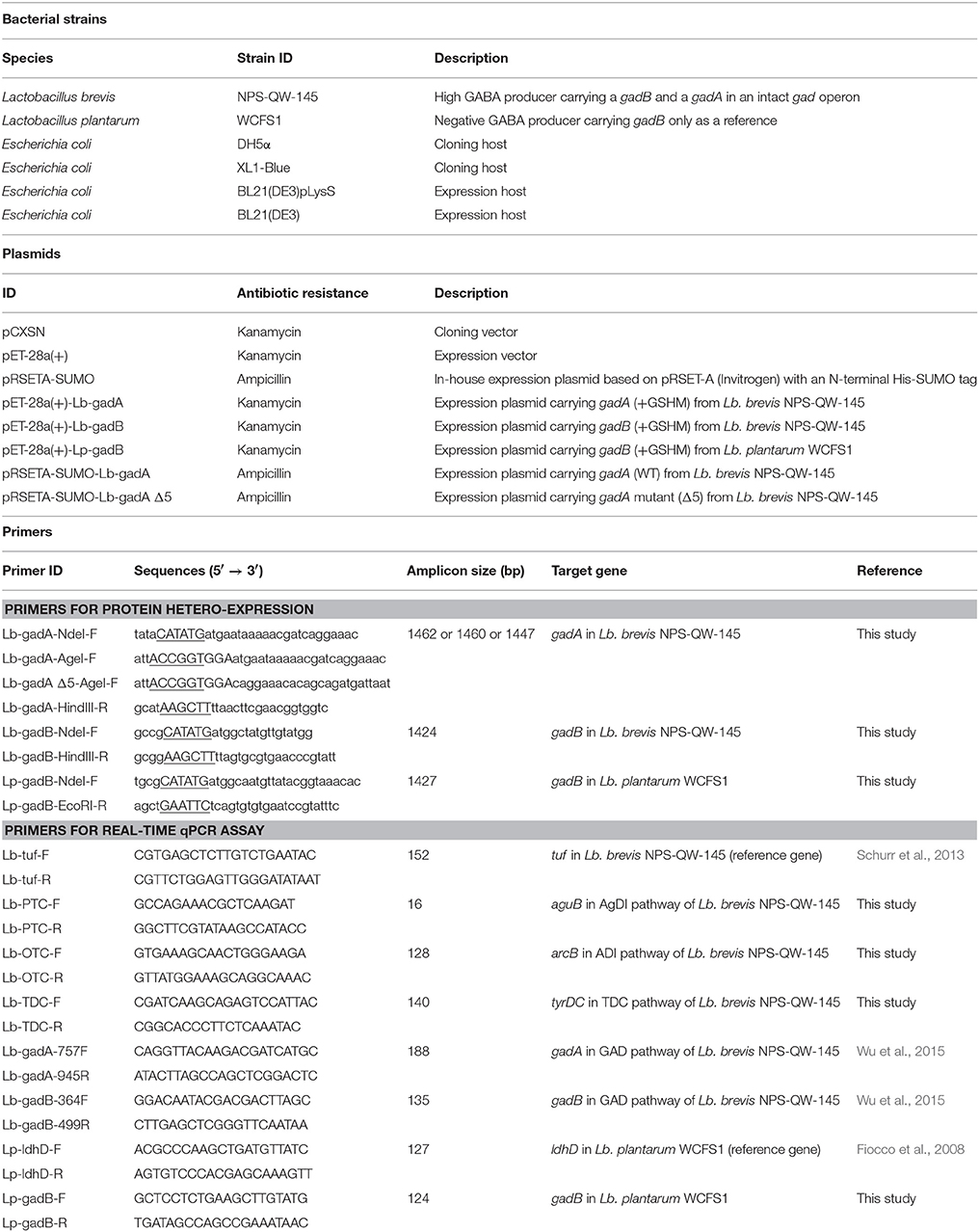
- 1School of Biological Sciences, The University of Hong Kong, Hong Kong, Hong Kong
- 2Department of Animal Science, University of Manitoba, Winnipeg, MB, Canada
- 3Department of Medical Microbiology, University of Manitoba, Winnipeg, MB, Canada
- 4Victoria University, Melbourne, VIC, Australia
Many strains of lactic acid bacteria (LAB) and bifidobacteria have exhibited strain-specific capacity to produce γ-aminobutyric acid (GABA) via their glutamic acid decarboxylase (GAD) system, which is one of amino acid-dependent acid resistance (AR) systems in bacteria. However, the linkage between bacterial AR and GABA production capacity has not been well established. Meanwhile, limited evidence has been provided to the global diversity of GABA-producing LAB and bifidobacteria, and their mechanisms of efficient GABA synthesis. In this study, genomic survey identified common distribution of gad operon-encoded GAD system in Lactobacillus brevis for its GABA production among varying species of LAB and bifidobacteria. Importantly, among four commonly distributed amino acid-dependent AR systems in Lb. brevis, its GAD system was a major contributor to maintain cytosolic pH homeostasis by consuming protons via GABA synthesis. This highlights that Lb. brevis applies GAD system as the main strategy against extracellular and intracellular acidification demonstrating its high capacity of GABA production. In addition, the abundant GadA retained its activity toward near-neutral pH (pH 5.5–6.5) of cytosolic acidity thus contributing to efficient GABA synthesis in Lb. brevis. This is the first global report illustrating species-specific characteristic and mechanism of efficient GABA synthesis in Lb. brevis.
Introduction
Many species of lactic acid bacteria (LAB) and bifidobacteria are of economic and health importance and have been extensively used for the production of yogurt, cheese, milk beverage, and fermented vegetables for thousands of years (Leroy and De Vuyst, 2004). These bacteria have been considered as potential probiotics for promoting human health (Hill et al., 2014). Interactions between ingested LAB or bifidobacteria and host are associated with immunomodulation (Bron et al., 2012; van Baarlen et al., 2013; Sivan et al., 2015), gut homeostasis maintenance (Gareau et al., 2010; Delzenne et al., 2011), and brain behavior (Tillisch et al., 2013; Möhle et al., 2016). Moreover, metabolites from these bacteria, such as lactate, have also been extensively evaluated for brain function and plasticity promotion (Herzog et al., 2013; Tang et al., 2014; Yang et al., 2014), bacteriocin for killing pathogens (Cotter et al., 2013; Martinez et al., 2013), exopolysaccharide as food texturing agent (Wu et al., 2014) and immune modulator (Fanning et al., 2012). In the past decades, γ-aminobutyric acid (GABA) has drawn a lot of attentions because GABA is the most abundant inhibitory neurotransmitter that maintains neuro functions of human central nervous system. Although GABA may not be able to cross human blood-brain barrier (Kuriyama and Sze, 1971; Boonstra et al., 2015), GABA approved as a food ingredient and its food carriers have shown anti-hypertensive and anti-depressant activities as two main functions to the host after oral administration (Diana et al., 2014; Wu and Shah, 2016). However, GABA content in natural animal and plant products is very low, thus solutions have been sought from microorganisms including LAB and bifidobacteria for their capability of producing GABA.
GABA synthesis via glutamate decarboxylation in bacteria has been associated with acid resistance (Su et al., 2011; De Biase and Pennacchietti, 2012; Teixeira et al., 2014). Glutamic acid decarboxylase (GAD) system encoded by the gad operon is responsible for glutamate decarboxylation and GABA secretion in bacteria and consists of two important elements—Glu/GABA antiporter GadC and GAD enzyme either GadA or GadB (Capitani et al., 2003). This system converts glutamate into GABA and while doing so consumes protons thus maintaining cytosolic pH homeostasis (Krulwich et al., 2011). Our recent review and another previous review summarized enormous reports presenting specific strains of LAB and bifidobacteria having varying capacities to produce GABA (Li and Cao, 2010; Wu and Shah, 2016). This suggests that the efficiency of GAD system in these bacteria depends on their activities of Glu/GABA antiporter and GAD enzyme. Thus, most studies have been carried out to characterize individual strains for their GABA production in varying conditions either controlled or natural fermentations (Li and Cao, 2010; Dhakal et al., 2012; Wu and Shah, 2015, 2016). Among these reported strains, we found that isolates of Lactobacillus brevis appear to be efficient for GABA production (Wu and Shah, 2016). Importantly, these GABA-producing Lb. brevis strains have different sources of origin such as human intestine, Korean kimchi and brewery, suggesting their varying genetic backgrounds but exhibiting similar GABA synthesis capacities (Wu and Shah, 2016).
Currently, hundreds of species of LAB and bifidobacteria that have been systematically identified, and their genome sequences have been deposited in public databases including DDBJ/EMBL/GenBank databases. Due to the importance and applications of LAB and bifidobacteria in food industry and human health, it is crucial to gain insights into global diversity of GABA-producing bifidobacteria and LAB mainly including Lactobacillus and Lactococcus. These bacteria are able to catabolize sugars to produce lactic acid efficiently (Leroy and De Vuyst, 2004), but they have also developed multiple acid resistance systems including F0F1-ATPase proton pump, sodium/proton antiporter, amino acid decarboxylation and deimination, alkali production and biofilm formation (Hutkins and Nannen, 1993; Zhao and Houry, 2010; Liu et al., 2015). Since multiple strategies as stated above in these bacteria are available to cope against acids, the linkage between bacterial acid resistance and GABA production capacity derived from GAD system has not been well established. Moreover, it is vital to unravel the mechanism of efficient GABA synthesis in these isolates especially in Lb. brevis. In the present study, Lb. brevis NPS-QW-145 (hereafter Lb. brevis 145) isolated previously from Korean kimchi was used as a model organism of high GABA-producing lactic acid bacterium (Wu and Shah, 2015). The chromosome of this organism has been completely sequenced in this study; genomic survey and biochemical toolkits have been carried out to illustrate efficient machinery of GABA production from this organism.
Materials and Methods
Bacterial Strains, Cultivation Conditions, and Genome Sequencing
GABA-producing Lb. brevis 145 and a reference strain, Lb. plantarum WCFS1, were cultivated in Lactobacilli MRS medium (BD Company, Franklin Lakes, NJ). Unless otherwise stated, Lactobacillus strains used in this study were anaerobically cultivated in the above medium statically. For cloning and protein expression purposes, E. coli strains as indicated in Table 1 were cultivated in Luria-Bertani (LB) medium with or without antibiotics supplemented to the medium. The model strain of high GABA-producing Lb. brevis 145 was completely sequenced in this study (please refer to Supplementary Materials and Methods for details). The NCBI accession number for the chromosome of Lb. brevis NPS-QW-145 is CP015398.
Genomic Survey on the Distribution of gad Operon and Genes Encoding Glutamate Decarboxylase in LAB and Bifidobacteria
Currently, there are more than a thousand of strains of LAB and bifidobacteria that have been sequenced and deposited in NCBI-GenBank database. Thus, genomic survey was carried out for the presence or absence of gad operon and genes encoding glutamate decarboxylase in most of the sequenced strains (all assembly levels; 890 strains) of Bifidobacterium and LAB including Lactobacillus, Lactococcus, Leuconostoc, Pedicoccus, Oenococcus, Weissella, and Streptococcus thermophilus released in NCBI-GenBank genome database. Only the species having at least two strains sequenced were included in this genomic survey. In addition, the distribution of acid resistance systems including F0F1-ATPase system, amino acid/cation:proton antiporters, glutamic acid decarboxylase (GAD) system, tyrosine decarboxylase (TDC) system, agmatine deiminase (AgDI) system, arginine deiminase (ADI) system and urea system in Lb. brevis was also surveyed in terms of presence or absence of above AR systems in the sequenced strains.
Extracellular pH (pHex) and Intracellular pH (pHin) Measurements
Extracellular pH (pHex) was measured directly with the pH meter. The fluorescent probe–5(6)-carboxyfluorescein diacetate N-succinimidyl ester (cFDA-SE; Thermo Fisher Scientific) was used to label bacterial cells for intracellular pH (pHin) measurement. There are several reports on the pHin measurement of bacteria suspended in citrate-based buffer (Siegumfeldt et al., 2000; Teixeira et al., 2014); however, this buffer did not allow the discrimination between cells equilibrated to pH between 3.5 and 5.0 thus limiting the detection range of cFDA-SE probe (Hansen et al., 2016). Thus, phosphate-buffered saline (PBS) was used to re-suspend bacterial cells for pHin measurement in the range of pH 3.5–7.0 in this study according to the previous studies (Breeuwer et al., 1996; Hansen et al., 2016). Briefly, the pHex of cultures was measured first, and bacterial cells were centrifuged, washed and re-suspended in PBS where its pH level was adjusted to the pHex of the cultures. The cell density was adjusted to an optical density (λ = 600) of 0.6–0.7, followed by the addition of both cFDA-SE and glucose to the final concentrations of 10 μM and 10 mM, respectively. The mixture was incubated in the dark at 37°C for 30 min. The stained cells were later harvested by centrifugation at 12,000 × g and 4°C for 5 min, and were re-suspended in the PBS buffer with the same pHex prior to the measurement.
Fluorescence intensities of the stained cells were measured in the Fluorescence Spectrophotometer F-7000 (Hitachi High-technologies, Shenzhen, China) at the excitation wavelengths of 488 nm (pH-sensitive) and 435 nm (pH-insensitive) by rapidly altering the monochromator between both wavelengths. The emission wavelength was started from 400 to 650 nm. Both excitation and emission slit width was 5 nm. For the calibration curve (pH 3.5–7.0) of Lb. brevis, the stained cells were suspended in PBS buffer having different pH values adjusted by hydrochloric acid (HCl). Valinomycin (0.2 mM in methanol; Sigma) and nigericin (0.2 mM in methanol; Sigma) were added to the final concentrations of 5 μM followed by incubation at 37°C for 10 min resulting in the equilibration of both potassium and proton ions across cell membrane. The equilibrated cells were later measure the same as described above.
Acid Resistance and Challenge Assays
Acid resistance assay was carried out based on previously method described (Seo et al., 2015). Briefly, cells of Lb. brevis 145 grown for 12 h (early stationary phase) in Lactobacilli MRS broth were inoculated into Lactobacilli MRS broth (pH 2.5) supplemented with or without 1 g/L of MSG, arginine, agmatine sulfate or tyrosine. The initial cell density after inoculation for acid challenge was 1.4 × 108 CFU/mL. Cells were incubated at 37°C for 2 h statically. Plate count method was then applied to assess the survival rates of Lb. brevis cells under above conditions. For acid challenge assay, the 3-h Lb. brevis cultures (lag phase, not acid-adapted cells) were centrifuged, washed and suspended in PBS buffer (pH 7.0), followed by cFDA-SE staining as described above. The stained cells were then placed in fluorescence spectrophotometer and their fluorescence emission intensities were measured every 5 min at 37°C. Acid challenge was achieved at the point of 5 min by adding hydrochloric acid that changed pHex from 7.0 to 3.5. After another 5 min, the stock solution (100 mM) of individual substrate (glutamate, arginine, agmatine, and tyrosine) was added to the final concentration of 10 mM in the solution. The changes in pHin were recorded accordingly as per the above description.
Growth of Lb. brevis 145 in Lactobacilli MRS Broth Containing Arginine, Glutamate, Agmatine, and Tyrosine
Cells of Lb. brevis 145 grown for 12 h (early stationary phase) in Lactobacilli MRS broth were inoculated into Lactobacilli MRS broth (pH 6.5) supplemented with or without the mixture of MSG, arginine, agmatine sulfate salt and tyrosine (1 g/L each; Sigma). However, tyrosine was partially dissolved in MRS broth (pH 6.5), and the insoluble tyrosine (solid form in the broth) was removed by membrane filtering (0.45 μm). The initial cell density after inoculation for acid challenge was 1.0 × 108 CFU/mL and the cells were incubated at 37°C for 36 h statically. Samples were collected at the time point of 0, 3, 6, 9, 12, 18, 24, and 36 h. Subsequently, measurements of acids, amino acids, amines and gene expression were carried out as discussed below. Plate count method was used to assess the cell viability, and pHin and pHex measurements were carried out as stated above.
Measurements of Acids Production
Acids in the culture broth were analyzed by HPLC (Model Shimadzu LC-2010A, Shimadzu Corporation, Kyoto, Japan) equipped with Aminex HPX-87H column (300 × 7.8 mm; Bio-Rad). Briefly, cell-free supernatants (diluted if necessary) after removal of bacterial cells and passing through Acrodisc® syringe filter with 0.20 μm Supor® membrane (Pall, Ann Arbor, MI, USA) were injecting into the HPLC system. Acids were eluted with 5 mM sulfuric acid with a flow of 0.60 mL/min. The temperature of the column was maintained at 50°C and the absorbance of the detector was set to 210 nm for detection.
Measurements of Amino Acids and Amines
Amino acids including glutamic acid, GABA, arginine, ornithine and tyrosine were separated and quantified as per the method we have previously developed (Wu and Shah, 2016). Briefly, cell-free supernatant was collected after centrifugation of fresh cultures. The derivatization was achieved by adding 200 μL acetonitrile, 200 μL NaHCO3 (1 M, pH 9.8 adjusted with NaOH), 300 μL distilled water and 100 μL dansyl chloride solution (40 g/L; dissolved in acetonitrile) to 100 μL supernatant (diluted if necessary), then the mixture was kept in an oven (40°C) for 60 min. After derivatization, 100 μL of 20% (v/v) acetic acid was added to stop the reaction and centrifuged at 10,000 × g and 20°C for 2 min. The supernatant was passed through a 0.22 μm membrane filter before HPLC analysis.
Amines including agmatine, putrescine, and tyramine in the supernatants were analyzed as previously described (Dugo et al., 2006). Briefly, 1.6 mL of 10 g/L dansyl chloride solution (dissolved in acetone) was added to 1.5 mL of the supernatant (diluted if necessary). The pH of the mixture was adjusted to pH 8.2–8.3 with 40 g/L of NaCO3 solution, then the mixture was heated in a water-bath (40°C) for 60 min in dark. After derivatization, acetone was removed under a stream of nitrogen and then the volume was made up to 5 mL with acetonitrile and centrifuged at 1000 × g and 20°C for 2 min. The supernatant was passed through a 0.22 μm membrane filter before HPLC analysis.
Dansyl amino acids and dansyl amines were separated and detected using a previously developed gradient elution mode (Wu et al., 2015) by HPLC (Model Shimadzu LC-2010A, Shimadzu) equipped with a Kromasil 5 μ 100A C18 column (250 × 4.6 mm; Phenomenex). All the standards of amino acids and amines were derivatized and analyzed as the same in the above procedures. The absorbance for dansyl-amino acids and dansyl-amines were recorded at 275 nm and 254 nm, respectively.
Total RNA Extraction and Real-Time Quantitative PCR Assay
Total RNA extraction using hot SDS/phenol method, DNase I treatment, cDNA synthesis, qPCR assay were carried out as per previously described (Wu et al., 2015). The expression of target genes (gadA, gadB, aguB, arcB, and tyrDC in Lb. brevis 145; gadB in Lb. plantarum WCFS1) listed in Table 1 was quantified by real-time qPCR assay. Reference genes including tuf in Lb. brevis and ldhD in Lb. plantarum listed in Table 1 were used to normalize the expression of target genes in both strains. The efficacy of qPCR amplification using each pair of primers was in the range of 90–110%. Comparative critical threshold method (2−ΔΔCt) was used to calculate the relative gene expression. The qPCR assay was performed in duplicates for each sample and independent experiments were carried out in triplicates.
Glutamate Decarboxylation Assay
For the procedure of cloning, hetero-expression and purification of GadA and GadB from Lb. brevis 145 and GadB from Lb. plantarum WCFS1, please refer to Supplementary Materials and Methods for details. The effects of pH (3.0–6.6) and temperature (30–90°C) on the purified GADs including Lb. plantarum GadB, Lb. brevis GadB, and Lb. brevis GadA and its mutants (Lb. brevis GadA Δ5 and Lb. brevis GadA +GSHM) were carried out. Low concentrations of PLP such as 20 μM is sufficient to activate the activities of both Lb. plantarum GadB and Lb. brevis GadB (Fan et al., 2012; Yu et al., 2012; Shin et al., 2014), thus the reaction mixture consisted of 5 μL of 1 M monosodium glutamate (MSG; Sigma), 5 μL of 2 mM pyridoxal 5′-phosphate (PLP; Sigma), 500 μL of McIlvaine (citrate-phosphate) buffer, and 10 μg of GAD. Previous studies have indicated the PLP- and sulfate ion-dependent activation of Lb. brevis GadA and its homologous Lb. zymae GadB (Ueno et al., 1997; Park et al., 2014); thus, for Lb. brevis GadA and its mutants, the reaction mixture contained 5 μL of 1 M MSG, 5 μL of 20 mM PLP, 500 μL of sulfate buffer (0.8 M sodium sulfate and 50 mM sodium acetate; different pH), and 10 μg of GAD. The kinetics of these enzymes was determined under their optimal pH and temperature, and the concentrations of substrate (glutamate) ranged from 0.9 to 36 mM.
Statistical Analysis
All presented data in the bar charts and tables correspond to means ± standard deviation. Significant difference was carried out by using IBM SPSS Statistics 20.0 version.
Results
Genomic Survey Identifies Common Distribution of gad operon in Lb. brevis
Currently, there have been two major pathways reported for GABA production either through the degradation of putrescine (Puu and ADC pathways) or via the decarboxylation of glutamate (GAD pathway) in bacteria (Figure 1A). However, GABA production from putrescine degradation is not common in LAB and bifidobacteria due to the absence of Puu and ADC pathways. Interestingly, genes (gadA and gadB) encoding glutamate decarboxylases (GadA and GadB) and an intact gad operon including gadR (regulator), gadA and gadC (Glu/GABA antiporter) have been identified in all of the sequenced strains of Lb. brevis having varying origins excluding the one of strain WK12 with incomplete (contig levels) genome sequence (Figure 1C). The location of gadB is far away from gad operon in Lb. brevis suggesting there may be two different gene regulations for gadA and gadB in this species. However, arrangement of gadA and gadC close to each other in the genome could ensure the timely co-regulation of transcription and translation of gadA and gadC for GABA production (De Biase and Pennacchietti, 2012). Moreover, phylogenetic analysis of four gene components (gadR, gadC, gadA, and gadB) indicated the presence of two isoforms of glutamate decarboxylases, GadA and GadB, in Lb. brevis; all of the four components from varying Lb. brevis strains are highly conserved in this species (Figure 1B). This implies that co-existence of gadA and gadB in Lb. brevis contributes to its GABA synthesizing capacity.
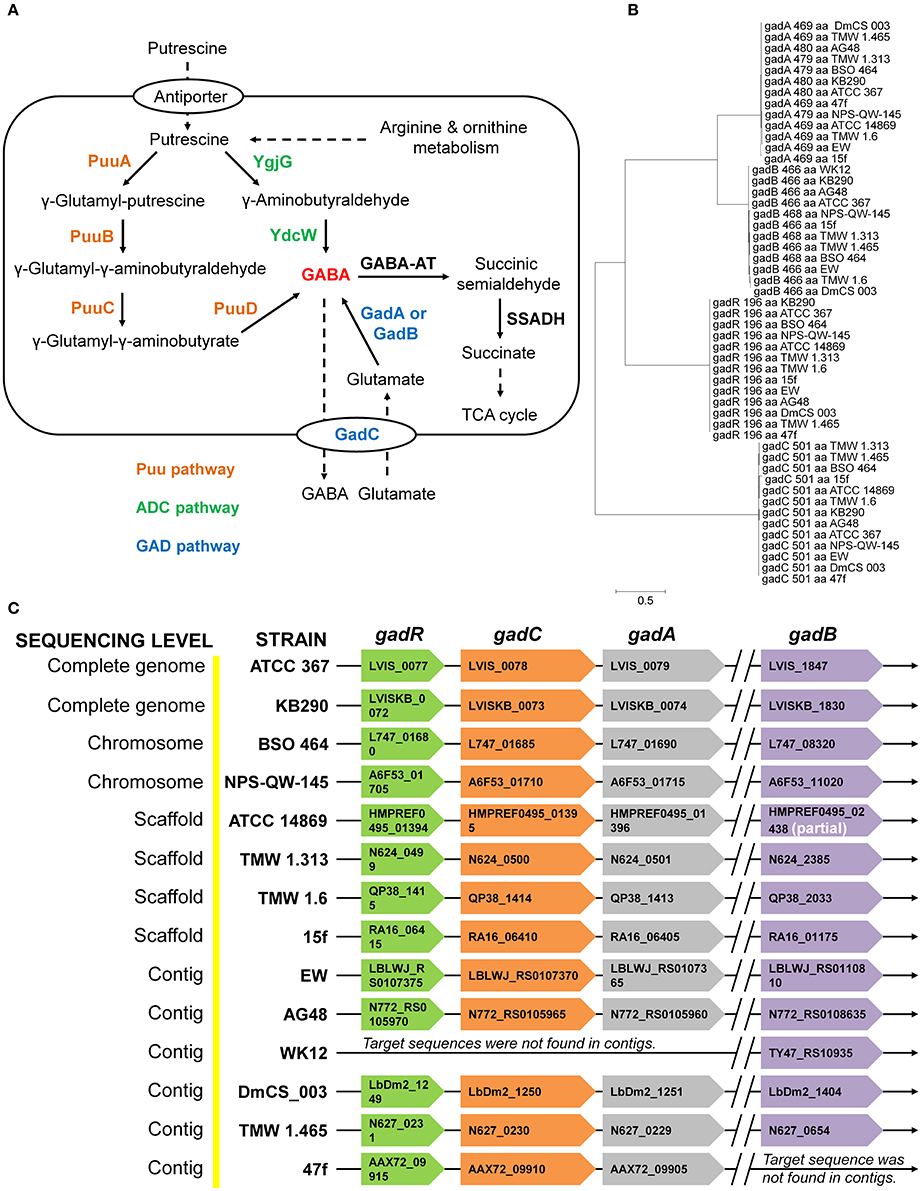
Figure 1. Common distribution and arrangement of gad operon and genes encoding glutamic acid decarboxylase in the genomes of Lb. brevis. (A) General GABA production in bacteria from GAD pathway and putrescine degradation pathways (Puu pathway and ADC pathway). (B) Phylogeny of amino acids sequences of four components in gad operon demonstrating two isoforms of glutamate decarboxylases and the highly-conserved four genetic components in Lb. brevis. (C) Common distribution of gad operon in all the sequenced strains of Lb. brevis. Denotations: GABA-AT, GABA aminotransferase; SSADH, succinic semialdehyde dehydrogenase; gadA, glutamate decarboxylase isoform A; gadB, glutamate decarboxylase isoform B, gadR, transcriptional regulator; gadC, Glu/GABA antiporter. The phylogenetic tree was generated from MEGA (version 6.0) after MUSCLE alignment of amino acids sequences of each component in gad operon. The length of each component (locus tag indicated) in gad operon is indicated in the braces. The chromosome of a model strain Lb. brevis NPS-QW-145 was completely sequenced in this study and its NCBI accession no. is CP015398. All gene loci and genome data were collected from NCBI genome database (genome assembly and annotation report) on 10 January 2016.
Global Diversity of GABA-Producing LAB and Bifidobacteria
In order to gain a global view on the distribution of gad operon and genes encoding glutamate decarboxylases in LAB and bifidobacteria, a genomic survey was carried out on the sequenced LAB and bifidobacteria species, of which at least two strains have been sequenced and deposited in GenBank database (accessed on 10 January 2016). As shown in Table 2, many strains carry gadA or gadB, but gadC is only present in the genomes of limited strains indicating their strain-specific characteristic of GABA production. Among gad operon-positive strains, Lb. brevis is the only one species that has close to 100% probability to carry the intact gad operon in their chromosomes. All the sequenced strains of B. dentium possessed gadBC operon in their chromosomes. However, there is very little information on GABA production from B. dentium with only gadB gene, whereas there are several reports on high GABA production from Lb. brevis with two gad genes (Li and Cao, 2010; Dhakal et al., 2012; Wu and Shah, 2016). Thus, the present study focused on only Lb. brevis for its efficient GABA synthesis. Moreover, it was found that other common reported GABA-producing species such as Lb. reuteri and Lc. lactis showed strain-specific GABA biosynthesis at the genetic level.
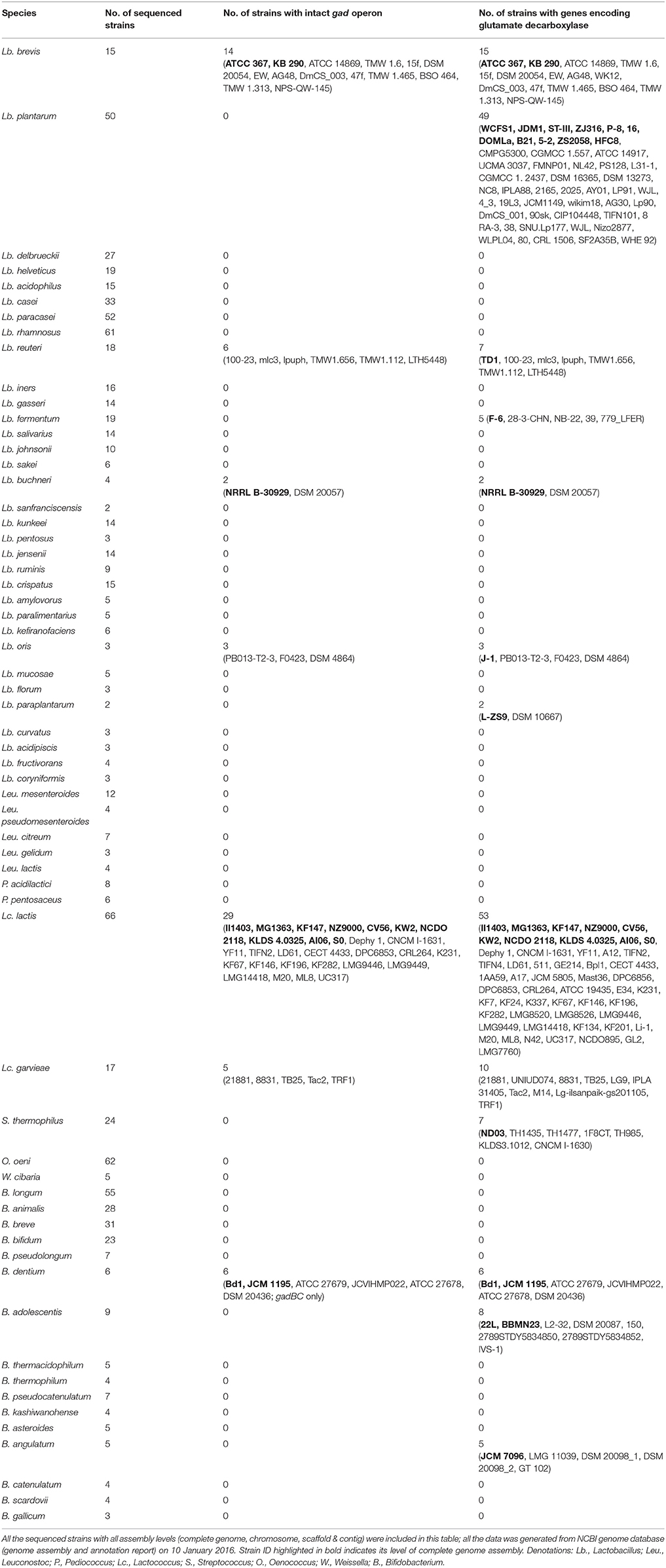
Table 2. Distribution of gad operon and genes encoding glutamate decarboxylases in the sequenced lactic acid bacteria and bifidobacteria.
Although, there are several reports on GABA-producing Lb. plantarum without sequencing their genomes (Li and Cao, 2010; Wu and Shah, 2016), in this study we found that gadC is absent in all the sequenced strains of Lb. plantarum (Table 2). Hence, a fermentation study was carried out using the sequenced Lb. plantarum WCFS1 as a model organism. Although, there was an incomplete pathway for GABA metabolism in this organism based on KEGG pathway (Figures S1A,B), its viable counts, gadB transcripts level, acid profiles and GABA production did not significantly change after glutamate supplementation to the medium (Figures S1C–H). Although gadB transcript was increased in the stationary phase suggesting an enhanced GABA synthesis, gadC in LAB is required and specific for exchanging extracellular glutamate with intracellular GABA (Figure S1D). Our cultivation experiment on Lb. plantarum WCFS1 complied with previous gene knock-out studies in other bacteria with intact gad operon that GadC is an important element for GABA production (Cotter et al., 2001; Lu et al., 2013).
Diverse and Constant Acid Resistance Systems in Lb. brevis
A genomic survey on the distribution of amino acid-dependent AR systems in all the sequenced strains of Lb. brevis was carried out. Except for universal amino acid-dependent AR systems including F0F1-ATPase system and amino acid/cation:proton antiporter, extra amino acid-dependent AR systems including GAD system, tyrosine decarboxylase (TDC) system, agmatine deiminase (AgDI) system and arginine deiminase (ADI) system are commonly present in all sequenced strains of Lb. brevis (Figure 2A and Table 3).
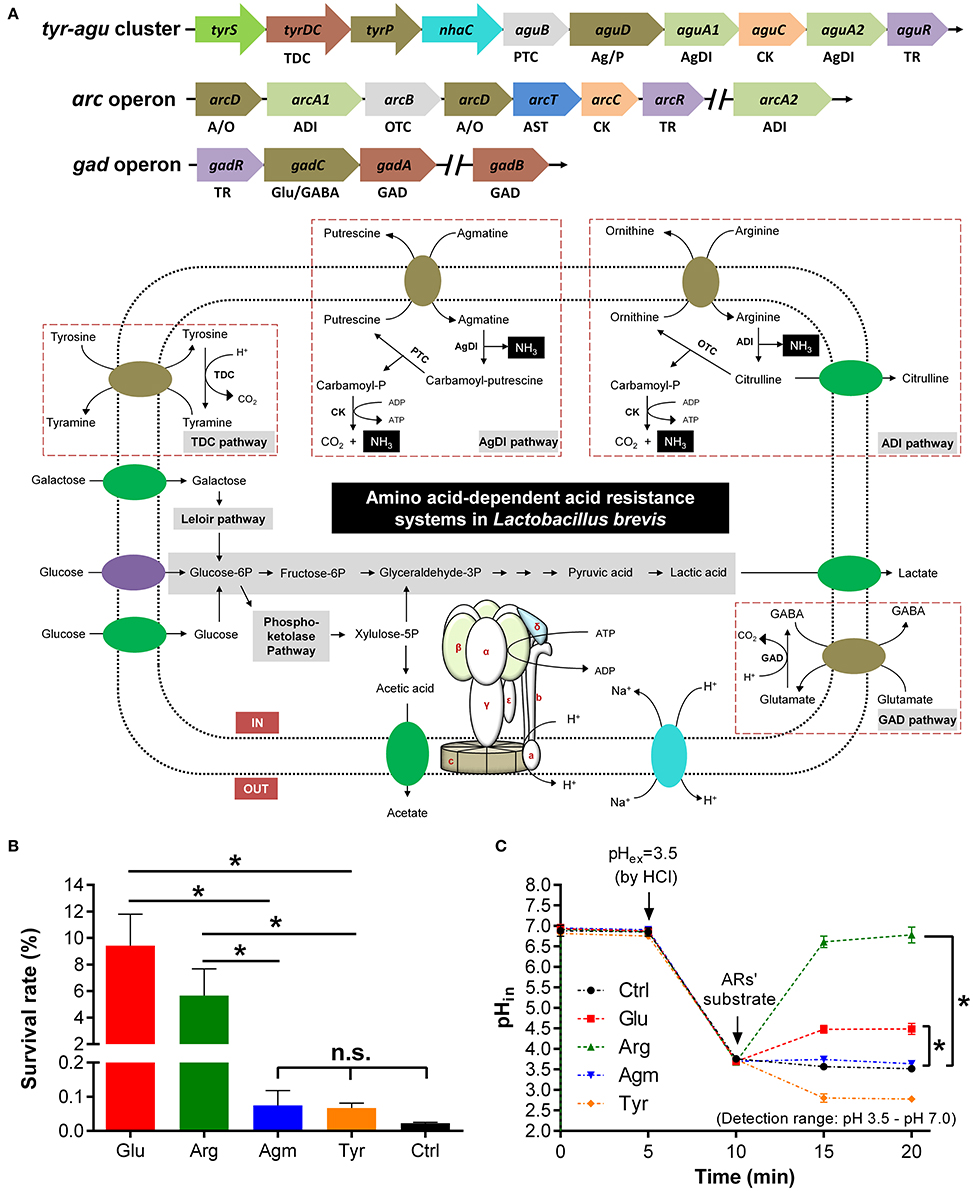
Figure 2. GAD system in Lb. brevis improves cell viability by maintaining intracellular pH homeostasis. (A) Carbohydrate metabolism and amino acid-dependent acid resistance (AR) systems in the model strains Lb. brevis 145. (B) Effect of amino acid-dependent ARs' substrates on survival rate of Lb. brevis cells (12-h cultures; acid-adapted cells) during acid resistance assay (37°C and 2-h incubation) carried out in Lactobacilli MRS medium (pH 2.5). (C) Effect of amino acid-dependent ARs' substrates on intracellular pH (pHin) of Lb. brevis cells (3-h cultures; non-acid-adapted cells) upon acid challenge tested at 37°C (extracellular pH–pHex decreased from pH 6.5 to pH 3.5. Glutamate, arginine and agmatine were dissolved in PBS buffer (pH 3.5) and tyrosine was dissolved in 0.1 M hydrochloric acid (HCl; after addition of tyrosine, pHin of the cell was out of detection range but was still calculated from the equation of standard curve (pHin = −0.1141 × + 1.4035 × RFU488/435 + 2.6307; R2 = 0.9849; pH range: 3.5–7.0) of pH and RFU488/435 (RFU, relative fluorescence units). Cells were suspended in phosphate-buffered saline but not citrate-based buffer for pHin measurements ranging from pH 3.5 to pH 7.0. Denotations: GAD, glutamate decarboxylase; TDC, tyrosine decarboxylase; PTC, putrescine carbamoyltransferase; OTC, ornithine carbamoyltransferase; ADI, arginine deiminase; AgDI, agmatine deiminase; CK, carbamate kinase; TR, transcriptional regulator; A/O, arginine/ornithine antiporter; Ag/P, agmatine/putrescine antiporter; Glu/GABA, glutamate/GABA antiporter. Experiments were performed in triplicates and data is presented as mean ± standard derivation (SD). *p < 0.05; n.s., not significant.
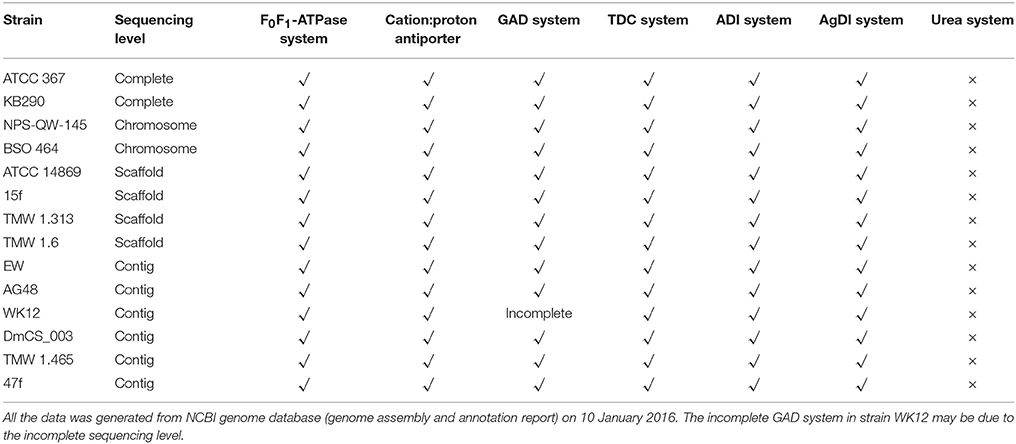
Table 3. Acid resistance systems in the sequenced Lactobacillus brevis based on NCBI annotated protein database.
Activation of GAD System in Lb. brevis Upon Challenge with Exogenous Acid
Although, the contribution of AR systems for acid resistance in other LAB such as Lb. reuteri has been characterized (Teixeira et al., 2014), individual contribution of above AR systems to resist exogenous acids in Lb. brevis has not been well demonstrated. In this study, it was found that both glutamate and arginine were more effective in increasing the survivability of Lb. brevis significantly (p < 0.05) when exposed to acidic condition (pH 2.5; HCl-adjusted) for 2 h than that by agmatine and tyrosine (Figure 2B). Moreover, both glutamate and arginine were also able to increase the intracellular pH of Lb. brevis significantly (p < 0.05) than agmatine and tyrosine after challenging cells with hydrochloric acid (Figure 2C). This suggests that glutamate and arginine increased cell viability of Lb. brevis by increasing intracellular pH of the cell thus maintaining its metabolic activity. However, the increased level of intracellular pH of Lb. brevis cells by arginine and glutamate differed greatly. This may be due to the stages of cells selected for acid resistance experiment (12-h culture; acid-adapted cells) and acid challenge assay (3-h cultures; non-acid-adapted cells). The stages of cells may affect the contents of key cytosolic enzymes including GadA, GadB, ADI, and OTC from the four amino acid-dependent AR systems (Figure 3D). In addition, the increase in the intracellular pH by glutamate may be explained by GABA synthesis that consumed cytosolic protons in Lb. brevis upon acid challenge. Since the function of GadC is activated under acidic conditions (Ma et al., 2012), GAD system in Lb. brevis may be activated by acid challenge.
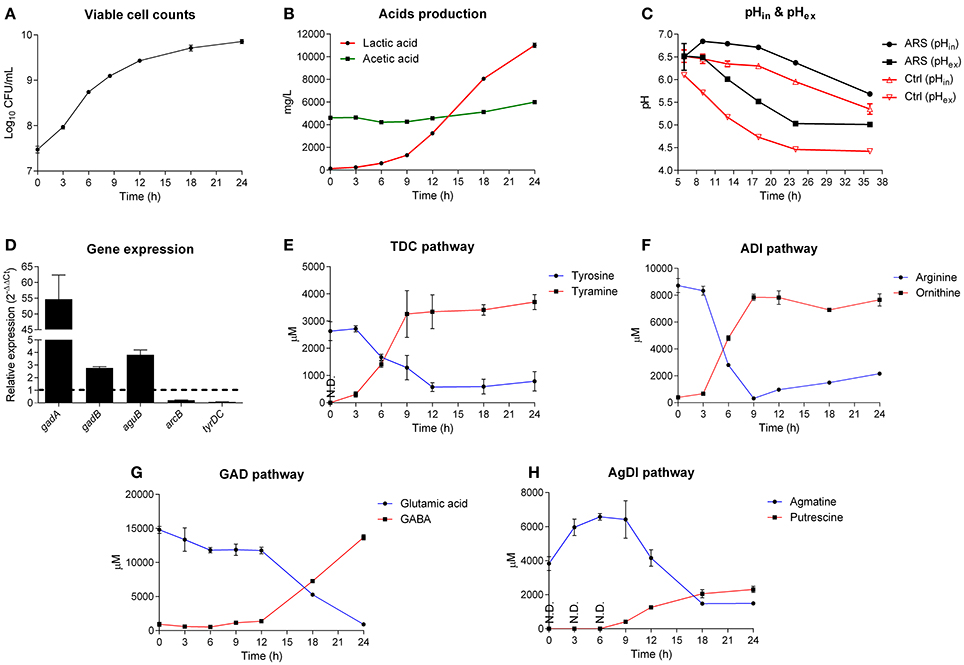
Figure 3. GAD system is a major contributor for acid resistance of Lb. brevis. (A) Growth curve of Lb. brevis in lactobacilli MRS medium supplemented with extra glutamate, arginine, agmatine and tyrosine. (B) Lactic acid and acetic acid production. (C) Intracellular pH (pHin; cFDA-SE as the probe) and extracellular pH (pHex) of Lb. brevis cells incubated in lactobacilli MRS medium with (ARS) or without (as control) amino acid-dependent ARs' substrates. (D) Relative gene expression of key genes of different amino acid-dependent AR systems in Lb. brevis at 18 h normalized to that at 6 h of cultivation. (E–H) Changes in the concentrations of the end products from each amino acid-dependent AR system. The experiment was carried out in triplicates and data was presented as mean ± standard derivation (SD).
GAD System in Lb. brevis Is a Major Contributor for Resisting Endogenous Acid
To understand the association between GAD system for endogenous acid (lactic acid) resistance and GABA production capacity of Lb. brevis, a 24-h course of cultivation experiment on Lb. brevis 145 in Lactobacilli MRS broth supplemented with four amino acid-dependent AR's substrates (glutamate, arginine, agmatine and tyrosine) was carried out. Firstly, it was observed that the AR's substrates were able to increase the intracellular pH of Lb. brevis cells by about 0.5 compared to that in Lb. brevis without the AR's substrates supplementation (Figures 3A–C). In addition, results indicate that Lb. brevis maintained its intracellular pH level in the near-neutral range (pH 5.5–6.5) or close to neutral range (pH 6.5–7.0) when extracellular pH decreased spontaneously by secreting protons during its lactic acid production (Figures 3B,C). Moreover, gene transcription of gadB and aguB increased by at least 2 fold whereas gadA mRNA was highly up-regulated by about 55 fold at the point of 18 h (stationary phase) when compared to that at 6 h (logarithmic phase) (Figure 3D). In combination with the higher rate of GABA production at 18 h than that at 6 h (Figure 3G), it appears that GAD system was more active in stationary phase than that in lag and logarithmic phases. In general, changes in the end metabolites of four amino acid-dependent AR systems also demonstrated that utilization of the AR's substrates varied with phases. Based on the metabolic data in Figures 3E–H and gene expression data in Figure 3D, TDC and ADI pathways were active in early cultivation (Figures 3E,F) but were down-regulated in late cultivation (Figure 3D), while GAD and AgDI pathways were more active in stationary phase (Figures 3G,H). Although a gene encoding lysine decarboxylase was found in Lb. brevis 145, the lysine content little changed suggesting an inactive state of this AR system (Figure S2).
The acid resistance and challenge assays may not be able to indicate actual contributions of AgDI and TDC pathways for resisting acids (Figures 2B,C); however, changes in the content of metabolites during different cultivation stages evidenced that all the four amino acid-dependent AR systems contributed to intracellular pH homeostasis (Figure 3C). Since GABA production from Lb. brevis was increased in the stationary and late stages, its GAD system played an important role in bacterial acid resistance because its lactic acid accumulated largely during late cultivation (Figure 3B).
GadA Supports GABA Synthesis in Lb. brevis toward a Weak pH Range
Two Gads, GadA and GadB, from Lb. brevis and a reference type I GadB from Lb. plantarum were hetero-expressed and purified for enzyme assay (Figure S3). Two mutants of Lb. brevis GadA (wild-type; WT), Lb. brevis GadA +GSHM (extra GSHM to N-terminus) and Lb. brevis GadA Δ5 (5 amino acids deleted from N-terminus), were constructed in this study. As shown in Figure 4A, Lb. brevis GadA (WT) exhibited a narrow activity spectrum toward the change of temperature, but still retained about 50% of the highest activity at 37°C, whereas Lb. brevis GadB and Lb. plantarum GadB were heat-stable enzymes. More interestingly, Lb. brevis GadB and Lb. plantarum GadB retained similar catalytic spectrum in the range of pH 3.0–5.5 while Lb. brevis GadA (WT) exhibited activity toward a weak acidity range (pH 5.5–6.6) (Figure 4B). The in vitro enzyme kinetic assay (Figure 4C and Table 4) demonstrated that catalytic efficiency of Lb. brevis GadA (WT) was largely lower than that of two GadB enzymes. However, these three Gads had a very similar Km value suggesting a similar level of substrate saturation (Table 4). Moreover, Lb. brevis GadA (WT) exhibited a significant (p < 0.05) activity than its two mutants with modifications to its N-terminus (Figure 4D). Previous gel filtration studies have indicated the tetramer state of Lb. brevis GadA (Hiraga et al., 2008), the monomer state of Lb. brevis GadB (Yu et al., 2012; Shi et al., 2014) while Lb. plantarum GadB function as dimers (Shin et al., 2014). This suggests that N-terminus of Lb. brevis GadA may be critical to the tetramer formation via incorporation to its N-terminus. The phylogenetic analysis of representative Gads from LAB and Bifidobacterium highlighted that Lb. brevis GadA was a type III group of Gads which differed largely from Lb. brevis GadB (sub-group 2) and Lb. plantarum GadB (sub-group 1) which were classified in type I Gads (Figure 4E). Overall, Lb. brevis, as a cell factory, exhibited GABA biosynthesizing capacity in a board range of acidity, especially in near-neutral pH range (pH 5.5–6.5).
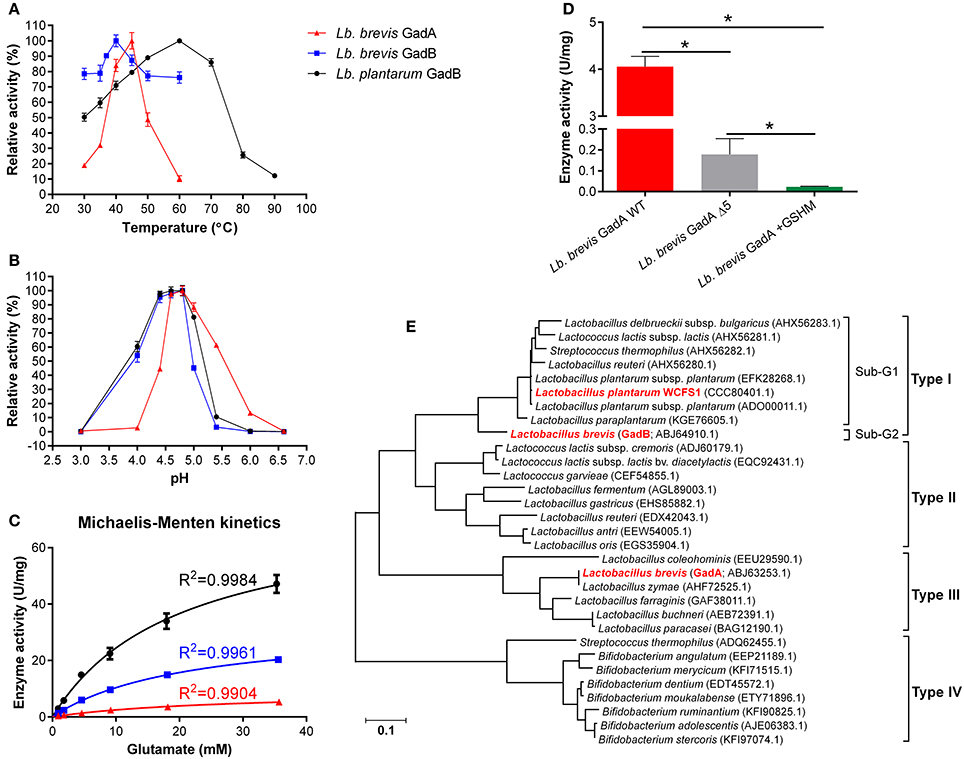
Figure 4. GadA supports GABA synthesis in Lb. brevis toward a weak pH range of cytosolic acidity. (A) Temperature effect on Gads tested under constant pH 4.8. (B) Acidity effect on Gads tested under constant 37°C. (C) Enzyme kinetics tested under their optimal pH and temperature. (D) Activities of wild-type Lb. brevis GadA and its mutants. (E) Phylogeny of representative Gads from LAB and Bifidobacterium. The well-characterized Lb. plantarum GadB was used as a reference Gad. The phylogenetic tree was generated from MEGA (version 6.0) after MUSCLE alignment of amino acids sequences of Gads. GraphPad Prism version 6.0 was used to generate kinetic curves of three Gads. The enzyme assay was carried out in duplicates and data is presented as mean ± standard derivation (SD). *p < 0.05.
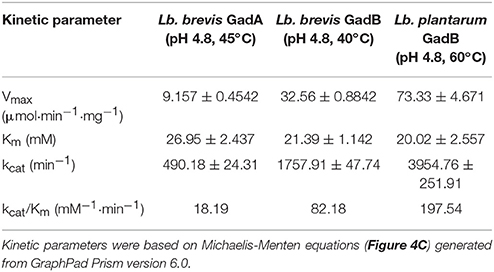
Table 4. Kinetic parameters of Lb. brevis GadA, Lb. brevis GadB and Lb. plantarum GadB under optimal conditions.
Discussion
GABA synthesis can also be derived from putrescine degradation process via either Puu pathway or arginine decarboxylase (ADC) pathway as shown in Figure 1A in some bacterial species such as Escherichia coli (Kurihara et al., 2008, 2010); however, based on our KEGG survey on arginine and proline metabolism map of complete sequenced bacterial strains, these two pathways are not commonly present in LAB and bifidobacteria (data not shown). Thus, GABA production from LAB and bifidobacteria is mainly derived from GAD pathway if it is present in them. Due to the important biofunctionalities of GABA and GABA-producing LAB and bifidobacteria, several studies have characterized GABA production from individual strains (Li and Cao, 2010; Dhakal et al., 2012; Diana et al., 2014; Wu and Shah, 2016). The diversity of GABA-producing LAB and bifidobacteria has been poorly understood either at genetic level or from experimental insights. Our genomic survey conducted on most of the sequenced strains of LAB and bifidobacteria released in GenBank has clearly indicated that the genetics of Lb. brevis differed with other species of LAB and bifidobacteria in terms of GABA production: (1) an intact gad operon, and (2) two genes (gadA & gadB) encoding glutamate decarboxylases in their genomes (Figure 1C and Table 2). Thus, GABA production from Lb. brevis appears to be species-specific based on our current genetic observation (Table 2) and previously reported GABA yields from various strains of Lb. brevis (Li and Cao, 2010; Dhakal et al., 2012; Wu and Shah, 2016). This differs with the strain-specificity of GABA biosynthesis in other species such as Lactococcus lactis and Lb. reuteri (Table 2). To our knowledge, this is the first global insights into genetic diversity of GABA producer among above food-grade bacteria.
It is clear that GAD system is one of AR systems in bacteria to cope against acidic environments thus allowing them to maintain metabolic potential and cell viability (De Biase and Pennacchietti, 2012). Multiple AR systems including universal and amino acids-based AR systems function in the bacteria, but the contribution of GAD system to bacterial acid resistance has not been clearly demonstrated so far. Since Lactobacillus genus including Lb. brevis is a high lactic acid producer (Leroy and De Vuyst, 2004), excessive intracellular protons from endogenous or exogenous acids can be neutralized or eliminated by their AR systems. Among the four amino acid-dependent AR systems that were commonly identified in Lb. brevis, our results indicate the major contribution of GAD system for acid resistance and challenge in this organism upon either short-term or long-term acidic exposure, and either extracellular or intracellular acidification (Figures 2, 3). For instance, high amount of lactic acid was produced and accumulated in the medium during stationary phase of Lb. brevis cultivation, this organism utilized the highly active GAD system that decarboxylated glutamate into GABA greatly at this stage contributing to the increase in cytosolic pH (Figure 3). This highlights that Lb. brevis, a lactic acid producer, applies GAD system as the main strategy for its acid resistance thus reflecting its high capacity of GABA production.
Considering high GABA production from Lb. brevis and its GAD system, it is crucial to understand the mechanism of efficient GABA synthesis for this phenomenon. Several studies have reported individual Lb. brevis GadB tested either in acetate buffer or citrate-phosphate buffer exhibiting its activity spectrum toward acidic conditions (pH 3.5–5.5) but not near-neutral pH (pH 5.5–6.5; Yu et al., 2012; Seo et al., 2013; Lin et al., 2014), but the activity of only one GadA from Lb. brevis IFO 12005 has been reported twice focusing on its activation of enzymatic activity during in vitro conditions (Ueno et al., 1997; Hiraga et al., 2008). However, these studies did not provide systematic and comparative insights into this decarboxylation machinery in Lb. brevis. In general, Lb. brevis GadA retained its activity from pH 4.0 to pH 6.6 in the sulfate buffer (Figure 4B). This showed similar activity spectrum, particularly in near-neutral pH range (pH 5.5–6.5), with another reported Lb. brevis IFO 12005 GadA tested in pyridine-HCl buffer after sulfate activation and enzyme purification via gel filtration (Ueno et al., 1997; Hiraga et al., 2008). The characteristic of Lb. brevis GadA differs greatly with Lb. brevis GadB since the former requires enzyme activation by high concentrations of sulfate (Hiraga et al., 2008). As for Lb. brevis GadB, our study and previous reports showed its active activity in a more acidic range (pH 3.0–5.5; Figure 4B; Yu et al., 2012; Seo et al., 2013). Taken together, GadA and GadB in Lb. brevis cells may help them synthesize GABA toward a broad range of intracellular pH (pH 3.0–6.6).
As shown in Figure 3C, intracellular pH of Lb. brevis cells during a standard 24-h course of cultivation in MRS medium was in the range of pH 5.8–6.5. This indicates that glutamate decarboxylation may be very slow due to limited activities of both Gads in this near-neutral pH range (pH 5.5–6.5). Moreover, pHin of Lb. brevis cells decreased in the late incubation (24–36 h) thus may increase GABA synthesis by the enhanced activities of both GadA and GadB. This may be able to explain why resting cells could be used for GABA production (Zhang et al., 2012). Moreover, transmembrane potential, which contributes to amino acid-dependent acid resistance in bacteria, could be increased by glutamate decarboxylation (Teixeira et al., 2014). The transmembrane ΔpH in the ARS group was less than that in the control after the incubation of 9 h (Figure 3C) indicating that transmembrane potential in ARS group may be higher than that in the control; this effect may be attributed by glutamate decarboxylation in Lb. brevis.
The tetramer formation of Lb. brevis GadA in vitro needs high content of sulfate ions which is not similar to that in the in vivo conditions (Hiraga et al., 2008). This may be due to the fact that in vitro catalytic activity does not fully represent in vivo situation of the enzyme, though in general it concurs to that in vivo in bacteria (Davidi et al., 2016). However, we observed that glutamate was almost decarboxylated into GABA within 24 h (Figure 3G), even at the supplementation level of 10 g/L of monosodium glutamate (Wu and Shah, 2015), and GadA showed similar spectrum of enzyme activity either in sulfate buffer (Figure 4D) or in pyridine-HCl buffer (Hiraga et al., 2008). This may suggest that the pH value rather than the composition of the buffer affects the activity of GadA greatly after the activation process for GadA.
The near-neutral intracellular pH of Lb. brevis cells limited activities of cytosolic Gads though GadA and GadB were highly regulated (Figures 3C,D, 4B). Thus, approaches to improve their activities in near-neutral pH (pH 5.5–6.5) would be of economic and biological importance. Two studies that applied mutation strategy to Lb. brevis GadB demonstrated that these mutants extended their activities toward near-neutral acidity (Yu et al., 2012; Shi et al., 2014). However, we observed the extended activity of GadA in near-neutral pH range (pH 5.5–6.5) supporting GABA synthesis in Lb. brevis offering a novel insight into the diversity of Gads from LAB and bifidobacteria (Figure 4B). Although detailed regulatory mechanism for GABA synthesis in Lb. brevis is not clear, the gadA mRNA transcript became more abundant than gadB mRNA transcript in Lb. brevis at the stationary phase; this may contribute to its high GABA production by this organism during late cultivation (Figures 3D,G). The classification of Gads from LAB and bifidobacteria also indicated that Lb. brevis GadA and GadB were unique Gads differing with other Gads (Figure 4E). In general, we concluded that Lb. brevis GadA, a novel type III Gad that may play a more important role than Lb. brevis GadB, exhibited its activity toward weak pH range (pH 5.5–7.0) of bacterial cytosolic acidity. This supports the efficient GABA synthesis in Lb. brevis as a microbial cell factory.
Apart from the contribution of GAD system to acid resistance during late cultivation of Lb. brevis, our results showed that its ADI system was active in early cultivation, specifically in lag and log phases (Figures 2, 3D,F). This implies that ADI pathway plays a key role in early acid resistance by alkali production.
In conclusion, GAD system was commonly distributed in Lb. brevis among various species of LAB and bifidobacteria, and was the major contributor for acid resistance thus exhibiting a high capacity of GABA production of the cell. GadA in GAD system played a key role in GABA synthesis via its extended activity toward a near-neutral pH range (pH 5.5–6.5) of cytosolic acidity of Lb. brevis cells. Further understanding on tetramer formation and structural insights of Lb. brevis GadA would be necessary to unravel the decarboxylation machinery in a weak acidic condition.
Author Contributions
QW and NS designed the study; HT and EK performed the draft genome sequencing of Lactobacillus brevis NPS-QW-145; QW performed the experiments; QW and YL analyzed the data; QW and NS wrote the manuscript.
Funding
This work is partially supported by Seed Funding Programme for Basic Research (project code: 201511159156), University Research Committee, The University of Hong Kong.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the approval from Prof. Hongzhe Sun for instrumental usage, and Dr. Xinming Yang and Ms. Ya Yang for their technical assistance in fluorescence spectrophotometry and gel-filtration chromatography at the Department of Chemistry of The University of Hong Kong.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00206/full#supplementary-material
References
Boonstra, E., de Kleijn, R., Colzato, L. S., Alkemade, A., Forstmann, B. U., and Nieuwenhuis, S. (2015). Neurotransmitters as food supplements: the effects of GABA on brain and behavior. Front. Psychol. 6:1520. doi: 10.3389/fpsyg.2015.01520
Breeuwer, P., Drocourt, J., Rombouts, F. M., and Abee, T. (1996). A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl. Environ. Microb. 62, 178–183.
Bron, P. A., van Baarlen, P., and Kleerebezem, M. (2012). Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 10, 66–78. doi: 10.1038/nrmicro2690
Capitani, G., De Biase, D., Aurizi, C., Gut, H., Bossa, F., and Grütter, M. G. (2003). Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 22, 4027–4037. doi: 10.1093/emboj/cdg403
Cotter, P. D., Gahan, C. G., and Hill, C. (2001). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. doi: 10.1046/j.1365-2958.2001.02398.x
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Davidi, D., Noor, E., Liebermeister, W., Bar-Even, A., Flamholz, A., Tummler, K., et al. (2016). Global characterization of in vivo enzyme catalytic rates and their correspondence to in vitro k(cat) measurements. Proc. Natl. Acad. Sci. U.S.A. 113, 3401–3406. doi: 10.1073/pnas.1514240113
De Biase, D., and Pennacchietti, E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86, 770–786. doi: 10.1111/mmi.12020
Delzenne, N. M., Neyrinck, A. M., Bäckhed, F., and Cani, P. D. (2011). Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 7, 639–646. doi: 10.1038/nrendo.2011.126
Dhakal, R., Bajpai, V. K., and Baek, K. H. (2012). Production of GABA (γ-aminobutyric acid) by microorganisms: a review. Braz. J. Microbiol. 43, 1230–1241. doi: 10.1590/S1517-83822012000400001
Diana, M., Quilez, J., and Rafecas, M. (2014). Gamma-aminobutyric acid as a bioactive compound in foods: a review. J. Funct. Foods 10, 407–420. doi: 10.1016/j.jff.2014.07.004
Dugo, G., Vilasi, F., La Torre, G. L., and Pellicano, T. M. (2006). Reverse phase HPLC/DAD determination of biogenic amines as dansyl derivatives in experimental red wines. Food Chem. 95, 672–676. doi: 10.1016/j.foodchem.2005.07.001
Fan, E., Huang, J., Hu, S., Mei, L., and Yu, K. (2012). Cloning, sequencing and expression of a glutamate decarboxylase gene from the GABA producing strain Lactobacillus brevis CGMCC 1306. Ann. Microbiol. 62, 689–698. doi: 10.1007/s13213-011-0307-5
Fanning, S., Hall, L. J., Cronin, M., Zomer, A., Macsharry, J., Goulding, D., et al. (2012). Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U.S.A. 109, 2108–2113. doi: 10.1073/pnas.1115621109
Fiocco, D., Crisetti, E., Capozzi, V., and Spano, G. (2008). Validation of an internal control gene to apply reverse transcription quantitative PCR to study heat, cold and ethanol stresses in Lactobacillus plantarum. World J. Microb. Biot. 24, 899–902. doi: 10.1007/s11274-007-9556-7
Gareau, M. G., Sherman, P. M., and Walker, W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastro. Hepat. 7, 503–514. doi: 10.1038/nrgastro.2010.117
Hansen, G., Johansen, C. L., Marten, G., Wilmes, J., Jespersen, L., and Arneborg, N. (2016). Influence of extracellular pH on growth, viability, cell size, acidification activity, and intracellular pH of Lactococcus lactis in batch fermentations. Appl. Microbiol. Biotechnol. 100, 5965–5976. doi: 10.1007/s00253-016-7454-3
Herzog, R. I., Jiang, L., Herman, P., Zhao, C., Sanganahalli, B. G., Mason, G. F., et al. (2013). Lactate preserves neuronal metabolism and function following antecedent recurrent hypoglycemia. J. Clin. Invest. 123, 1988–1998. doi: 10.1172/JCI65105
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro. Hepat. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hiraga, K., Ueno, Y., and Oda, K. (2008). Glutamate decarboxylase from Lactobacillus brevis: activation by ammonium sulfate. Biosci. Biotech. Bioch. 72, 1299–1306. doi: 10.1271/bbb.70782
Hutkins, R. W., and Nannen, N. L. (1993). pH homeostasis in lactic acid bacteria. J. Dairy Sci. 76, 2354–2365. doi: 10.3168/jds.S0022-0302(93)77573-6
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kurihara, S., Kato, K., Asada, K., Kumagai, H., and Suzuki, H. (2010). A putrescine-inducible pathway comprising PuuE-YneI in which gamma-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192, 4582–4591. doi: 10.1128/JB.00308-10
Kurihara, S., Oda, S., Tsuboi, Y., Kim, H. G., Oshida, M., Kumagai, H., et al. (2008). Gamma-glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 283, 19981–19990. doi: 10.1074/jbc.M800133200
Kuriyama, K., and Sze, P. Y. (1971). Blood-brain barrier to H3-gamma-aminobutyric acid in normal and amino oxyacetic acid-treated animals. Neuropharmacology 10, 103–108. doi: 10.1016/0028-3908(71)90013-X
Leroy, F., and De Vuyst, L. (2004). Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Tech. 15, 67–78. doi: 10.1016/j.tifs.2003.09.004
Li, H., and Cao, Y. (2010). Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 39, 1107–1116. doi: 10.1007/s00726-010-0582-7
Lin, L., Hu, S., Yu, K., Huang, J., Yao, S. J., Lei, Y. L., et al. (2014). Enhancing the activity of glutamate decarboxylase from Lactobacillus brevis by directed evolution. Chinese J. Chem. Eng. 22, 1322–1327. doi: 10.1016/j.cjche.2014.09.025
Liu, Y., Tang, H., Lin, Z., and Xu, P. (2015). Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol. Adv. 33, 1484–1492. doi: 10.1016/j.biotechadv.2015.06.001
Lu, P., Ma, D., Chen, Y., Guo, Y., Chen, G. Q., Deng, H., et al. (2013). L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 23, 635–644. doi: 10.1038/cr.2013.13
Ma, D., Lu, P., Yan, C., Fan, C., Yin, P., Wang, J., et al. (2012). Structure and mechanism of a glutamate-GABA antiporter. Nature 483, 632–U161. doi: 10.1038/nature10917
Martinez, F. A., Balciunas, E. M., Converti, A., Cotter, P. D., and de Souza Oliveira, R. P. (2013). Bacteriocin production by Bifidobacterium spp. A review. Biotechnol. Adv. 31, 482–488. doi: 10.1016/j.biotechadv.2013.01.010
Möhle, L., Mattei, D., Heimesaat, M. M., Bereswill, S., Fischer, A., Alutis, M., et al. (2016). Ly6Chi monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep 15, 1945–1956. doi: 10.1016/j.celrep.2016.04.074
Park, J. Y., Jeong, S. J., and Kim, J. H. (2014). Characterization of a glutamate decarboxylase (GAD) gene from Lactobacillus zymae. Biotechnol. Lett. 36, 1791–1799. doi: 10.1007/s10529-014-1539-9
Schurr, B. C., Behr, J., and Vogel, R. F. (2013). Role of the GAD system in hop tolerance of Lactobacillus brevis. Eur. Food Res. Technol. 237, 199–207. doi: 10.1007/s00217-013-1980-3
Seo, M. J., Nam, Y. D., Lee, S. Y., Park, S. L., Yi, S. H., and Lim, S. I. (2013). Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing gamma-aminobutyric acid. Biosci. Biotech. Bioch. 77, 853–856. doi: 10.1271/bbb.120785
Seo, S. W., Kim, D., O'brien, E. J., Szubin, R., and Palsson, B. O. (2015). Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 6:7970. doi: 10.1038/ncomms8970
Shi, F., Xie, Y., Jiang, J., Wang, N., Li, Y., and Wang, X. (2014). Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzyme Microb. Tech. 61–62, 35–43. doi: 10.1016/j.enzmictec.2014.04.012
Shin, S. M., Kim, H., Joo, Y., Lee, S. J., Lee, Y. J., Lee, S. J., et al. (2014). Characterization of glutamate decarboxylase from Lactobacillus plantarum and its C-terminal function for the pH dependence of activity. J. Agr. Food Chem. 62, 12186–12193. doi: 10.1021/jf504656h
Siegumfeldt, H., Björn Rechinger, K., and Jakobsen, M. (2000). Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microb. 66, 2330–2335. doi: 10.1128/AEM.66.6.2330-2335.2000
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi: 10.1126/science.aac4255
Su, M. S., Schlicht, S., and Gänzle, M. G. (2011). Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 10(Suppl 1):S8. doi: 10.1186/1475-2859-10-S1-S8
Tang, F., Lane, S., Korsak, A., Paton, J. F., Gourine, A. V., Kasparov, S., et al. (2014). Lactate-mediated glia-neuronal signalling in the mammalian brain. Nat. Commun. 5:3284. doi: 10.1038/ncomms4284
Teixeira, J. S., Seeras, A., Sanchez-Maldonado, A. F., Zhang, C., Su, M. S., and Gänzle, M. G. (2014). Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 42, 172–180. doi: 10.1016/j.fm.2014.03.015
Tillisch, K., Labus, J., Kilpatrick, L., Jiang, Z., Stains, J., Ebrat, B., et al. (2013). Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 144, 1394–1401.e4. doi: 10.1053/j.gastro.2013.02.043
Ueno, Y., Hayakawa, K., Takahashi, S., and Oda, K. (1997). Purification and characterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotech. Bioch. 61, 1168–1171. doi: 10.1271/bbb.61.1168
van Baarlen, P., Wells, J. M., and Kleerebezem, M. (2013). Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 34, 208–215. doi: 10.1016/j.it.2013.01.005
Wu, Q., Law, Y. S., and Shah, N. P. (2015). Dairy Streptococcus thermophilus improves cell viability of Lactobacillus brevis NPS-QW-145 and its gamma-aminobutyric acid biosynthesis ability in milk. Sci. Rep. 5:12885. doi: 10.1038/srep12885
Wu, Q., and Shah, N. P. (2015). Gas release-based prescreening combined with reversed-phase HPLC quantitation for efficient selection of high-gamma-aminobutyric acid (GABA)-producing lactic acid bacteria. J. Dairy Sci. 98, 790–797. doi: 10.3168/jds.2014-8808
Wu, Q., and Shah, N. P. (2016). High γ-aminobutyric acid production from lactic acid bacteria: emphasis on Lactobacillus brevis as a functional dairy starter. Crit. Rev. Food Sci. Nutr. doi: 10.1080/10408398.2016.1147418. [Epub ahead of print].
Wu, Q., Tun, H. M., Leung, F. C., and Shah, N. P. (2014). Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Sci. Rep. 4:4974. doi: 10.1038/srep04974
Yang, J. Y., Ruchti, E., Petit, J. M., Jourdain, P., Grenningloh, G., Allaman, I., et al. (2014). Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc. Natl. Acad. Sci. U.S.A. 111, 12228–12233. doi: 10.1073/pnas.1322912111
Yu, K., Lin, L., Hu, S., Huang, J., and Mei, L. (2012). C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH. Enzyme Microb. Tech. 50, 263–269. doi: 10.1016/j.enzmictec.2012.01.010
Zhang, Y., Song, L., Gao, Q., Yu, S. M., Li, L., and Gao, N. F. (2012). The two-step biotransformation of monosodium glutamate to GABA by Lactobacillus brevis growing and resting cells. Appl. Microbiol. Biot. 94, 1619–1627. doi: 10.1007/s00253-012-3868-8
Keywords: genomic survey, Lactobacillus brevis, γ-aminobutyric acid (GABA), glutamic acid decarboxylase, acid resistance
Citation: Wu Q, Tun HM, Law Y-S, Khafipour E and Shah NP (2017) Common Distribution of gad Operon in Lactobacillus brevis and its GadA Contributes to Efficient GABA Synthesis toward Cytosolic Near-Neutral pH. Front. Microbiol. 8:206. doi: 10.3389/fmicb.2017.00206
Received: 10 November 2016; Accepted: 30 January 2017;
Published: 14 February 2017.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Shu-Wei Marcia Su, Pennsylvania State University, USASergio I. Martinez-Monteagudo, South Dakota State University, USA
Copyright © 2017 Wu, Tun, Law, Khafipour and Shah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nagendra P. Shah, bnBzaGFoQGhrdS5oaw==
 Qinglong Wu
Qinglong Wu Hein Min Tun
Hein Min Tun Yee-Song Law
Yee-Song Law Ehsan Khafipour
Ehsan Khafipour Nagendra P. Shah1,4*
Nagendra P. Shah1,4*