Abstract
Background and objective:
This pilot study aimed to identify potential blood DNA methylation (BDM) biomarker genes for the diagnosis of liver fibrosis in non-alcoholic fatty liver disease (NAFLD).
Methods:
We included a total of 16 NAFLD patients with significant (SLF, liver fibrosis stage ≥ 2) and 16 patients with non-significant liver fibrosis (NSLF, fibrosis stages 0–1). The association between BDM and liver fibrosis was analyzed. Genes were selected based on a stepwise-filtering with CpG islands containing significant differentially methylated probes.
Results:
The two groups of patients were distinguishable through both t-distributed stochastic neighbor embedding (t-SNE) analysis and unsupervised hierarchical clustering analysis based on their BDM status. BDM levels were significantly higher in the NSLF group than in the SLF group. The methylation levels in the island and shelf regions were also significantly higher in the NSLF group, as well as the methylation levels in the first exon, 3′-untranslated region, body, ExonBnd, non-intergenic region, transcription start site (TSS)1500, and TSS200 regions (all p < 0.05). BDM status was associated with greater histological liver fibrosis, but not with age, sex, or other histological features of NAFLD (p < 0.05). The methylation levels of the hypomethylated CpG island region of CISTR, IFT140, and RGS14 genes were increased in the NSLF group compared to the SLF group (all p < 0.05).
Conclusion:
BDM may stratify NAFLD patients with significant and non-significant liver fibrosis. The CISTR, IFT140, and RGS14 genes are potential novel candidate BDM biomarkers for liver fibrosis and these pilot data suggest further work on BDM biomarkers is warranted.
Introduction
Non-alcoholic fatty liver disease (NAFLD) has become a public health problem, due to its high prevalence globally (1), and there is now evidence that it is a multisystem disease (2) increasing risk of multiple cardiometabolic disorders, such as cardiovascular disease (3) and type 2 diabetes (4). NAFLD is a complex multifactorial disease involving genetic, metabolic and environmental factors. Over the past decade, research into pathogenesis, diagnosis, and treatment has revealed different aspects of NAFLD, challenging the definition of clinical practice and the accuracy of treatment strategies. Recently, several international experts reached a consensus that the old definition of NAFLD does not reflect current knowledge, suggesting that metabolic dysfunction-associated fatty liver disease (MAFLD) is a more appropriate definition (5–7).
Liver fibrosis, which is typically characterized by the deposition of excess extracellular matrix and loss of hepatocyte function, is frequently associated with NAFLD. Studies have revealed that the severity of liver fibrosis is the strongest prognostic factor for all-cause mortality and the development of liver-related complications in people with NAFLD (8–11). The risk of liver-related mortality also increases exponentially with the severity of liver fibrosis (12). NAFLD patients with significant liver fibrosis (SLF, i.e., those with F stage ≥ 2) are at higher risk of liver-related mortality than those with non-significant liver fibrosis (NSLF; F stage 0–1) (12). Therefore, appropriate interventions should be performed in a timely manner to prevent or delay the development of end-stage liver disease associated with liver fibrosis (4). Currently, liver biopsy is the gold standard for the diagnosis of NAFLD and assessment of the severity of liver fibrosis. However, this procedure is invasive, expensive, and potentially associated with some acute complications. There is, therefore, an unmet need for developing non-invasive and cost-effective tests for diagnosing and staging NAFLD, especially for distinguishing between patients with SLF and those with NSLF (13–19).
DNA methylation adds a methyl group to cytosine in the CpG dinucleotide and is one of the major epigenetic forms of DNA modification. DNA methylation often causes transcriptional silencing of gene expression and alterations in chromosomal instability, cell cycle regulation, viability, differentiation, apoptosis, and signaling pathways (20, 21). Methylated circulating and tissue DNA have emerged as biomarkers or therapeutic targets in many chronic diseases, such as prostate cancer (22), bladder cancer (23), pulmonary fibrosis (24), Alzheimer’s disease (25), and NAFLD (26, 27). A previous study revealed that hypomethylation of DNA in early-stage liver fibrosis is critical for the development and progression of liver fibrosis (28). Hepatic stellate cells are activated by methylation changes (resulting in excessive accumulation of extracellular matrix), which are the hallmark of liver fibrosis (29–32). Plasma DNA methylation of peroxisome proliferator-activated receptor (PPAR-γ) can potentially be used to non-invasively stratify liver fibrosis severity in NAFLD (33). Some key methylated CpG sites from peripheral blood leukocytes might also be used as serum biomarkers to differentiate patients with simple steatosis from those with non-alcoholic steatohepatitis (NASH) (34). A recent study by Lai et al. revealed that patients with NAFLD had decreased hepatic levels of global DNA methylation, a parameter that decreases in parallel with increasing hepatic inflammation grade and disease progression (35). However, to date, blood DNA methylation biomarker genes for the diagnosis of liver fibrosis in NAFLD remain to be identified.
In this pilot study, we investigated the association between blood DNA methylation and advanced or mild liver fibrosis, and identified potential blood DNA methylation biomarker genes for the diagnosis of liver fibrosis in patients with biopsy-confirmed NAFLD. Our findings may provide clues for the development of non-invasive tests to diagnose NAFLD-related liver fibrosis.
Materials and Methods
Patients and Samples
Patients with biopsy-proven NAFLD were recruited as described in our previous study (36). These patients were recruited at the First Affiliated Hospital of Wenzhou Medical University from December 2016 to January 2018. Specifically, for this study, we randomly selected 16 patients with high liver fibrosis and 16 patients with low liver fibrosis by liver histological staging. The presence of fibrosis stage ≥ 2 was defined as presence of significant liver fibrosis (SLF), whereas fibrosis stages 0–1 were defined as non-significant (NSLF). The two groups of NAFLD patients were well matched for age and sex. For each patient, a complete medical history was obtained, the blood sample was collected and immediately stored at −80°C until use, and a liver biopsy was performed. Patients who had a prior history of significant alcohol consumption, viral hepatitis, autoimmune hepatitis, and other known causes of chronic liver diseases, or those with incomplete data were excluded from the study (36, 37).
The diagnosis of NAFLD was based on histological abnormalities in the liver biopsy examined with hematoxylin-eosin and Masson’s trichrome staining. All stained biopsy samples were analyzed by two experienced liver pathologists, who were blinded to the clinical and laboratory data of participants. All histologic features were scored according to the NASH Clinical Research Network classification system (38), and the fibrosis stage (1–4) was estimated for each NAFLD specimen (39). The fibrosis stage was scored as follows: no fibrosis, stage 0, perisinusoidal or portal fibrosis, stage 1; perisinusoidal and portal/periportal fibrosis, stage 2; bridging fibrosis, stage 3; highly suspicious or definite cirrhosis, stage 4 (39). The presence of fibrosis stage ≥ 2 was defined as SLF, whereas fibrosis stages 0–1 were defined as NSLF (36, 37).
This study was registered in the Chinese Clinical Trial Registry (ChiCTR-EOC-17013562). The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University (2016-246, 1 December 2016). Written informed consent was obtained from each participant.
DNA Extraction and Illumina 850K (EPIC) DNA Methylation Array
Genomic DNA from fresh-frozen blood samples was isolated using the Magnetic Universal Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. The Infinium Methylation EPIC BeadChips (Illumina, San Diego, CA, United States) was used to detect bisulfite-converted genomic DNA by hybridization. After scanning (iScan, Illumina), the raw intensity data were preprocessed, normalized, and analyzed with the ChAMP package (2.14.0) (40). Cross-reactive probes, probes with single nucleotide polymorphisms (SNPs) (41), and probes on X or Y chromosomes, as well as all probes with incomplete and non-significant detection P-values (P > 0.01) were removed. The resulting dataset of autosomal probes was used for subsequent analyses.
Differential Methylation Analysis
Beta (β) values were calculated to denote the methylation levels at CpG sites. The β value 0.0 represents no methylation and 1.0 represent 100% methylation (42). Mean β values were calculated for moderate to severe liver fibrosis (i.e., the SLF group) and mild or no liver fibrosis (i.e., the NSLF group). The Δβ value was the result of the mean β value of cases minus that of controls. A positive Δβ value indicated relative hypermethylation, and a negative value denoted hypomethylation in blood DNA. The limma (3.40.2) package was used to analyze differential methylation (43). Differentially methylated probes (DMPs) were generated and identified after comparing the mean β values between the NSLF and SLF groups for a particular CpG site. The cut-off threshold for significant DMPs was a Benjamini-Hochberg-adjusted p-value for multiple comparisons <0.05 and |Δβ| > 0.1.
t-Distributed Stochastic Neighbor Embedding and Clustering Analysis
To examine whether the blood DNA methylation status was associated with the severity of liver fibrosis, the t-SNE analysis and unsupervised hierarchical clustering analysis of the overall blood DNA methylation status of NAFLD patients with increasing liver fibrosis were performed using the pheatmap package in R V4.1.0.
Distribution Analysis of Differential Methylated Probes
Differentially methylated probes were classified into different groups according to their distributions of CpG island regions and gene regions. The expected counts were calculated with the probes that remained after filtering. A multinomial goodness-of-fit Chi squared test was performed to analyze the significance of differences between groups. The blood DNA methylation levels and the distributions of DMPs in CpG island regions and gene regions in the SLF and NSLF groups were compared.
Enrichment of Differentially Methylated Probe Genes in Gene Ontology Terms and Kyoto Encyclopedia of Genes and Genomes Pathways
The genes that contained DMPs were first mapped and identified. The enrichment of these DMP genes in GO terms and KEGG pathways was analyzed using the DAVID bioinformatics tool (44).
Identification of Candidate Biomarkers From Blood Differentially Methylated Genes for Liver Fibrosis
To identify potential biomarkers from differentially methylated genes for liver fibrosis in the blood, DMPs found at the CpG sites were filtered via the following sequential steps: (1) selection of DMPs with a mean blood DNA methylation level greater than 0.7 that represents extremely high methylation, or less than 0.3 that represents extremely low methylation, because extremely high or low DNA methylation levels are more likely to be associated with abnormal gene expression; (2) selection of DMPs in non-intergenic region (IGR); (3) island and shelf regions in CpG islands; and (4) after applying the above-mentioned filters, the resultant DMPs were mapped to genes. Genes with at least two DMPs were considered candidate biomarkers for liver fibrosis from blood differentially methylated genes. Their methylation levels in the SLF and NSLF groups were subsequently compared.
Statistical Analysis
Statistical analyses of all data were performed with R (Version 4.1.0). Probes showing significant differences in DNA methylation levels between the NSLF and SLF groups of NAFLD patients were identified using the limma package. A Benjamini-Hochberg-adjusted p-value < 0.05 and |Δβ| > 0.1 for multiple comparisons was considered to be statistically significant.
Results
Clinical Information for Patients With Liver Fibrosis
A total of 32 selected patients with biopsy-proven NAFLD and varying levels of liver fibrosis were divided into two groups: the SLF group with high liver fibrosis (n = 16) and the NSLF group with low liver fibrosis (n = 16). As shown in Table 1, the two groups of patients were well matched for age and sex. No significant differences were found in main biochemical parameters, except for higher HbA1c levels in the SLF group, as well as in other histology features of NASH (steatosis, lobular inflammation, or ballooning).
TABLE 1
| Levels of liver fibrosis | High (n = 16) | Low (n = 16) | P-value | |
| Age (years) | 32.31 ± 8.20 | 33.75 ± 6.82 | 0.59 | |
| Sex (n, %) | Men | 13 (81.2) | 14 (87.5) | 1.00 |
| Women | 3 (18.8) | 2 (12.5) | ||
| Body mass index (kg/m2) | 28.72 ± 3.9 | 27.31 ± 3.1 | 0.27 | |
| Waist circumference (cm) | 95.50 ± 9.6 | 92.56 ± 6.9 | 0.33 | |
| Fibrosis stage (n, %) | 0 | 0 (0.0) | 3 (18.8) | <0.001* |
| 1 | 0 (0.0) | 13 (81.2) | ||
| 2 | 12 (75.0) | 0 (0.0) | ||
| 3 | 3 (18.8) | 0 (0.0) | ||
| 4 | 1 (6.2) | 0 (0.0) | ||
| NAS steatosis (n, %) | 0 | 1 (6.2) | 1 (6.2) | 0.20 |
| 1 | 6 (37.5) | 4 (25.0) | ||
| 2 | 1 (6.2) | 6 (37.5) | ||
| 3 | 8 (50.0) | 5 (31.2) | ||
| NAS hepatocellular ballooning (n, %) | 0 | 1 (6.2) | 2 (12.5) | 0.72 |
| 1 | 9 (56.2) | 7 (43.8) | ||
| 2 | 6 (37.5) | 7 (43.8) | ||
| NAS lobular inflammation (n, %) | 1 | 11 (68.8) | 12 (75.0) | 1.00 |
| 2 | 5 (31.2) | 4 (25.0) | ||
| Glucose (mmol/L) | 4.84 ± 0.63 | 5.36 ± 0.97 | 0.08 | |
| HbA1c (%) | 6.20 ± 1.36 | 5.41 ± 0.27 | 0.03* | |
| Insulin (pmol/L) | 122.25 ± 63.09 | 290.18 ± 492.64 | 0.19 | |
| C-Peptide (pmol/L) | 1055.47 ± 370.62 | 1498.88 ± 1339.00 | 0.21 | |
| Total cholesterol (mmol/L) | 5.13 ± 0.90 | 5.03 ± 1.31 | 0.80 | |
| Triglycerides (mmol/L) | 2.06 ± 1.00 | 2.51 ± 1.51 | 0.33 | |
| HDL-cholesterol (mmol/L) | 1.02 ± 0.10 | 0.99 ± 0.17 | 0.45 | |
| LDL-cholesterol (mmol/L) | 3.21 ± 0.87 | 3.06 ± 0.97 | 0.64 | |
| Total bilirubin (μmol/L) | 19.12 ± 8.11 | 16.31 ± 6.45 | 0.29 | |
| Albumin (g/L) | 47.67 ± 3.12 | 48.58 ± 3.58 | 0.45 | |
| Alanine aminotransferase (U/L) | 107.75 ± 86.28 | 160.44 ± 156.93 | 0.25 | |
| Aspartate aminotransferase (U/L) | 83.56 ± 73.71 | 75.56 ± 54.80 | 0.73 | |
| Alkaline phosphatase (U/L) | 81.69 ± 16.52 | 89.75 ± 20.20 | 0.23 | |
| Gamma-glutamyltransferase (U/L) | 77.94 ± 59.23 | 79.62 ± 39.62 | 0.93 | |
| Serum creatinine (μmol/L) | 65.44 ± 12.72 | 67.50 ± 12.85 | 0.65 | |
| eGFR (ml/min/1.73 m2) | 127.45 ± 16.14 | 125.84 ± 14.48 | 0.77 | |
| Uric acid (μmol/L) | 464.19 ± 109.70 | 448.62 ± 120.35 | 0.71 |
Main clinical and biochemical characteristics of patients with biopsy-proven NAFLD stratified by high or low levels of liver fibrosis.
The Blood DNA Methylation Status in the Significant Liver Fibrosis and Non-significant Liver Fibrosis Groups
To examine whether the blood DNA methylation status was associated with greater liver fibrosis, we performed the t-SNE analysis and unsupervised hierarchical clustering analysis of the overall blood DNA methylation status amongst the two groups of NAFLD patients. Our results show that the SLF and NSLF groups were significantly different through both the t-SNE analysis (Figure 1A) and the unsupervised hierarchical clustering analysis (Figure 1B) based on the blood overall DNA methylation status.
FIGURE 1

The blood DNA methylation status in NAFLD patients with different levels of liver fibrosis. The SLF and NSLF groups of NAFLD patients were stratified with the t-SNE analysis (A) and the unsupervised hierarchical clustering analysis (B) based on the overall blood DNA methylation status. (C) The methylation levels (i.e., the beta value) of blood DNA in the SLF and NSLF groups. (D,E) Comparison of the methylation levels of different CpG island regions (D) and different gene regions (E) of blood DNA in the SLF and NSLF groups. SLF, significant liver fibrosis; NSLF, non-significant liver fibrosis.
The blood DNA methylation levels (i.e., the beta value) (Figure 1C) and the methylation levels in the island and shelf regions (Figure 1D) were significantly higher in the NSLF group than in the SLF group. The methylation levels in the first exon, 3′-untranslated region (UTR), body, ExonBnd, IGR, transcription start site (TSS)1500, and TSS200 regions were also significantly higher in the NSLF group Figure 1E). Conversely, there were no significant differences in the open sea and shore regions (Figure 1D) or the 5′-UTR region (Figure 1E) between the two groups of patients.
Blood Differentially Methylated Probes in Patients With Diagnosed Liver Fibrosis
We identified a total of 233 DMPs in blood samples from our NAFLD patients with varying levels of liver fibrosis (Figure 2A). Both hypomethylated and hypermethylated DMPs were significantly enriched in the CpG island (Figure 2B) and gene regions (Figure 2C). Hypomethylated DMPs were over-represented in the island and shelf regions (Figure 2B), as well as in the first exon, 3′-UTR, 5′-UTR, and TSS200 regions (Figure 2C). Conversely, hypermethylated DMPs were markedly under-represented in the first exon and 3′-UTR regions (Figure 2C), and hypermethylated DMPs were over-represented in the body, IGR, and TSS1500 regions (Figure 2C).
FIGURE 2
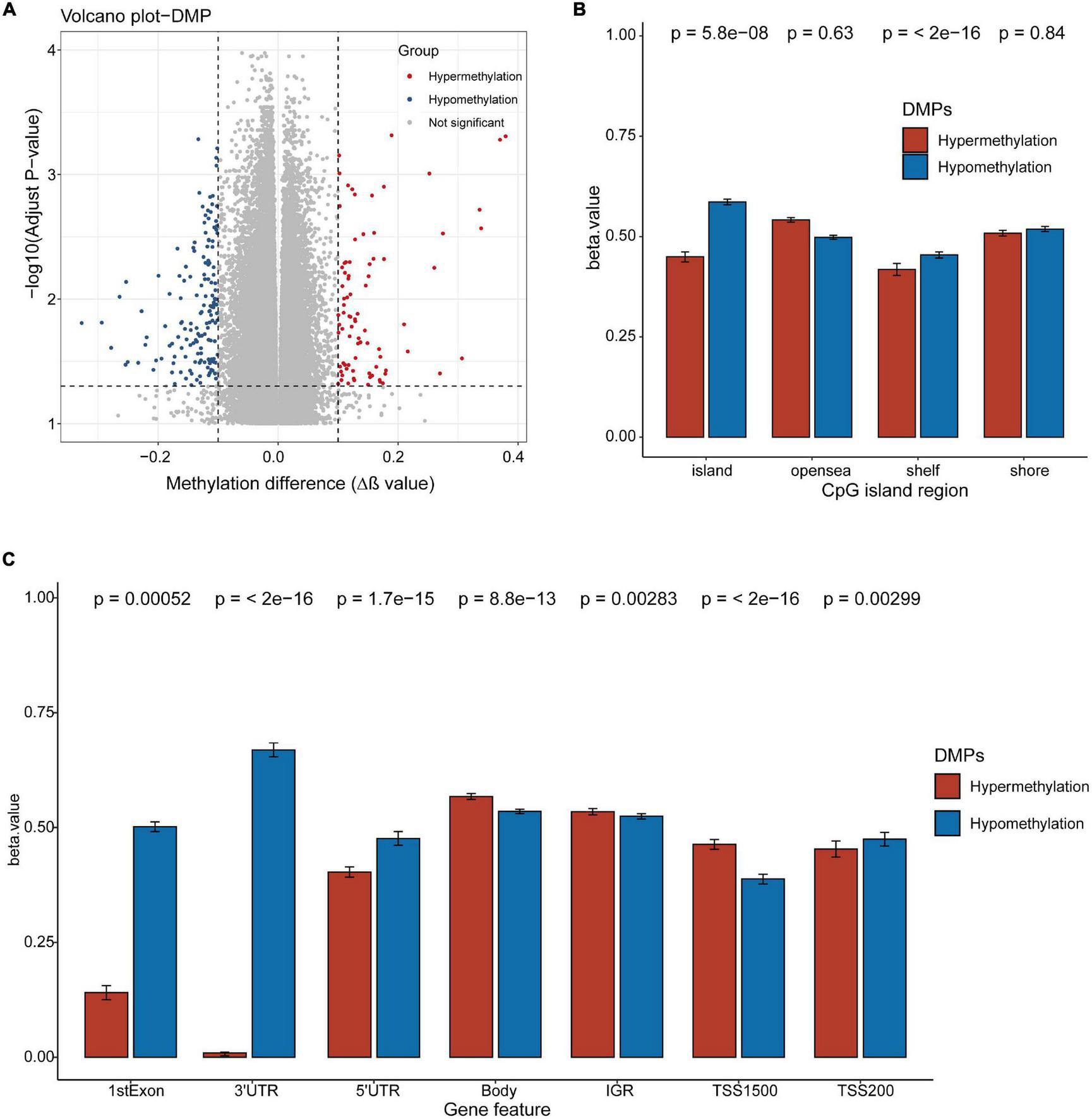
Differentially methylated probes detected in the blood DNA of NAFLD patients with diagnosed liver fibrosis. (A) DNA methylation sites (probes) in the blood DNA of NAFLD patients with diagnosed liver fibrosis. Dots, DNA methylation probes. The cutoff values for differentially methylated probes (DMPs): adjusted p < 0.05 and |Δβ| > 0.1. Red dots, hypermethylated DMPs; blue dots, hypomethylated DMPs. (B,C) The DMPs in CpG island regions (B) and gene regions (C). hypermethylated DMPs (red), and hypomethylated DMPs (blue). P-values were calculated with the Chi-squared tests.
Enrichment of Differentially Methylated Probe Genes in Gene Ontology Terms and Kyoto Encyclopedia of Genes and Genomes Pathways
We mapped the genes that hosted DMPs and analyzed the enrichment of DMP genes in GO terms and KEGG pathways. Our results show that hemophilic and cell-cell adhesion, as well as muscle cell differentiation and development, were the top enriched GO biological process terms (Figure 3A). Additionally, superoxide-generating nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activator activity, 5S rRNA binding, phospholipid binding, and phosphatidylinositol-related binding activity were the top enriched GO molecular function terms (Figure 3B). Under GO cellular components, inhibitory synapse, basement membrane, A band, actin filament, cilia-related structures, extracellular matrix (ECM), and neuromuscular junction were the top enriched terms (Figure 3C). Finally, vascular smooth muscle contraction, inflammatory mediator regulation of transient receptor potential (TRP) channels, signaling pathways such as Rap1, Apelin, Hippo, and vascular endothelial growth factor (VEGF), and amino and nucleotide sugar metabolism were the top enriched KEGG pathways (Figure 3D).
FIGURE 3
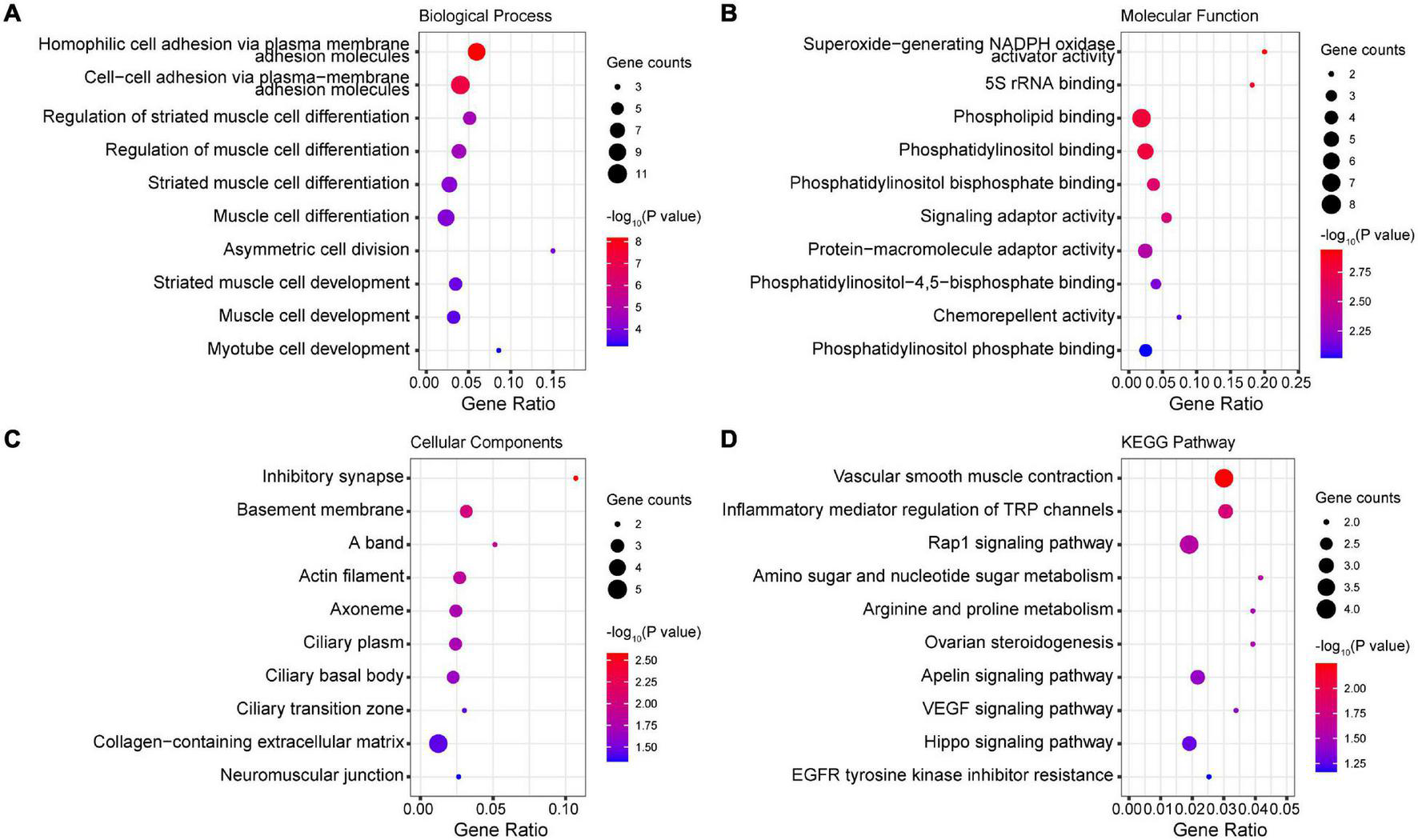
Enrichment of DMP genes in GO terms and KEGG pathways. (A) Top enriched GO biological process terms. (B) Top enriched GO molecular function terms. (C) Top enriched GO cellular components terms. (D) Top enriched KEGG pathways.
Association of Blood DNA Methylation With Liver Fibrosis
We examined the association of blood DNA methylation with liver fibrosis and other clinical features using the unsupervised hierarchical clustering analysis. Our results show that the blood DNA methylation status was significantly associated with greater severity of liver fibrosis, but not with age, sex, and other histologic features of NASH (lobular inflammation, hepatocellular ballooning, or steatosis) (Figure 4A).
FIGURE 4
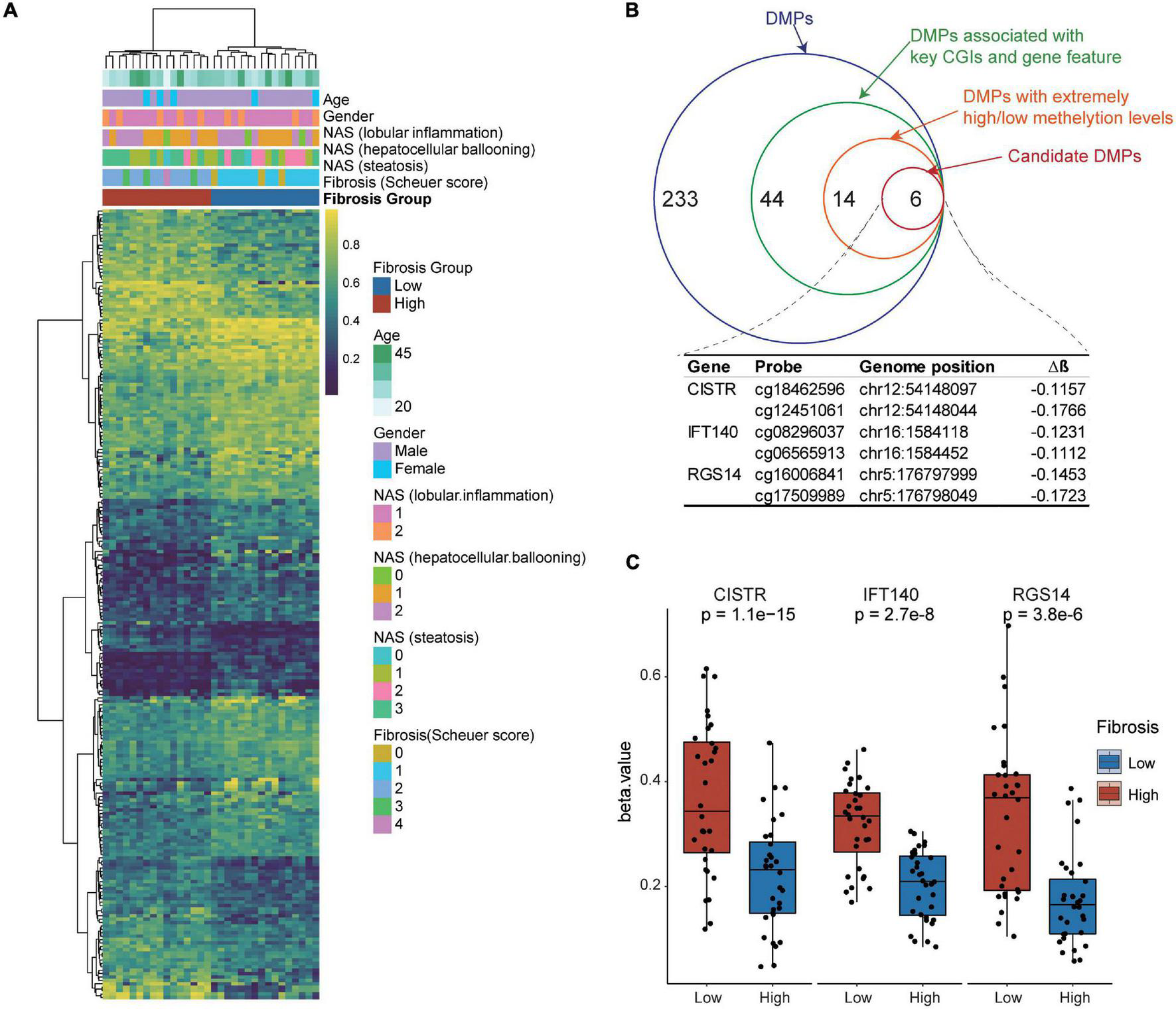
Identification of candidate biomarkers from blood differentially methylated genes for liver fibrosis. (A) Association of DNA methylation and clinical features with liver fibrosis. The unsupervised hierarchical cluster analysis of DMPs was performed. DMPs in CpG islands are presented in the rows and independent samples are presented in the columns. The colors in the cells represent methylation level. Blue, unmethylated; red, fully methylated. (B) Step-wise selection of candidate DMPs for liver fibrosis. Three genes, i.e., CISTR, IFT140, and RGS14, each harboring at least two DMPs were finally selected. (C) The methylation levels in the CpG island regions of blood CISTR, IFT140, and RGS14 genes in the NSLF group were significantly higher than those in the SLF group.
Identification of Candidate Biomarkers From Blood Differentially Methylated Genes for Liver Fibrosis
To identify candidate biomarkers from blood differentially methylated genes for liver fibrosis, we performed a step-wise data-filtering in terms of mean methylation, gene features, CpG islands, and number of DMPs in the genes. We identified a total of six hypomethylated DMPs mapped to the chondrogenesis-associated transcript (CISTR), intraflagellar transport complex A (IFT140), and regulator of G-protein signaling 14 (RGS14) genes (Figure 4B). The methylation levels in the CpG island regions of the CISTR, IFT140, and RGS14 genes in the NSLF group were significantly higher than those in the SLF group (Figure 4C).
Discussion
In this pilot study involving Chinese individuals with biopsy-proven NAFLD, we showed for the first time that histologically classified SLF and NSLF patient groups can be accurately distinguished via the t-SNE analysis and unsupervised hierarchical clustering analysis, based on their blood overall DNA methylation status. The blood DNA methylation levels and the methylation levels in the island and shelf regions were significantly greater in the NSLF group than in the SLF group, but they did not differ between men and women, and were not associated with age, NAS lobular inflammation, NAS hepatocellular ballooning, or NAS steatosis. Consistent with previous results presented by Murphy et al. human genes in the livers with advanced fibrosis were generally hypomethylated relative to those in the livers with mild fibrosis (45). It is known that DNA methylation is a critical process for modulating the activity of hepatic stellate cells, which are essential for fibrogenesis (29–31, 46); so consistent with previous studies, our findings suggest that blood DNA methylation can differentiate NAFLD patients with SLF and NSLF (34, 35). Additionally, we have confirmed and extended previous findings showing that the DNA methylation levels in the island and shelf regions were higher in the NSLF group. We have also shown that the methylation levels in the first exon, 3′-UTR, body, ExonBnd, IGR, TSS1500, and TSS200 regions were significantly higher in the NSLF group. These data suggest that more severe liver fibrosis is characterized by lower levels of blood DNA methylation, which is in agreement with the results from the studies by Komatsu et al. (28) and Lai et al. (35). Thus, it is likely that blood DNA methylation is a potential biomarker for NAFLD-related liver fibrosis and for stratification of SLF and NSLF.
Non-alcoholic fatty liver disease development is influenced by genetic susceptibility and heritability explains inter-individual and ethnic differences of NAFLD development and progression to fibrosis (47). Romeo et al. documented an interplay between the patatin-like phospholipase domain containing 3 (PNPLA3) variant I148M and advanced fibrosis in NASH patients (48). Another gene found by genome-wide association studies to be a risk factor for NAFLD, is the trans-membrane 6 superfamily member 2 (TM6SF2), non-synonymous variant rs58542926 (49). Dongiovanni et al. showed that carriers of this genetic variant had lower plasma lipids and more severe steatosis and fibrosis than non-carriers (50). Other genetic variants potentially implicated in NAFLD development and progression were neurocan (NCAN-rs2228603), protein phosphatase 1, regulatory (inhibitor) subunit 3B (PPP1R3B-rs4240624), glucokinase regulator (GCKR-rs780094), and lysophospholipase-like 1 (LYPLAL1-rs12137855) (51).
Previous studies have also shown that plasma DNA methylation of PPAR-γ can potentially be used for non-invasive detection of liver fibrosis severity in NAFLD (33). Additionally, peripheral blood leukocyte ACSL4, CRLS1, CTP1A, SIGIRR, SSBP1, and ZNF622 genes containing DMPs might be used as serum biomarkers to stratify NAFLD patients into groups with simple hepatic steatosis and NASH (34). In the current study, we found that the CISTR, IFT140, and RGS14 genes were hypomethylated in the island and shelf regions of the CpG island. The methylation levels of blood CISTR, IFT140, and RGS14 genes were also significantly higher in the NSLF group than in the SLF group. The hypomethylation status of these genes in NAFLD patients with liver fibrosis was consistent with the findings by Komatsu et al. (28) and Lai et al. (35). Therefore, the CISTR, IFT140, and RGS14 genes may be potential novel candidate blood methylation biomarkers for the diagnosis of liver fibrosis in NAFLD.
The RGS14 gene encodes a multi-functional nucleocytoplasmic shuttling signaling protein involved in modulating microtubule dynamics and spindle formation (46). RGS14 acts as a scaffolding protein that integrates the G protein, Ras/extracellular signal-regulated kinase (ERK), and calcium/calmodulin signaling pathways essential for spine plasticity and cell signaling (52); RGS14 may also reduce hepatic ischemia–reperfusion injury mainly through its interaction with TAK1 and the JNK/p38 signaling axis (53). IFT140 is a key component of IFT complex A, which is responsible for retrograde transportation in cilia. IFT140 is essential in promoting dentin formation and repair, (54) as well as chondrogenic and osteogenic differentiation during bone development (55). The distinctive monoallelic phenotype causes liver cysts (56). The CISTR gene is evolutionarily conserved and involved in mesenchymal and prechondrogenic development (57). The roles of the CISTR, IFT140, and RGS14 genes in the onset of liver fibrosis in NAFLD patients and their roles in the blood and the relationship with liver fibrosis remain unclear. Therefore, these genes require further validation as potential candidate blood methylation biomarkers for the diagnosis of NAFLD-related liver fibrosis through in-depth progression, limiting or promoting its progression (58). The challenge now is to better understand epigenetic mechanisms and their interactions; these experimental studies could provide further insights into the pathogenesis of chronic liver disease (59). Our analysis has shown that genes containing DMPs in the blood DNA of patients with NAFLD are enriched in hemophilic and cell-cell adhesion, muscle cell differentiation and development under GO biological process terms. Additionally, superoxide-generating NADPH oxidase activator activity, 5S rRNA binding, phospholipid binding, and phosphatidylinositol-related binding activity were enriched under GO molecular function terms. These genes were also enriched in inhibitory synapse, basement membrane, A band, actin filament, ciliary related structures, ECM, and neuromuscular junction under GO cellular components terms. Vascular smooth muscle contraction, inflammatory mediator regulation of TRP channels, signaling pathways such as Rap1, Apelin, Hippo, and VEGF, and amino and nucleotide sugar metabolism were also affected according to KEGG pathway analysis. Collectively, these findings suggest that the blood DNA methylation biomarker genes that are modified with NAFLD-related liver fibrosis are multi-functional genes.
Our pilot study has some important limitations. Firstly, the sample size of the study was small and these findings need to be further validated by larger studies in other ethnic groups. In addition, some in vitro cell and in vivo animal experiments are now ongoing to further study whether the corresponding gene methylation plays a role in the development and progression of liver fibrosis. Secondly, our study design was cross-sectional and cannot prove causality. Since the DNA methylation profiles of hemostatic genes may not necessarily correlate between liver tissue and peripheral blood in individuals (60), the genes hosting DMPs in the blood DNA of these patients need further investigation.
Conclusion
The results of our pilot study show that blood DNA methylation markers may allow identification of NAFLD patients with SLF and NSLF. The CISTR, IFT140, and RGS14 genes are likely novel candidate blood methylation biomarkers for the diagnosis of liver fibrosis in NAFLD. Genes such as CISTR, IFT140, and RGS14 that contain DMPs in the blood DNA of NAFLD patients may provide clues to investigate the role of DNA methylation in the progression of liver fibrosis. Our findings provide new insights into the development of better non-invasive tests for diagnosing liver fibrosis in NAFLD. Further studies are needed to validate these results in independent cohorts in different ethnic groups.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The data presented in the study are deposited at https://doi.org/10.5281/zenodo.6240813.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee in Clinical Research (ECCR) of The First Affiliated Hospital of Wenzhou Medical University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Q-FS, W-YL, YL, and M-HZ: study concept and design. P-WZ, H-LM, O-YH, LH, and GL: acquisition of data. Y-YL: pathology analysis. L-JT: drafting of the manuscript. GT and CB: critical revision. M-JW: statistical analysis. M-HZ and J-GD: study supervision. All authors contributed to the manuscript for important intellectual content and approved the submission.
Funding
This work was funded by grants from the National Natural Science Foundation of China (82070588), High Level Creative Talents from Department of Public Health in Zhejiang Province (S2032102600032), and Project of New Century 551 Talent Nurturing in Wenzhou. GT was supported in part by grants from the School of Medicine, University of Verona, Verona, Italy. CB was supported in part by the NIHR Southampton Biomedical Research Centre (IS-BRC-20004), United Kingdom.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- NAFLD
non-alcoholic fatty liver disease
- DMPs
differentially methylated probes
- SLF
significant liver fibrosis
- NSLF
non-significant liver fibrosis
- t-SNE
t-distributed stochastic neighbor embedding
- UTR
untranslated region
- IGR
intergenic region
- TSS
transcription start site
- NAS
NAFLD activity score
- PPAR-γ
peroxisome proliferator-activated receptor
- NASH
non-alcoholic steatohepatitis
- SNPs
single nucleotide polymorphisms
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NADPH
nicotinamide adenine dinucleotide phosphate
- ECM
extracellular matrix
- TRP
transient receptor potential
- VEGF
vascular endothelial growth factor
- ERK
extracellular signal-regulated kinase.
References
1.
Zheng KI Fan JG Shi JP Wong VW Eslam M George J et al From NAFLD to MAFLD: a “redefining” moment for fatty liver disease. Chin Med J. (2020) 133:2271–3. 10.1097/CM9.0000000000000981
2.
Byrne CD Targher G. NAFLD: a multisystem disease. J Hepatol. (2015) 62:S47–64.
3.
Mantovani A Csermely A Petracca G Beatrice G Corey KE Simon TG et al Non-alcoholic fatty liver disease and risk of fatal and non-fatal cardiovascular events: an updated systematic review and meta-analysis. Lancet Gastroenterol Hepatol. (2021) 6:903–13. 10.1016/S2468-1253(21)00308-3
4.
Chalasani N Younossi Z Lavine JE Charlton M Cusi K Rinella M et al The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American association for the study of liver diseases. Hepatology. (2018) 67:328–57.
5.
Eslam M Newsome PN Sarin SK Anstee QM Targher G Romero-Gomez M et al A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J Hepatol. (2020) 73:202–9.
6.
Xian YX Weng JP Xu F. MAFLD vs. NAFLD: shared features and potential changes in epidemiology, pathophysiology, diagnosis, and pharmacotherapy. Chin Med J. (2020) 134:8–19. 10.1097/CM9.0000000000001263
7.
Nan Y An J Bao J Chen H Chen Y Ding H et al The Chinese society of hepatology position statement on the redefinition of fatty liver disease. J Hepatol. (2021) 75:454–61.
8.
Hagström H Nasr P Ekstedt M Hammar U Stål P Hultcrantz R et al Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. (2017) 67:1265–73. 10.1016/j.jhep.2017.07.027
9.
Ekstedt M Hagström H Nasr P Fredrikson M Stål P Kechagias S et al Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. (2015) 61:1547–54. 10.1002/hep.27368
10.
Vilar-Gomez E Calzadilla-Bertot L Wai-Sun Wong V Castellanos M Aller-de la Fuente R Metwally M et al Fibrosis severity as a determinant of cause-specific mortality in patients with advanced nonalcoholic fatty liver disease: a multi-national cohort study. Gastroenterology. (2018) 155:443–57.e17.
11.
Angulo P Kleiner DE Dam-Larsen S Adams LA Bjornsson ES Charatcharoenwitthaya P et al Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. (2015) 149:389–97.e10. 10.1053/j.gastro.2015.04.043
12.
Dulai PS Singh S Patel J Soni M Prokop LJ Younossi Z et al Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. (2017) 65:1557–65.
13.
Angulo P Hui JM Marchesini G Bugianesi E George J Farrell GC et al The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. (2007) 45:846–54. 10.1002/hep.21496
14.
Harrison SA Oliver D Arnold HL Gogia S Neuschwander-Tetri BA. Development and validation of a simple NAFLD clinical scoring system for identifying patients without advanced disease. Gut. (2008) 57:1441–7. 10.1136/gut.2007.146019
15.
Heyens LJM Busschots D Koek GH Robaeys G Francque S. Liver fibrosis in non-alcoholic fatty liver disease: from liver biopsy to non-invasive biomarkers in diagnosis and treatment. Front Med. (2021) 8:615978. 10.3389/fmed.2021.615978
16.
Önnerhag K Hartman H Nilsson PM Lindgren S. Non-invasive fibrosis scoring systems can predict future metabolic complications and overall mortality in non-alcoholic fatty liver disease (NAFLD). Scand J Gastroenterol. (2019) 54:328–34. 10.1080/00365521.2019.1583366
17.
Rios RS Zheng KI Targher G Byrne CD Zheng MH. Non-invasive fibrosis assessment in non-alcoholic fatty liver disease. Chin Med J. (2020) 133:2743–5. 10.1097/cm9.0000000000000989
18.
Zhou YJ Zheng KI Targher G Byrne CD Zheng MH. Non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis. Lancet Gastroenterol Hepatol. (2021) 6:9–10.
19.
Zhou YJ Wong VW Zheng MH. Consensus scoring systems for nonalcoholic fatty liver disease: an unmet clinical need. Hepatobiliary Surg Nutr. (2021) 10:388–90. 10.21037/hbsn-21-80
20.
Dhar GA Saha S Mitra P Nag Chaudhuri R. DNA methylation and regulation of gene expression: guardian of our health. Nucleus. (2021) 64:259–70. 10.1007/s13237-021-00367-y
21.
Sriraman A Debnath TK Xhemalce B Miller KM. Making it or breaking it: DNA methylation and genome integrity. Essays Biochem. (2020) 64:687–703. 10.1042/EBC20200009
22.
Wu L Yang Y Guo X Shu XO Cai Q Shu X et al An integrative multi-omics analysis to identify candidate DNA methylation biomarkers related to prostate cancer risk. Nat Commun. (2020) 11:3905. 10.1038/s41467-020-17673-9
23.
Kandimalla R van Tilborg AA Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nat Rev Urol. (2013) 10:327–35. 10.1038/nrurol.2013.89
24.
Zhou S Wang X Gao H Zeng Y. DNA methylation in pulmonary fibrosis. Adv Exp Med Biol. (2020) 1255:51–62. 10.1007/978-981-15-4494-1_4
25.
Wei X Zhang L Zeng Y. DNA methylation in Alzheimer’s disease: in brain and peripheral blood. Mech Ageing Dev. (2020) 191:111319. 10.1016/j.mad.2020.111319
26.
Hyun J Jung Y. DNA methylation in nonalcoholic fatty liver disease. Int J Mol Sci. (2020) 21:8138.
27.
Ma J Nano J Ding J Zheng Y Hennein R Liu C et al DNA methylation signature of hepatic fat reveals a potential causal pathway for nonalcoholic fatty liver disease. Diabetes. (2019) 68:1073–83. 10.2337/DB18-1193
28.
Komatsu Y Waku T Iwasaki N Ono W Yamaguchi C Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med Genomics. (2012) 5:5. 10.1186/1755-8794-5-5
29.
Bian EB Huang C Wang H Wu BM Zhang L Lv XW et al DNA methylation: new therapeutic implications for hepatic fibrosis. Cell Signal. (2013) 25:355–8. 10.1016/j.cellsig.2012.10.007
30.
Yang JJ Tao H Huang C Shi KH Ma TT Bian EB et al DNA methylation and MeCP2 regulation of PTCH1 expression during rats hepatic fibrosis. Cell Signal. (2013) 25:1202–11. 10.1016/j.cellsig.2013.01.005
31.
Bian EB Zhao B Huang C Wang H Meng XM Wu BM et al New advances of DNA methylation in liver fibrosis, with special emphasis on the crosstalk between microRNAs and DNA methylation machinery. Cell Signal. (2013) 25:1837–44. 10.1016/j.cellsig.2013.05.017
32.
Zeybel M Hardy T Robinson SM Fox C Anstee QM Ness T et al Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenet. (2015) 7:25. 10.1186/s13148-015-0056-6
33.
Hardy T Zeybel M Day CP Dipper C Masson S McPherson S et al Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. (2017) 66:1321–8. 10.1136/gutjnl-2016-311526
34.
Wu J Zhang R Shen F Yang R Zhou D Cao H et al Altered DNA methylation sites in peripheral blood leukocytes from patients with simple steatosis and nonalcoholic steatohepatitis (NASH). Med Sci Monit. (2018) 24:6946–67. 10.12659/MSM.909747
35.
Lai Z Chen J Ding C Wong K Chen X Pu L et al Association of hepatic global DNA methylation and serum one-carbon metabolites with histological severity in patients with NAFLD. Obesity (Silver Spring Md). (2020) 28:197–205. 10.1002/oby.22667
36.
Zhou YJ Ye FZ Li YY Pan XY Chen YX Wu XX et al Individualized risk prediction of significant fibrosis in non-alcoholic fatty liver disease using a novel nomogram. United Eur Gastroenterol J. (2019) 7:1124–34. 10.1177/2050640619868352
37.
Sun DQ Zheng KI Xu G Ma HL Zhang HY Pan XY et al PNPLA3 rs738409 is associated with renal glomerular and tubular injury in NAFLD patients with persistently normal ALT levels. Liver Int. (2020) 40:107–19. 10.1111/liv.14251
38.
Kleiner DE Brunt EM Van Natta M Behling C Contos MJ Cummings OW et al Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology (2005) 41:1313–21. 10.1002/hep.20701
39.
Stål P. Liver fibrosis in non-alcoholic fatty liver disease – diagnostic challenge with prognostic significance. World J Gastroenterol. (2015) 21:11077–87. 10.3748/wjg.v21.i39.11077
40.
Morris TJ Butcher LM Feber A Teschendorff AE Chakravarthy AR Wojdacz TK et al ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. (2014) 30:428–30. 10.1093/bioinformatics/btt684
41.
Zhou W Laird PW Shen H. Comprehensive characterization, annotation and innovative use of infinium DNA methylation BeadChip probes. Nucleic Acids Res. (2017) 45:e22. 10.1093/nar/gkw967
42.
Du P Zhang X Huang CC Jafari N Kibbe WA Hou L et al Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics (2010) 11:587. 10.1186/1471-2105-11-587
43.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. (2004) 3:Article3.
44.
Huang da W Sherman BT Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. 10.1038/nprot.2008.211
45.
Murphy SK Yang H Moylan CA Pang H Dellinger A Abdelmalek MF et al Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. (2013) 145:1076–87. 10.1053/j.gastro.2013.07.047
46.
Martin-McCaffrey L Willard FS Pajak A Dagnino L Siderovski DP D’Souza SJ. RGS14 is a microtubule-associated protein. Cell Cycle. (2005) 4:953–60. 10.4161/cc.4.7.1787
47.
Macaluso FS Maida M Petta S. Genetic background in nonalcoholic fatty liver disease: a comprehensive review. World J Gastroenterol. (2015) 21:11088–111. 10.3748/wjg.v21.i39.11088
48.
Romeo S Kozlitina J Xing C Pertsemlidis A Cox D Pennacchio LA et al Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. (2008) 40:1461–5. 10.1038/ng.257
49.
Kozlitina J Smagris E Stender S Nordestgaard BG Zhou HH Tybjærg-Hansen A et al Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. (2014) 46:352–6. 10.1038/ng.2901
50.
Dongiovanni P Petta S Maglio C Fracanzani AL Pipitone R Mozzi E et al Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. (2015) 61:506–14. 10.1002/hep.27490
51.
Speliotes EK Yerges-Armstrong LM Wu J Hernaez R Kim LJ Palmer CD et al Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. (2011) 7:e1001324. 10.1371/journal.pgen.1001324
52.
Harbin NH Bramlett SN Montanez-Miranda C Terzioglu G Hepler JR. RGS14 regulation of post-synaptic signaling and spine plasticity in brain. Int J Mol Sci. (2021) 22:6823. 10.3390/ijms22136823
53.
Zhang JK Ding MJ Liu H Shi JH Wang ZH Wen PH et al Regulator of G-protein signaling 14 protects the liver from ischemia-reperfusion injury by suppressing TGF-β-activated kinase 1 activation. Hepatology. (2022) 75:338–52. 10.1002/hep.32133
54.
Li G Liu M Zhang S Wan H Zhang Q Yue R et al Essential role of IFT140 in promoting dentinogenesis. J Dent Res. (2018) 97:423–31. 10.1177/0022034517741283
55.
Zhang C Zhang S Sun Y. Expression of IFT140 during bone development. J Histochem Cytochem. (2019) 67:723–34. 10.1369/0022155419859357
56.
Senum SR Li YSM Benson KA Joli G Olinger E Lavu S et al Monoallelic IFT140 pathogenic variants are an important cause of the autosomal dominant polycystic kidney-spectrum phenotype. Am J Hum Genet. (2022) 109:136–56. 10.1016/j.ajhg.2021.11.016
57.
Maass PG Rump A Schulz H Stricker S Schulze L Platzer K et al A misplaced lncRNA causes brachydactyly in humans. J Clin Investig. (2012) 122:3990–4002. 10.1172/JCI65508
58.
Zeybel M Mann DA Mann J. Epigenetic modifications as new targets for liver disease therapies. J Hepatol. (2013) 59:1349–53. 10.1016/j.jhep.2013.05.039
59.
Zeybel M Vatansever S Hardy T Sarı AA Cakalağaoğlu F Avcı A et al DNA methylation profiling identifies novel markers of progression in hepatitis B-related chronic liver disease. Clin Epigenet. (2016) 8:48. 10.1186/s13148-016-0218-1
60.
Olsson Lindvall M Angerfors A Andersson B Nilsson S Davila Lopez M Hansson L et al Comparison of DNA methylation profiles of hemostatic genes between liver tissue and peripheral blood within individuals. Thromb Haemost. (2021) 121:573–83. 10.1055/s-0040-1720980
Summary
Keywords
NAFLD, liver fibrosis, blood DNA methylation, biomarker, MAFLD
Citation
Sun Q-F, Tang L-J, Wang M-J, Zhu P-W, Li Y-Y, Ma H-L, Huang O-Y, Hong L, Li G, Byrne CD, Targher G, Liu W-Y, Lu Y, Ding J-G and Zheng M-H (2022) Potential Blood DNA Methylation Biomarker Genes for Diagnosis of Liver Fibrosis in Patients With Biopsy-Proven Non-alcoholic Fatty Liver Disease. Front. Med. 9:864570. doi: 10.3389/fmed.2022.864570
Received
28 January 2022
Accepted
22 February 2022
Published
31 March 2022
Volume
9 - 2022
Edited by
Yueyong Zhu, First Affiliated Hospital of Fujian Medical University, China
Reviewed by
Enqiang Chen, Sichuan University, China; Yu-Chen Fan, Shandong University, China
Updates
Copyright
© 2022 Sun, Tang, Wang, Zhu, Li, Ma, Huang, Hong, Li, Byrne, Targher, Liu, Lu, Ding and Zheng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming-Hua Zheng, zhengmh@wmu.edu.cnJi-Guang Ding, djg5011@163.com
†These authors have contributed equally to this work
This article was submitted to Gastroenterology, a section of the journal Frontiers in Medicine
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.