- Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Key Laboratory of Dairy Products Processing, Ministry of Agriculture, Inner Mongolia Agricultural University, Hohhot, China
Amoxicillin is one of the most commonly prescribed antibiotics for bacterial infections and gastrointestinal disorders. To investigate the adaptation of Lactobacillus (L.) casei Zhang to amoxicillin stress, an iTRAQ-based comparative proteomic analysis was performed to compare the protein profiles between the parental L. casei Zhang and its amoxicillin-resistant descendent strains. Our results revealed a significant increase in the relative expression of 38 proteins (>2.0-folds, P < 0.05), while the relative expression of 34 proteins significantly decreased (<−2.0-folds, P < 0.05). The amoxicillin-resistant descendent strain exhibited marked alterations in carbohydrate and amino acid metabolism. Moreover, certain components involving in membrane metabolism were activated. The differences in the proteomic profiles between the two strains might explain the enhanced stress resistance of the adapted bacteria.
Introduction
Antibiotic resistance is a global health problem. Particularly, many antibiotics have become less effective due to rapid bacterial adaptive evolution facilitated by the frequent use of antibiotics in medicine and agriculture (Arias and Murray, 2015). Meanwhile, the increased use of antibiotics can also introduce a selective pressure which leads to the development of multi-resistance characteristics in some of the bacterial populations (Chen and Jiang, 2014). Owing to significant clinical concerns, many previous studies have investigated the relationship between antibiotic resistance and genome stability of pathogenic bacteria, especially when environmental antibiotic selective pressure is present (Andersson, 2006). Meanwhile, it is crucial to develop new strategies to prevent and control the spread of antibiotic resistance (Normark and Normark, 2002). One successful strategy is to minimize the use of antibiotics by including alternative and/or adjunct treatments for certain diseases (Schultz and Haas, 2011). For example, the combined use of probiotics and antibiotics was clinically effective in treating gastrointestinal disorders (Wright et al., 2015). Thus, there is growing interest in applying probiotics to improve human health and in clinical practice (Reid, 2017).
One concern, however, is the potential risk of evolutionary adaptation of probiotics toward antibiotic resistance after prolonged drug exposure (Perreten et al., 1997). Since most published studies have focused only on pathogenic bacteria, there are insufficient data to assess the safe use of probiotic bacteria in clinical practice. Although horizontal gene transfer is a major mechanism that had shaped the antibiotic resistance pattern of probiotics bacteria in evolution, genome point mutations cannot be neglected (Woodford and Ellington, 2007). Some lactobacilli strains have been shown to gain antibiotic resistance via point mutations (Curragh and Colllns, 1992). On the other hand, by a whole-genome resequencing approach, our earlier work monitored the genetic changes of Lactobacillus casei Zhang during long-term culture in an antibiotics-containing medium; and we found that, unlike pathogenic bacteria, the accumulation of de novo mutations occurred only initially but not after an extended period of antibiotics selection (Wang et al., 2017). However, mechanistic changes occurring at the functional level remain uncharacterized.
In a previous long-term propagation experiment, our laboratory isolated an L. casei Zhang descendent (L. casei Zhang-A-600) that had elevated resistance to amoxicillin (Wang et al., 2017). Amoxicillin is one of the most commonly prescribed antibiotics used for treating bacterial infections and gastrointestinal disorders (Kabbani et al., 2017; Zerbetto De Palma et al., 2017). It kills bacteria by inhibiting the process of cell wall synthesis. Although amoxicillin resistance is not yet considered as a serious clinical concern, several amoxicillin-tolerant strains have been isolated from gastric biopsy specimens of patients (van Zwet et al., 1998); thus, its clinical significance should not be neglected. The present work hypothesized that the amoxicillin-resistant descendent adapted to antibiotics stress via modulating cellular protein expression. Comparative proteomics analysis is an efficient tool to reveal functional differences between wild-type and mutant cells or between cells cultivated under different conditions (Wang et al., 2013). Thus, an iTRAQ-based proteomic analysis was performed to elucidate the resistant phenotype of the descendent at a global protein expression level.
Materials and Methods
Bacterial Isolates and Growth
The L. casei Zhang descendent strain (L. casei Zhang-A-600) was more resistant to amoxicillin resistance with a minimum inhibitory concentration (MIC) of 8 μg/mL (vs. 2 μg/mL for the parental strain; Wang et al., 2017). The resistant strain was isolated from long-term propagation of the parental strain in lactic acid bacteria (LAB) susceptibility test medium broth (LSM), consisting of 90% Iso-sensitest medium (IST; OXOID, CM0473) and 10% MRS (Klare et al., 2005), supplemented with 0.5 μg/mL amoxicillin (Wang et al., 2017). At this antibiotics concentration, the bacterial growth was suppressed by 50%. The growth of bacteria (optical density, pH, and viable counts) was monitored every 2 h (from 0 to 30 h of cultivation). All experiments were performed in triplicate.
Sample Preparation
To minimize the interfering effect between the antimicrobial compound and the growth medium components (Klare et al., 2005), the minimal growth medium, LSM, was chosen for the current study. For proteomic analysis, the parental and descendent L. casei Zhang cells were collected after 24 h of cultivation in amoxicillin-containing LSM (0.5 μg/mL of amoxicillin). In each case, 4 biological replicates of samples were prepared. The culture conditions used here were the same as our previous study which aimed to characterize the adaptation of the resistant strain at the genomic level (Wang et al., 2017). Briefly, the bacterial cells were pelleted by centrifugation and washed 4 times with phosphate buffered saline (PBS). One milliliter of lysis buffer (7 M urea, 4% SDS, and 1x protease inhibitor cocktail) was added to each sample. The mixtures were then sonicated on ice and spun at 13,000 rpm for 10 min at 4°C. The supernatant of each sample was separately collected.
Protein Digestion and iTRAQ Labeling
The protein concentration of the supernatants was estimated by the bicinchoninic acid protein assay. One hundred microgram of protein of each sample was adjusted to a final volume of 100 μL with 100 mM triethylammonium bicarbonate (TEAB), followed by adding 5 μL of 200 mM DTT and incubating at 55°C for 1 h. Afterwards, 5 μL of iodoacetamide (375 mM) was added to each sample, followed by 30 min incubation in dark at room temperature. Then the protein was precipitated with ice-cold acetone and redissolved in TEAB (20 μL). Proteins were digested with sequence-grade modified trypsin (Promega, Madison, WI) and labeled using the iTRAQ reagents kit. The labeled samples were combined, desalted (Sep-Pak C18 SPE column, Waters, Milford, MA), and vacuum dried.
High pH Reverse Phase Separation
Phase separation was performed as described by Gilar with some modifications (Gilar et al., 2005). Briefly, the peptide mixture was redissolved in buffer A (buffer A: 10 mM ammonium formate in water, pH 10.0, adjusted with ammonium hydroxide). The dissolved peptide mixtures were then fractionated by high pH separation using the Aquity UPLC system (Waters Corporation, Milford, MA) connected to a reverse phase column (BEH C18 column, 2.1 × 150 mm, 1.7 μm, 300 Å, Waters Corporation, Milford, MA). A linear gradient, starting from 0% B to 45% B in 35 min (B: 10 mM ammonium formate in 90% ACN, pH 10.0, adjusted with ammonium hydroxide), was used in the high pH separation. The column flow rate and temperature were maintained at 250 μL/min and 45°C, respectively. Sixteen fractions were separately collected and dried in a vacuum concentrator.
Low pH Nano-HPLC-MS/MS Analysis
The fractions were redissolved in a solvent composed of solvents C and D (C: 0.1% formic acid in water; D: 0.1% formic acid in ACN), separated by nano LC and analyzed by on-line electrospray tandem mass spectrometry. The experiments were performed on a Nano Aquity UPLC system (Waters Corporation, Milford, MA) connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive) (Thermo Fisher Scientific, Bremen, Germany) with an online nano-electrospray ion source. Eight microliters of each peptide sample were loaded onto the trap column (Thermo Scientific Acclaim PepMap C18, 100 μm × 2 cm), with a flow of 10 μl/min for 3 min, to be separated on a 75 μm × 25 cm Acclaim PepMap C18 analytical column. A linear gradient, from 5% D to 30% D in 95 min, was used. The column was re-equilibrated at initial conditions for 15 min. The column flow rate and temperature were maintained at 300 μL/min and 45°C, respectively. An electrospray voltage of 2.0 kV was applied to the mass spectrometer inlet.
The Q-Exactive mass spectrometer was operated in the data-dependent mode, switching automatically between MS and MS/MS acquisition. Survey full-scan MS spectra (m/z 350–1,600) of mass resolution of 70K were acquired, followed by 15 sequential higher-energy collisional dissociation (HCD) MS/MS scans of 17.5K resolution. In all cases, one 30-s dynamic exclusion micro-scan was recorded. The MS/MS fixed first mass was set to 100.
Database Searching
Tandem mass spectra were extracted by ProteoWizard (version 3.0.5126; Thermo Fisher Scientific) using the Proteome Discoverer software (version 1.4.0.288; Thermo Fisher Scientific). All MS/MS samples were analyzed using Mascot (version 2.3; Matrix Science, London, UK), which was used to search the NCBI database (Taxonomy: Lactobacillus casei Zhang, 2804 entries; trypsin digestion; 0.050 Da fragment ion mass tolerance and 10.0 PPM parent ion tolerance). Moreover, in the Mascot search, cysteine carbamidomethylation and iTRAQ 8plex of lysine and the n-terminus were opted as fixed modifications, while methionine oxidation and iTRAQ 8plex of tyrosine were specified as variable modifications.
Quantitative Data Analysis
Statistical analyses were performed following the recommendations of Predrag Radivojac and Olga Vitek (Radivojac and Vitek, 2012). The percolator algorithm of <1% was used to control the false discovery rate. Only unique peptides were quantified. Experimental biases were corrected by normalization with the protein median. The minimum number of observed proteins was 1000. Statistical analysis was performed under the R environment (Student's t-tests, p < 0.05 was considered statistically significant). A cut-off level of 2.0-fold change was applied to select differentially expressed proteins; and only those showing a consistent expression change in all 4 biological replicates were considered as differentially expressed proteins. They were functionally assigned by the clusters of orthologous genes (COGs) and the Kyoto Encyclopedia of Genes and Genomes databases (Tatusov et al., 1997).
Construction and Analysis of Gene Disruption Mutants
Two genes, LCAZH_0490 and LCAZH_0521, were selected as target candidates to be genetically disrupted. They putatively encoded an OmpR family DNA-binding response regulator and a surface-associated protein, respectively. The plasmids and primers used for constructing the gene disruption mutants are listed in Table 1.
The gene disruption mutants were constructed using a cre-lox-based system (Lambert et al., 2007). To disrupt the LCAZH_0490 gene, the upstream and downstream flanking regions of the LCAZH_0490 gene were amplified by PCR by two primer pairs (0490upF and 0490upR; 0490downF and 0490downR) using the genomic DNA of L. casei Zhang-A-600 as template. These fragments were then cloned between the Xho I or Pme I and Ecol53 KI or Bgl II restriction sites of the suicide vector pNZ5319 to create the recombinant mutagenesis vector, pNZ5319-0490 Up-Down, which was introduced into L. casei Zhang-A-600 by electroporation. Colonies harboring the anticipated inserts in the desired orientation were identified by the combined use of the primers 85, 87, and an insert-specific primer. Chloramphenicol-resistant transformants were selected and replica plated to check for erythromycin-sensitive clones. Candidate double-crossover mutant clones were first analyzed by PCR (with the primer pairs, catR and catF, EryintF, and EryintR), followed by verifying the correct integration of the 0490::lox66-P32-cat-lox71 cassette into the genome using the primer combination of 0490upF or catR and catF or 0490downR. In order to excise the P32-cat selectable marker cassette, the cre expression plasmid, pMSPcre, was transformed into the 0490::lox66-P32-cat-lox71 gene replacement mutant. Erythromycin-resistant and chloramphenicol-sensitive colonies were selected by another round of replica plating. The cre-mediated recombination and correct excision of the P32-cat cassette were confirmed by PCR using primers spanning the recombination locus (0490upF and 0490downR). The pMSPcre vector was cured from L. casei Zhang-A-600 Δ0490 colonies by growth without erythromycin selection pressure. This plasmid was constructed from pMSP3535 (provided by Professor Jian Kong, Shandong University, unpublished). Similar procedures were used to inactivate the LCAZH_0521 gene in L. casei Zhang-A-600.
The amoxicillin-resistant phenotype of the gene disrupted mutants were evaluated by determining their MICs of amoxicillin (Guo et al., 2017). Moreover, the OD value at the time of observing the MIC by witness was determined to quantify the growth performance of the wild-type and mutant strains. The Student's t-test at a confidence level of 0.05 was used to evaluate any difference in the growth performance between different strains.
Nucleotide Sequence Accession Number
The gene disruption plasmids were designed and constructed based on the genome sequence of L. casei Zhang (GenBank database accession number: CP001084.2). The sequences of gene disruption regions of the mutant strains, Zhang-A-600-delta-0490 and Zhang-A-600-delta-0521, have been deposited to the GenBank database under the accession numbers MG021089 and MG021090.
Results
Growth of L. casei Zhang-A-600 and the Parental Strains
The growth characteristics (viable counts, pH, and OD values) of the parental L. casei Zhang strain and the amoxicillin-resistant Zhang-A-600 descendent were monitored. The growth performance of L. casei Zhang-A-600 was better than that of the parental strain when amoxicillin was present in the culture medium (Figures 1A–C). Both the maximum viable count and cell density of L. casei Zhang-A-600 were significantly higher (P < 0.05) than that of the parental strain, which were 6.3 × 108 CFU/mL and 1.36, respectively. The L. casei Zhang-A-600 culture entered stationary phase at 12 h (vs. 10 h for the parental strain; Figure 1A). Moreover, the pH of the L. casei Zhang-A-600 culture dropped much faster than that of the parental strain (Figure 1B).
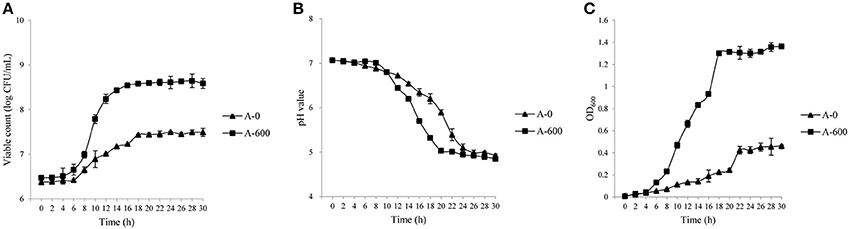
Figure 1. Growth of Lactobacillus casei strains in lactic acid bacteria susceptibility test medium broth (LSM) supplemented with amoxicillin. Changes in the viable counts (A), pH (B), and OD600 (C) were monitored over 30 h. The parental and the amoxicillin-resistant (Zhang-A-600) strains are represented by “A-0” and “A-600,” respectively. Error bars represent standard deviation.
Differentially Expressed Proteins Identified in L. casei Zhang-A-600
When L. casei Zhang-A-600 was grown in the presence of amoxicillin, the expression of 38 proteins significantly increased (>2.0-folds, P < 0.05) compared with the parental cells (Table 2). Most of these proteins could be assigned to specific COG functional categories.
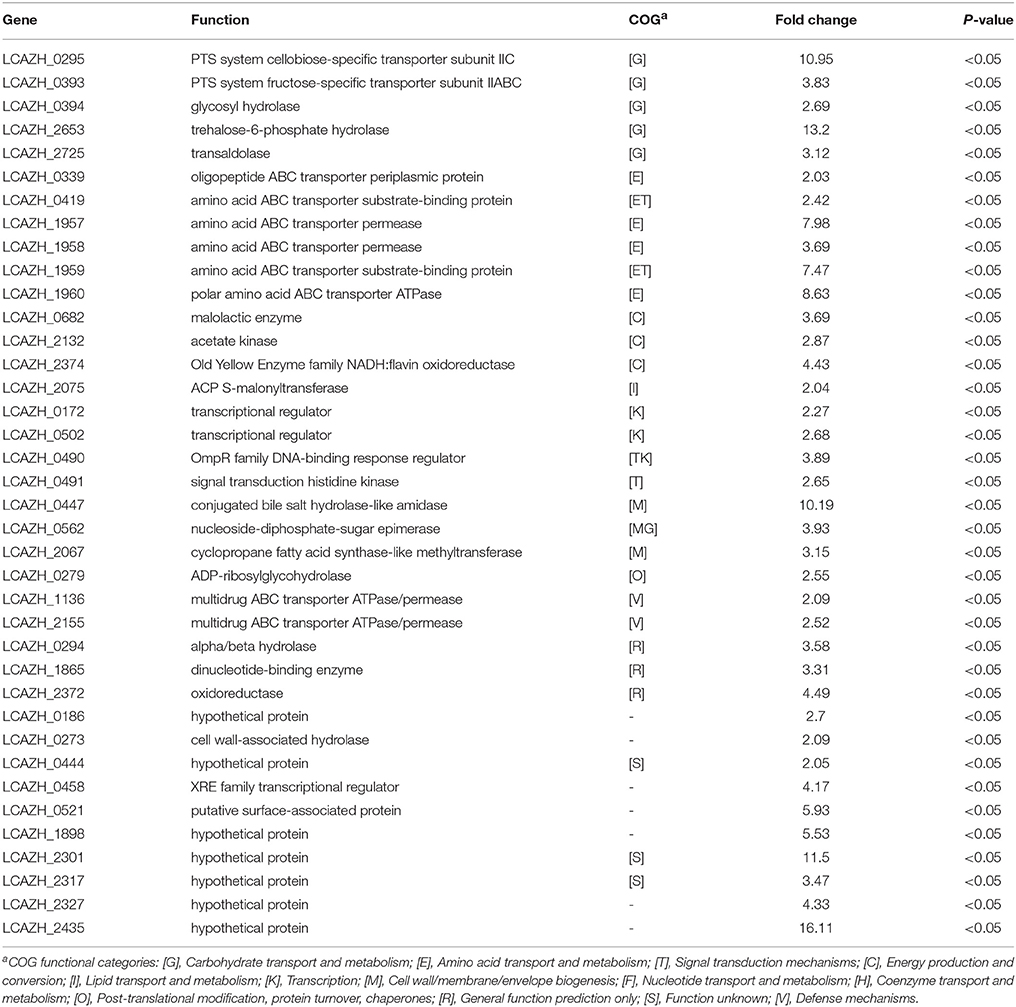
Table 2. Up-regulated Lactobacillus casei Zhang-A-600 proteins in comparison with the parental strain.
Six of the highly expressed proteins (15.8%) were involved in amino acid transport and metabolism (E), including a transporter component for oligopeptides (LCAZH_0339), an amino acid ABC transporter substrate-binding protein (LCAZH_0419), as well as a set of proteins responsible for amino acid transport (LCAZH_1957–LCAZH_1960). Five other highly expressed proteins (12.8%) were associated with carbohydrate transport and metabolism (G), namely the PTS system cellobiose-specific transporter subunit IIC (LCAZH_0295), the PTS system fructose-specific transporter subunit IIABC (LCAZH_0393), a trehalose-6-phosphate hydrolase (LCAZH_2653), and a transaldolase (LCAZH_2725).
Some of the highly expressed proteins fell into the COG classes T and M, which were connected with cellular stress response. These included the OmpR family DNA-binding response regulator (LCAZH_0490), a signal transduction histidine kinase (LCAZH_0491), a conjugated bile salt hydrolase-like amidase (LCAZH_0447), a nucleoside-diphosphate-sugar epimerase (LCAZH_0562), and a cyclopropane fatty acid synthase-like methyltransferase (LCAZH_2067). The functions of several other differentially expressed proteins were unknown.
In contrast to the spectrum of highly expressed proteins, the majority of the lowly expressed proteins (35.3%) belonged to the COG class G (Table 3). Among them, 4 putative proteins, namely the PTS system cellobiose-specific transporter subunits IIA and IIB (LCAZH_2637, LCAZH_2638), a triosephosphate isomerase (LCAZH_2697), and a fructose/tagatose bisphosphate aldolase (LCAZH_2698), were encoded by genes located within an operon-like structure in the L. casei Zhang genome. Two other lowly expressed proteins were assigned to the COG class V, i.e., the antimicrobial peptide ABC transporter permease and ATPase (LCAZH_1927, LCAZH_1928).
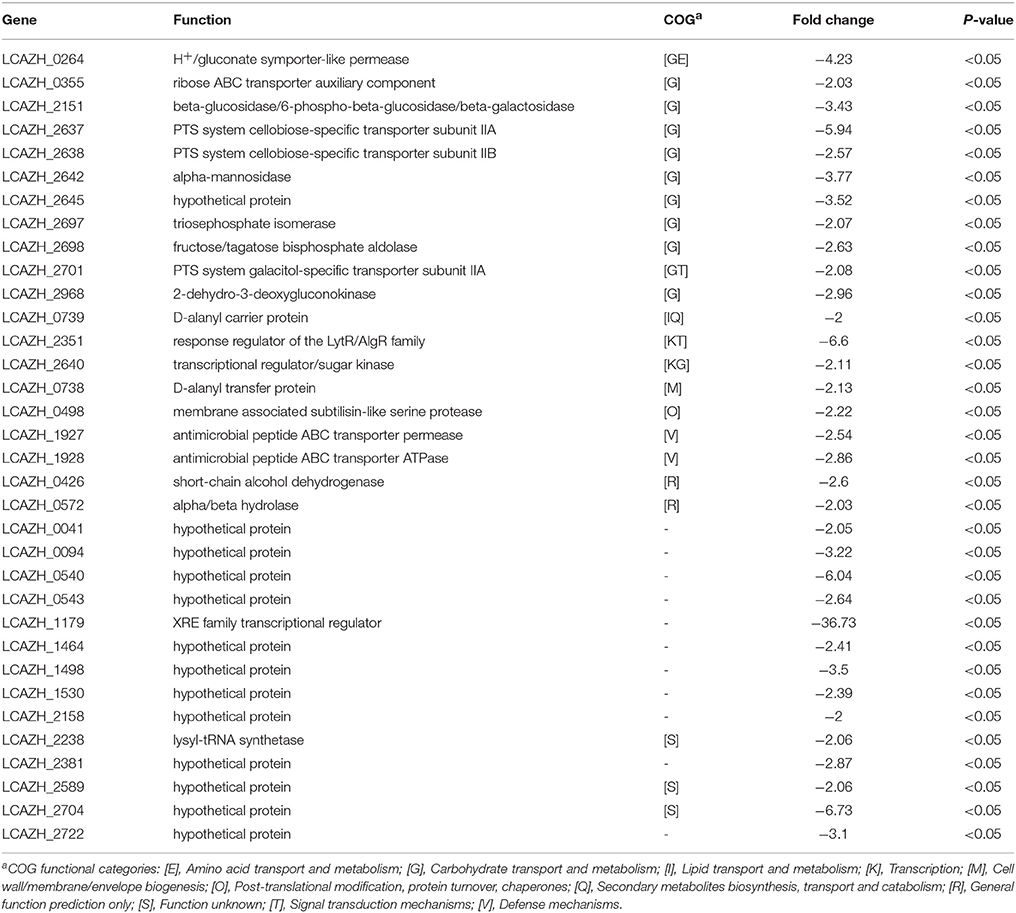
Table 3. Down-regulated Lactobacillus casei Zhang-A-600 proteins in comparison with the parental strain.
Amoxicillin-Resistant Phenotype of the Mutants
The target knockout genes, LCAZH_0490 and LCAZH_0521, were selected because of their predicted molecular functions in stress response. No significant difference (P > 0.05) was observed in the MICs of amoxicillin between the gene disruption mutants and the wild-type, L. casei Zhang-A-600. However, the mutants, particularly Δ0490 that lacked the response regulator, grew slower than L. casei Zhang-A-600 in the presence of amoxicillin. As shown in Figure 2, the OD of the L. casei Zhang-A-600 culture was 1.31-fold higher (P < 0.05) than that of the mutant Δ0490 cultivated in LSM with 4 μg/mL amoxicillin.
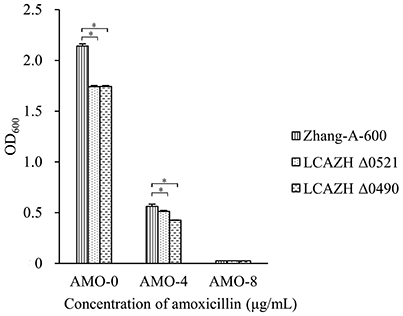
Figure 2. Effect of amoxicillin concentration on the OD600 of the culture medium of the wild-type (Zhang-A-600) and the mutants (LCAZH Δ0490 and LCAZH Δ0521). Error bars represent standard deviation. Asterisks indicate the level of statistical significance (“*” represents P < 0.05); no asterisk indicates no significance (P > 0.05).
Discussion
The combined use of antibiotics and probiotics has recently been shown to improve the eradication rate for certain infections (Kafshdooz et al., 2017). However, there is yet insufficient data to assess the safe use of probiotic bacteria in clinical practice. Particularly, the antibiotics-induced adaptation responses of probiotics are not well characterized. Previously, our laboratory isolated an amoxicillin-resistant L. casei Zhang strain in a long-term antibiotics-driven evolution experiment. Here, we aimed to investigate the mechanisms of amoxicillin resistance of this strain using a comparative proteomics approach.
The amoxicillin-resistant isolate had altered carbohydrate metabolism. Although glucose was the main carbon source in the culture medium used in the experiment, several proteins involving in carbohydrate metabolism, including beta-glucoside metabolism (LCAZH_0295), fructose utilization (LCAZH_0393 and LCAZH_2725), and trehalose utilization (LCAZH_2653), were highly expressed compared with the parental strain. In contrast, the expression of some glycolysis- and gluconeogenesis-associated proteins (LCAZH_2697 and LCAZH_2698) decreased, reflecting an altered substrate requirement for the antibiotics-resistant strain. Interestingly, the expression of 1 component (LCAZH_0295) of the PTS system cellobiose-specific transporter subunit increased, while the relative abundance of two other components (LCAZH_2637 and LCAZH_2638) of the same transporter decreased. It is hard to explain the inconsistent changes between the individual transporter components. However, since PTS systems are involved in regulating gene expression, we speculate that the divergent expression of these components was associated with cellular protection against environmental stressors via modulation of gene expression (Nascimento et al., 2004).
The switching of carbon utilization from glucose to other substrates often happens when the growth environment turns acidic. For example, this was observed at the start of the late growth phase of L. casei Zhang when it was grown in cow milk and soy milk (Wang et al., 2012a,b). In addition, the growth medium used in this study was a minimal medium that was suboptimal for the growth of lactobacilli, which might have enhanced the switching of carbon utilization. The medium contained hydrolyzed casein, which could potentially serve as an alternative carbohydrate source (Williams et al., 2000). Two other proteins, an acetate kinase (LCAZH_2132) and a malolactic enzyme (LCAZH_0682), were possibly modulated by the carbon source switching as well. The former protein catalyzes the formation of acetyl phosphate from acetate and generates ATP, while the latter one catalyzes the production of L-latate and CO2 from L-malate via decarboxylation (Poolman et al., 1991; Puri et al., 2014). Likewise, the lack of mono-/di-carbohydrates, citrates, and amino acids in the minimal growth medium could have been another factor contributing to the carbon source switching in acidic environment. These survival strategies and acid-tolerant mechanisms are well documented (Behr et al., 2007).
Amino acids are essential for bacterial growth, and the modulation of amino acids metabolism is another strategy that helps increase bacterial stress tolerance. In order to survive, L. casei Zhang cells have to acquire adequate amino acids from the direct growth environment. To aid efficient acquisition of amino acids, LAB usually possess an effective proteolytic enzyme system that generally consists of multiple cell surface-associated proteinases, transport systems, and peptidases. Before any peptides or amino acids can be translocated to the cytoplasm, proteins would first need to be broken down by the bacterial proteinases (Zhang et al., 2015). Overall speaking, the expression of individual protein components of the proteolytic enzyme system was not induced except 1 oligopeptide ABC transporter periplasmic protein (LCAZH_0339), which might be necessary for oligopeptide uptake. This may suggest that the consecutive expression of the proteolytic proteins was enough to support the bacterial growth until the deceleration phase. One interesting observation regarding the amino acid metabolism was the increased expression of the protein clustered LCAZH_1957–LCAZH_1960. This is a putative transporter for polar amino acids, although its substrates are yet to be identified. This finding may suggest that polar amino acids are important in coping with environmental antibiotics stress. Moreover, the disruption of an amino acid permease-coding gene in L. acidophilus greatly increased the acid and bile sensitivity of the mutant (Azcarate-Peril et al., 2004).
Typically, the two component systems (TCS) consist of a sensor kinase and a response regulator; and they together play crucial roles in facilitating bacterial adaptation to environmental changes (El-Sharoud, 2005). The genome of L. casei encodes a relatively high number of TCS (Zhang et al., 2010), allowing cells to monitor their direct environment and respond rapidly to external stimuli, including chemical changes, acid, bile, and salt stresses (Landete et al., 2010; Alcántara et al., 2011; Revilla-Guarinos et al., 2013). Two component systems also confer adaptive antibiotic resistance to the species Pseudomonas aeruginosa and Enterococcus faecalis (Fernández et al., 2010; Hancock and Perego, 2012). In particular, the TCS of Pseudomonas aeruginosa activates a lipopolysaccharide modification operon that confers antibiotic resistance to the bacteria. The amoxicillin-resistant L. casei Zhang strain was found to have an increased expression in one TCS pair (LCAZH_0490 and LCAZH_0491). However, no significant difference was noted in the MIC of amoxicillin between the TCS-disrupted mutant and the wild-type strain, suggesting that this TCS might not contribute directly to the amoxicillin-resistant phenotype. Alternatively, the accumulated mutations in the antibiotics-resistant strain might have bypass the effect of TCS inactivation (Wang et al., 2017).
Cell surface is the interface between the bacterial cell and the environment when LAB confront situations of adversity. Acid and hypersaline stresses could cause alterations in the fatty acid metabolism of L. casei (Machado et al., 2004; Wu et al., 2014). Similar to our previous work, the expression of a cyclopropane fatty acid synthase-like methyltransferase (LCAZH_2067) and an ACP S-malonyltransferase (LCAZH_2075) increased in the antibiotics-resistant strain. These two proteins participate in fatty acid biosynthesis. During fatty acid biosynthesis, the cyclopropane fatty acid synthase-like methyltransferase catalyzes the addition of a methylene residue across the cis double bond of C16:1n(9), C18:1n(9), or C18:1n(11) unsaturated fatty acids to form an unsaturated cyclopropane derivative; and the ACP S-malonyltransferase catalyzes the formation of malonyl-ACP (Payne et al., 2001). The activation of these enzymes could be a part of the membrane adaptation to the surrounding environment (Wang et al., 2017). Meanwhile, we also observed an increased expression in a WxL domain (IPR027994)-containing surface-associated protein (LCAZH_0521). Some WxL domain-containing proteins can interact with cell wall peptidoglycan and are responsive to stress (Brinster et al., 2007). However, no significant phenotypic change was observed in the gene disruption mutant LCAZH Δ0521. Further inspection of the genome of L. casei Zhang revealed two other WxL domain-containing proteins (LCAZH_0527 and LCAZH_0529); and whether they serve any compensatory role in the mutant LCAZH Δ0521 remains to be further explored.
Conclusion
In summary, we compared the proteomes of a resistant L. casei Zhang strain isolated in an amoxicillin-driven evolution experiment and its parental line. The resistant descendent strain exhibited alterations in the carbohydrate, amino acid, and membrane metabolism. These metabolic adaptations might have enhanced the cell survival in response to the stressors. Interestingly, a TCS was found to be associated with the experimental evolution. However, further experiments are required to confirm its role in antibiotic resistance in probiotic bacteria.
Author Contributions
WenZ and HZ designed the study. WenZ, L-YK, and JW wrote the manuscript. JW, HG, CC, WeiZ, and L-YK performed the experiments. WenZ and JW analyzed the data. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 31660457) and the Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (Grant no. NJYT-17-B05). We would like to thank Prof. Jian Kong of Shangdong University for providing the plasmid pMSPcre.
References
Alcántara, C., Revilla-Guarinos, A., and Zúñiga, M. (2011). Influence of two-component signal transduction systems of Lactobacillus casei BL23 on tolerance to stress conditions. Appl. Environ. Microbiol. 77, 1516–1519. doi: 10.1128/AEM.02176-10
Andersson, D. I. (2006). The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9, 461–465. doi: 10.1016/j.mib.2006.07.002
Arias, C. A., and Murray, B. E. (2015). A new antibiotic and the evolution of resistance. N. Engl. J. Med. 372, 1168–1170. doi: 10.1056/NEJMcibr1500292
Azcarate-Peril, M. A., Altermann, E., Hoover-Fitzula, R. L., Cano, R. J., and Klaenhammer, T. R. (2004). Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70, 5315–5322. doi: 10.1128/AEM.70.9.5315-5322.2004
Behr, J., Israel, L., Gänzle, M. G., and Vogel, R. F. (2007). Proteomic approach for characterization of hop-inducible proteins in Lactobacillus brevis. Appl. Environ. Microbiol. 73, 3300–3306. doi: 10.1128/AEM.00124-07
Brinster, S., Furlan, S., and Serror, P. (2007). C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189, 1244–1253. doi: 10.1128/JB.00773-06
Chen, Z., and Jiang, X. (2014). Microbiological safety of chicken litter or chicken litter-based organic fertilizers: a review. Agriculture 4, 1–29. doi: 10.3390/agriculture4010001
Curragh, H. J., and Colllns, M. A. (1992). High levels of spontaneous drug resistance in Lactobacillus. J. Appl. Bacteriol. 73, 31–36. doi: 10.1111/j.1365-2672.1992.tb04965.x
El-Sharoud, W. M. (2005). Two-component signal transduction systems as key players in stress responses of lactic acid bacteria. Sci. Prog. 88, 203–228. doi: 10.3184/003685005783238381
Fernández, L., Gooderham, W. J., Bains, M., Mcphee, J. B., Wiegand, I., and Hancock, R. E. (2010). Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54, 3372–3382. doi: 10.1128/AAC.00242-10
Gilar, M., Olivova, P., Daly, A. E., and Gebler, J. C. (2005). Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J. Sep. Sci. 28, 1694–1703. doi: 10.1002/jssc.200500116
Guo, H., Pan, L., Li, L., Lu, J., Kwok, L., Menghe, B., et al. (2017). Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 82, 724–730. doi: 10.1111/1750-3841.13645
Hancock, L., and Perego, M. (2012). Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184, 5189–5825. doi: 10.1128/JB.184.21.5819-5825.2002
Kabbani, T. A., Pallav, K., Dowd, S. E., Villafuerte-Galvez, J., Vanga, R. R., Castillo, N. E., et al. (2017). Prospective randomized controlled study on the effects of Saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes. 8, 17–32. doi: 10.1080/19490976.2016.1267890
Kafshdooz, T., Akbarzadeh, A., Majdi Seghinsara, A., Pourhassan, M., Nasrabadi, H. T., and Milani, M. (2017). Role of probiotics in managing of Helicobacter Pylori infection: a review. Drug Res. (Stuttg) 67, 88–93. doi: 10.1055/s-0042-116441
Klare, I., Konstabel, C., Müller-Bertling, S., Reissbrodt, R., Huys, G., Vancanneyt, M., et al. (2005). Evaluation of new broth media for microdilution antibiotic susceptibility testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl. Environ. Microbiol. 71, 8982–8986. doi: 10.1128/AEM.71.12.8982-8986.2005
Lambert, J. M., Bongers, R. S., and Kleerebezem, M. (2007). Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73, 1126–1135. doi: 10.1128/AEM.01473-06
Landete, J. M., García-Haro, L., Blasco, A., Manzanares, P., Berbegal, C., Monedero, V., et al. (2010). Requirement of the Lactobacillus casei MaeKR two-component system for L-malic acid utilization via a malic enzyme pathway. Appl. Environ. Microbiol. 76, 84–95. doi: 10.1128/AEM.02145-09
Machado, M. C., López, C. S., Heras, H., and Rivas, E. A. (2004). Osmotic response in Lactobacillus casei ATCC 393: biochemical and biophysical characteristics of membrane. Arch. Biochem. Biophys. 422, 61–70. doi: 10.1016/j.abb.2003.11.001
Nascimento, M. M., Lemos, J. A., Abranches, J., Gonçalves, R. B., and Burne, R. A. (2004). Adaptive acid tolerance response of Streptococcus sobrinus. J. Bacteriol. 186, 6383–6390. doi: 10.1128/JB.186.19.6383-6390.2004
Normark, B. H., and Normark, S. (2002). Evolution and spread of antibiotic resistance. J. Intern. Med. 252, 91–106. doi: 10.1046/j.1365-2796.2002.01026.x
Payne, D. J., Warren, P. V., Holmes, D. J., Ji, Y., and Lonsdale, J. T. (2001). Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today 6, 537–544. doi: 10.1016/S1359-6446(01)01774-3
Perreten, V., Schwarz, F., Cresta, L., Boeglin, M., Dasen, G., and Teuber, M. (1997). Antibiotic resistance spread in food. Nature 389, 801–802. doi: 10.1038/39767
Poolman, B., Molenaar, D., Smid, E. J., Ubbink, T., Abee, T., Renault, P. P., et al. (1991). Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173, 6030–6037. doi: 10.1128/jb.173.19.6030-6037.1991
Puri, P., Goel, A., Bochynska, A., and Poolman, B. (2014). Regulation of acetate kinase isozymes and its importance for mixed-acid fermentation in Lactococcus lactis. J. Bacteriol. 196, 1386–1393. doi: 10.1128/JB.01277-13
Radivojac, P., and Vitek, O. (2012). Statistical mass spectrometry-based proteomics. BMC Bioinformatics 13:S1. doi: 10.1186/1471-2105-13-S16-S1
Reid, G. (2017). Probiotic use in an infectious disease setting. Expert Rev. Anti Infect. Ther. 1, 1–7. doi: 10.1080/14787210.2017.1300061
Revilla-Guarinos, A., Gebhard, S., Alcántara, C., Staron, A., Mascher, T., and Zúñiga, M. (2013). Characterization of a regulatory network of peptide antibiotic detoxification modules in Lactobacillus casei BL23. Appl. Environ. Microbiol. 79, 3160–3170. doi: 10.1128/AEM.00178-13
Schultz, M. J., and Haas, L. E. (2011). Antibiotics or probiotics as preventive measures against ventilator-associated pneumonia: a literature review. Crit Care 15:R18. doi: 10.1186/cc9963
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
van Zwet, A. A., Vandenbroucke-Grauls, C. M., Thijs, J. C., Van Der Wouden, E. J., Gerrits, M. M., and Kusters, J. G. (1998). Stable amoxicillin resistance in Helicobacter pylori. Lancet 352:1595. doi: 10.1016/S0140-6736(98)00064-6
Wang, J. C., Zhang, W. Y., Zhong, Z., Wei, A. B., Bao, Q. H., Zhang, Y., et al. (2012a). Gene expression profile of probiotic Lactobacillus casei Zhang during the late stage of milk fermentation. Food Control. 25, 321–327. doi: 10.1016/j.foodcont.2011.10.036
Wang, J. C., Zhang, W. Y., Zhong, Z., Wei, A. B., Bao, Q. H., Zhang, Y., et al. (2012b). Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk. J. Ind. Microbiol. Biotechnol. 39, 191–206. doi: 10.1007/s10295-011-1015-7
Wang, J., Dong, X., Shao, Y., Guo, H., Pan, L., Hui, W., et al. (2017). Genome adaptive evolution of Lactobacillus casei under long-term antibiotic selection pressures. BMC Genomics 18:320. doi: 10.1186/s12864-017-3710-x
Wang, J., Wu, R., Zhang, W., Sun, Z., Zhao, W., and Zhang, H. (2013). Proteomic comparison of the probiotic bacterium Lactobacillus casei Zhang cultivated in milk and soy milk. J. Dairy Sci. 96, 5603–5624. doi: 10.3168/jds.2013-6927
Williams, A. G., Withers, S. E., and Banks, J. M. (2000). Energy sourcs of non-starter lactic acid bacteria isolated from Cheddar cheese. Int. Dairy J. 10, 17–23. doi: 10.1016/S0958-6946(00)00027-3
Woodford, N., and Ellington, M. J. (2007). The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 13, 5–18. doi: 10.1111/j.1469-0691.2006.01492.x
Wright, K., Wright, H., and Murray, M. (2015). Probiotic treatment for the prevention of antibiotic-associated diarrhoea in geriatric patients: a multicentre randomised controlled pilot study. Australas. J. Ageing 34, 38–42. doi: 10.1111/ajag.12116
Wu, C., He, G., and Zhang, J. (2014). Physiological and proteomic analysis of Lactobacillus casei in response to acid adaptation. J. Ind. Microbiol. Biotechnol. 41, 1533–1540. doi: 10.1007/s10295-014-1487-3
Zerbetto De Palma, G., Mendiondo, N., Wonaga, A., Viola, L., Ibarra, D., Campitelli, E., et al. (2017). Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from buenos aires City. Microb. Drug Resist. 23, 351–358. doi: 10.1089/mdr.2015.0361
Zhang, W. Y., Chen, Y. F., Zhao, W. J., Kwok, L. Y., and Zhang, H. P. (2015). Gene expression of proteolytic system of Lactobacillus helveticus H9 during milk fermentation. Ann. Microbiol. 65, 1171–1175. doi: 10.1007/s13213-014-0902-3
Keywords: Lactobacillus casei Zhang, proteomic analysis, amoxicillin, adaptive evolution, stress resistance
Citation: Wang J, Guo H, Cao C, Zhao W, Kwok L-Y, Zhang H and Zhang W (2018) Characterization of the Adaptive Amoxicillin Resistance of Lactobacillus casei Zhang by Proteomic Analysis. Front. Microbiol. 9:292. doi: 10.3389/fmicb.2018.00292
Received: 24 June 2017; Accepted: 07 February 2018;
Published: 20 February 2018.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Weili Liang, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, ChinaZhao Chen, University of California, Davis, United States
Copyright © 2018 Wang, Guo, Cao, Zhao, Kwok, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenyi Zhang, zhangwenyizi@163.com
 Jicheng Wang
Jicheng Wang Lai-Yu Kwok
Lai-Yu Kwok Heping Zhang
Heping Zhang Wenyi Zhang
Wenyi Zhang