G, Perkhofer S, Haas H, Schrettl M, Wurzner R, Dierich MP, et al. Potential basis for amphotericin B resistance in Aspergillus terreus. Antimicrob Agents Chemother. (2008) 52:1553–5. doi: 10.1128/AAC.01280-07
219. Liang T, Chen W, Yang X, Wang Q, Wan Z, Li R, et al. The elevated endogenous reactive oxygen species contribute to the sensitivity of the amphotericin B-resistant isolate of Aspergillus flavus to triazoles and echinocandins. Front Microbiol. (2021) 12:680749. doi: 10.3389/fmicb.2021.680749
220. Ashu EE, Korfanty GA, Samarasinghe H, Pum N, You M, Yamamura D, et al. Widespread amphotericin B-resistant strains of Aspergillus fumigatus in Hamilton, Canada. Infect Drug Resist. (2018) 11:1549–55. doi: 10.2147/IDR.S170952
221. Rudramurthy SM, Paul RA, Chakrabarti A, Mouton JW, Meis JF. Invasive aspergillosis by Aspergillus flavus: epidemiology, diagnosis, antifungal resistance, and management. J Fungi (Basel). (2019) 5:55. doi: 10.3390/jof5030055
222. Vermes A, Guchelaar HJ, Dankert J. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J Antimicrob Chemother. (2000) 46:171–9. doi: 10.1093/jac/46.2.171
223. Gsaller F, Furukawa T, Carr PD, Rash B, Jochl C, Bertuzzi M, et al. Mechanistic basis of pH-dependent 5-flucytosine resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. (2018) 62:e02593–17. doi: 10.1128/AAC.02593-17
224. Vahedi-Shahandashti R, Lass-Florl C. Novel antifungal agents and their activity against Aspergillus species. J Fungi (Basel). (2020) 6:213. doi: 10.3390/jof6040213
225. du Pre S, Beckmann N, Almeida MC, Sibley GEM, Law D, Brand AC, et al. Effect of the novel antifungal drug F901318 (olorofim) on growth and viability of Aspergillus fumigatus. Antimicrob Agents Chemother. (2018) 62:e231–18. doi: 10.1128/AAC.00231-18
226. Sandison T, Ong V, Lee J, Thye D. Safety and pharmacokinetics of CD101 IV, a novel echinocandin, in healthy adults. Antimicrob Agents Chemother. (2017) 61:e01627–16. doi: 10.1128/AAC.01627-16
227. Jallow S, Govender NP. Ibrexafungerp: a first-in-class oral triterpenoid glucan synthase inhibitor. J Fungi (Basel). (2021) 7:163. doi: 10.3390/jof7030163
228. Borroto-Esoda K, Barat S, Angulo D, Holden K, Warn P. SCY-078 demonstrates significant antifungal activity in a murine model of invasive aspergillosis. Open Forum Infect Dis. (2017) 4(Suppl. 1):S472. doi: 10.1093/ofid/ofx163.1207
229. Nakamura I, Ohsumi K, Takeda S, Katsumata K, Matsumoto S, Akamatsu S, et al. ASP2397 is a novel natural compound that exhibits rapid and potent fungicidal activity against Aspergillus species through a specific transporter. Antimicrob Agents Chemother. (2019) 63:e02689–18. doi: 10.1128/AAC.02689-18
230. Scholthof KB. The disease triangle: pathogens, the environment and society. Nat Rev Microbiol. (2007) 5:152–6. doi: 10.1038/nrmicro1596
231. Bernardo-Cravo AP, Schmeller DS, Chatzinotas A, Vredenburg VT, Loyau A. Environmental factors and host microbiomes shape host-pathogen dynamics. Trends Parasitol. (2020) 36:616–33. doi: 10.1016/j.pt.2020.04.010
232. Misslinger M, Hortschansky P, Brakhage AA, Haas H. Fungal iron homeostasis with a focus on Aspergillus fumigatus. Biochim Biophys Acta Mol Cell Res. (2021) 1868:118885. doi: 10.1016/j.bbamcr.2020.118885
233. Aznar A, Dellagi A. New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J Exp Bot. (2015) 66:3001–10. doi: 10.1093/jxb/erv155
234. Bairwa G, Hee Jung W, Kronstad JW. Iron acquisition in fungal pathogens of humans. Metallomics. (2017) 9:215–27. doi: 10.1039/c6mt00301j
235. Blatzer M, Schrettl M, Sarg B, Lindner HH, Pfaller K, Haas H. SidL, an Aspergillus fumigatus transacetylase involved in biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Appl Environ Microbiol. (2011) 77:4959–66. doi: 10.1128/AEM.00182-11
236. Haas H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat Prod Rep. (2014) 31:1266–76. doi: 10.1039/c4np00071d
237. Liu H, Xu W, Bruno VM, Phan QT, Solis NV, Woolford CA, et al. Determining Aspergillus fumigatus transcription factor expression and function during invasion of the mammalian lung. PLoS Pathog. (2021) 17:e1009235. doi: 10.1371/journal.ppat.1009235
238. Matthaiou EI, Sass G, Stevens DA, Hsu JL. Iron: an essential nutrient for Aspergillus fumigatus and a fulcrum for pathogenesis. Curr Opin Infect Dis. (2018) 31:506–11. doi: 10.1097/QCO.0000000000000487
239. Leal SM Jr, Roy S, Vareechon C, Carrion S, Clark H, Lopez-Berges MS, et al. Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. (2013) 9:e1003436. doi: 10.1371/journal.ppat.1003436
240. Kramer J, Ozkaya O, Kummerli R. Bacterial siderophores in community and host interactions. Nat Rev Microbiol. (2020) 18:152–63. doi: 10.1038/s41579-019-0284-4
241. Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. (2012) 10:525–37. doi: 10.1038/nrmicro2836
242. Healy C, Munoz-Wolf N, Strydom J, Faherty L, Williams NC, Kenny S, et al. Nutritional immunity: the impact of metals on lung immune cells and the airway microbiome during chronic respiratory disease. Respir Res. (2021) 22:133. doi: 10.1186/s12931-021-01722-y
243. Seyoum Y, Baye K, Humblot C. Iron homeostasis in host and gut bacteria – a complex interrelationship. Gut Microbes. (2021) 13:1–19. doi: 10.1080/19490976.2021.1874855
244. Bastos RW, Rossato L, Valero C, Lagrou K, Colombo AL, Goldman GH. Potential of gallium as an antifungal agent. Front Cell Infect Microbiol. (2019) 9:414. doi: 10.3389/fcimb.2019.00414
245. Zhao J, Yu W. Interaction between Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis. PeerJ. (2018) 6:e5931. doi: 10.7717/peerj.5931
246. Margalit A, Carolan JC, Kavanagh K. Bacterial interactions with Aspergillus fumigatus in the immunocompromised lung. Microorganisms. (2021) 9:435. doi: 10.3390/microorganisms9020435
247. Rouanet A, Bolca S, Bru A, Claes I, Cvejic H, Girgis H, et al. Live biotherapeutic products, a road map for safety assessment. Front Med (Lausanne). (2020) 7:237. doi: 10.3389/fmed.2020.00237
248. Yildiz S, Pereira Bonifacio Lopes JP, Berge M, Gonzalez-Ruiz V, Baud D, Kloehn J, et al. Respiratory tissue-associated commensal bacteria offer therapeutic potential against pneumococcal colonization. Elife. (2020) 9:e53581. doi: 10.7554/eLife.53581
249. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. (2015) 517:205–8. doi: 10.1038/nature13828
250. Mazzola M. Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie Van Leeuwenhoek. (2002) 81:557–64. doi: 10.1023/a:1020557523557
251. Mendes R, Kruijt M, De Bruijn I, Dekkers E, Van Der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. (2011) 332:1097–100. doi: 10.1126/science.1203980
252. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. (2012) 336:1255–62. doi: 10.1126/science.1224203
253. Pepper JW, Rosenfeld S. The emerging medical ecology of the human gut microbiome. Trends Ecol Evol. (2012) 27:381–4. doi: 10.1016/j.tree.2012.03.002
254. Martens K, Pugin B, De Boeck I, Spacova I, Steelant B, Seys SF, et al. Probiotics for the airways: potential to improve epithelial and immune homeostasis. Allergy. (2018) 73:1954–63. doi: 10.1111/all.13495
255. De Boeck I, van den Broek MFL, Allonsius CN, Spacova I, Wittouck S, Martens K, et al. Lactobacilli have a niche in the human nose. Cell Rep. (2020) 31:107674. doi: 10.1016/j.celrep.2020.107674
256. De Rudder C, Garcia-Timermans C, De Boeck I, Lebeer S, Van de Wiele T, Calatayud Arroyo M. Lacticaseibacillus casei AMBR2 modulates the epithelial barrier function and immune response in a donor-derived nasal microbiota manner. Sci Rep. (2020) 10:16939. doi: 10.1038/s41598-020-73857-9
257. Cho DY, Skinner D, Lim DJ, McLemore JG, Koch CG, Zhang S, et al. The impact of Lactococcus lactis (probiotic nasal rinse) co-culture on growth of patient-derived strains of Pseudomonas aeruginosa. Int Forum Allergy Rhinol. (2020) 10:444–9. doi: 10.1002/alr.22521
Keywords: chronic respiratory disease, microbiome, mycobiome, aspergillosis, live biotherapeutic products, disease management, environmental interference
Citation: Palmieri F, Koutsokera A, Bernasconi E, Junier P, von Garnier C and Ubags N (2022) Recent Advances in Fungal Infections: From Lung Ecology to Therapeutic Strategies With a Focus on Aspergillus spp.. Front. Med. 9:832510. doi: 10.3389/fmed.2022.832510
Received: 09 December 2021; Accepted: 22 February 2022;
Published: 21 March 2022.
Edited by:
Mehdi Razzaghi-Abyaneh, Pasteur Institute of Iran (PII), IranReviewed by:
Koshika Yadava, Vector Biopharma AG, SwitzerlandSarah Sze Wah Wong, Institut Pasteur, France
Copyright © 2022 Palmieri, Koutsokera, Bernasconi, Junier, von Garnier and Ubags. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabio Palmieri, ZmFiaW8ucGFsbWllcmlAdW5pbmUuY2g=; Niki Ubags, bmlraS51YmFnc0BjaHV2LmNo
 Fabio Palmieri
Fabio Palmieri Angela Koutsokera
Angela Koutsokera Eric Bernasconi
Eric Bernasconi Pilar Junier
Pilar Junier Christophe von Garnier
Christophe von Garnier Niki Ubags
Niki Ubags