- Medical Equipment Department, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, Shandong, China
Gastrointestinal tumors pose a significant clinical challenge due to their high heterogeneity and the difficulties in early diagnosis. The article systematically reviews the latest advances in multi-omics technologies in gastrointestinal tumor research, focusing on their contributions to early screening, biomarker discovery, and treatment optimization. Genomics reveals genetic characteristics and heterogeneity of tumors; transcriptomics helps identify molecular subtypes and potential therapeutic targets; proteomics provides important information on core proteins and the immune microenvironment; and metabolomics offers promising biomarkers for early diagnosis. Furthermore, emerging fields such as epigenomics, metagenomics, and lipidomics, through the construction of multi-scale frameworks, have opened new paths for molecular subtyping and targeted therapy. By integrating these multi-dimensional data, multi-omics integration enables a panoramic dissection of driver mutations, dynamic signaling pathways, and metabolic-immune interactions. However, challenges such as data heterogeneity, insufficient algorithm generalization, and high costs limit clinical translation. In the future, the integration of single-cell multi-omics, artificial intelligence, and deep learning technologies with multi-omics may offer more efficient strategies for the precise diagnosis and personalized treatment of gastrointestinal tumors.
1 Introduction
Gastrointestinal tumors, including gastric cancer (GC), colorectal cancer (CRC), and esophageal cancer, are among the leading causes of cancer-related mortality worldwide, with over 5 million new cases and nearly 3.5 million deaths annually, accounting for more than 25% of the global cancer burden (1). Despite progress in conventional treatments such as surgery, chemotherapy, and targeted therapies, patient prognosis remains limited by two core challenges: tumor heterogeneity and the lack of reliable early diagnostic biomarkers (2).
Tumor heterogeneity manifests as spatiotemporal dynamics in molecular features. For instance, approximately 50% of GC patients are diagnosed at advanced stages, and the 5-year survival rate for metastatic CRC is below 15% (3). This clinical dilemma stems from the interplay of multilayered biological processes, including genomic instability, epigenetic dysregulation, metabolic reprogramming, and immune microenvironment remodeling (4–6). Traditional single-omics studies (e.g., genomics or proteomics) can reveal changes at specific molecular levels but struggle to elucidate the synergistic mechanisms driving tumor initiation and progression through multi-omics interaction networks (7, 8). For example, KRAS mutations require transcriptomic analysis to uncover their regulatory effects on the MAPK/ERK pathway (9, 10), while metabolomics can further clarify mutation-induced metabolic phenotypes such as the Warburg effect (11, 12).
The integrated application of multi-omics technologies offers systemic solutions to address critical bottlenecks in gastrointestinal tumor research (13, 14). By harmonizing multi-dimensional data from genomics, transcriptomics, proteomics, and metabolomics, integrated multi-omics reveals driver mutations, signaling pathways, and metabolic-immune crosstalk, offering systemic insights (15, 16). For example, in CRC, whole-exome sequencing (WES) revealed that APC gene deletion activates the Wnt/β-catenin pathway (17), while metabolomics further demonstrated that this pathway drives glutamine metabolic reprogramming through the upregulation of glutamine synthetase (18). Such cross-omics integration not only overcomes the limitations of single-marker analyses (e.g., HER2 protein overexpression requiring validation by ERBB2 gene amplification) but also enables dynamic tracking of therapeutic resistance through liquid biopsy multi-omics (e.g., ctDNA mutations combined with exosomal PD-L1 protein) (19). For instance, in metastatic CRC, combined detection of KRAS G12D mutations and exosomal EGFR phosphorylation levels predicts cetuximab resistance 12 weeks in advance (20).
In recent years, the deep integration of artificial intelligence (AI) with multi-omics has revolutionized precision medicine (21, 22). Machine learning algorithms, such as deep residual networks (ResNet-101), analyze heterogeneous multi-omics datasets to identify potential biomarkers and construct prognostic models (23, 24). For instance, the deep residual network (ResNet-101) integrated multi-omics data from CRC to build an mass spectrometry imaging (MSI) status prediction model, achieving an AUC of 0.93 (95% CI: 0.917–0.948) in 10,452 samples, and maintaining an AUC of 0.89 (95% CI: 0.866–0.914) in an independent external validation cohort, significantly outperforming traditional PCR testing (AUC = 0.85) (25). Single-cell spatial multi-omics technologies [e.g., single-cell RNA sequencing (scRNA-seq) combined with desorption electrospray ionization mass spectrometry imaging (DESI-MSI)] dissect cellular heterogeneity and metabolic-immune interaction networks within the tumor microenvironment (26). These approaches have uncovered metabolic-immunoregulatory features of cancer stem cell subpopulations, such as CD133+ cells secreting IL-6 (>35 pg/ml) to polarize M2 macrophages (CD206+ proportion increased from 12% to 54%, p < 0.001) and suppress CD8+ T cell infiltration via spatial lactate gradients (>5 mmol/mm2) (27, 28). These discoveries provide novel pathways for developing CAR-T therapies (e.g., dual-targeting CD133/IL-6R) and metabolic intervention strategies (e.g., LDHA inhibitors) (29, 30).
This essay attempts to methodically review the development of multi-omics technologies in gastrointestinal tumor research, with a focus on their functions in early screening, biomarker discovery, and treatment optimization, as well as the use of methodological references for the clinical translation of multi-omics-based precision diagnostic and therapeutic strategies.
2 Multi-omics technologies
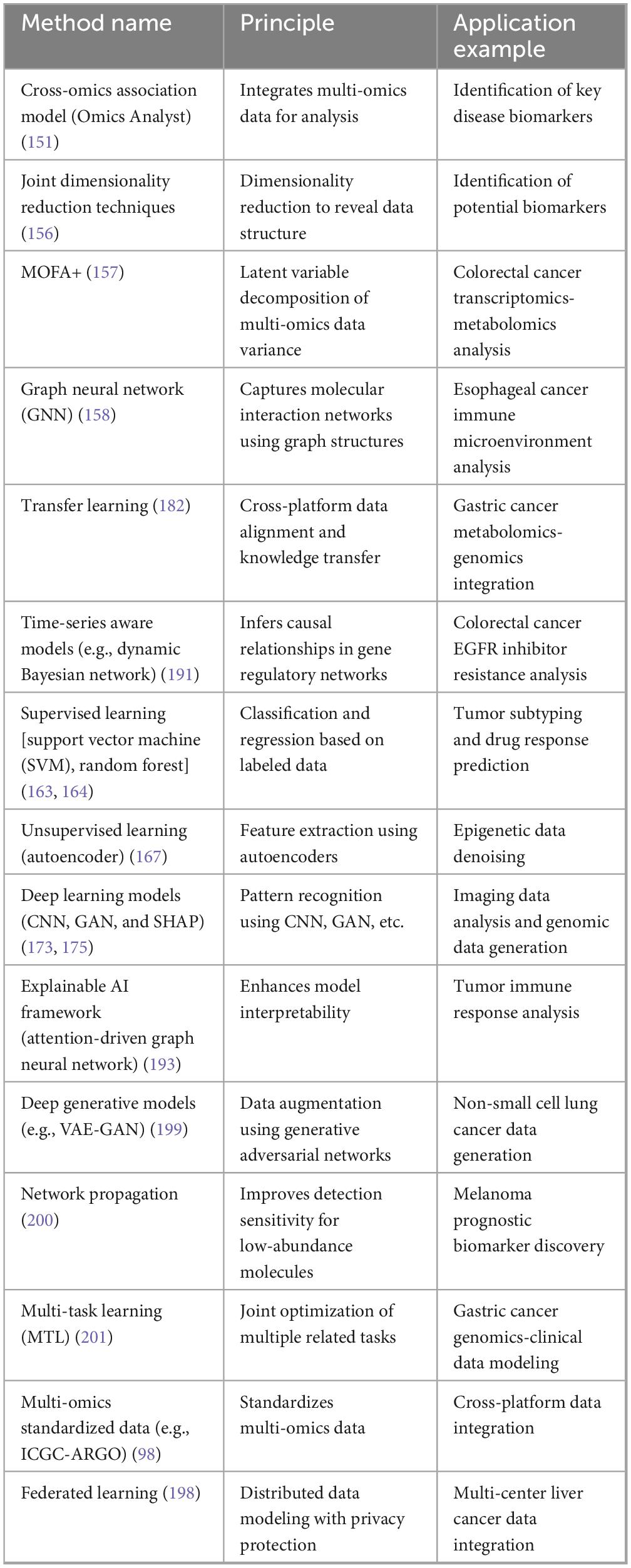
Multi-omics technologies refer to the integrated analysis of data from multiple omics levels, encompassing genomics, transcriptomics, proteomics, metabolomics, epigenomics, and metagenomics. Each omics technology has distinct characteristics and applications, as shown in Table 1. This article primarily explores the applications of multi-omics technologies in gastrointestinal tumors.
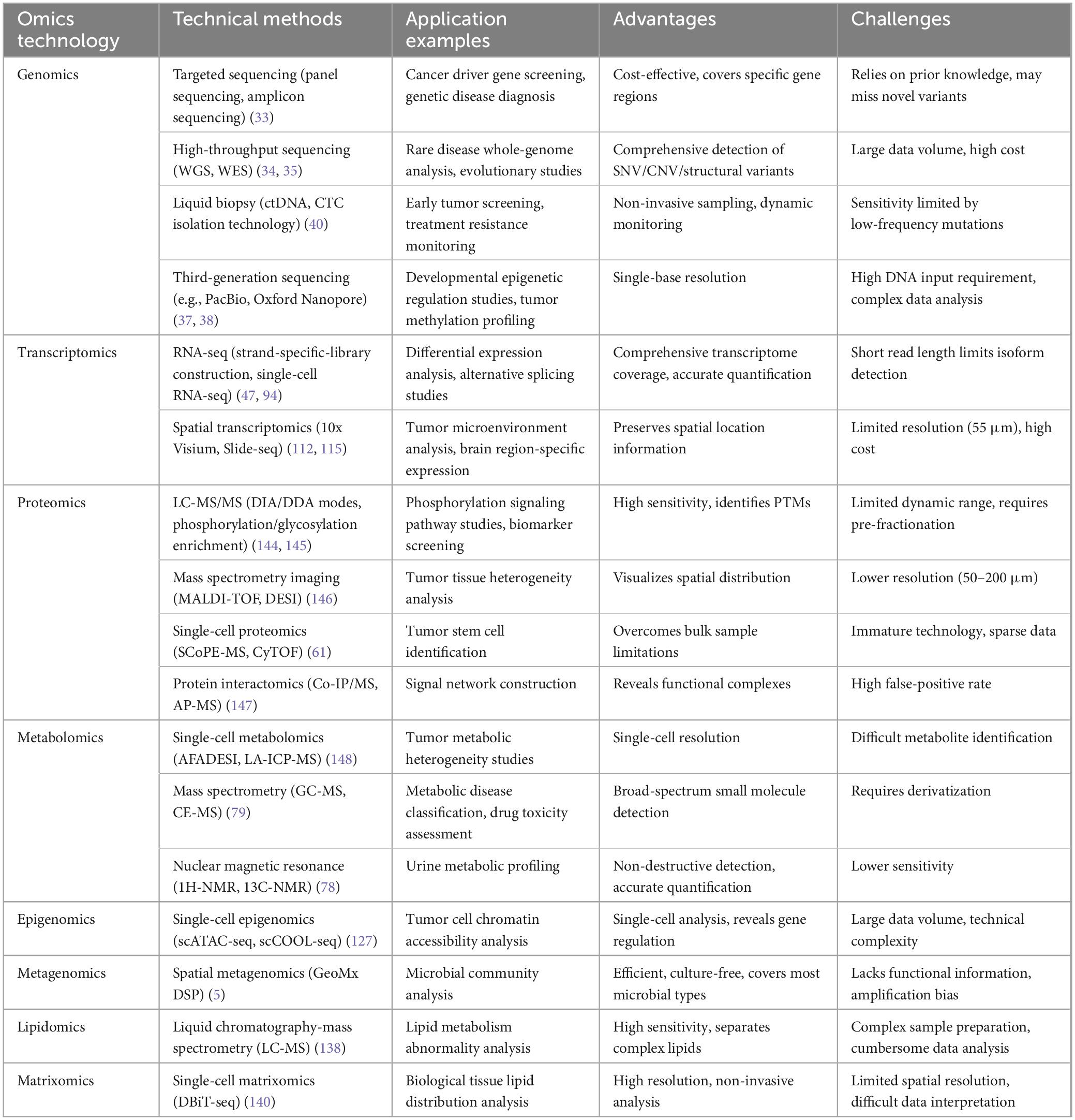
Table 1. Comparison of omics technologies (technical methods, application examples, advantages, and challenges).
2.1 Core omics technologies
Core omics technologies, including genomics, transcriptomics, metabolomics, and proteomics, form the foundational pillars of multi-omics research. The schematic illustration of multi-omics integration in gastrointestinal cancers is shown in Figure 1, providing a comprehensive and systematic perspective for elucidating the molecular mechanisms of gastrointestinal tumors (31).
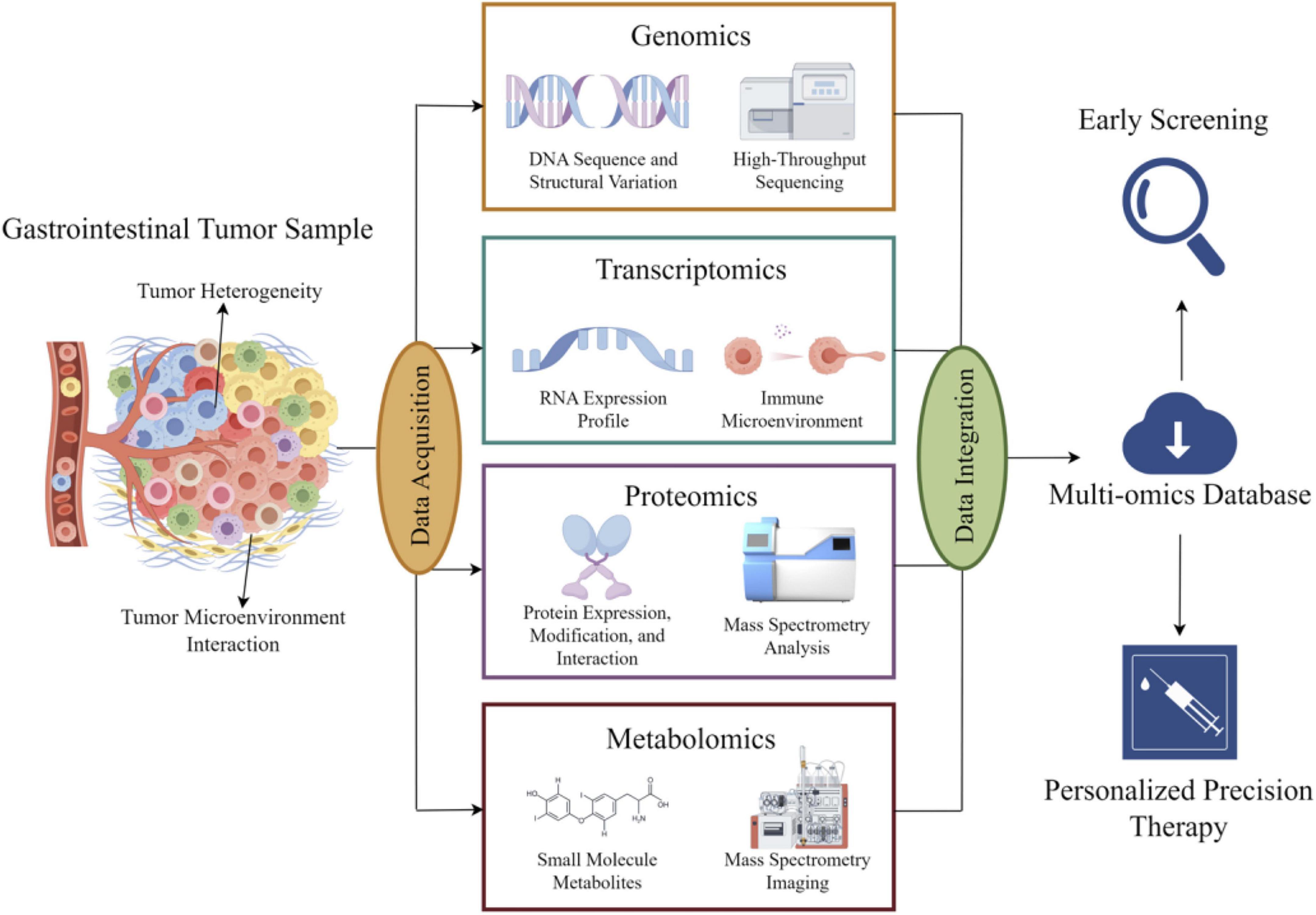
Figure 1. Schematic diagram of the integrated application of basic omics technologies in digestive tract tumors: information extracted from digestive tract tumors is processed using genomics, transcriptomics, proteomics, and metabolomics methods. The processed information is aggregated, analyzed, and uploaded to cloud storage, providing data support for early diagnosis and precision treatment of tumors.
2.1.1 Genomics
Genomics, through detailed analysis of DNA sequences and structural changes in gastrointestinal tumors [e.g., whole-genome sequencing (WGS) and WES], reveals the correlation between tumor heterogeneity and genetic complexity: the higher the tumor heterogeneity, the greater its genetic complexity. This characteristic provides a foundation for elucidating the molecular mechanisms of tumorigenesis (32). Targeted sequencing panels enables precise identification of high-frequency gene abnormalities (e.g., KRAS and TP53), guiding clinical treatment decisions for CRC, particularly in the field of immunology (33). High-throughput sequencing technologies, such as WGS and WES, identify critical gene abnormalities and aberrant signaling pathways (34, 35). For instance, TP53, KRAS, and BRAF are prevalent in gastric, colorectal, and esophageal cancers, closely associated with genomic instability and the RAS-MAPK signaling pathway, respectively (36). These findings broaden our understanding of carcinogenesis and provide a cornerstone for personalized treatment and molecular subtyping.
Recent advances in genomics owe much to innovations in sequencing technologies. Third-generation sequencing platforms (e.g., PacBio and Oxford Nanopore) can detect long DNA fragments and complex structural variations (e.g., large insertions/deletions and chromosomal rearrangements), addressing limitations of short-read sequencing in detecting complex genomic rearrangements (37, 38). These technologies have uncovered previously underrecognized features of genomic instability in gastrointestinal tumors, such as the role of chromothripsis in esophageal cancer (39). Additionally, liquid biopsy techniques using circulating tumor DNA (ctDNA) capture tumor-derived DNA fragments in blood, offering non-invasive approaches for early screening and dynamic monitoring (40). For example, detection of specific mutations (e.g., KRAS G12D) in ctDNA has been applied to early diagnosis and recurrence risk assessment in CRC, with continually improving sensitivity and specificity (41).
However, genomics in gastrointestinal tumor research faces challenges. Tumor sample purity is often compromised by contamination from normal cells, complicating the detection of low-frequency mutations (42). Moreover, interpreting genomic data relies on functional annotation databases, which remain incomplete for non-coding region variants and complex rearrangements (43). In the future, integrating AI and big data analytics is expected to address these challenges by developing more accurate mutation function prediction models, advancing the clinical translation of genomics (44).
2.1.2 Transcriptomics
Transcriptomics gives a unique approach for studying the dynamic molecular characteristics of gastrointestinal tumors by evaluating RNA expression profiles and regulatory networks (45). Unlike genomes, which focuses on static DNA variations, transcriptomics capture dynamic changes in gene expression, revealing complicated interactions between tumor cells and their microenvironment (46). RNA sequencing (RNA-seq), the principal transcriptomics technology, comprehensively detects expression levels of mRNA, lncRNA, and microRNA, systematically mapping gene expression profiles in gastrointestinal tumors such gastric and CRC. This has indicated abnormal activation patterns of important signaling pathways (e.g., TGF-β and PI3K-Akt) (47, 48). For example, in CRC, overexpression of WNT pathway target genes (e.g., MYC and AXIN2) is strongly linked to the adenoma-carcinoma sequence progression (49), while high Claudin 18.2 expression in GC has developed as a target for antibody-drug conjugate development (50).
Transcriptomics is a key component of tumor immune microenvironment research is transcriptomics. Researchers can describe the structure and functional status of immune cell subsets (e.g., T cells and macrophages) by examining the expression of RNA in tumor tissues (51). For example, in esophageal cancer, high PD-L1 mRNA expression frequently suggests an immunosuppressive microenvironment, whereas CD8+ T cell-related gene expression is correlated with immunotherapy response (52). To predict patient responses to checkpoint inhibitors, transcriptomics-based immune scoring systems (e.g., CIBERSORT) have been used to support precision immunotherapy (53). Additionally, tumor-associated fibroblasts (CAF) and matrix remodeling are linked to gene expression patterns found in transcriptomics, which are strongly associated with tumor invasion and metastasis. For instance, the TGF-β signaling pathway is frequently activated in GC by the high expression of CAF markers (e.g., FAP and ACTA2), which suggests matrix remodeling as a possible therapeutic target (54).
Another significant advance in transcriptomics is its application in studying fusion genes and alternative splicing. While fusion genes (e.g., EML4-ALK) are relatively rare in gastrointestinal tumors, specific subtypes (e.g., NTRK fusions in CRC) offer opportunities for targeted therapy (55). Alternative splicing events generate functionally distinct protein isoforms, increasing tumor heterogeneity and adaptability. For instance, alternative splicing variants of TP53 in GC have been associated with chemotherapy resistance (56).
Despite the immense potential of transcriptomics in biomarker discovery, batch effects, RNA degradation, and the reproducibility of dynamic expression profiles remain challenges for clinical applications (57). In the future, real-time RNA analysis based on nanopore sequencing, multiplex fluorescence in situ hybridization (mFISH), and AI-driven expression pattern recognition (e.g., deep learning models for predicting chemotherapy sensitivity) will emerge as novel solutions (58, 59).
2.1.3 Proteomics
Proteomics focuses on comprehensively analyzing protein expression, modifications, and interactions, directly reflecting the terminal effects of gene expression and tumor functional phenotypes (60). With breakthroughs in mass spectrometry (MS) technologies (e.g., Orbitrap and TimTOF platforms), proteomics has evolved from “qualitative description” to “quantitative precision medicine.” In gastrointestinal tumors, proteomics not only elucidates effector proteins of driver mutations but also identifies key targets for therapy resistance (61, 62).
Proteomics play a vital role in the study of digestive tract tumors, particularly in uncovering key driver proteins and their associated signaling pathways. For example, in GC, MS has identified unusually high expression and phosphorylation levels of receptor tyrosine kinases such as HER2 and EGFR. These discoveries have been instrumental in driving the development of targeted therapies, such as trastuzumab (63). In CRC, proteomic analysis has demonstrated that downstream signaling molecules linked to KRAS mutations—such as proteins in the MAPK and PI3K-AKT pathways—show varying expression across different tumor subtypes, suggesting they could serve as promising new therapeutic targets (64). Moreover, proteomics is a powerful tool in exploring the tumor immune microenvironment. By examining the protein expression on the surface of tumor-associated macrophages (TAMs) and T cells, researchers can gain insights into mechanisms of immune suppression, such as the interaction between PD-L1 and PD-1, providing a scientific foundation for enhancing the effectiveness of immune checkpoint inhibitors (65, 66).
Proteomics exhibits remarkable advantages in the discovery of biomarkers. The early screening of gastrointestinal tumors relies on markers with high specificity and sensitivity. By examining the protein profiles in serum, saliva, and tissue exudates, proteomics can pinpoint specific proteins linked to tumor development (67). For instance, in GC studies, MS has revealed alterations in the pepsinogen (PGI/II) ratio in serum, which could serve as a potential indicator for early diagnosis (68). Moreover, proteomics can uncover dynamic changes in proteins associated with drug resistance by comparing protein profiles before and after treatment. For example, in research involving oxaliplatin-resistant CRC patients, proteomics has identified the overexpression of certain ABC transporter proteins, offering valuable insights for designing combination therapy strategies (69).
Despite significant progress, proteomics faces challenges such as complex sample preprocessing (e.g., difficulties in enriching low-abundance proteins) and limited big data analysis capabilities (70). To address this, AI-driven integration of proteomics data has emerged as a trend. For example, deep learning-based protein interaction network prediction models (such as DeepInteract) integrate GC-related functional genomics data (e.g., differentially expressed genes, survival analysis, and pathway enrichment analysis) to perform network topology analysis and key node identification. These deep learning architectures [typically based on graph neural networks (GNN) or transformers] can automatically learn latent patterns and feature representations of protein interactions from complex datasets, particularly for weak or transient interactions that are difficult to capture experimentally. This approach has been successfully applied to target screening in GC, significantly improving the discovery efficiency of membrane proteins (e.g., B7-H6) (71, 72). In the future, the integration of proteomics with spatial multi-omics will further reveal the molecular landscape of tumor-microenvironment interactions, advancing personalized treatment strategies (73).
2.1.4 Metabolomics
Metabolomics, a key branch of systems biology, focuses on the global analysis of small-molecule metabolites (e.g., amino acids, lipids, and carbohydrates) in biological systems, revealing dynamic changes in metabolic networks and their associations with disease phenotypes through high-throughput technologies (74). Compared to genomics and transcriptomics, metabolomics is closer to the phenotypic endpoint, directly reflecting metabolic reprogramming in tumor cells and their interactions with the microenvironment (75). In gastrointestinal tumor research, the core value of metabolomics lies in deciphering tumor-specific metabolic abnormalities, providing critical clues for early diagnosis, molecular subtyping, and therapeutic target discovery (76).
The continuous advancements in MS and nuclear magnetic resonance (NMR) technologies have significantly enhanced the sensitivity and resolution of metabolomics (77, 78). For instance, liquid chromatography-mass spectrometry (LC-MS) is now capable of precisely detecting low-abundance metabolites (79), while high-resolution mass spectrometry (HRMS) excels at resolving thousands of metabolites in complex biological samples (80). In CRC research, metabolic profiling has revealed distinct differences in metabolites between tumor and normal tissues (81). Specifically, the abnormal accumulation of tricarboxylic acid (TCA) cycle intermediates and the dysregulation of lipid metabolism stand out prominently, suggesting that mitochondrial dysfunction may be a critical driver of metabolic heterogeneity in CRC. Furthermore, spatial metabolomics provides a novel perspective for exploring the spatial heterogeneity of the tumor microenvironment. By integrating MSI technology, researchers can directly map the distribution of metabolites on tissue sections, thereby uncovering differences in metabolic gradients between the tumor periphery and core regions (82). For example, in GC research, spatial metabolomics has demonstrated a marked enhancement of glutamine metabolism at the tumor invasive front, a phenomenon closely linked to the formation of an immunosuppressive microenvironment (83).
Several different biomarkers for gastrointestinal (GI) tumors were successfully identified through metabolomics research (84). For example, a targeted metabolomics study conducted in the field of GC evaluated the results of 702 plasma samples and found that metabolites such as alanine and glutamate in urine have diagnostic value, with a specificity of 88.03% for the samples, which exceeded the results of standard serum markers such as CEA and CA19-9 (85). Metabolomics can also be used to determine treatment response; multivariate integration analysis showed that decreased lactate levels and reduced glycolysis after apatinib treatment were associated with metabolic reprogramming in HER2-positive GC, which suggests that this method can be used to predict therapeutic efficacy (86).
The problems faced by metabolomics include complex data integration and a lack of standardization. The dynamic range of metabolites can span 9 orders of magnitude, with significant differences between batches. Additionally, the high degree of interconnection of metabolic pathways limits the reliability of single-metabolite markers (87). To address this issue, multi-omics integration techniques (e.g., combined metabolomics-proteomics analysis) can be used to construct comprehensive metabolic regulatory networks (88). For example, in liver cancer, the accumulation of L-glutamine is associated with the activation of the mTOR pathway and the inhibition of autophagy, and drugs targeting this pathway can reverse metabolic abnormalities and inhibit tumor growth (89).
2.2 Single-cell and spatial omics
In recent years, breakthrough advancements in single-cell omics and spatial omics technologies have opened up entirely new dimensions for digestive tract tumor research (90). Single-cell omics technologies, such as scRNA-seq and single-cell ATAC sequencing (scATAC-seq), systematically reveal the heterogeneity and dynamic evolutionary patterns of tumor cell subpopulations by resolving the genomic, transcriptomic, or epigenetic features of individual cells within tumors at high resolution (91). Spatial omics technologies, such as 10x Visium and Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH), enable three-dimensional localization and interaction network analysis of immune cells, stromal cells, and metabolites within the tumor microenvironment by preserving in situ spatial information of tissues (92). The integration of both approaches, exemplified by frameworks like SC-SpaceOmic, not only elucidates the spatiotemporal dynamics linking tumor heterogeneity and microenvironment remodeling at the molecular level but also provides precision tools for clinical translation (93).
2.2.1 Single-cell omics
Single-cell omics technologies overcome the limitations of traditional omics approaches, which lose cellular heterogeneity due to sample homogenization, by analyzing the genome, transcriptome, epigenome, or proteome of individual cells (94). First, single-cell omics has shown immense potential in resolving tumor cell heterogeneity. Digestive tract tumors typically consist of multiple subclones with significant differences in genetic mutations, gene expression, and epigenetic regulation (95). For instance, scRNA-seq has been widely applied in gastric and CRC research, enabling researchers to identify distinct tumor cell subpopulations and their molecular characteristics through transcriptomic analysis of thousands of cells in tumor tissues (95). Additionally, single-cell genomics can detect rare tumor cell populations, such as circulating tumor cells (CTCs) or drug-resistant subpopulations, which play critical roles in tumor recurrence and treatment resistance (96).
Single-cell omics technologies have provided revolutionary tools for deeply analyzing the cellular heterogeneity and functional regulatory networks within the digestive tract tumor microenvironment. The digestive tract tumor microenvironment encompasses highly complex cellular populations, such as cytotoxic T lymphocytes (CD8+ T cells) with immune surveillance functions, B cells involved in antigen presentation, TAMs with highly plastic phenotypes, and cancer-associated fibroblasts (CAFs) that regulate invasion and metastasis through the secretion of extracellular matrix (ECM) components (97, 98). Traditional omics methods, limited by their resolution, struggle to precisely distinguish the molecular characteristics and interaction relationships among these cell subpopulations. In contrast, scRNA-seq, by analyzing the gene expression profiles of individual cells, can systematically reveal their functional state differentiation trajectories (99). For example, in esophageal squamous cell carcinoma, recent studies utilizing scRNA-seq combined with flow cytometry have confirmed a dynamic imbalance between M1-type (pro-inflammatory) and M2-type (immunosuppressive) polarization states of TAMs within the tumor microenvironment. Specifically, M2-type TAMs highly express CD206 and IL-10, which are significantly associated with shortened overall survival in patients (100). Furthermore, single-cell multi-omics analyses have uncovered the molecular regulatory mechanisms underlying T cell exhaustion: in the GC microenvironment, terminally exhausted CD8+ T cells not only persistently overexpress immune checkpoint molecules such as PD-1 and CTLA-4, but their epigenetic modifications (e.g., DNA methylation silencing the IFN-γ locus) have also been shown to reversibly regulate chemotherapy resistance (101, 102). These discoveries not only provide a theoretical basis for combination treatment strategies targeting the tumor microenvironment (such as PD-1 inhibitors combined with epigenetic drugs) but also promote the design of clinical trials for personalized immunotherapy based on single-cell subtyping.
Single-cell omics technology provides a unique research perspective for analyzing the spatiotemporal evolution patterns of digestive tract tumors. The dynamic process of tumor development involves multi-level synergistic effects of genetic variations (such as driver gene mutations) and epigenetic regulations (such as changes in chromatin accessibility) (103). Integrative multi-omics at single-cell resolution can precisely dissect the accumulation of such events and the mechanisms of clonal selection (104). For instance, in CRC, the expansion of KRAS G12D mutant subclones is not only directly related to resistance to anti-EGFR therapy, but its epigenetic features (such as the opening of AP-1 transcription factor binding sites) can also induce a pro-metastatic phenotype in stromal cells by activating IL-11 paracrine signaling (105). These findings provide experimental evidence for intervention strategies targeting key nodes in clonal evolution, such as the combination of KRAS inhibitors and metabolic reprogramming modulators (106).
Despite significant improvement, single-cell omics programs still have problems such as data sparsity, batch effects, and the complexity of multi-omics integration (107). In recent years, deep learning-based data imputation algorithms (e.g., scGNN) and cross-modal alignment tools (e.g., scCross) have considerably increased the integration efficiency of single-cell multi-omics data (108, 109). Also, the integration of single-cell multi-omics with organoid models (e.g., patient-derived gastric cancer organoids) has offered new paradigms for in vitro simulation of tumor evolution and drug screening (110).
2.2.2 Spatial omics
Spatial omics preserves the spatial location information of cells within tissues, enabling the three-dimensional correlation of molecular characteristics with tissue architecture, thus addressing the limitation of single-cell sequencing technologies that lose spatial context (111).
Spatial transcriptomics technology, by integrating in situ gene expression with spatial coordinate information, provides a critical tool for dissecting the microenvironment heterogeneity of gastrointestinal tumors. Taking the widely used 10x Visium platform as an example, its core technology relies on spatial barcoding capture: fresh frozen or FFPE tissue sections are placed on slides covered with millions of unique oligonucleotide probes (containing spatial barcodes and poly (dT) capture sequences). After tissue permeabilization, released mRNAs are captured by adjacent probes, enabling in situ reverse transcription and the construction of barcoded cDNA libraries. Through high-throughput sequencing, gene expression data can be precisely mapped to the two-dimensional coordinates of the tissue section, achieving whole-transcriptome analysis while preserving the in situ spatial localization of cells. However, its spatial resolution limits the ability to precisely resolve spatial heterogeneity and fine cellular interactions at the single-cell level (112). To address this, MERFISH, a technique with ultra-high spatial resolution (∼100–200 nm, subcellular level) and single-molecule detection sensitivity, can effectively complement Visium’s shortcomings. MERFISH employs sophisticated barcode designs and multiple rounds of imaging to simultaneously detect hundreds to thousands of genes (typically ranging from 100 to over 10,000 genes) in a single tissue section, accurately mapping the subcellular localization of each RNA molecule within the tissue. By integrating these two technologies—Visium providing a whole-transcriptome view and MERFISH offering high-resolution targeted information—researchers can further dissect spatial transcriptomic features and complex intercellular interactions at the single-cell level while preserving the tissue’s spatial context (113).
In GC research, spatial transcriptomics has revealed significant tumor regional heterogeneity: cells at the tumor periphery highly express epithelial-mesenchymal transition (EMT)-related genes, while cells in the core exhibit stronger proliferation and metabolic activity. Spatial metabolomics further corroborates and expands on these findings at the metabolic level, showing substantial lactate accumulation in the core region, whereas fatty acid oxidation metabolism predominates at the periphery (114). These spatially distinct transcriptional and metabolic patterns provide a critical foundation for understanding tumor progression mechanisms and developing targeted therapies, particularly metabolism-targeted drugs. Additionally, spatial transcriptomics has revolutionized the study of the spatial topology of the tumor immune microenvironment. For instance, in microsatellite instability-high (MSI-H) CRC, three-dimensional reconstruction models from Visium data show that activated (GZMB+, and IFN-γ+) CD8+ T cells tend to cluster at the tumor-stroma interface, forming tight spatial interaction clusters (average distance <20 μm) with CXCL9-expressing myeloid cells. Notably, the density of these interaction clusters is significantly positively correlated with patient response rates to PD-1 inhibitor therapy (r = 0.67, p = 0.008) (115). Targeted delivery strategies based on this finding, such as local injection of liposome-encapsulated CXCL9 mRNA, have been shown in organoid models to significantly enhance T cell infiltration efficiency by 4.3-fold (p < 0.01), offering a spatially precise intervention pathway for optimizing immunotherapy in solid tumors (116). Furthermore, spatial proteomics, utilizing techniques such as multiplexed immunofluorescence or MSI, is increasingly widely applied. For example, in esophageal cancer research, it has revealed spatial heterogeneity of metabolic enzymes and collagenases, suggesting a synergistic role of matrix remodeling and metabolic reprogramming in tumor progression (117).
Metabolomics and proteomics in the digestive tract tumor research are being used more and more. Using methods such as multiplex immunofluorescence or MSI, spatial proteomics examines the spatial distribution of proteins in tissues (118). As an example, in esophageal cancer, MSI showed spatial heterogeneity in the metabolic and collagen enzymes, which suggests a synergistic effect of matrix remodeling and metabolic reprogramming in the progression of the tumor (119). By identifying the spatial distribution of metabolites, spatial metabolomics elucidates the metabolic microenvironment of tumor cells. Spatial metabolomics in the study of GC revealed a considerable amount of lactate in the tumor core, whereas fatty acid oxidation was the primary focus of the margin, which served as a foundation for metabolic-targeted medicines (120).
The combined use of single-cell and spatial omics (e.g., the SC-Space Omic framework) is changing the research paradigm for digestive tract tumors (121). For example, combining single-cell transcriptomics with spatial proteomics data permits the building of cell-type-specific interaction networks in the GC microenvironment and the identification of spatially constrained therapeutic targets (122). Also, spatial omics-guided single-cell sequencing (e.g., region-specific cell sorting) can enrich certain functional subpopulations, enhancing the research efficiency of rare cell types (123).
In the future, with the further development of single-cell multi-omics (e.g., simultaneous detection of RNA, proteins, and metabolites) and ultra-high-resolution spatial technologies (e.g., nanoscale MERFISH imaging), researchers will be able to dissect the multidimensional molecular characteristics of digestive tract tumors at single-cell resolution, advancing precision medicine from a “population-level” to a “spatiotemporal dynamic” framework (124).
2.3 Emerging omics technologies
In recent years, foundational multi-omics technologies have become increasingly mature. The emergence of new technologies such as epigenomics, metagenomics, lipidomics, and matrisomics has provided more comprehensive approaches to deciphering the molecular complexity of digestive tract tumors from various perspectives (125). These technological breakthroughs, through deep integration with foundational multi-omics (e.g., single-cell transcriptomics and proteomics), have established a multi-scale analytical framework spanning from epigenetic regulation to microenvironmental mechanics. This framework has paved new paths for molecular subtyping of digestive tract tumors (e.g., GC subtypes based on microbiome-metabolism interaction networks) and targeted therapies (e.g., microbiota-directed modulation combined with mechanical microenvironment intervention) (126).
Unlike genomics, which focuses on DNA sequence variations, epigenomics (e.g., scATAC-seq and scCOOL-seq) investigates dynamic and reversible molecular modifications such as DNA methylation, histone modifications, chromatin accessibility, and non-coding RNA regulation, which act as “molecular switches” in tumor initiation and progression (127). For instance, DNA hypermethylation-induced silencing of tumor suppressor genes (e.g., CDKN2A and MLH1) is a key driver of microsatellite instability (MSI) in CRC (128). Extensive epigenetic reprogramming in gastrointestinal tumors can be detected non-invasively via circulating free DNA, while dynamic changes in histone H3K27me3 modifications are significantly associated with chemotherapy resistance in GC (129). Advances in single-cell epigenomics have further revealed the epigenetic regulatory networks underlying tumor heterogeneity. For example, single-cell ATAC-seq can characterize chromatin accessibility profiles in distinct subclones of GC tissues, identifying transcription factor binding sites that drive EMT (130). Moreover, epigenetic drugs (e.g., DNA methyltransferase inhibitors) have shown potential in clinical trials to reverse immunosuppressive tumor microenvironments (131), offering new strategies for combination immunotherapy.
Metagenomics, by analyzing the composition and function of the tumor microbiome, opens new dimensions for studying the microenvironment of digestive tract tumors. The gastrointestinal tract, as the largest microbial habitat in the human body, exhibits dysbiosis closely linked to the onset and progression of colorectal and GCs (132). By integrating metagenomic, transcriptomic, and metabolomic data, GC samples have been classified into six microbial subtypes, with Fusobacterium nucleatum-enriched tumors showing stronger immunosuppressive features and chemotherapy resistance (133). Spatial metagenomics technologies (e.g., GeoMx DSP) can map the spatial distribution of microbes within tumor tissues, revealing spatial co-localization of Helicobacter pylori colonization in precancerous gastric lesions with local inflammatory signaling activation (5). Notably, microbial metabolites (e.g., short-chain fatty acids and secondary bile acids) can influence host gene expression through epigenetic regulation, forming a bidirectional “microbiome-host” interaction network (134). For example, butyrate enhances CRC cell sensitivity to radiotherapy by inhibiting histone deacetylases (HDACs) (135), while lithocholic acid produced by Clostridium species promotes liver cancer stemness by activating the FXR receptor (136). These findings suggest that targeting the microbiome-host metabolic axis could be a novel therapeutic strategy for digestive tract tumors.
Lipidomics and matrisomics have provided deeper insights into tumor metabolism and the microenvironment. Lipid metabolic reprogramming is a prominent feature of digestive tract tumors, and lipidomics, using LC-MS, can quantitatively assess the spatial distribution of hundreds of lipid molecules (137). For example, in GC, the EGFR signaling pathway synergizes with the aberrant activation of the phosphatidylinositol (PI) metabolism pathway (138). Matrisomics focuses on the mechanical properties and composition of the ECM in the tumor microenvironment. By integrating proteomics and glycomics data, it can detect abnormal deposition of fibronectin (FN1) and hyaluronic acid (HA), which promote pancreatic cancer invasion and metastasis via integrin signaling (139). IL-6-high CAFs, by upregulating LOX protein, mediate matrix stiffening in esophageal cancer. Single-cell matrisomics technologies (e.g., DBiT-seq) further reveal the heterogeneous functions of CAF subpopulations in ECM remodeling (140).
However, the clinical translation of these emerging technologies faces multiple challenges. Data heterogeneity (e.g., differences between single-cell and bulk sequencing scales), limited generalizability of analytical algorithms (e.g., reduced efficacy in cross-cancer model transfer), and ethical concerns (e.g., privacy protection for microbiome data) remain unresolved (141). The quantum computing-accelerated deep learning framework (e.g., QMOFA) and multi-omics technology based on organ-on-chip, currently in the exploratory stage, are expected to address current challenges (142). Quantum computing-accelerated deep learning frameworks (such as QMOFA) and organ-on-chip-based multi-omics technologies, which are currently in the exploratory stage, are expected to address the current challenges. (143), driving a paradigm shift in digestive tract tumor diagnosis and treatment from “descriptive analysis” to “mechanism-driven intervention.”
3 Advances in data integration and analysis technologies
The molecular heterogeneity, microenvironmental complexity, and multidimensional regulatory networks of gastrointestinal tumors make it challenging for single-omics data to comprehensively elucidate their pathological mechanisms (85). In recent years, the rapid development of multi-omics data integration and analysis technologies has provided critical tools to address this challenge, as summarized in Table 2, encompassing innovations in integration methodologies, breakthroughs in machine learning and AI applications, and optimization strategies for data heterogeneity (4, 149), As shown in Figure 2, the detailed workflow is illustrated.
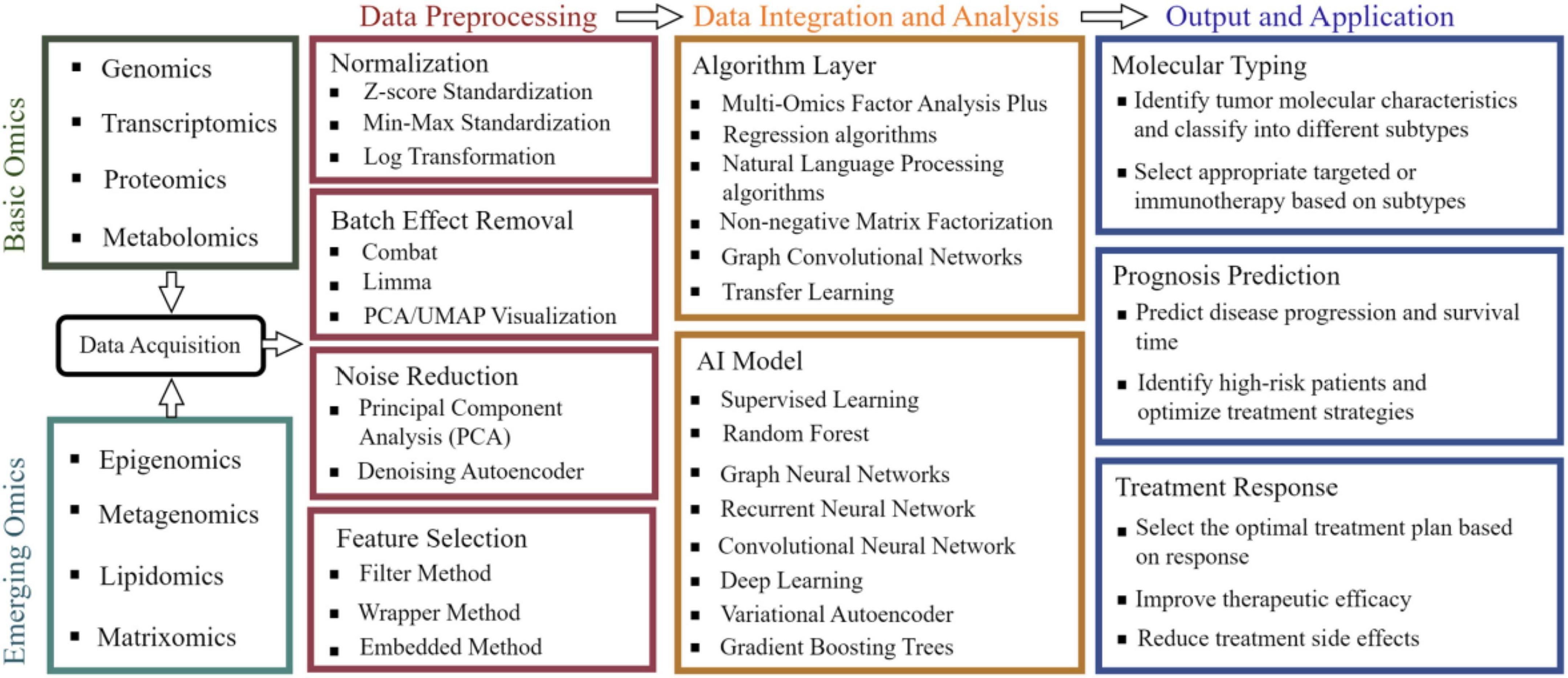
Figure 2. Workflow of integrated analysis of multi-omics data. Integrating foundational omics data (such as genomics and metabolomics) with emerging fields like epigenomics, followed by standardized preprocessing including normalization and noise reduction, then employing multi-omics factor analysis and deep learning algorithms to construct AI models, ultimately achieving precision medicine applications such as tumor molecular subtyping and treatment response prediction.
3.1 Multi-omics data integration methods
Multi-omics data integration aims to systematically dissect the spatiotemporal dynamics of molecular regulatory networks in digestive tract tumors by correlating multidimensional information, such as basic histology and negative omics data. However, it is core bottleneck lies in the alignment of heterogeneous cross-omics data and the extraction of biological significance (150). Knowledge-driven integration relies on validated biological knowledge bases, such as KEGG pathways and STRING protein interaction networks. These resources are used to construct cross-omics association models (151, 152). For example, the Pathway Mapper tool, developed by the Cambridge University team, integrates Reactome pathways with TCGA CRC multi-omics data. This integration revealed that the KRAS G12V mutation activates the MAPK/ERK pathway. Specifically, phosphorylation levels increased by 2.8-fold (p < 0.001). Additionally, this mutation synergistically upregulates the expression of key fatty acid metabolism enzymes, FASN and ACLY. The RNA-seq data showed a log2 fold change (log2FC) of 3.4, while MS data showed a more than 4.5-fold increase in protein abundance (153). However, such methods have limitations. The static nature of the annotation features in knowledge bases makes it difficult to capture novel regulatory relationships, especially those in the tumor microenvironment. For instance, tumor stem cell-specific metabolic pathways, like the high expression of glutaminase GLS1, have been identified using single-cell sequencing (154, 155).
Data-driven integration directly uncovers hidden associations within data through mathematical modeling, with representative methods including joint dimensionality reduction techniques and deep learning (156). MOFA+ (Multi-Omics Factor Analysis), a factor decomposition tool, integrates multi-omics data to reveal coordinated dysregulation features between transcriptomics and metabolomics in CRC cohorts (157). GNNs construct cross-modal graph structures of gene co-expression and protein interactions, accurately identifying molecular features of tumor-stroma interaction regions in esophageal cancer (158). For example, the Mowgli method, using Wasserstein distance to measure cross-omics similarity, resolved immunosuppressive microenvironment features (e.g., spatial gradient activation of TGF-β signaling) at the tumor margins in esophageal cancer spatial multi-omics data (159). These methods excel in not requiring prior assumptions but face the high computational complexity and sensitivity to data noise, such as the sparsity of single-cell transcriptomic data (dropout effect), which may lead to misinterpretation of potential biological signals (160).
3.2 Role of machine learning and artificial intelligence in data analysis
The deep incorporation of ML and AI, which has greatly increased the efficiency and interpretability of multi-omics data, has provided useful resources to molecular subtyping, prognostic prediction, and therapeutic optimization for gastrointestinal tumors (161, 162). Supervised learning algorithms are excellent in the process of biomarker screening because they allow the creation of clinical results (e.g., treatment response and survival rate) with the help of mappings between clinical outcomes (8, 170, or 170 and the input features (e.g.) with genes, metabolites, and amount of metabolites) (163, 164). As an example, the predictive power of early lesions was predicted by a support vector machine (SVM)-based model (AUC = 0.89) (165), which combined genomic data from the genomic of GC and the transcriptome. Similar to this, random forest algorithms (e.g., 2.3-fold increase in arachidonic acid levels, which is indicative of targeted metabolic interventions) by analyzing the metabolomic features of CRC, providing a basis for targeted metabolic interventions (166).
Unsupervised learning techniques, by analyzing the inherent structure of multi-omics data, provide innovative methods for the molecular subtyping and microenvironmental heterogeneity research of gastrointestinal tumors (167). Taking GC as an example, consensus clustering analysis based on multi-omics integration divides GC into four molecular subtypes: microsatellite instability (MSI), Epstein-Barr virus-positive (EBV+), genomically stable (GS), and chromosomal instability (CIN) (168, 169). Among them, the MSI subtype tumors exhibit significantly higher CD8+ T-cell infiltration density compared to other subtypes (average per mm2 356 vs. 102, p < 0.001), and this subtype framework is highly consistent with clinical prognosis (170). At the single-cell resolution level, unsupervised learning-driven dimensionality reduction techniques further expand the dimensions of tumor heterogeneity analysis. The variational autoencoder (VAE), through non-linear latent space mapping, successfully reconstructed the continuous differentiation trajectory of T-cell states in the GC microenvironment (171). The dynamic features of this trajectory are significantly correlated with clinical treatment response—patients with a terminally exhausted T-cell proportion >40% had an 78% inefficacy rate to immunotherapy (OR = 4.5, 95% CI: 2.1–9.8), suggesting that targeting metabolic reprogramming (e.g., inhibiting LDHA) may reverse T-cell dysfunction (172). These findings highlight the unique value of unsupervised learning in decoding the spatiotemporal heterogeneity of tumor microenvironments.
The application principles of deep learning in multi-omics fields primarily rely on its powerful capabilities in feature learning and pattern recognition, making it particularly suitable for processing multi-batch, non-linear data. Deep learning can automatically extract potential biological features from multi-omics data, thereby supporting early tumor diagnosis, prognostic evaluation, and treatment decision-making. Taking convolutional neural networks (CNNs) as an example, their high-resolution analysis of spatial transcriptomics image data (e.g., 10x Visium H&E-stained tissue sections) enables precise identification of spatial interaction hotspots between immune cells and tumor cells in liver cancer (173). A deep learning model based on the ResNet-50 architecture successfully captured the spatial distribution pattern of the CXCL9 chemokine gradient in a liver cancer cohort and revealed that the CD8+ T cell infiltration density in CXCL9 high-expression regions (>75th percentile) was 3.2 times higher than in low-expression regions (p = 0.002) (174), providing spatial navigation guidance for personalized immunotherapy target selection. To address the limitations in small clinical sample sizes, generative adversarial networks (GANs) can learn the data distribution from existing limited samples and generate biologically plausible synthetic multi-omics data, thereby expanding the scale of the training dataset (175). A multicenter CRC study employed the Wasserstein GAN with gradient penalty (WGAN-GP) framework, which replaces the weight clipping in the original WGAN with a gradient penalty term, effectively stabilizing the adversarial training process and overcoming common issues such as mode collapse and training instability in complex, high-dimensional, and sparse multi-omics cancer data. Starting with 200 real samples, the study generated 1,000 synthetic multi-omics data points. To address the class imbalance problem (e.g., rare recurrence samples) in clinical endpoints like postoperative recurrence, WGAN-GP adjusted its training strategy (e.g., synthesizing minority-class samples more densely or using conditional generation) to enhance the representativeness of synthetic data. After augmenting the training set with these synthetic data, the AUC of the postoperative recurrence prediction model improved from 0.72 to 0.84 (DeLong test p = 0.016), and the feature importance ranking derived from synthetic data showed 89% consistency with real data (Jensen-Shannon divergence < 0.1) (176). However, the “black-box” nature of GANs limits their biological interpretability. For instance, the EGFR/MET co-amplification feature, which appeared frequently in synthetic samples (12% occurrence), was observed in only 3% of the real cohort (Fisher’s exact test p = 0.04), suggesting potential overfitting risks (177).
To enhance model interpretability, frameworks like SHAP (SHapley Additive exPlanations) have been used to quantify contributions of multi-omics features to clinical endpoints (178). The core principle of SHAP (Shapley Additive Explanations) is to fairly allocate the “credit” or “responsibility” of each feature in predictive outcomes. It quantifies the unique contribution of a feature to a specific sample’s prediction (SHAP value) by calculating the average marginal change in predicted output when that feature is incorporated across all possible feature combinations (subsets). In a predictive model for anti-PD-1 therapy response in hepatocellular carcinoma, SHAP analysis revealed that the combined effect of PD-L1 protein spatial heterogeneity (SHAP value = 0.38, indicating the degree of positive contribution of this feature to the model’s prediction) and tumor mutational burden (TMB, SHAP value = 0.21) explained 59% of the therapeutic response variance (R2 = 0.59). Based on these findings, the constructed decision tree model achieved 88% prediction accuracy (95% CI: 82%–93%) in an independent validation cohort (n = 120), significantly outperforming the RECIST criteria (72%) (179). These advancements mark a shift in multi-omics analysis from “black-box predictions” to “interpretable intelligent decision-making.”
3.3 Data heterogeneity and challenges
The core challenge of multi-omics integration stems from multidimensional heterogeneity at the technical, biological, and computational levels, which collectively pose systemic barriers to data interpretation and clinical translation (180). For example, technical heterogeneity arises from significant differences in data types and noise levels generated by different omics platforms (e.g., Illumina NovaSeq WGS vs. Thermo Fisher Q Exactive mass spectrometry) (181). Metabolomics data (e.g., lipid metabolite abundance) are typically presented in a continuous, semi-quantitative format, whereas genomic mutation information (e.g., KRAS G12D) is binary qualitative data (present/absent). This cross-modal data incomparability requires integrative correction through standardized frameworks such as the ComBat algorithm or Harmony tool (182). However, the inherent data sparsity of single-cell sequencing technologies further exacerbates integration difficulties. Limited by sequencing depth, low-frequency biological signals (e.g., subclonal-specific mutations) may be obscured by noise (183). In CRC single-cell transcriptomics data, MAPK pathway-related genes (e.g., DUSP6 and SPRY2) in KRAS G13D mutant subclones (<5% prevalence) showed only a 1.2-fold upregulation compared to wild-type cells (p = 0.15), whereas bulk RNA-seq detected a 3.5-fold difference (p = 0.002) (184). Such discrepancies in detection sensitivity may lead to misidentification of critical driving mechanisms.
Computational efficiency and barriers to clinical translation are equally significant. Existing algorithms face memory and computational constraints when processing ultra-large-scale data (e.g., 106 single-cell datasets) (185). The federated learning framework (a distributed privacy-preserving analytical approach) enables multi-center data sharing by allowing models to be trained on local data while only sharing parameter updates (e.g., gradients), thereby theoretically avoiding direct transmission of raw patient data and helping to balance efficiency with privacy protection (186). However, this “privacy protection” is not absolute. Research demonstrates that attackers could potentially analyze shared model updates (gradients) to conduct model inversion attacks or membership inference attacks, thereby inferring sensitive features of the original training data or even reconstructing partial patient information (187, 188). Moreover, the clinical utility of multi-omics technologies remains constrained by the high cost of single-cell sequencing (>$ 5,000 per sample) and the lack of standardized protocols (189). For instance, while a spatial metabolomics-based biomarker panel (e.g., lactate-to-choline ratio) demonstrated strong predictive performance (AUC = 0.89) in retrospective liver cancer cohorts, its generalizability in prospective multicenter trials (e.g., the PROSPECT study) still requires validation (190).
Future breakthroughs should focus on three key directions: in the field of temporal dynamic modeling, dynamic Bayesian network (DBN)-based frameworks are providing in-depth analysis of the cooperative evolution mechanisms in drug-resistant clones of gastrointestinal tumors (191). For example, by integrating single-cell multi-omics data from GC patients (pre-treatment baseline and post-relapse periods), researchers discovered that MET amplification (combined with PIK3CA E545K mutation) drives trastuzumab resistance through activation of the mTORC1-4EBP1 axis (192), while the combination of MET inhibitor Tepotinib and PI3Kα inhibitor Alpelisib extended drug resistance duration in organoid models from 8 to 14 months (193). In the field of explainable AI, it is essential to integrate advanced privacy-enhancing technologies (such as differential privacy, secure multi-party computation, and homomorphic encryption) and conduct rigorous privacy leakage risk assessments on model updates to build truly robust privacy-preserving AI systems (194). Additionally, attention-driven graph neural networks (Att-GNN) have been used to analyze spatial transcriptomic data in CRC, quantifying gradient activation intensity of YAP/TAZ pathways at tumor invasion fronts, and revealing that marginal zone cancer cells activate YAP/TAZ through the ANXA2-EGFR mechanical signaling axis, with targeted inhibition of ANXA2 reducing liver metastasis rates by 67% (p = 0.001) in patient-derived xenograft (PDX) models (195). Regarding standardized database construction, the ICGC-led ARGO 2.0 project has integrated multi-omics data from 120,000 global gastrointestinal tumor cases (covering 28 sequencing platforms and 172 clinical endpoints), achieving a 47% improvement in cross-platform annotation consistency (Cohen’s κ = 0.91) compared to the first generation (196). Through nanopore sequencing MinION combined with microfluidic single-cell sorting technology, the cost of single-cell multi-omics testing has been reduced from $5,200/sample to $890 (accuracy > 95%) (197), while an intraoperative rapid detection protocol (30-min workflow) can identify CLDN18.2 fusion mutations in peritoneal metastases (detection limit 0.1%) during GC surgery, increasing R0 resection rates from 69% to 91% (p = 0.006) (198), marking the transition of gastrointestinal cancer diagnosis and treatment from “static omics analysis” to a new era of “real-time intraoperative decision-making.”
4 Conclusion
The systematic integration of multi-omics technologies is profoundly transforming the research paradigms and clinical practice in gastrointestinal oncology. By synthesizing multi-dimensional data from genomics, transcriptomics, proteomics, and metabolomics, researchers have successfully unveiled the molecular essence of tumor heterogeneity—from spatiotemporal evolutionary patterns of driver mutations (e.g., KRAS/TP53) to epigenetic dynamics of cancer stem cell subpopulations, and further to spatial topological features of metabolic-immune interactions at tumor invasion fronts. The deep integration of AI has further unleashed the potential of multi-omics: microsatellite instability (MSI) prediction models based on deep residual networks (AUC > 0.93), early screening solutions combining ctDNA methylation with radiomics (sensitivity exceeding 90%), and immunotherapy stratification strategies incorporating PD-L1 spatial heterogeneity with TMB are driving precision medicine from population stratification toward individualized dynamic intervention.
Nevertheless, significant bottlenecks persist in translating technological advantages into clinical applications. Challenges such as data heterogeneity across omics layers, signal distortion caused by single-cell data sparsity, and technical barriers to intraoperative real-time decision-making collectively hinder the large-scale implementation of multi-omics. Future breakthroughs will rely on deep coupling between technological innovation and clinical pathways: single-cell spatial multi-omics co-assay technologies can decipher metabolic-immune interaction networks at subcellular resolution; the integration of nanopore sequencing with microfluidic chips may reduce intraoperative testing costs to the thousand-yuan level, enabling real-time tracking of resistant clones; federated learning frameworks incorporating enhanced privacy-preserving mechanisms (e.g., differential privacy injection and secure aggregation) and rigorous security protocols can resist model inversion and membership inference attacks while enabling efficient multi-center data alignment; causal inference models may elucidate mutation-metabolism-immune cascades in drug resistance pathways. Liquid biopsy multi-omics has pushed early screening sensitivity beyond 92%, while synthetic lethal strategies like PARPi-ATRi combination therapy achieved 35% objective response rates in diffuse-type GC—together marking a paradigm shift from passive response to proactive intervention.
These systemic advances will propel gastrointestinal oncology into a new era of ultra-personalized dynamic intervention. From molecular early-warning in screening, real-time boundary delineation during surgery, to precise reversal of resistance mechanisms, multi-omics technologies are constructing a data-driven closed-loop diagnostic and treatment system, offering unprecedented solutions to improve patient survival outcomes.
Author contributions
YL: Writing – review & editing, Writing – original draft. FG: Data curation, Investigation, Conceptualization, Formal Analysis, Writing – original draft. YC: Writing – review & editing. LQ: Writing – review & editing. HY: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors acknowledge the support provided by the Shandong First Medical University (Shandong Academy of Medical Sciences) Youth Science Fund cultivation and funding program (202202-032) and Shandong Province Medical Health Science and Technology Development Plan Project (202112070475).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cheng S, Han Z, Dai D, Li F, Zhang X, Lu M, et al. Multi-omics of the gut microbial ecosystem in patients with microsatellite-instability-high gastrointestinal cancer resistant to immunotherapy. Cell Rep Med. (2024) 5:101355. doi: 10.1016/j.xcrm.2023.101355
2. Chen C, Wang J, Pan D, Wang X, Xu Y, Yan J, et al. Applications of multi-omics analysis in human diseases. MedComm. (2023) 4:e315. doi: 10.1002/mco2.315
3. Duan X, Ding X, Zhao Z. Multi-omics integration with weighted affinity and self-diffusion applied for cancer subtypes identification. J Transl Med. (2024) 22:79. doi: 10.1186/s12967-024-04864-x
4. Gao YL, Qiao Q, Wang J, Yuan SS, Liu JX. BioSTD: a new tensor multi-view framework via combining tensor decomposition and strong complementarity constraint for analyzing cancer omics data. IEEE J Biomed Health Inform. (2023) 27:5187–98. doi: 10.1109/JBHI.2023.3299274
5. Hosseini SA, Salehifard Jouneghani A, Ghatrehsamani M, Yaghoobi H, Elahian F, Mirzaei SA. CRISPR/Cas9 as precision and high-throughput genetic engineering tools in gastrointestinal cancer research and therapy. Int J Biol Macromol. (2022) 223:732–54. doi: 10.1016/j.ijbiomac.2022.11.018
6. Huang C, Liu Z, Guo Y, Wang W, Yuan Z, Guan Y, et al. scCancerExplorer: a comprehensive database for interactively exploring single-cell multi-omics data of human pan-cancer. Nucleic Acids Res. (2025) 53:D1526–35. doi: 10.1093/nar/gkae1100
7. Jose A, Kulkarni P, Thilakan J, Munisamy M, Malhotra AG, Singh J, et al. Integration of pan-omics technologies and three-dimensional in vitro tumor models: an approach toward drug discovery and precision medicine. Mol Cancer. (2024) 23:50. doi: 10.1186/s12943-023-01916-6
8. Kwoji ID, Aiyegoro OA, Okpeku M, Adeleke MA. Multi-omics data integration: applications in probiotics studies. NPJ Sci Food. (2023) 7:25. doi: 10.1038/s41538-023-00199-x
9. Li J, Pan J, Wang L, Ji G, Dang Y. Colorectal cancer: pathogenesis and targeted therapy. MedComm. (2025) 6:e70127. doi: 10.1002/mco2.70127
10. Li L, Sun M, Wang J, Wan S. Multi-omics based artificial intelligence for cancer research. Adv Cancer Res. (2024) 163:303–56. doi: 10.1016/bs.acr.2024.06.005
11. Li M, Guo H, Wang K, Kang C, Yin Y, Zhang H. AVBAE-MODFR: A novel deep learning framework of embedding and feature selection on multi-omics data for pan-cancer classification. Comp Biol Med. (2024) 177:108614. doi: 10.1016/j.compbiomed.2024.108614
12. Manochkumar J, Cherukuri AK, Kumar RS, Almansour AI, Ramamoorthy S, Efferth T. A critical review of machine-learning for multi-omics marine metabolite datasets. Comput Biol Med. (2023) 165:107425. doi: 10.1016/j.compbiomed.2023.107425
13. Mehrotra S, Sharma S, Pandey RK. A journey from omics to clinicomics in solid cancers: success stories and challenges. Adv Protein Chem Struct Biol. (2024) 139:89–139. doi: 10.1016/bs.apcsb.2023.11.008
14. Piroozkhah M, Gholinezhad Y, Piroozkhah M, Shams E, Nazemalhosseini-Mojarad E. The molecular mechanism of actions and clinical utilities of tumor infiltrating lymphocytes in gastrointestinal cancers: a comprehensive review and future prospects toward personalized medicine. Front Immunol. (2023) 14:1298891. doi: 10.3389/fimmu.2023.1298891
15. Regner MJ, Garcia-Recio S, Thennavan A, Wisniewska K, Mendez-Giraldez R, Felsheim B, et al. Defining the regulatory logic of breast cancer using single-cell epigenetic and transcriptome profiling. Cell Genom. (2025) 5:100765. doi: 10.1016/j.xgen.2025.100765
16. Sun H, Zhang C, Zheng Y, Liu C, Wang X, Cong X. Glutamine deficiency promotes recurrence and metastasis in colorectal cancer through enhancing epithelial-mesenchymal transition. J Transl Med. (2022) 20:330. doi: 10.1186/s12967-022-03523-3
17. Tang LY, Spezia M, Chen T, Shin JH, Wang F, Stappenbeck F, et al. Oxysterol derivatives Oxy186 and Oxy210 inhibit WNT signaling in non-small cell lung cancer. Cell Biosci. (2022) 12:119. doi: 10.1186/s13578-022-00857-9
18. Wang FA, Zhuang Z, Gao F, He R, Zhang S, Wang L, et al. TMO-Net: an explainable pretrained multi-omics model for multi-task learning in oncology. Genome Biol. (2024) 25:149. doi: 10.1186/s13059-024-03293-9
19. Xiao S, Lin H, Wang C, Wang S, Rajapakse JC. Graph neural networks with multiple prior knowledge for multi-omics data analysis. IEEE J Biomed Health Inform. (2023) 27:4591–600. doi: 10.1109/JBHI.2023.3284794
20. Xiao Y, Bi M, Guo H, Li M. Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis. EBioMedicine. (2022) 79:104001. doi: 10.1016/j.ebiom.2022.104001
21. Yan W, Chen Y, Hu G, Shi T, Liu X, Li J, et al. MiR-200/183 family-mediated module biomarker for gastric cancer progression: an AI-assisted bioinformatics method with experimental functional survey. J Transl Med. (2023) 21:163. doi: 10.1186/s12967-023-04010-z
22. Yang C, Ou Y, Zhou Q, Liang Y, Li W, Chen Y, et al. Methionine orchestrates the metabolism vulnerability in cisplatin resistant bladder cancer microenvironment. Cell Death Dis. (2023) 14:525. doi: 10.1038/s41419-023-06050-1
23. Yang M, Yang H, Ji L, Hu X, Tian G, Wang B, et al. A multi-omics machine learning framework in predicting the survival of colorectal cancer patients. Comp Biol Med. (2022) 146:105516. doi: 10.1016/j.compbiomed.2022.105516
24. Yang Z, Guan F, Bronk L, Zhao L. Multi-omics approaches for biomarker discovery in predicting the response of esophageal cancer to neoadjuvant therapy: a multidimensional perspective. Pharmacol Ther. (2024) 254:108591. doi: 10.1016/j.pharmthera.2024.108591
25. Ye B, Li Z, Wang Q. A novel artificial intelligence network to assess the prognosis of gastrointestinal cancer to immunotherapy based on genetic mutation features. Front Immunol. (2024) 15:1428529. doi: 10.3389/fimmu.2024.1428529
26. Zafari N, Bathaei P, Velayati M, Khojasteh-Leylakoohi F, Khazaei M, Fiuji H, et al. Integrated analysis of multi-omics data for the discovery of biomarkers and therapeutic targets for colorectal cancer. Comp Biol Med. (2023) 155:106639. doi: 10.1016/j.compbiomed.2023.106639
27. Zhang N, Kandalai S, Zhou X, Hossain F, Zheng Q. Applying multi-omics toward tumor microbiome research. iMeta. (2023) 2:e73. doi: 10.1002/imt2.73
28. Zhang R, Hu M, Chen HN, Wang X, Xia Z, Liu Y, et al. Phenotypic heterogeneity analysis of APC-mutant colon cancer by proteomics and phosphoproteomics identifies RAI14 as a key prognostic determinant in East Asians and Westerners. Mol Cell Proteomics. (2023) 22:100532. doi: 10.1016/j.mcpro.2023.100532
29. Borah K, Das HS, Budhathoki RK, Aurangzeb K, Mallik S. DOMSCNet: a deep learning model for the classification of stomach cancer using multi-layer omics data. Brief Bioinform. (2025) 26:bbaf115. doi: 10.1093/bib/bbaf115
30. Yan Y, Shen S, Li J, Su L, Wang B, Zhang J, et al. Cross-omics strategies and personalised options for lung cancer immunotherapy. Front Immunol. (2024) 15:1471409. doi: 10.3389/fimmu.2024.1471409
31. Minciuna CE, Tanase M, Manuc TE, Tudor S, Herlea V, Dragomir MP, et al. The seen and the unseen: molecular classification and image based-analysis of gastrointestinal cancers. Comput Struct Biotechnol J. (2022) 20:5065–75. doi: 10.1016/j.csbj.2022.09.010
32. Datta J, Bianchi A, De Castro Silva I, Deshpande NU, Cao LL, Mehra S, et al. Distinct mechanisms of innate and adaptive immune regulation underlie poor oncologic outcomes associated with KRAS-TP53 co-alteration in pancreatic cancer. Oncogene. (2022) 41:3640–54. doi: 10.1038/s41388-022-02368-w
33. Pataky RE, Peacock S, Bryan S, Sadatsafavi M, Regier DA. Using genomic heterogeneity to inform therapeutic decisions for metastatic colorectal cancer: an application of the value of heterogeneity framework. Appl Health Econ Health Policy. (2024) 23:441–52. doi: 10.1007/s40258-024-00926-9
34. Takeuchi Y, Yoshida K, Halik A, Kunitz A, Suzuki H, Kakiuchi N, et al. The landscape of genetic aberrations in myxofibrosarcoma. Int J Cancer. (2022) 151:565–77. doi: 10.1002/ijc.34051
35. Yang H, Xiao X, Zeng L, Zeng H, Zheng Y, Wang J, et al. Integrating cfDNA liquid biopsy and organoid-based drug screening reveals PI3K signaling as a promising therapeutic target in colorectal cancer. J Transl Med. (2024) 22:132. doi: 10.1186/s12967-023-04675-6
36. Xie F, Luo S, Liu D, Lu X, Wang M, Liu X, et al. Genomic and transcriptomic landscape of human gastrointestinal stromal tumors. Nat Commun. (2024) 15:9495. doi: 10.1038/s41467-024-53821-1
37. Zhan T, Betge J, Schulte N, Dreikhausen L, Hirth M, Li M, et al. Digestive cancers: mechanisms, therapeutics and management. Signal Trans Target Ther. (2025) 10:24. doi: 10.1038/s41392-024-02097-4
38. Kim DE, Oh HJ, Kim HJ, Kim YB, Kim ST, Yim H. Synergistic two-step inhibition approach using a combination of trametinib and onvansertib in KRAS and TP53-mutated colorectal adenocarcinoma. Biomed Pharmacother. (2025) 182:117796. doi: 10.1016/j.biopha.2024.117796
39. Nadeem U, Drapkin BJ. Atypical small cell lung cancer: a new malignancy characterized by chromothripsis, carcinoid tumors, and wild-type RB1 and TP53. Cancer Discov. (2025) 15:8–10. doi: 10.1158/2159-8290.CD-24-1494
40. White MG, Zeineddine MA, Fallon EA, Zeineddine FA, Dansby J, Chowdhury S, et al. The landscape of ctDNA in appendiceal adenocarcinoma. Clin Cancer Res. (2025) 31:551–60. doi: 10.1158/1078-0432.CCR-24-2474
41. Choi Y, Dharia NV, Jun T, Chang J, Royer-Joo S, Yau KK, et al. Circulating tumor DNA dynamics reveal KRAS G12C mutation heterogeneity and response to treatment with the KRAS G12C inhibitor divarasib in solid tumors. Clin Cancer Res. (2024) 30:3788–97. doi: 10.1158/1078-0432.CCR-24-0255
42. Zhao Y, O’Keefe CM, Hsieh K, Cope L, Joyce SC, Pisanic TR, et al. Multiplex digital methylation-specific PCR for noninvasive screening of lung cancer. Adv Sci. (2023) 10:e2206518. doi: 10.1002/advs.202206518
43. Kolesnikov A, Cook D, Nattestad M, Brambrink L, McNulty B, Gorzynski J, et al. Local read haplotagging enables accurate long-read small variant calling. Nat Commun. (2024) 15:5907. doi: 10.1038/s41467-024-50079-5
44. Zhou S, Xie Y, Feng X, Li Y, Shen L, Chen Y. Artificial intelligence in gastrointestinal cancer research: image learning advances and applications. Cancer Lett. (2025) 614:217555. doi: 10.1016/j.canlet.2025.217555
45. de Back TR, van Hooff SR, Sommeijer DW, Vermeulen L. Transcriptomic subtyping of gastrointestinal malignancies. Trends Cancer. (2024) 10:842–56. doi: 10.1016/j.trecan.2024.06.007
46. Oliver AJ, Huang N, Bartolome-Casado R, Li R, Koplev S, Nilsen HR, et al. Single-cell integration reveals metaplasia in inflammatory gut diseases. Nature. (2024) 635:699–707. doi: 10.1038/s41586-024-07571-1
47. Liu Z, Li L, Liu L, Zhu Z, Yu Y, Zhan S, et al. PDK3 drives colorectal carcinogenesis and immune evasion and is a therapeutic target for boosting immunotherapy. Am J Cancer Res. (2024) 14:3117–29. doi: 10.62347/QWKZ3078
48. Tang Q, Markby GR, MacNair AJ, Tang K, Tkacz M, Parys M, et al. TGF-β-induced PI3K/AKT/mTOR pathway controls myofibroblast differentiation and secretory phenotype of valvular interstitial cells through the modulation of cellular senescence in a naturally occurring in vitro canine model of myxomatous mitral valve disease. Cell Proliferat. (2023) 56:e13435. doi: 10.1111/cpr.13435
49. Zhong C, Xie Z, Zeng LH, Yuan C, Duan S. MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Front Immunol. (2022) 13:855078. doi: 10.3389/fimmu.2022.855078
50. Yu X, Ou J, Wang L, Li Z, Ren Y, Xie L, et al. Gut microbiota modulate CD8+ T cell immunity in gastric cancer through Butyrate/GPR109A/HOPX. Gut Microbes. (2024) 16:2307542. doi: 10.1080/19490976.2024.2307542
51. Chida K, Kawazoe A, Suzuki T, Kawazu M, Ueno T, Takenouchi K, et al. Transcriptomic profiling of MSI-H/dMMR gastrointestinal tumors to identify determinants of responsiveness to anti-PD-1 therapy. Clin Cancer Res. (2022) 28:2110–7. doi: 10.1158/1078-0432.CCR-22-0041
52. Xie Y, Lv Z, Wang Y, Ma J, Wei X, Zheng G, et al. Study on the efficacy of IFN-γ- and sPD-1-overexpressing BMSCs in enhancing immune effects for the treatment of lung adenocarcinoma. Front Immunol. (2025) 16:1554467. doi: 10.3389/fimmu.2025.1554467
53. Wu F, Jiang T, Chen G, Huang Y, Zhou J, Lin L, et al. Multiplexed imaging of tumor immune microenvironmental markers in locally advanced or metastatic non-small-cell lung cancer characterizes the features of response to PD-1 blockade plus chemotherapy. Cancer Commun. (2022) 42:1331–46. doi: 10.1002/cac2.12383
54. Wang X, Lu L, Hong X, Wu L, Yang C, Wang Y, et al. Cell-intrinsic PD-L1 ablation sustains effector CD8+ T cell responses and promotes antitumor T cell therapy. Cell Rep. (2024) 43:113712. doi: 10.1016/j.celrep.2024.113712
55. Sampson J, Ju HM, Zhang N, Yeoh S, Choi J, Bayliss R. Targeting ERBB3 and AKT to overcome adaptive resistance in EML4-ALK-driven non-small cell lung cancer. Cell Death Dis. (2024) 15:912. doi: 10.1038/s41419-024-07272-7
56. Wang Z, Li Y, Yang J, Sun Y, He Y, Wang Y, et al. CircCFL1 promotes TNBC stemness and immunoescape via deacetylation-mediated c-Myc deubiquitylation to facilitate mutant TP53 transcription. Adv Sci. (2024) 11:e2404628. doi: 10.1002/advs.202404628
57. Hsiehchen D, Bucheit L, Yang D, Beg MS, Lim M, Lee SS, et al. Genetic features and therapeutic relevance of emergent circulating tumor DNA alterations in refractory non-colorectal gastrointestinal cancers. Nat Commun. (2022) 13:7477. doi: 10.1038/s41467-022-35144-1
58. Huang CJ, Liu GT, Yeh YC, Chung SY, Chang YC, Chiang NJ, et al. Construction of hot tumor classification models in gastrointestinal cancers. J Transl Med. (2025) 23:218. doi: 10.1186/s12967-025-06230-x
59. Abebe JS, Verstraten R, Depledge DP. Nanopore-based detection of viral RNA modifications. mBio. (2022) 13:e0370221. doi: 10.1128/mbio.03702-21
60. Sun M, Tong Y, Yuan W, Wang Y, Pu Y, Huang W, et al. Proteomic characterization identifies clinically relevant subgroups of gastrointestinal stromal tumors. Gastroenterology. (2024) 166:450–65.e33. doi: 10.1053/j.gastro.2023.11.284
61. Lv D, Li D, Cai Y, Guo J, Chu S, Yu J, et al. CancerProteome: a resource to functionally decipher the proteome landscape in cancer. Nucleic Acids Res. (2024) 52:D1155–62. doi: 10.1093/nar/gkad824
62. Creighton CJ. Clinical proteomics towards multiomics in cancer. Mass Spectr Rev. (2024) 43:1255–69. doi: 10.1002/mas.21827
63. Li Y, Xu C, Wang B, Xu F, Ma F, Qu Y, et al. Proteomic characterization of gastric cancer response to chemotherapy and targeted therapy reveals new therapeutic strategies. Nat Commun. (2022) 13:5723. doi: 10.1038/s41467-022-33282-0
64. Maruyama K, Shimizu Y, Nomura Y, Oh-Hara T, Takahashi Y, Nagayama S, et al. Mechanisms of KRAS inhibitor resistance in KRAS-mutant colorectal cancer harboring Her2 amplification and aberrant KRAS localization. NPJ Precis Oncol. (2025) 9:4. doi: 10.1038/s41698-024-00793-6
65. Chen C, Liu Y, Li Q, Zhang Z, Luo M, Liu Y, et al. The genetic, pharmacogenomic, and immune landscapes associated with protein expression across human cancers. Cancer Res. (2023) 83:3673–80. doi: 10.1158/0008-5472.CAN-23-0758
66. Song WM, Elmas A, Farias R, Xu P, Zhou X, Hopkins B, et al. Multiscale protein networks systematically identify aberrant protein interactions and oncogenic regulators in seven cancer types. J Hematol Oncol. (2023) 16:120. doi: 10.1186/s13045-023-01517-2
67. Fan H, Li X, Li ZW, Zheng NR, Cao LH, Liu ZC, et al. Urine proteomic signatures predicting the progression from premalignancy to malignant gastric cancer. EBioMedicine. (2022) 86:104340. doi: 10.1016/j.ebiom.2022.104340
68. Shang C, Zhang Y, Wang Y, Zhao W, Sun X, Dong X, et al. Role of ITGB2 protein structure and molecular mechanism in precancerous lesions of gastric cancer: influencing the occurrence and development of cancer through the CXCL1-CXCR2 axis. Int J Biol Macromol. (2025) 296:139772. doi: 10.1016/j.ijbiomac.2025.139772
69. Liu YF, Feng ZQ, Chu TH, Yi B, Liu J, Yu H, et al. Andrographolide sensitizes KRAS-mutant colorectal cancer cells to cetuximab by inhibiting the EGFR/AKT and PDGFRβ/AKT signaling pathways. Phytomedicine. (2024) 126:155462. doi: 10.1016/j.phymed.2024.155462
70. Cui M, Cheng C, Zhang L. High-throughput proteomics: a methodological mini-review. Lab Invest. (2022) 102:1170–81. doi: 10.1038/s41374-022-00830-7
71. Ferreira A, Pereira F, Reis C, Oliveira MJ, Sousa MJ, Preto A. Crucial role of oncogenic KRAS mutations in apoptosis and autophagy regulation: therapeutic implications. Cells. (2022) 11:2183. doi: 10.3390/cells11142183
72. Sun Z, Liu L, Chen J. Targeting non-histone methylation in gastrointestinal cancers: from biology to clinic. Eur J Med Chem. (2024) 278:116802. doi: 10.1016/j.ejmech.2024.116802
73. Joshi SK, Piehowski P, Liu T, Gosline SJC, McDermott JE, Druker BJ, et al. Mass spectrometry-based proteogenomics: new therapeutic opportunities for precision medicine. Annu Rev Pharmacol Toxicol. (2024) 64:455–79. doi: 10.1146/annurev-pharmtox-022723-113921
74. Liu S, Locasale JW. Delineating a role for methionine metabolism in colorectal cancer. Cancer Res. (2023) 83:3833–4. doi: 10.1158/0008-5472.CAN-23-3169
75. Shang Z, Ma Z, Wu E, Chen X, Tuo B, Li T, et al. Effect of metabolic reprogramming on the immune microenvironment in gastric cancer. Biomed Pharmacother. (2024) 170:116030. doi: 10.1016/j.biopha.2023.116030
76. Chen Y, Wang B, Zhao Y, Shao X, Wang M, Ma F, et al. Metabolomic machine learning predictor for diagnosis and prognosis of gastric cancer. Nat Commun. (2024) 15:1657. doi: 10.1038/s41467-024-46043-y
77. Cerrato A, Aita SE, Biancolillo A, Laganà A, Marini F, Montone CM, et al. An untargeted analytical workflow based on Kendrick mass defect filtering reveals dysregulations in acylcarnitines in prostate cancer tissue. Anal Chim Acta. (2024) 1307:342574. doi: 10.1016/j.aca.2024.342574
78. Mao T, Qin F, Zhang M, Li J, Li J, Lai M. Elevated serum β-hydroxybutyrate, a circulating ketone metabolite, accelerates colorectal cancer proliferation and metastasis via ACAT1. Oncogene. (2023) 42:1889–99. doi: 10.1038/s41388-023-02700-y
79. Zhao H, Zhao H, Wang J, Ren J, Yao J, Li Y, et al. Bovine omasum-inspired interfacial carbon-based nanocomposite for saliva metabolic screening of gastric cancer. Anal Chem. (2023) 95:11296–305. doi: 10.1021/acs.analchem.3c01358
80. Wang J, Kunzke T, Prade VM, Shen J, Buck A, Feuchtinger A, et al. Spatial metabolomics identifies distinct tumor-specific subtypes in gastric cancer patients. Clin Cancer Res. (2022) 28:2865–77. doi: 10.1158/1078-0432.CCR-21-4383
81. Du Y, Ding X, Ye Y. The spatial multi-omics revolution in cancer therapy: precision redefined. Cell Rep. Medicine. (2024) 5:101740. doi: 10.1016/j.xcrm.2024.101740
82. Liu X, Tao L, Jiang X, Qu X, Duan W, Yu J, et al. Isoporous membrane mediated imprinting mass spectrometry imaging for spatially-resolved metabolomics and rapid histopathological diagnosis. Small Methods. (2024) 8:e2301644. doi: 10.1002/smtd.202301644
83. Chi J, Shu J, Li M, Mudappathi R, Jin Y, Lewis F, et al. Artificial intelligence in metabolomics: a current review. Trends Anal Chem. (2024) 178:117852. doi: 10.1016/j.trac.2024.117852
84. Zhao H, Zhang Z, Liu H, Ma M, Sun P, Zhao Y, et al. Multi-omics perspective: mechanisms of gastrointestinal injury repair. Burns Trauma. (2025) 13:tkae057. doi: 10.1093/burnst/tkae057
85. Zhao G, Wang Y, Wang S, Li N. Comprehensive multi-omics analysis provides biological insights and therapeutic strategies for small-cell lung cancer. MedComm. (2024) 5:e569. doi: 10.1002/mco2.569
86. Turocy T, Crawford JM. Bacterial small molecule metabolites implicated in gastrointestinal cancer development. Nat Rev Microbiol. (2025) 23:106–21. doi: 10.1038/s41579-024-01103-4
87. Zhao J, Zhao X, Yu J, Gao S, Zhang M, Yang T, et al. A multi-platform metabolomics reveals possible biomarkers for the early-stage esophageal squamous cell carcinoma. Anal Chim Acta. (2022) 1220:340038. doi: 10.1016/j.aca.2022.340038
88. Yang S, Wang Z, Liu Y, Zhang X, Zhang H, Wang Z, et al. Dual mass spectrometry imaging and spatial metabolomics to investigate the metabolism and nephrotoxicity of nitidine chloride. J Pharm Anal. (2024) 14:100944. doi: 10.1016/j.jpha.2024.01.012
89. Lim J, Park C, Kim M, Kim H, Kim J, Lee DS. Advances in single-cell omics and multiomics for high-resolution molecular profiling. Exp Mol Med. (2024) 56:515–26. doi: 10.1038/s12276-024-01186-2
90. He J, Shen J, Luo W, Han Z, Xie F, Pang T, et al. Research progress on application of single-cell TCR/BCR sequencing technology to the tumor immune microenvironment, autoimmune diseases, and infectious diseases. Front Immunol. (2022) 13:969808. doi: 10.3389/fimmu.2022.969808
91. Liu X, Peng T, Xu M, Lin S, Hu B, Chu T, et al. Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications. J Hematol Oncol. (2024) 17:72. doi: 10.1186/s13045-024-01596-9
92. Xie B, Gao D, Zhou B, Chen S, Wang L. New discoveries in the field of metabolism by applying single-cell and spatial omics. J Pharm Anal. (2023) 13:711–25. doi: 10.1016/j.jpha.2023.06.002
93. Kiessling P, Kuppe C. Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases. Genome Med. (2024) 16:14. doi: 10.1186/s13073-024-01282-y
94. Wang B, Song B, Li Y, Zhao Q, Tan B. Mapping spatial heterogeneity in gastric cancer microenvironment. Biomed Pharmacother. (2024) 172:116317. doi: 10.1016/j.biopha.2024.116317
95. Ma H, Srivastava S, Ho SWT, Xu C, Lian BSX, Ong X, et al. Spatially resolved tumor ecosystems and cell states in gastric adenocarcinoma progression and evolution. Cancer Discov. (2025) 15:767–92. doi: 10.1158/2159-8290.CD-24-0605
96. Wu X, Yang X, Dai Y, Zhao Z, Zhu J, Guo H, et al. Single-cell sequencing to multi-omics: technologies and applications. Biomark Res. (2024) 12:110. doi: 10.1186/s40364-024-00643-4
97. Chu X, Tian Y, Lv C. Decoding the spatiotemporal heterogeneity of tumor-associated macrophages. Mol. Cancer. (2024) 23:150. doi: 10.1186/s12943-024-02064-1
98. Zhou W, Su M, Jiang T, Yang Q, Sun Q, Xu K, et al. SORC: an integrated spatial omics resource in cancer. Nucleic Acids Res. (2024) 52:D1429–37. doi: 10.1093/nar/gkad820
99. Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. (2023) 24:695–713. doi: 10.1038/s41580-023-00615-w
100. Ma RY, Black A, Qian BZ. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. (2022) 43:546–63. doi: 10.1016/j.it.2022.04.008
101. Pozniak J, Pedri D, Landeloos E, Van Herck Y, Antoranz A, Vanwynsberghe L, et al. A TCF4-dependent gene regulatory network confers resistance to immunotherapy in melanoma. Cell. (2024) 187:166–83.e25. doi: 10.1016/j.cell.2023.11.037
102. Wang X, Almet AA, Nie Q. The promising application of cell-cell interaction analysis in cancer from single-cell and spatial transcriptomics. Semin Cancer Biol. (2023) 95:42–51. doi: 10.1016/j.semcancer.2023.07.001
103. Yu X, Liu R, Gao W, Wang X, Zhang Y. Single-cell omics traces the heterogeneity of prostate cancer cells and the tumor microenvironment. Cell Mol Biol Lett. (2023) 28:38. doi: 10.1186/s11658-023-00450-z
104. Matsushita Y, Noguchi A, Ono W, Ono N. Multi-omics analysis in developmental bone biology. Jpn Dent Sci Rev. (2023) 59:412–20. doi: 10.1016/j.jdsr.2023.10.006
105. Wu X, Yan H, Qiu M, Qu X, Wang J, Xu S, et al. Comprehensive characterization of tumor microenvironment in colorectal cancer via molecular analysis. eLife. (2023) 12:e86032. doi: 10.7554/eLife.86032
106. Fu Y, Tao J, Gu Y, Liu Y, Qiu J, Su D, et al. Multiomics integration reveals NETosis heterogeneity and TLR2 as a prognostic biomarker in pancreatic cancer. NPJ Precis Oncol. (2024) 8:109. doi: 10.1038/s41698-024-00586-x
107. Ge S, Sun S, Xu H, Cheng Q, Ren Z. Deep learning in single-cell and spatial transcriptomics data analysis: advances and challenges from a data science perspective. Brief Bioinform. (2025) 26:bbaf136. doi: 10.1093/bib/bbaf136
108. Liu J, Ma J, Wen J, Zhou X. A cell cycle-aware network for data integration and label transferring of single-cell RNA-Seq and ATAC-Seq. Adv Sci. (2024) 11:e2401815. doi: 10.1002/advs.202401815
109. Conrad T, Altmüller J. Single cell- and spatial ’omics revolutionize physiology. Acta Physiol. (2022) 235:e13848. doi: 10.1111/apha.13848
110. Yue SSK, Tong Y, Siu HC, Ho SL, Law SYK, Tsui WY, et al. Divergent lineage trajectories and genetic landscapes in human gastric intestinal metaplasia organoids associated with early neoplastic progression. Gut. (2025) 74:522–38. doi: 10.1136/gutjnl-2024-332594
111. To A, Yu Z, Sugimura R. Recent advancement in the spatial immuno-oncology. Semin Cell Dev Biol. (2025) 166:22–8. doi: 10.1016/j.semcdb.2024.12.003
112. Baul S, Tanvir Ahmed K, Jiang Q, Wang G, Li Q, Yong J, et al. Integrating spatial transcriptomics and bulk RNA-seq: predicting gene expression with enhanced resolution through graph attention networks. Brief Bioinform. (2024) 25:bbae316. doi: 10.1093/bib/bbae316
113. Fangma Y, Liu M, Liao J, Chen Z, Zheng Y. Dissecting the brain with spatially resolved multi-omics. J Pharm Anal. (2023) 13:694–710. doi: 10.1016/j.jpha.2023.04.003
114. Wu Z, Trevino AE, Wu E, Swanson K, Kim HJ, D’Angio HB, et al. Graph deep learning for the characterization of tumour microenvironments from spatial protein profiles in tissue specimens. Nat Biomed Eng. (2022) 6:1435–48. doi: 10.1038/s41551-022-00951-w
115. Li J, Li L, Liang W, Li L, Wang R, Wang Z, et al. Spatial multi-omics analysis of metabolic heterogeneity in zebrafish exposed to microcystin-LR and its disinfection byproducts. Water Res. (2025) 280:123599. doi: 10.1016/j.watres.2025.123599
116. Hu Y, Jia H, Cui H, Song J. Application of spatial omics in the cardiovascular system. Research. (2025) 8:0628. doi: 10.34133/research.0628
117. Walker CR, Angelo M. Insights and opportunity costs in applying spatial biology to study the tumor microenvironment. Cancer Discov. (2024) 14:707–10. doi: 10.1158/2159-8290.CD-24-0348
118. Wang R, Hastings WJ, Saliba JG, Bao D, Huang Y, Maity S, et al. Applications of nanotechnology for spatial omics: biological structures and functions at nanoscale resolution. ACS Nano. (2025) 19:73–100. doi: 10.1021/acsnano.4c11505
119. Godfrey TM, Shanneik Y, Zhang W, Tran T, Verbeeck N, Patterson NH, et al. Integrating ambient ionization mass spectrometry imaging and spatial transcriptomics on the same cancer tissues to identify gene-metabolite correlations. Angew Chem Int Ed. (2025) 10:e202502028. doi: 10.1002/anie.202502028
120. Tu C, Kulasinghe A, Barbour A, Souza-Fonseca-Guimaraes F. Leveraging spatial omics for the development of precision sarcoma treatments. Trends Pharmacol Sci. (2024) 45:134–44. doi: 10.1016/j.tips.2023.12.006
121. Yang P, Jin K, Yao Y, Jin L, Shao X, Li C, et al. Spatial integration of multi-omics single-cell data with SIMO. Nat. Commun. (2025) 16:1265. doi: 10.1038/s41467-025-56523-4
122. Hui T, Zhou J, Yao M, Xie Y, Zeng H. Advances in spatial omics technologies. Small Methods. (2025) 9:e2401171. doi: 10.1002/smtd.202401171
123. Whittle JR, Kriel J, Fatunla OE, Lu T, Moffet JJD, Spiteri M, et al. Spatial omics shed light on the tumour organisation of glioblastoma. Semin Cell Dev Biol. (2025) 167:1–9. doi: 10.1016/j.semcdb.2024.12.006
124. Zhang Y, Lee RY, Tan CW, Guo X, Yim WW, Lim JC, et al. Spatial omics techniques and data analysis for cancer immunotherapy applications. Curr Opin Biotechnol. (2024) 87:103111. doi: 10.1016/j.copbio.2024.103111
125. Hong J, Jin HJ, Choi MR, Lim DW, Park JE, Kim YS, et al. Matrisomics: beyond the extracellular matrix for unveiling tumor microenvironment. Biochim Biophys Acta Rev Cancer. (2024) 1879:189178. doi: 10.1016/j.bbcan.2024.189178
126. Lancaster SM, Lee-McMullen B, Abbott CW, Quijada JV, Hornburg D, Park H, et al. Global, distinctive, and personal changes in molecular and microbial profiles by specific fibers in humans. Cell Host Microbe. (2022) 30:848–62. doi: 10.1016/j.chom.2022.03.036
127. Yu Z, Lv Y, Su C, Lu W, Zhang R, Li J, et al. Integrative single-cell analysis reveals transcriptional and epigenetic regulatory features of clear cell renal cell carcinoma. Cancer Res. (2023) 83:700–19. doi: 10.1158/0008-5472.CAN-22-2224
128. Pan X, Li X, Sun J, Xiong Z, Hu H, Ning S, et al. Enhancer methylation dynamics drive core transcriptional regulatory circuitry in pan-cancer. Oncogene. (2022) 41:3474–84. doi: 10.1038/s41388-022-02359-x
129. Zhao M, Xue G, He B, Deng J, Wang T, Zhong Y, et al. A multiomics dataset of paired CT image and plasma cell-free DNA end motif for patients with pulmonary nodules. Sci Data. (2025) 12:545. doi: 10.1038/s41597-025-04912-1
130. Bender KJ, Wang Y, Zhai CY, Saenz Z, Wang A, Neumann EK. Sample preparation method for MALDI mass spectrometry imaging of fresh-frozen spines. Anal Chem. (2023) 95:17337–46. doi: 10.1021/acs.analchem.3c03672
131. Xiao N, Liu H, Zhang C, Chen H, Li Y, Yang Y, et al. Applications of single-cell analysis in immunotherapy for lung cancer: current progress, new challenges and expectations. J Adv Res. (2024):462–4. doi: 10.1016/j.jare.2024.10.008
132. Yew WC, Young GR, Nelson A, Cheung W, Stewart CJ, Bridge SH, et al. The core phageome and its interrelationship with preterm human milk lipids. Cell Rep. (2023) 42:113373. doi: 10.1016/j.celrep.2023.113373
133. Hemmati MA, Monemi M, Asli S, Mohammadi S, Foroozanmehr B, Haghmorad D, et al. Using new technologies to analyze gut microbiota and predict cancer risk. Cells. (2024) 13:1987. doi: 10.3390/cells13231987
134. Iş Ö, Min Y, Wang X, Oatman SR, Abraham Daniel A, Ertekin-Taner N. Multi layered omics approaches reveal glia specific alterations in Alzheimer’s disease: a systematic review and future prospects. Glia. (2025) 73:539–73. doi: 10.1002/glia.24652
135. He Y, Ling Y, Zhang Z, Mertens RT, Cao Q, Xu X, et al. Butyrate reverses ferroptosis resistance in colorectal cancer by inducing c-Fos-dependent xCT suppression. Redox Biol. (2023) 65:102822. doi: 10.1016/j.redox.2023.102822
136. Li Z, Shen Y, Xin J, Xu X, Ding Q, Chen W, et al. Cryptotanshinone alleviates radiation-induced lung fibrosis via modulation of gut microbiota and bile acid metabolism. Phytother Res. (2023) 37:4557–71. doi: 10.1002/ptr.7926
137. Chen X, Cao S, Tao L, Yan R, Cao S, Hao J, et al. Establishment of MS LOC platform and its pilot application in clinical lipidomics. Talanta. (2025) 285:127314. doi: 10.1016/j.talanta.2024.127314
138. Turk A, Čeh E, Calin GA, Kunej T. Multiple omics levels of chronic lymphocytic leukemia. Cell Death Discov. (2024) 10:293. doi: 10.1038/s41420-024-02068-2
139. Yu W, Chen G, Yan J, Wang X, Zhu Y, Zhu L. Single-cell sequencing analysis reveals gastric cancer microenvironment cells respond vastly different to oxidative stress. J Transl Med. (2022) 20:250. doi: 10.1186/s12967-022-03411-w
140. Huang J, Tsang WY, Li ZH, Guan XY. The origin, differentiation, and functions of cancer-associated fibroblasts in gastrointestinal cancer. Cell Mol Gastroenterol Hepatol. (2023) 16:503–11. doi: 10.1016/j.jcmgh.2023.07.001
141. Yan M, Andersen TO, Pope PB, Yu Z. Probing the eukaryotic microbes of ruminants with a deep-learning classifier and comprehensive protein databases. Genome Res. (2025) 35:368–78. doi: 10.1101/gr.279825.124
142. Che G, Yin J, Wang W, Luo Y, Chen Y, Yu X, et al. Circumventing drug resistance in gastric cancer: a spatial multi-omics exploration of chemo and immuno-therapeutic response dynamics. Drug Resist Updat. (2024) 74:101080. doi: 10.1016/j.drup.2024.101080
143. Babu M, Snyder M. Multi-omics profiling for health. Mol Cell Proteomics. (2023) 22:100561. doi: 10.1016/j.mcpro.2023.100561
144. Tian X, Ou G, Hu S, Wang C, Han F, Gao L. Integrated network pharmacology and experimental verification to explore the molecular mechanism of Jingxin Zhidong formula for treating Tic disorder. J Ethnopharmacol. (2023) 305:116114. doi: 10.1016/j.jep.2022.116114
145. Gattu AK, Tanzer M, Yaron-Barir TM, Johnson JL, Jayavelu AK, Pan H, et al. Cell-intrinsic insulin signaling defects in human iPS cell-derived hepatocytes in type 2 diabetes. J Clin Invest. (2025) 135:e183513. doi: 10.1172/JCI183513
146. Arias-Hidalgo C, Juanes-Velasco P, Landeira-Viñuela A, García-Vaquero ML, Montalvillo E, Góngora R, et al. Single-cell proteomics: the critical role of nanotechnology. Int J Mol Sci. (2022) 23:6707. doi: 10.3390/ijms23126707
147. Serrano LR, Pelin A, Arrey TN, Damoc NE, Richards AL, Zhou Y, et al. Affinity purification mass spectrometry on the orbitrap-astral mass spectrometer enables high-throughput protein-protein interaction mapping. J Proteome Res. (2025) 24:2006–16. doi: 10.1021/acs.jproteome.4c01040
148. Lores-Padín A, Fernández B, García M, González-Iglesias H, Pereiro R. Real matrix-matched standards for quantitative bioimaging of cytosolic proteins in individual cells using metal nanoclusters as immunoprobes-label: a case study using laser ablation ICP-MS detection. Anal Chim Acta. (2022) 1221:340128. doi: 10.1016/j.aca.2022.340128
149. Li M, Chen Z, Deng S, Wang L, Yu X. MOSDNET: a multi-omics classification framework using simplified multi-view deep discriminant representation learning and dynamic edge GCN with multi-task learning. Comput Biol Med. (2024) 181:109040. doi: 10.1016/j.compbiomed.2024.109040
150. Zhong C, Xie T, Chen L, Zhong X, Li X, Cai X, et al. Immune depletion of the methylated phenotype of colon cancer is closely related to resistance to immune checkpoint inhibitors. Front Immunol. (2022) 13:983636. doi: 10.3389/fimmu.2022.983636
151. Bao Y, Chen Y, Lin L, Li J, Liu X, Wang G, et al. GutUDB: a comprehensive multiomics database for intestinal diseases. iMeta. (2024) 3:e195. doi: 10.1002/imt2.195
152. Lv T, Zhang Y, Liu J, Kang Q, Liu L. Multi-omics integration for both single-cell and spatially resolved data based on dual-path graph attention auto-encoder. Brief Bioinform. (2024) 25:bbae450. doi: 10.1093/bib/bbae450
153. Zhang SL, Cheng LS, Zhang ZY, Sun HT, Li JJ. Untangling determinants of gut microbiota and tumor immunologic status through a multi-omics approach in colorectal cancer. Pharmacol Res. (2023) 188:106633. doi: 10.1016/j.phrs.2022.106633
154. Zhao P, Feng Y, Wu J, Zhu J, Yang J, Ma X, et al. Efficient Sample Preparation System for Multi-Omics Analysis via Single Cell Mass Spectrometry. Anal Chem. (2023) 95:7212–9. doi: 10.1021/acs.analchem.2c05728
155. Palcau AC, Pulito C, De Pascale V, Casadei L, Valerio M, Sacconi A, et al. CircPVT1 weakens miR-33a-5p unleashing the c-MYC/GLS1 metabolic axis in breast cancer. J Exp Clin Cancer Res. (2025) 44:100. doi: 10.1186/s13046-025-03355-1
156. Sharma A, Debik J, Naume B, Ohnstad HO, Oslo Breast Cancer Consortium (Osbreac), Bathen TF, et al. Comprehensive multi-omics analysis of breast cancer reveals distinct long-term prognostic subtypes. Oncogenesis. (2024) 13:22. doi: 10.1038/s41389-024-00521-6
157. Massimino L, Barchi A, Mandarino FV, Spanò S, Lamparelli LA, Vespa E, et al. A multi-omic analysis reveals the esophageal dysbiosis as the predominant trait of eosinophilic esophagitis. J Transl Med. (2023) 21:46. doi: 10.1186/s12967-023-03898-x
158. Lee M, Kim PJ, Joe H, Kim HG. Gene-centric multi-omics integration with convolutional encoders for cancer drug response prediction. Comput Biol Med. (2022) 151:106192. doi: 10.1016/j.compbiomed.2022.106192
159. He X, Liu X, Zuo F, Shi H, Jing J. Artificial intelligence-based multi-omics analysis fuels cancer precision medicine. Semin Cancer Biol. (2023) 88:187–200. doi: 10.1016/j.semcancer.2022.12.009
160. Gao H, Zhang B, Liu L, Li S, Gao X, Yu B. A universal framework for single-cell multi-omics data integration with graph convolutional networks. Brief Bioinform. (2023) 24:bbad081. doi: 10.1093/bib/bbad081
161. Corti C, Cobanaj M, Dee EC, Criscitiello C, Tolaney SM, Celi LA, et al. Artificial intelligence in cancer research and precision medicine: applications, limitations and priorities to drive transformation in the delivery of equitable and unbiased care. Cancer Treat Rev. (2023) 112:102498. doi: 10.1016/j.ctrv.2022.102498
162. Cui K, Yao S, Liu B, Sun S, Gong L, Li Q, et al. A novel high-risk subpopulation identified by CTSL and ZBTB7B in gastric cancer. Br J Cancer. (2022) 127:1450–60. doi: 10.1038/s41416-022-01936-x
163. Xing W, Li M, Liu Y, Liu X, Li Y, Yang Y, et al. Multi-omics graph knowledge representation for pneumonia prognostic prediction. IEEE J Biomed Health Inform. (2025) 29:3021–34. doi: 10.1109/JBHI.2024.3488735
164. Strudwick J, Gardiner LJ, Denning-James K, Haiminen N, Evans A, Kelly J, et al. AutoXAI4Omics: an automated explainable AI tool for omics and tabular data. Brief Bioinform. (2024) 26:bbae593. doi: 10.1093/bib/bbae593
165. Li J, Li L, You P, Wei Y, Xu B. Towards artificial intelligence to multi-omics characterization of tumor heterogeneity in esophageal cancer. Semin Cancer Biol. (2023) 91:35–49. doi: 10.1016/j.semcancer.2023.02.009
166. Rozera T, Pasolli E, Segata N, Ianiro G. Machine learning and artificial intelligence in the multi-omics approach to gut microbiota. Gastroenterology. (2025). doi: 10.1053/j.gastro.2025.02.035
167. Li Z, Katz S, Saccenti E, Fardo DW, Claes P, Martins Dos, et al. Novel multi-omics deconfounding variational autoencoders can obtain meaningful disease subtyping. Brief Bioinform. (2024) 25:bbae512. doi: 10.1093/bib/bbae512
168. Wei Y, Li L, Zhao X, Yang H, Sa J, Cao H, et al. Cancer subtyping with heterogeneous multi-omics data via hierarchical multi-kernel learning. Brief Bioinform. (2023) 24:bbac488. doi: 10.1093/bib/bbac488
169. Li G, Zhou Z, Wang Z, Wang Z. Assessing Epstein-Barr virus in gastric cancer: clinicopathological features and prognostic implications. Infect Agent Cancer. (2023) 18:11. doi: 10.1186/s13027-023-00489-9
170. Athaya T, Ripan RC, Li X, Hu H. Multimodal deep learning approaches for single-cell multi-omics data integration. Brief Bioinform. (2023) 24:bbad313.
171. Singhania D, Rahaman R, Yao A. C2F-TCN: a framework for semi- and fully-supervised temporal action segmentation. IEEE Trans Pattern Anal Mach Intell. (2023) 45:11484–501. doi: 10.1109/TPAMI.2023.3284080
172. Fu Y, Wang J, Liu C, Liao K, Gao X, Tang R, et al. Glycogen synthase kinase 3 controls T-cell exhaustion by regulating NFAT activation. Cell Mol Immunol. (2023) 20:1127–39. doi: 10.1038/s41423-023-01075-0
173. Anteghini M, Gualdi F, Oliva B. How did we get there? AI applications to biological networks and sequences. Comput Biol Med. (2025) 190:110064. doi: 10.1016/j.compbiomed.2025.110064
174. Hao D, Li Q, Feng QX, Qi L, Liu XS, Arefan D, et al. SurvivalCNN: a deep learning-based method for gastric cancer survival prediction using radiological imaging data and clinicopathological variables. Artif Intell Med. (2022) 134: 102424. doi: 10.1016/j.artmed.2022.102424
175. Feng K, Zeng H, Zhao Y, Kong SG, Bu Y. Unsupervised spectral demosaicing with lightweight spectral attention networks. IEEE Trans Image Process. (2024) 33:1655–69. doi: 10.1109/TIP.2024.3364064
176. Shen WX, Liu Y, Chen Y, Zeng X, Tan Y, Jiang YY, et al. AggMapNet: enhanced and explainable low-sample omics deep learning with feature-aggregated multi-channel networks. Nucleic Acids Res. (2022) 50:e45. doi: 10.1093/nar/gkac010
177. Iacucci M, Santacroce G, Yasuharu M, Ghosh S. Artificial intelligence-driven personalized medicine: transforming clinical practice in inflammatory bowel disease. Gastroenterology. (2025) 10:S0016-5085(25)00494-9. doi: 10.1053/j.gastro.2025.03.005
178. Lin M, Guo J, Gu Z, Tang W, Tao H, You S, et al. Machine learning and multi-omics integration: advancing cardiovascular translational research and clinical practice. J Transl Med. (2025) 23:388. doi: 10.1186/s12967-025-06425-2
179. Ricciuti B, Wang X, Alessi JV, Rizvi H, Mahadevan NR, Li YY, et al. Association of high tumor mutation burden in non-small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol. (2022) 8:1160–8. doi: 10.1001/jamaoncol.2022.1981
180. Mok JH, Joo M, Cho S, Duong VA, Song H, Park JM, et al. Optimizing MS-based multi-omics: comparative analysis of protein, metabolite, and lipid extraction techniques. Metabolites. (2024) 14:34. doi: 10.3390/metabo14010034
181. Zhang J, Ren H, Jiang Z, Chen Z, Yang Z, Matsubara Y, et al. Strategic multi-omics data integration via multi-level feature contrasting and matching. IEEE Trans Nanobiosci. (2024) 23:579–90. doi: 10.1109/TNB.2024.3456797
182. Hernández-Lemus E, Ochoa S. Methods for multi-omic data integration in cancer research. Front Genet. (2024) 15:1425456. doi: 10.3389/fgene.2024.1425456
183. Yang X, Mann KK, Wu H, Ding J. scCross: a deep generative model for unifying single-cell multi-omics with seamless integration, cross-modal generation, and in silico exploration. Genome Biol. (2024) 25:198. doi: 10.1186/s13059-024-03338-z
184. Chiesa S, Mangraviti A, Martini M, Cenci T, Mazzarella C, Gaudino S, et al. Clinical and NGS predictors of response to regorafenib in recurrent glioblastoma. Sci Rep. (2022) 12:16265. doi: 10.1038/s41598-022-20417-y
185. Miao BB, Dong W, Gu YX, Han ZF, Luo X, Ke CH, et al. OmicsSuite: a customized and pipelined suite for analysis and visualization of multi-omics big data. Hortic Res. (2023) 10:uhad195. doi: 10.1093/hr/uhad195
186. Mangas-Florencio L, Herrero-Gómez A, Eills J, Azagra M, Batlló-Rius M, Marco-Rius I. A DIY bioreactor for in situ metabolic tracking in 3D cell models via hyperpolarized 13C NMR spectroscopy. Anal Chem. (2025) 97:1594–602. doi: 10.1021/acs.analchem.4c04183
187. Luchkina E, Waxman S. Talking about the absent and the abstract: referential communication in language and gesture. Perspect Psychol Sci. (2024) 19:978–92.
188. Yu C, Meng Z, Zhang W, Lei L, Ni J, Zhang K, et al. Secure and efficient federated learning against model poisoning attacks in horizontal and vertical data partitioning. IEEE Trans Neural Netw Learn Syst. (2025) 36:10913–27. doi: 10.1109/TNNLS.2024.3486028
189. Mani S, Lalani SR, Pammi M. Genomics and multiomics in the age of precision medicine. Pediatr Res. (2025) 97:1399–410. doi: 10.1038/s41390-025-04021-0
190. Yang T, Zhang N, Yang N. Single-cell sequencing in diabetic retinopathy: progress and prospects. J Transl Med. (2025) 23:49. doi: 10.1186/s12967-024-06066-x
191. Matsuoka T, Yashiro M. Bioinformatics analysis and validation of potential markers associated with prediction and prognosis of gastric cancer. Int J Mol Sci. (2024) 25:5880. doi: 10.3390/ijms25115880
192. Tamiya A, Osuga M, Harada D, Isa SI, Taniguchi Y, Nakamura K, et al. Mechanisms of resistance and correlation between pre-treatment co-alterations and prognosis to osimertinib in chemo-naïve advanced non-small cell lung cancer. Lung Cancer. (2024) 195:107917. doi: 10.1016/j.lungcan.2024.107917
193. Liu X, Mei W, Zhang P, Zeng C. PIK3CA mutation as an acquired resistance driver to EGFR-TKIs in non-small cell lung cancer: clinical challenges and opportunities. Pharmacol Res. (2024) 202:107123. doi: 10.1016/j.phrs.2024.107123
194. Malmin A, Thomseth VM, Førland PT, Khan AZ, Hetland HB, Chen X, et al. Associations between serial intravitreal injections and dry eye. Ophthalmology. (2023) 130:509–15. doi: 10.1016/j.ophtha.2023.01.009
195. Yang W, Zhang M, Zhang TX, Liu JH, Hao MW, Yan X, et al. YAP/TAZ mediates resistance to KRAS inhibitors through inhibiting proapoptosis and activating the SLC7A5/mTOR axis. JCI Insight. (2024) 9:e178535. doi: 10.1172/jci.insight.178535
196. Sato K, Faraji F, Cervantes-Villagrana RD, Wu X, Koshizuka K, Ishikawa T, et al. Targeting YAP/TAZ-TEAD signaling as a therapeutic approach in head and neck squamous cell carcinoma. Cancer Lett. (2025) 612:217467.
197. Li T, Gu Z, Zhou G. Expanding the dimensions of single-cell multi-omics to include mitochondrial DNA through fixation and permeabilization. Anal Chem. (2025) 97:5908–13. doi: 10.1021/acs.analchem.4c06777
198. Milite S, Caravagna G, Sottoriva A. MIDAA: deep archetypal analysis for interpretable multi-omic data integration based on biological principles. Genome Biol. (2025) 26:90. doi: 10.1186/s13059-025-03530-9
199. Ivanisevic T, Sewduth RN. Multi-omics integration for the design of novel therapies and the identification of novel biomarkers. Proteomes. (2023) 11:34. doi: 10.3390/proteomes11040034
200. Wang Q, He M, Guo L, Chai H. AFEI: adaptive optimized vertical federated learning for heterogeneous multi-omics data integration. Brief Bioinform. (2023) 24:bbad269. doi: 10.1093/bib/bbad269
Keywords: gastrointestinal tumors, multi-omics technologies, single-cell genomics, early screening, biomarkers, treatment optimization
Citation: Liu Y, Gao F, Cheng Y, Qi L and Yu H (2025) Applications and advances of multi-omics technologies in gastrointestinal tumors. Front. Med. 12:1630788. doi: 10.3389/fmed.2025.1630788
Received: 18 May 2025; Accepted: 07 July 2025;
Published: 23 July 2025.
Edited by:
Dongxu Wang, Qilu Hospital, Shandong University, ChinaReviewed by:
Junyu Long, Peking Union Medical College Hospital (CAMS), ChinaJinyang Huang, Hefei University of Technology, China
Qinghe Wu, National University of Singapore, Singapore
Copyright © 2025 Liu, Gao, Cheng, Qi and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haining Yu, eXVoYWluaW5nQHNkZm11LmVkdS5jbg==
†These authors share first authorship
 Yuqing Liu
Yuqing Liu Feng Gao†
Feng Gao† Yang Cheng
Yang Cheng