- 1Department of Veterinary Internal Medicine, Tottori University, Tottori, Japan
- 2Laboratory of Veterinary Microbiology, Nippon Veterinary and Life Science University, Tokyo, Japan
- 3Laboratory of Food Microbiology and Food Safety, Rakuno Gakuen University, Hokkaido, Japan
- 4Miyamoto Animal Hospital, Yamaguchi, Japan
- 5Sanritsu Zelkova Veterinary Laboratory, Kanagawa, Japan
- 6Monoris Co., Ltd., Tokyo, Japan
The emergence of antimicrobial resistance in Klebsiella spp., including resistance to extended-spectrum cephalosporins (ESC) and fluoroquinolones, is of great concern in both human and veterinary medicine. In this study, we investigated the prevalence of antimicrobial resistance in a total of 103 Klebsiella spp. isolates, consisting of Klebsiella pneumoniae complex (KP, n = 89) and K. oxytoca (KO, n = 14) from clinical specimens of dogs and cats in Japan. Furthermore, we characterized the resistance mechanisms, including extended-spectrum β-lactamase (ESBL), plasmid-mediated AmpC β-lactamase (PABL), and plasmid-mediated quinolone resistance (PMQR); and assessed genetic relatedness of ESC-resistant Klebsiella spp. strains by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). Antimicrobial susceptibility testing demonstrated that resistance rates to ampicillin, cephalothin, enrofloxacin, ciprofloxacin, trimethoprim/sulfamethoxazole, cefotaxime, gentamicin, tetracycline, chloramphenicol, amoxicillin-clavulanic acid, and cefmetazole were 98.1, 37.9, 37.9, 35.9, 35.0, 34.0, 31.1, 30.1, 28.2, 14.6, and 6.8%, respectively. Phenotypic testing detected ESBLs and/or AmpC β-lactamases in 31 of 89 (34.8%) KP isolates, but not in KO isolates. Resistances to 5 of the 12 antimicrobials tested, as well as the three PMQRs [qnrB, qnrS, and aac(6′)-Ib-cr], were detected significantly more frequently in ESBL-producing KP, than in non-ESBL-producing KP and KO. The most frequent ESBL was CTX-M-15 (n = 13), followed by CTX-M-14 (n = 7), CTX-M-55 (n = 6), SHV-2 (n = 5), CTX-M-2 (n = 2), and CTX-M-3 (n = 2). Based on the rpoB phylogeny, all ESBL-producing strains were identified as K. pneumoniae, except for one CTX-M-14-producing strain, which was identified as K. quasipneumoniae. All of AmpC β-lactamase positive isolates (n = 6) harbored DHA-1, one of the PABLs. Based on MLST and PFGE analysis, ST15 KP clones producing CTX-M-2, CTX-M-15, CTX-M-55, and/or SHV-2, as well as KP clones of ST1844-CTX-M-55, ST655-CTX-M-14, and ST307-CTX-M-15, were detected in one or several hospitals. Surprisingly, specific clones were detected in different patients at an interval of many months. These results suggest that multidrug-resistant ESBL-producing KP were clonally disseminated among companion animals via not only direct but also indirect transmission. This is the first report on large-scale monitoring of antimicrobial-resistant Klebsiella spp. isolates from companion animals in Japan.
Introduction
Members of the genus Klebsiella, belonging to the Enterobacteriaceae, are gram-negative bacilli that inhabit freshwater environments including surface water, sewage and soil, as well as the mucosal surfaces of mammals (Podschun and Ullmann, 1998). Klebsiella pneumoniae is the most medically-important species in the genus and is responsible, together with K. oxytoca (KO), for nosocomial infections in humans (Podschun and Ullmann, 1998). Strains classically identified as K. pneumoniae have been previously subdivided into phylogroups named KpI, KpII, and KpIII (Brisse et al., 2014). Recently, it has been proposed that these phylogroups be redesignated as distinct species, K. pneumoniae (KpI), K. quasipneumoniae (KpII), and K. variicola (KpIII) (Holt et al., 2015), which are collectively called K. pneumoniae complex (KP). In companion animals, Klebsiella spp. have been demonstrated to cause infections such as urinary tract infections (Ling et al., 2001), pyometra (Stone et al., 1988), upper respiratory infections (Adler et al., 2007), and septicemia (Roberts et al., 2000).
The emergence of antimicrobial resistance in Klebsiella spp. isolates is of great concern worldwide in human medicine (Lynch et al., 2013). It increases the risk of antimicrobial treatment failure not only in humans but also in companion animals. Similarly, the emergence of antimicrobial-resistant bacteria in companion animals may have important human public health consequences if isolates are transmitted to humans by their pets (Guardabassi et al., 2004; Lloyd, 2007). Understanding the prevalence of antimicrobial resistance among Klebsiella spp. isolates is thus important both from veterinary medicine and public health perspectives.
Resistance to extended-spectrum cephalosporins (ESC) and fluoroquinolones in gram-negative bacteria, including Klebsiella spp., is of particular concern (Paterson, 2006). ESC resistance is mainly associated with the production of plasmid-mediated extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases (PABLs), and carbapenemases (Kenneth, 2010; Rubin and Pitout, 2014). Although fluoroquinolone resistance is mainly acquired by modification of their target enzymes, it may also involve acquisition of plasmid-mediated quinolone resistance (PMQR) determinants (Fàbrega et al., 2005; Guillard et al., 2015). PMQRs determine relatively small increases in quinolone resistance, but these changes are sufficient to mediate natural selection of mutants that have higher levels of resistance (Strahilevitz et al., 2009). In recent years, these important resistance mechanisms in Klebsiella spp. isolates from companion animals have been well documented in several European countries, including Germany (Stolle et al., 2013; Ewers et al., 2014), Italy (Donati et al., 2014), France (Haenni et al., 2012; Poirel et al., 2013), Spain (Hidalgo et al., 2013), and Switzerland (Wohlwend et al., 2015). However, the status of emerging antimicrobial resistance in Klebsiella spp. in companion animals remains unknown in many other countries, including Japan.
The aim of the present study was to investigate the prevalence of antimicrobial resistance, and provide molecular characterization of ESC resistance and PMQRs in Klebsiella spp. isolates from clinical specimens from dogs and cats visited to different veterinary hospitals throughout Japan. A further aim was to assess the epidemiological relatedness of ESC-resistant Klebsiella spp. strains.
Materials and Methods
Bacterial Isolates
A total of 103 Klebsiella spp. isolates, consisting of 89 KP and 14 KO, were obtained from clinical specimens collected from dogs (n = 78) and cats (n = 25) that visited veterinary hospitals between 2003 and 2015. These hospitals were located at the following 15 prefectures in Japan: Hokkaido, Fukui, Gunma, Ibaraki, Saitama, Tokyo, Chiba, Kanagawa, Nagano, Aichi, Osaka, Hyogo, Tottori, Yamaguchi, and Fukuoka prefectures. The specimens were isolated from various anatomical sites, assessed as being sites of bacterial infection by clinical veterinarians, including the urinary tract (n = 48), skin (n = 11), genitals (n = 9), and respiratory organs (n = 9), ears (n = 7), and digestive organs (n = 6); as well as pus from unspecified locations (n = 8), and body fluids including ascites (n = 4). One specimen was of unknown anatomical origin. The details of Klebsiella spp. isolates used in this study are shown in Supplementary Table 1. No information was available regarding previous antimicrobial treatment of the dogs and cats. This study was carried out in accordance with the recommendations of Guidelines for Proper Conduct of Animal Experiments, Science Council of Japan. All enrolled animals received the best practice veterinary care, and the owners granted informed consent. Bacterial identification was conducted by growth status on CHROMagar orientation medium (Ohkusu, 2000), using the API 20E kit (SYSMEX bioMérieux Co., Ltd., Tokyo, Japan). Because KP and KO often present similar biochemical patterns (Chander et al., 2011), the species-specific PCR was further carried out for confirmation of bacterial species, as previously reported (Chander et al., 2011). All confirmed Klebsiella spp. isolates were stored at −80°C in 10% skim milk.
Antimicrobial Susceptibility Testing
Susceptibilities to ampicillin (AMP), amoxicillin-clavulanic acid (ACV), cephalothin (CPL), cefmetazole (CMZ), cefotaxime (CTX), meropenem (MPM), tetracycline (TET), gentamicin (GEN), chloramphenicol (CHL), trimethoprim/sulfamethoxazole (TMS), ciprofloxacin (CIP), and enrofloxacin (ENR) were determined. Susceptibility testing was conducted using the agar dilution method, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2013a). The results obtained were interpreted as per the criteria contained within CLSI guidelines (CLSI, 2013b,c). Escherichia coli ATCC 25922 was used as a control strain.
ESC-resistant (i.e., CTX MIC ≥ 2 μg/mL) strains were screened for production of ESBLs and AmpC-type β-lactamases using a commercial combination disk kit (MAST Diagnostics, Co. Ltd., UK), which can separately detect the two type of β-lactamases by comparing the inhibition zones of the cefpodoxime (CPD) disk to the inhibition zones of each of the CPD plus inhibitor (i.e., ESBL inhibitor, AmpC inhibitor, or both) disks.
Detection of PMQR Genes
Genomic DNA from each of the isolates was prepared by suspending several colonies in 0.5 ml of water and boiling for 10 min. These samples were used as templates for further genetic analyses. All isolates were screened for eight PMQR genes [i.e., qnrA, qnrB, qnrC, qnrD, qnrS, qepA, aac(6′)-Ib-cr, and oqxAB genes] using multiplex PCR (Ciesielczuk et al., 2013). Positive genes were confirmed subsequently by single PCR. Randomly selected PCR products of oqxAB and all of the products of the other PMQRs were directionally sequenced with the same primers, for confirmation.
Characterization of β-Lactamase Genes in ESC-Resistant Klebsiella spp. Strains
All of the ESC-resistant strains were screened for class A β-lactamase genes (i.e., blaTEM and blaSHV), which were identified using PCR and DNA sequencing, as previously reported (Kojima et al., 2005). In the isolates with ambiguous sequences of blaSHV, indicating that they have both chromosomal and plasmid blaSHV genes (Haanperä et al., 2008), the two blaSHV genes were separately identified, as previously described (Lee et al., 2006). In ESBL-positive strains, the CTX-M-type β-lactamase genes were detected using multiplex PCR (Xu et al., 2007); for the positive isolates, the genes were amplified and sequenced to identify CTX-M subtypes using group-specific PCR primers (Kojima et al., 2005; Shibata et al., 2006). In AmpC-positive strains, PABL genes (i.e., ACC, FOX, MOX, DHA, CIT, and EBC groups) were screened by multiplex PCR (Pérez-Pérez and Hanson, 2002), amplified and then bi-directionally sequenced using specific primers (Yan et al., 2002).
Multilocus Sequence Typing and Phylogenetic Analysis of ESC-Resistant Klebsiella spp. Strains
For ESC-resistant KP strains, multilocus sequence typing (MLST) with seven genes (i.e., gapA, infB, mdh, pgi, phoE, rpoB, and tonB) was carried out according to the protocol on the Institut Pasteur website (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). New alleles and STs were submitted to the MLST website and new ST numbers were assigned. eBURST v3 analysis (http://eburst.mlst.net/v3/instructions/) was performed to identify clonal complexes (CCs), defined as groups of two or more independent isolates sharing identical alleles at six or more loci (Ewers et al., 2014). Following Breurec et al. (2013), we further split the STs into clonal groups (CGs) mapping within the large central CC.
In addition, the phylogenetic relationship of all ESC-resistant strains was investigated based on the rpoB gene sequence (Brisse et al., 2014).
Pulsed-Field Gel Electrophoresis of ESBL-Producing Klebsiella spp. Strains
Pulsed-field gel electrophoresis (PFGE) was performed on ESBL-producing KP (ESBL-KP) strains, as previously described (Herschleb et al., 2007). DNA embedded in agarose was digested with XbaI (Takara Bio, Inc., Japan) and then electrophoresed using CHEF DRIII (Bio-Rad, Hercules, CA, USA). PFGE profiles were digitized for analysis using BioNumerics software (version 5.10; Applied Maths, TX, USA). All fragment sizes within the gel were normalized using the molecular weight method. A similarity matrix was calculated using the Dice coefficient, and cluster analysis was performed using the UPGMA algorithm. A cluster was defined based on a similarity cut-off of 80% with 1.0% optimization and 1.0% band tolerance.
Statistical Analysis
Prevalence of antimicrobial resistance and PMQR genes between the three groups (i.e., ESBL-KP and non-ESBL-KP, and KO strains) was compared using the Fisher's exact test. A P value lower than 0.05 was considered significant. The Bonferroni correction for multiple comparisons was applied, lowering the threshold for significance to a value of P ≤ 0.0167.
Results
Antimicrobial Susceptibilities of Klebsiella spp. Isolates
The numbers of isolates with resistance to AMP, CPL, ENR, CIP, TMS, CTX, GEN, TET, CHL, ACV, and CMZ were 101 (98.1%), 39 (37.9%), 39 (37.9%), 37 (35.9%), 36 (35.0%), 35 (34.0%), 32 (31.1%), 31 (30.1%), 29 (28.2%), 15 (14.6%), and 7 (6.8%), respectively, in 103 Klebsiella spp. clinical isolates (Supplementary Table 1). All of the isolates tested had low MIC for MEM (≤0.031 to 0.063 μg/mL).
Based on MIC of CTX, ESC resistance was detected in 35 KP isolates but not in KO isolates. The combination disk screening test revealed that 31 of 35 (88.6%) ESC-resistant KP strains produced ESBLs with or without AmpC β-lactamases, whereas the remaining four isolates produced AmpC β-lactamases but not ESBLs. Resistances against TET, GEN, TMS, CIP, and ENR, as well as cephalosporins including CPL and CTX, were detected significantly more frequently in ESBL-KP strains, than in non-ESBL-KP and KO strains (P < 0.0167, Table 1).
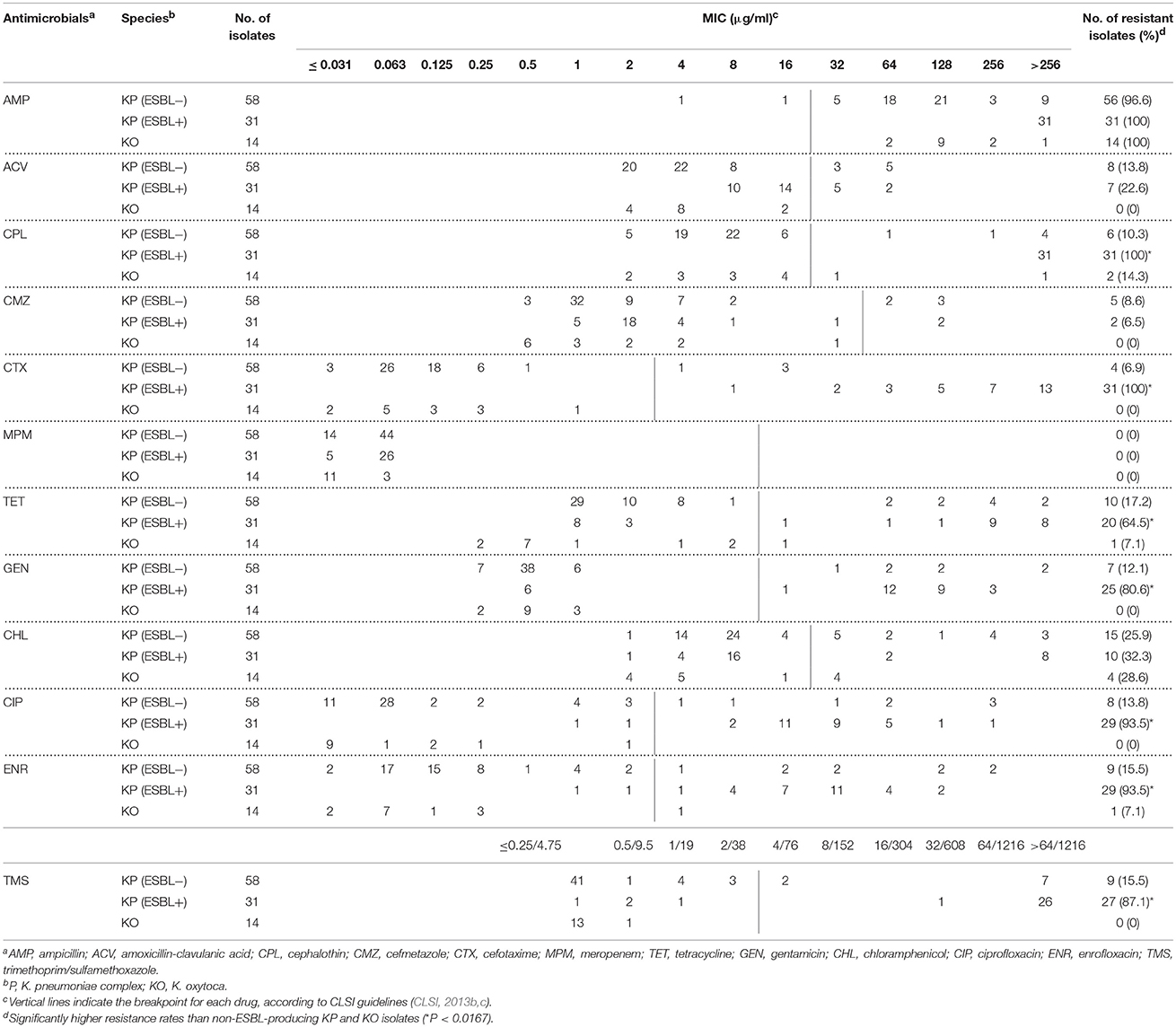
Table 1. MIC distribution and resistance rates among Klebsiella spp. isolates from dogs and cats in Japan.
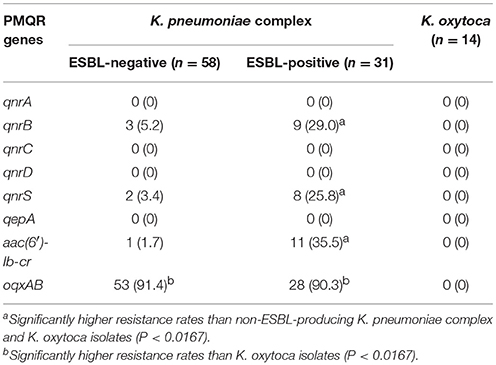
Prevalence of PMQR Genes in Klebsiella spp. Isolates
Of the eight PMQRs tested, qnrB, qnrS, aac(6′)-Ib-cr, and oqxAB genes were detected in KP clinical isolates, whereas none of the PMQR genes were detected in KO isolates (Table 2). The qnrB, qnrS, and aac(6′)-Ib-cr genes were significantly more prevalent in ESBL-KP strains than in non-ESBL-KP strains (respectively 29.0 vs. 5.2, 25.8 vs. 3.4, and 35.5 vs. 1.7%, P < 0.0167). However, the oqxAB gene was detected in approximately 90% of isolates of both ESBL-KP and non-ESBL-KP strains: there was no significant difference in prevalence of the gene between the strains (P > 0.05).
Prevalence of β-Lactamases among ESC-Resistant Klebsiella spp. Strains
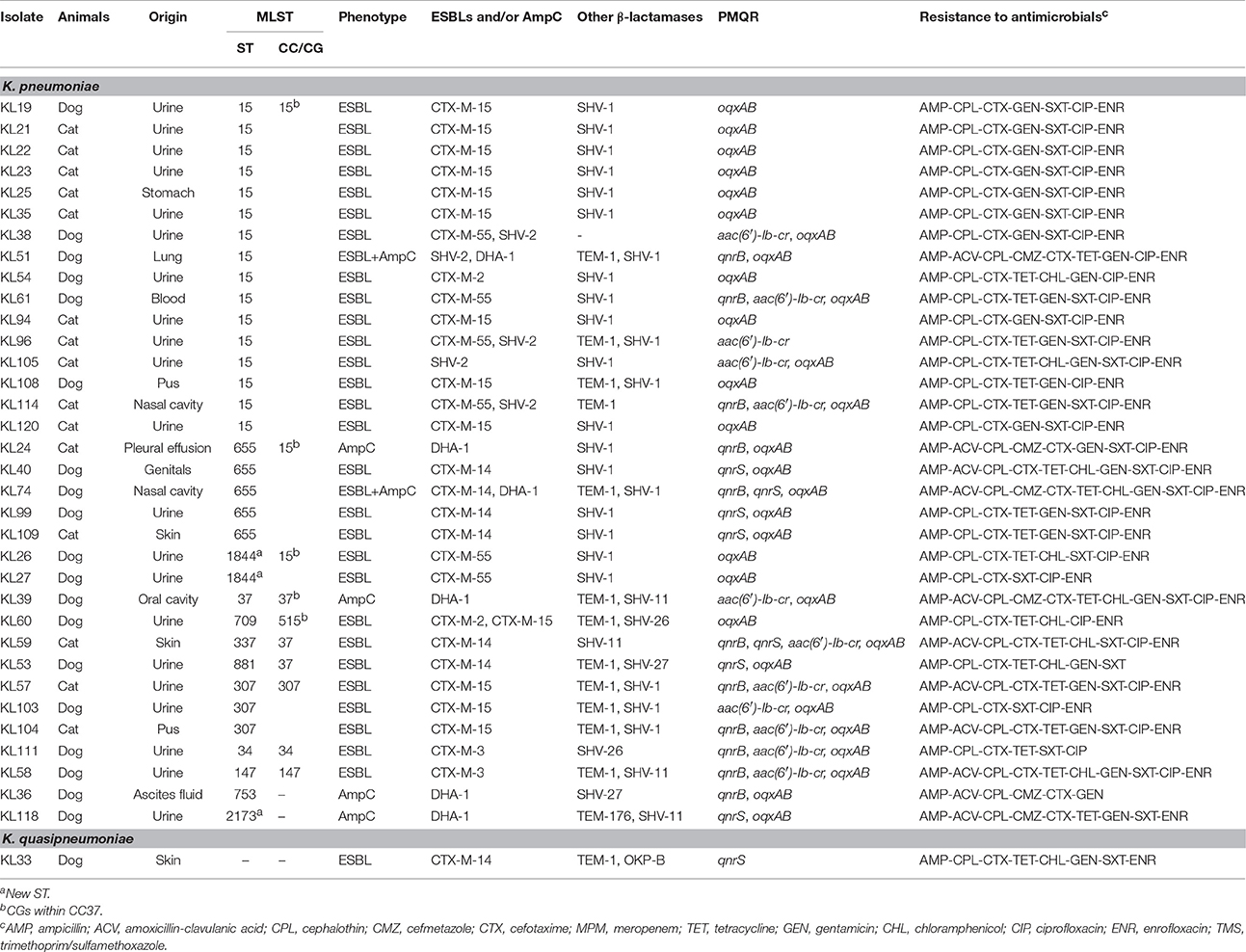
Table 3 shows the detailed characteristics of 35 ESC-resistant clinical strains. Twenty-seven and four ESC-resistant strains harbored one and two ESBL genes, respectively. Of the ESBL genes detected in this study, CTX-M-15 was the most prevalent (n = 13), followed by CTX-M-14 (n = 7), CTX-M-55 (n = 6), SHV-2 (n = 5), CTX-M-2 (n = 2), and CTX-M-3 (n = 2). All of the six AmpC-positive strains (n = 6), two of which were also positive for ESBLs, harbored DHA-1, one of the PABLs. As for β-lactamases other than ESBLs and PABLs, SHV-1 and TEM-1 were prevalent (n = 24 and 13, respectively); other minor β-lactamases including SHV-11, SHV-26, SHV-27, OKP-B, and TEM-176 were also detected (n = 4, 2, 2, 1, and 1, respectively). Three ESC-resistant strains harbored two types of SHV genes (i.e., SHV-1 and SHV-2).
Phylogenetic Relationship of ESC-Resistant Klebsiella spp. Strains
The rpoB sequences of 35 ESC-resistant KP clinical strains were compared with those of seven type strains of Klebsiella genus (Supplementary Figure 1). The sequence of rpoB allele 1, which was identified in 29 strains, differed from those of the three type strains (K. pneumoniae subsp. pneumoniae ATCC 13883, K. pneumoniae subsp. ozaenae ATCC 11296, and K. pneumoniae sub sp. rhinoscleromatis CIP52-210) by 1 nucleotide (99.8% similarity). The sequence of rpoB allele 4, which was identified in four strains, was identical with those of K. pneumoniae subsp. pneumoniae ATCC 13883 and K. pneumoniae subsp. ozaenae ATCC 11296, whereas the rpoB sequence of the KL111 strain (allele 7) differed from these two subspecies by 1 nucleotide (99.8% similarity). The rpoB sequence of the KL33 strain (allele 13) was identical with that of K. quasipneumoniae subsp. similipneumoniae 07A044T.
As the result of phylogenetic analysis, all ESC-resistant strains fell within the K. pneumoniae cluster (KpI), except for the KL33 strain, which fell within the K. quasipneumoniae cluster (KpII).
MLST Typing of ESC-Resistant Klebsiella spp. Strains
As shown in Table 3, 34 ESC-resistant K. pneumoniae clinical isolates investigated by MLST were assigned to 13 Sequence Types (STs): ST15 (allelic profile 1-1-1-1-1-1-1, n = 16), ST655 (1-1-1-1-1-1-23, n = 5), ST307 (4-1-2-52-1-1-7, n = 3), ST1844 (1-1-1-1-1-1-297, n = 2), ST34 (2-3-6-1-9-7-4, n = 1), ST37 (2-9-2-1-13-1-16, n = 1), ST147 (3-4-6-1-7-4-38, n = 1), ST337 (2-1-11-1-1-1-13, n = 1), ST709 (1-1-1-1-1-1-4, n = 1), ST753 (14-1-2-1-135-4-12, n = 1), ST881 (2-68-1-1-10-4-13, n = 1), and ST2173 (4-6-19-13-1-4-22, n = 1). Figure 1 illustrates a population snapshot by eBURST analysis for our collection, against 2250 previously-reported STs obtained from the MLST database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html: accessed on 4 February 2016). Seven of the thirteen STs generated in our collection were placed into CC37. In this CC, ST15 and its single locus variants (SLVs, ST655 and ST1844) were included in CG15; ST37 and ST709 were included in CG37 and CG515, respectively. Of non-CC37 STs, ST34, ST147, and ST307 were defined as predicted founders of their respective CCs.
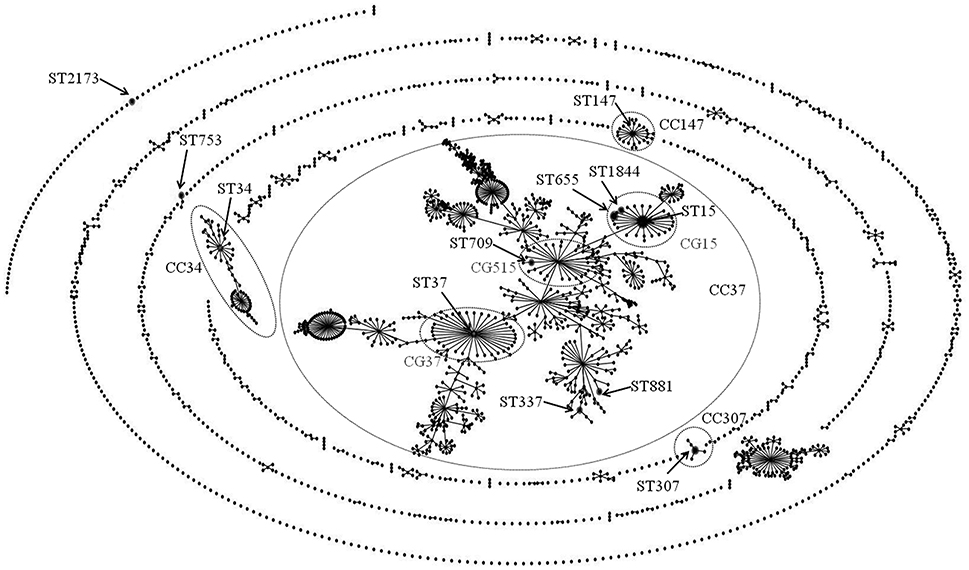
Figure 1. Population snapshot by eBURST analysis for ESC-resistant K. pneumoniae strains against the entire K. pneumoniae MLST database. The STs identified in this study are labeled with arrows. The names of the CCs are based on the ST assigned as the founder genotype. The relative size of the circles indicates the prevalence of STs and lines between STs connect SLVs. The straggly CC in the center of the picture was further separated into CGs, characterized as subsets of this complex (Breurec et al., 2013).
PFGE Analysis of ESBL-Producing Klebsiella spp. Strains
In the PFGE analysis, ESBL-KP clinical strains formed five distinct clusters (Figure 2). Cluster I consisted of ST15 strains producing CTX-M-2, CTX-M-55, and/or SHV-2 obtained from the same or different veterinary hospitals in Tokyo and the neighboring prefectures (Supplementary Figure 2), except for the KL38 strain. Clusters II and III contained two ST1844-CTX-M-55 strains and four ST655-CTX-M-15 strains, respectively, obtained from several hospitals located mainly in Tokyo prefecture. Cluster IV consisted of eight ST15-CTX-M-15 strains obtained from two hospitals located in Aichi prefecture. Cluster V contained two ST307-CTX-M-15 strains obtained from different prefectures. Clusters I, III, and IV each contained strains detected at an interval of several months. The remaining strains were independent of the clusters.
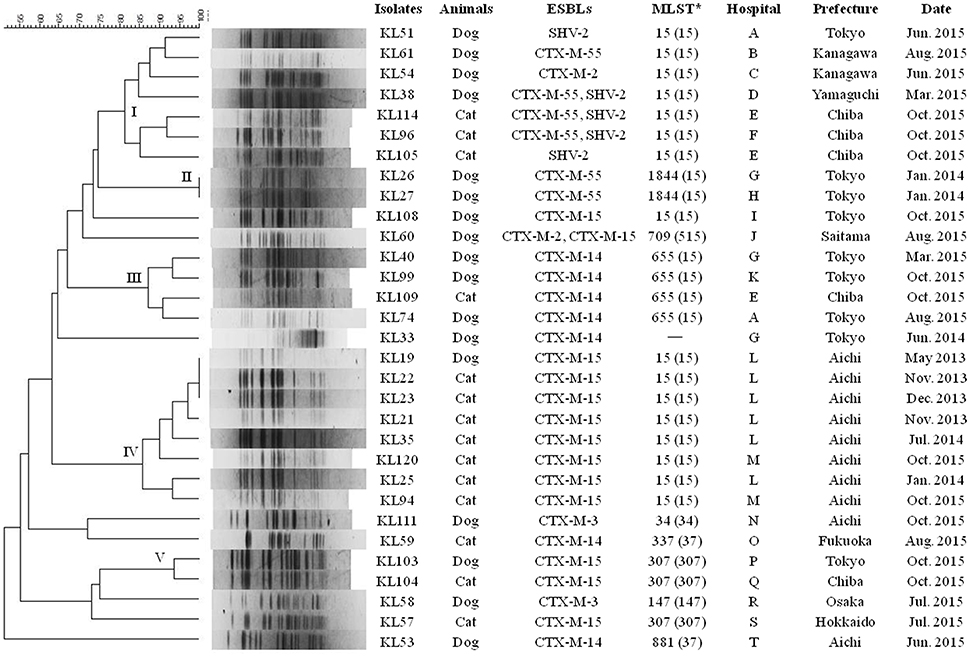
Figure 2. PFGE profiles of 31 ESBL-producing Klebsiella spp. strains from dogs and cats in Japan. The numbers embedded in the phylogenetic tree mean clusters. *The CG or CC of each ST is given in parenthesis. ST was not assigned to KL33 strain because this strain was identified as K. quasipneumoniae.
Discussion
There have been few reports on the prevalence of antimicrobial resistance in overall populations of Klebsiella spp. clinical isolates from companion animals, although numerous investigations focusing on the strains with resistance to cephalosporins and/or fluoroquinolones have been carried out. This study demonstrated that more than 30% of Klebsiella spp. isolates in our collection exhibited resistance to CPL, CTX, TET, GEN, TMS, CIP, or ENR, in addition to extremely high prevalence of AMP resistance, possibly due to chromosomal β-lactamases (Hæggman et al., 2004). On the other hand, all of the Klebsiella spp. isolates exhibited low MICs against MEM (≤0.063 μg/ml), which were much lower than the screening cut-offs for the detection of carbapenemases proposed by both CLSI (2013c) and European Committee on Antimicrobial Susceptibility Testing (Hrabák et al., 2014). The resistance rates in our collection were higher than those of KP as human respiratory pathogen in Japan, in which resistance rates against 35 of 36 tested antimicrobials were less than 10% (Yanagihara et al., 2015). Likewise, lower resistance rates (i.e., less than 20% in most tested antimicrobials) were found in 17 German canine and feline Klebsiella spp. isolates (Grobbel et al., 2007). We have previously found similar inter-country differences in prevalence of antimicrobial resistance in other pathogens from companion animals (Harada et al., 2012a,b, 2014). Systematic inter-country evaluation of Klebsiella spp. isolates from companion animals would provide a better understanding of country-specific trends in antimicrobial resistance.
We found higher prevalence of ESBLs in KP clinical isolates (31/89, 34.8%), compared with those from companion animals in Italy (15/70, 21.4%, Donati et al., 2014), and in Germany and other European countries (84/1112, 7.6%, Ewers et al., 2014). In addition, this carriage rate of ESBLs was extremely higher than that in human isolates in Japan (8/484, 1.6%, Sato et al., 2015). These data suggest that the risk of ESBL carriage is relatively high in KP clinical isolates from companion animals in Japan. In this study, ESBL-KP strains frequently exhibited resistance against not only cephalosporins but also the other classes of antimicrobials. This finding indicates a strong inclination toward multidrug resistance in ESBL-KP, similar to KP human isolates (Hyle et al., 2005). This high prevalence of multidrug resistance may contribute to the selection and persistence of ESBL-KP in clinical settings (Coque et al., 2008), and should be taken into account when treating companion animals with KP infections. Genetic analysis of β-lactamases revealed extremely high prevalence of CTX-M-type β-lactamases, especially CTX-M-15, in ESC-resistant KP clinical strains, similar to previous studies (Donati et al., 2014; Ewers et al., 2014). CTX-M-3 was also identified in this study, which has been reported in KO isolates in France (Ewers et al., 2014). As for ESBLs detected in KP human strains in Japan, CTX-M-14 was predominant, whereas CTX-M-15 was in the minority (Nagasaka et al., 2015). To the best of our knowledge, CTX-M-2, CTX-M-14, and CTX-M-55 were first detected in Klebsiella spp. isolates from companion animals.
As for β-lactamases other than ESBLs, all of the identified AmpC-type β-lactamases were DHA-1 in ESC-resistant KP clinical strains, in accordance with previous reports of KP isolates from humans (Nagasaka et al., 2015). Likewise, Wohlwend et al. (2015) have reported that DHA-1 is mainly responsible for ESC resistance in KP isolates from companion animals in a veterinary hospital in Switzerland. SHV-type β-lactamases, including one kind of ESBL (i.e., SHV-2), were detected in nearly all of the ESC-resistant KP strains. Furthermore, OKP-type β-lactamase, closely related to SHV-type, was firstly identified in Klebsiella spp. isolates from companion animals. The OKP-type enzymes have been specifically found in the phylogenetic group KpII of K. pneumoniae (Hæggman et al., 2004; Fevre et al., 2005), namely, K. quasipneumoniae. In fact, the OKP-B-positive strain (KL33) was identified as K. quasipneumoniae based on the rpoB phylogeny. It might be necessary to investigate the significance of K. quasipneumoniae as pathogen and antimicrobial resistance reservoirs in companion animals in the future.
The MLST analysis revealed that ST15 was the most common among ESBL-KP clinical strains from companion animals in Japan. In particular, ST15 clone was identified more frequently in cats than in dogs, in contrast to the other STs, indicating the spread of ST15 clone among cats has a role in its prevalence. Previous studies in Italy and France have found that ST101 and ST274 clones were predominant in ESBL-KP from companion animals (Poirel et al., 2013; Donati et al., 2014). These data indicate that the predominant ST in ESBL-KP varies by country. In this study, we found predominance of ST15-CTX-M-15, a well-known international clone of KP (Ewers et al., 2014), which was also detected in France (Haenni et al., 2012) and Germany (Ewers et al., 2014). Based on the PFGE analysis, this clone was disseminated among different patients visiting the same hospital (cluster IV). This result strongly suggests nosocomial infections of ESBL-KP clone, and similar findings have previously been reported (Haenni et al., 2012; Poirel et al., 2013; Ewers et al., 2014). The PFGE analysis also identified the other ST15 clone (cluster I). This clone consisted of strains that produce various ESBLs and were obtained from several hospitals. Thus, we identified not only intra- but also inter-hospital dissemination of ST15 ESBL-KP clones among companion animals. In addition to ST15 clone, we found that several ESBL-KP clones are spread among different hospitals, most of which were located in the same or neighboring prefecture. Surprisingly, several clones were repeatedly identified at an interval of many months, implying that ESBL-KP clones can be maintained inside or outside of hospitals for a long period. Therefore, the dissemination of ESBL-KP clones among companion animals may occur not only via direct spread from animal to animal, but also via indirect transmission from potential reservoirs and sources in the environment, as often seen in human medicine (Hendrik et al., 2015). Our data emphasizes the need for infection control in hospitals and in the community to prevent dissemination of ESBL-KP clones among companion animals.
Nagasaka et al. (2015) carried out MLST analysis for ESC-resistant KP strains from human patients in Japan. They reported a high prevalence of CG11, CG17, and CG37 with a minority of the other STs. In our collection, however, CG15, an international clonal group (Breurec et al., 2013), was predominant. On other hand, we confirmed the presence of several STs in common with human isolates (i.e., ST37 and ST147) as a minor population. These findings suggest that transmission of ESC-resistant KP between companion animals and humans may occur but is relatively uncommon in Japan. Poirel et al. (2013) have shown that CTX-M-15 genes in KP isolates from companion animals were located on a different plasmid from that of human isolates, in France, and indicated that these KP isolates evolved separately from the human reservoir. In contrast, the German study by Ewers et al. (2014) indicated that ST15-CTX-M-15 KP strains were shared by humans and animals. Hidalgo et al. (2013) firstly identified IncR plasmids, which have often been associated with human isolates, in KP isolates from dogs and cats. Further studies would be needed to clarify whether animals can act as a reservoir of antimicrobial-resistant KP, including ESBL-KP, for humans.
Of the detected PMQRs in this study, oqxAB is detected in nearly all of KP clinical isolates but not in KO isolates. This result can be explained by the fact that the PMQR is a chromosomally-encoded gene in KP (Yuan et al., 2012). As for other PMQRs, aac(6′)-Ib-cr, qnrB, and qnrS were relatively prevalent in our collection, and similar findings were confirmed in ESBL-KP isolates from human in Japan (Nagasaka et al., 2015). In contrast, the latter two PMQRs have hardly been detected in ESBL-KP isolates from companion animals in European countries (Donati et al., 2014; Ewers et al., 2014). These findings may suggest local spread of PMQRs among companion animals and humans in Japan. Furthermore, these PMQRs were more prevalent in ESBL-KP strains than non-ESBL-KP and KO isolates. This implies an epidemiological link between genes of PMQRs and ESBLs (Cantón and Coque, 2006; Strahilevitz et al., 2009). Such a link may explain the higher rates of fluoroquinolone resistance in ESBL-KP clinical strains, compared with non-ESBL-KP strains, as seen in this study.
In conclusion, we carried out the first large-scale monitoring of antimicrobial resistance in Klebsiella spp. clinical isolates from companion animals in Japan. Our data demonstrated a high prevalence of multidrug-resistant ESBL-KP strains, most of which harbored PMQRs. In addition, ESBL-KP strains were predominantly identified as to the ST15-CTX-M-15 clone, and scarcely contained human-related ST clone. Epidemiological data may suggest that ESBL-KP isolates are disseminated clonally, via intrahospital and interhospital transmission. Overall, Klebsiella spp. isolates in our collection exhibited higher rates of antimicrobial resistance and ESBL carriage than human clinical isolates in Japan, but the similarity of STs and ESBL types between isolates from human and companion animals were less common. We strongly believe that ESBL-KP poses a serious threat of antimicrobial resistance to companion animal medicine, especially in Japan, although the public health risk of ESBL-KP isolates from companion animals remains to be assessed.
Author Contributions
KH designed and coordinated the study, performed genetic analysis including detection of resistance genes and MLST analysis, and wrote the draft of manuscript. KH, TS, YM, and KK carried out the identification and antimicrobial susceptibility testing of Klebsiella spp. TS and MU conducted PFGE analysis. YK (MAH), TM, YT, AO, and YK (NVLU) participated in the collection of Klebsiella spp. isolates. All authors read and approved the final manuscript.
Funding
This work was financially supported by JSPS KAKENHI Grant Number 24780299.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Noriyuki Kitadai (DS Pharma Animal Health Co., Ltd.) for providing Klebsiella spp. isolates from companion animals. We thank the team of curators of the Institut Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr/.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01021
Supplementary Table 1. The details of Klebsiella spp. isolates used in this study. aAMP, ampicillin; ACV, amoxicillin-clavulanic acid; CPL, cephalothin; CMZ, cefmetazole; CTX, cefotaxime; MPM, meropenem; TET, tetracycline; GEN, gentamicin; CHL, chloramphenicol; TMS, trimethoprim/sulfamethoxazole; CIP, ciprofloxacin; ENR, enrofloxacin. R, resistant.
Supplementary Figure 1. Phylogenetic relationship based on the rpoB gene sequence. The tree was obtained with the neighbor-joining method with Kimura's two-parameter distance. Strain name is indicated for each sequence. The values at the nodes correspond to the bootstrap values obtained after 1000 replicates (Brisse et al., 2014).
Supplementary Figure 2. Geographical distribution of 31 ESBL-producing Klebsiella spp. strains detected in this study in Japan. Bold type denotes prefecture names. The numbers of strains are shown in parentheses.
References
Adler, K., Radeloff, I., Stephan, B., Greife, H., and Hellmann, K. (2007). Bacteriological and virological status in upper respiratory tract infections of cats (cat common cold complex). Berl. Münch. Tierärztl. 120, 120–125.
Breurec, S., Guessennd, N., and Timinouni, M. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major town: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
Brisse, S., Passet, V., and Grimont, P. A. (2014). Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 64, 3146–3152. doi: 10.1099/ijs.0.062737-0
Cantón, R., and Coque, T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Chander, Y., Ramakrishnan, M. A., Jindal, N., Hanson, K., and Goyal, S. M. (2011). Differentiation of Klebsiella pneumoniae and K. oxytoca by multiplex polymerase chain reaction. Intern. J. App. Res. Vet. Med. 9, 138–142.
Ciesielczuk, H., Hornsey, M., Choi, V., Woodford, N., and Wareham, D. W. (2013). Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 62, 1823–1827. doi: 10.1099/jmm.0.064428-0
Clinical Laboratory Standards Institute (CLSI) (2013a). Performance Standards for Antimicrobial Disk and Dilution Suscptibility Tests for Bacteria Isolated From Animals. Approved Standard-Fourth Edn. CLSI document VET01-A4, Wayne, PA, USA.
Clinical Laboratory Standards Institute (CLSI) (2013b). Performance Standards for Antimicrobial Disk and Dilution Suscptibility Tests for Bacteria Isolated From Animals. Second Informational Supplement. CLSI document VET01-S2, Wayne, PA, USA.
Clinical Laboratory Standards Institute (CLSI) (2013c). Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. CLSI document M100-S20, Wayne, PA, USA.
Coque, T. M., Baquero, F., and Cantón, R. (2008). Increasing prevalence of ESBL-producing enterobacteriaceae in Europe. Euro. Surveill. 13, 573–583.
Donati, V., Feltrin, F., Hendriksen, R. S., Svendsen, C. A., Cordaro, G., García-Fernández, A., et al. (2014). Extended-spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistance in Klebsiella spp. from companion animals in Italy. PLoS ONE 9:e90564. doi: 10.1371/journal.pone.0090564
Ewers, C., Stamm, I., Pfeifer, Y., Wieler, L. H., Kopp, P. A., Schønning, K., et al. (2014). Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J. Antimicrob. Chemother. 69, 2676–2680. doi: 10.1093/jac/dku217
Fàbrega, A., Madurga, S., Giralt, E., and Vila, J. (2005). Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2, 40–61. doi: 10.1111/j.1751-7915.2008.00063.x
Fevre, C., Passet, V., Weill, F. X., Grimont, P. A., and Brisse, S. (2005). Variants of the Klebsiella pneumoniae OKP chromosomal beta-lactamase are divided into two main groups, OKP-A and OKP-B. Antimicrob. Agents Chemother. 49, 5149–5152. doi: 10.1128/AAC.49.12.5149-5152.2005
Grobbel, M., Lübke-Becker, A., Alesík, E., Schwarz, S., Wallmann, J., Werckenthin, C., et al. (2007). Antimicrobial susceptiblity of Klebsiella spp. and Proteus spp. from various organ systems of horses, dogs and cats as determined in the BfT-GermVet monitoring program 2004-2006. Berl. Münch. Tierärztl. Wochenschr. 120, 402–411.
Guardabassi, L., Schwarz, S., and Lloyd, D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54, 321–332. doi: 10.1093/jac/dkh332
Guillard, T., de Jong, A., Limelette, A., Lebreil, A. L., Madoux, J., and de Champs, C. (2015). Characterization of quinolone resistance mechanisms in Enterobacteriaceae recovered from diseased companion animals in Europe. Vet. Microbiol. doi: 10.1016/j.vetmic.2015.11.033. [Epub ahead of print].
Haanperä, M., Forssten, S. D., Huovinen, P., and Jalava, J. (2008). Typing of SHV extended-spectrum β-lactamases by pyrosequencing in Klebsiella pneumoniae strains with chromosomal SHV β-lactamase. Antimicrob. Agents Chemother. 52, 2632–2635. doi: 10.1128/AAC.01259-07
Hæggman, S., Löfdahl, S., Paauw, A., Verhoef, J., and Brisse, S. (2004). Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 2400–2408. doi: 10.1128/AAC.48.7.2400-2408.2004
Haenni, M., Ponsin, C., Métayer, V., Médaille, C., and Madec, J. (2012). Veterianary hospital-acquired infections in pets with a ciprofloxacain-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. J. Antimicrob. Chemother. 67, 770–771. doi: 10.1093/jac/dkr527
Harada, K., Arima, S., Niina, A., Kataoka, Y., and Takahashi, T. (2012a). Characterization of Pseudomonas aeruginosa isolates from dogs and cats in Japan: current status of antimicrobial resistance and prevailing resistance mechanisms. Microbiol. Immunol. 56, 123–127. doi: 10.1111/j.1348-0421.2011.00416.x
Harada, K., Niina, A., Nakai, Y., Kataoka, Y., and Takahashi, T. (2012b). Prevalence of antimicrobial resistance in relation to virulence genes and phylogenetic origins among urogenital Escherichia coli isolates from dogs and cats in Japan. Am. J. Vet. Res. 73, 409–417. doi: 10.2460/ajvr.73.3.409
Harada, K., Niina, A., Shimizu, T., Mukai, Y., Kuwajima, K., Miyamoto, T., et al. (2014). Phenotypic and molecular characterization of antimicrobial resistance in Proteus mirabilis isolates from dogs. J. Med. Microbiol. 63, 1561–1567. doi: 10.1099/jmm.0.081539-0
Hendrik, T. C., Voor in't Holt, A. F., and Vos, M. C. (2015). Cilnical and molecular epidemiology of extended-spectrum beta-lactamase-producing Klebsiella spp.: a systematic review and meta-analyses. PLoS ONE 10:e0140754. doi: 10.1371/journal.pone.0140754
Herschleb, J., Ananiev, G., and Schwartz, D. C. (2007). Pulsed-field gel electrophoresis. Nat. Protoc. 2, 677–684. doi: 10.1038/nprot.2007.94
Hidalgo, L., Gutierrez, B., Ovejero, C. M., Carrilero, L., Matrat, S., Saba, C. K., et al. (2013). Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and Qnr4. Antimicrob. Agents Chemother. 57, 4532–4534. doi: 10.1128/AAC.00491-13
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Hrabák, J., Chudáèková, E., and Papagiannitsis, C. C. (2014). Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin. Microbiol. Infect. 20, 839–853. doi: 10.1111/1469-0691.12678
Hyle, E. P., Lipworth, A. D., Zaoutis, T. E., Nachamkin, I., Fishman, N. O., Bilker, W. B., et al. (2005). Risk factors for increasing multidrug resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella species. Clin. Infect. Dis. 40, 1317–1324. doi: 10.1086/429239
Kenneth, S. T. (2010). Extended-spectrum β-lactamases, AmpC, and carbapenemase issues. J. Clin. Microbiol. 48, 1019–1025. doi: 10.1128/JCM.00219-10
Kojima, A., Ishii, Y., Ishihara, K., Esaki, H., Asai, T., Oda, C., et al. (2005). Extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49, 3533–3537. doi: 10.1128/AAC.49.8.3533-3537.2005
Lee, Y. H., Cho, B., Bae, I. K., Chang, C. L., and Jeong, S. H. (2006). Klebsiella pneumoniae strains carrying the chromosomal SHV-11 β-lactamase gene produce the plasmid-mediated SHV-12 extended-spectrum β-lactamase more frequently than those carrying the chromosomal SHV-1 β-lactamase gene. J. Antimicrob. Chemother. 57, 1259–1261. doi: 10.1093/jac/dkl115
Ling, G. V., Norris, C. R., Franti, C. E., Eisele, P. H., Johnson, D. L., Ruby, A. L., et al. (2001). Interrelations of organism prevalence, specimen collection method, and host age, sex, and breed among 8,354 canine urinary tract infections (1969-1995). J. Vet. Intern. Med. 15, 341–347.
Lloyd, D. H. (2007). Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 45(Suppl. 2), S148–S152. doi: 10.1086/519254
Lynch, J. P. III, Clark, N. M., and Zhanel, G. G. (2013). Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended-spectrum β-lactamases and carbapenemases). Expert Opin. Pharmacother. 14, 199–210. doi: 10.1517/14656566.2013.763030
Nagasaka, Y., Kimura, K., Yamada, K., Wachino, J., Jin, W., Notake, S., et al. (2015). Genetic profiles of fluoroquinolone-nonsusceptible Klebsiella pneumoniae among cephalosporin-resistant K. pneumoniae. Microb. Drug Resist. 21, 224–233. doi: 10.1089/mdr.2014.0150
Ohkusu, K. (2000). Cost-effective and rapid presumptive identification of gram-negative bacilli in routine urine, pus and stool culture: evaluation of the use of CHROMagar orientation medium in conjunction with biochemical tests. J. Clin. Microbiol. 38, 4586–4592.
Paterson, D. L. (2006). Resistance in gram-negative bacteria: enterobacteriaceae. Am. J. Infect. Control. 34(5 Suppl. 1), S20–S28. doi: 10.1016/j.ajic.2006.05.238
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex-PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epideimiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Poirel, L., Nordmann, P., Ducroz, S., Boulouis, H. J., Arné, P., and Millemann, Y. (2013). Extended-spectrum β-lactamase CTX-M-15-producing Klebsiella pneumoniae of sequence type ST274 in companion animals. Antimicrob. Agents Chemother. 57, 2372–2375. doi: 10.1128/AAC.02622-12
Roberts, D. E., McClain, H. M., Hansen, D. S., Currin, P., and Howerth, E. W. (2000). An outbreak of Klebsiella pneumoniae infection in dogs with severe enteritis and septicaemia. J. Vet. Diagn. Invest. 12, 168–173. doi: 10.1177/104063870001200215
Rubin, J. E., and Pitout, J. D. (2014). Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet. Microbiol. 170, 10–18. doi: 10.1016/j.vetmic.2014.01.017
Sato, T., Hara, T., Horiyama, T., Kanazawa, S., Yamaguchi, T., and Maki, H. (2015). Mechanism of resistance and antibacterial susceptibiilty in extended-spectrum β-lactamase phenotype Klebsiella pneumoniae and Klebsiella oxytoca isolated between 2000 and 2010 in Japan. J. Med. Microbiol. 64, 538–543. doi: 10.1099/jmm.0.000057
Shibata, N., Kurokawa, H., Doi, Y., Yagi, T., Yamane, K., Wachino, J., et al. (2006). PCR classification of CTX-M-type β-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob. Agents Chemother. 50, 791–795. doi: 10.1128/AAC.50.2.791-795.2006
Stolle, I., Prenger-Berninghoff, E., Stamm, I., Scheufen, S., Hassdenteufel, E., Guenther, S., et al. (2013). Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 68, 2802–2808. doi: 10.1093/jac/dkt259
Stone, E. A., Littman, M. P., Robertson, J. L., and Bovée, K. C. (1988). Renal dysfunction in dogs with pyometra. J. Am. Vet. Med. Assoc. 193, 457–464.
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Wohlwend, N., Endimiani, A., Francey, T., and Perreten, V. (2015). Third-generation-cephalosporin-resistant Klebsiella pneumoniae isolates from humans and companion animals in Switzerland: spread of a DHA-producing sequence type 11 clone in a veterinary setting. Antimicrob. Agents Chemother. 59, 2949–2955. doi: 10.1128/AAC.04408-14
Xu, L., Ensor, V., Gossain, S., Nye, K., and Hawkey, P. (2007). Rapid and simple detection of blaCTX−M genes by multiplex PCR assay. J. Med. Microbiol. 54, 1183–1187. doi: 10.1099/jmm.0.46160-0
Yan, J. J., Ko, W. C., Jung, Y. C., Chuang, C. L., and Wu, J. J. (2002). Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 β-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40, 3121–3126. doi: 10.1128/JCM.40.9.3121-3126.2002
Yanagihara, K., Kadota, J., Aoki, N., Matsumoto, T., Yoshida, M., Yagisawa, M., et al. (2015). Nationwide surveillance of bacterial respiratory pathogens conducted by the surveillance committee of Japanese Society of Chemotherapy, the Japanese Association for Infectious Diseases, and the Japanese Society for Clinical Microbiology in 2010: general view of the pathogens' antibacterial susceptibility. J. Infect. Chemother. 21, 410–420. doi: 10.1016/j.jiac.2015.02.008
Keywords: Klebsiella spp., dogs, cats, extended-spectrum β-lactamases, multidrug resistance, clonal dissemination
Citation: Harada K, Shimizu T, Mukai Y, Kuwajima K, Sato T, Usui M, Tamura Y, Kimura Y, Miyamoto T, Tsuyuki Y, Ohki A and Kataoka Y (2016) Phenotypic and Molecular Characterization of Antimicrobial Resistance in Klebsiella spp. Isolates from Companion Animals in Japan: Clonal Dissemination of Multidrug-Resistant Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae. Front. Microbiol. 7:1021. doi: 10.3389/fmicb.2016.01021
Received: 22 February 2016; Accepted: 15 June 2016;
Published: 29 June 2016.
Edited by:
Teresa M. Coque, Hospital Universitario Ramón y Cajal, SpainReviewed by:
Angela Novais, Faculdade de Farmácia da Universidade do Porto, PortugalBruno Gonzalez-Zorn, Universidad Compultense de Madrid, Spain
Copyright © 2016 Harada, Shimizu, Mukai, Kuwajima, Sato, Usui, Tamura, Kimura, Miyamoto, Tsuyuki, Ohki and Kataoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuki Harada, ay1oYXJhZGFAbXVzZXMudG90dG9yaS11LmFjLmpw
 Kazuki Harada
Kazuki Harada Takae Shimizu1
Takae Shimizu1 Tomomi Sato
Tomomi Sato Masaru Usui
Masaru Usui Asami Ohki
Asami Ohki