- 1Jiangsu Key Laboratory of Zoonosis, Yangzhou University, Yangzhou, China
- 2Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 3Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agri-Food Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
- 4Joint International Research Laboratory of Agriculture and Agri-Product Safety of the Ministry of Education, Yangzhou University, Yangzhou, China
Salmonella enterica serovar Gallinarum biovars Pullorum (S. Pullorum) and Gallinarum (S. Gallinarum) can result in pullorum disease and fowl typhoid in avian species, respectively, and cause considerable economic losses in poultry in many developing countries. Conventional Salmonella serotyping is a time-consuming, labor-intensive and expensive process, and the two biovars cannot be distinguished using the traditional serological method. In this study, we developed a rapid and reliable one-step multiplex polymerase chain reaction (PCR) assay to simultaneously identify and discriminate the biovars Pullorum and Gallinarum. The multiplex PCR method focused on three specific genes, stn, I137_08605 and ratA. Based on bioinformatics analysis, we found that gene I137_08605 was present only in S. Pullorum and S. Gallinarum, and a region of difference in ratA was deleted only in S. Pullorum after comparison with that of S. Gallinarum and other Salmonella serovars. Three pairs of primers specific for the three genes were designed for the multiplex PCR system and their selectivity and sensitivity were determined. The multiplex PCR results showed that S. Pullorum and S. Gallinarum could be identified and discriminated accurately from all tested strains including 124 strains of various Salmonella serovars and 42 strains of different non-Salmonella pathogens. In addition, this multiplex PCR assay could detect a minimum genomic DNA concentration of 67.4 pg/μL, and 100 colony forming units. The efficiency of the multiplex PCR was evaluated by detecting natural-occurring Salmonella isolates from a chicken farm. The results demonstrated that the established multiplex PCR was able to identify S. Gallinarum and S. Pullorum individually, with results being consistent with traditional serotyping and biochemical testing. These results demonstrated that a highly accurate and simple biovar-specific multiplex PCR assay could be performed for the rapid identification and discrimination of Salmonella biovars Gallinarum and Pullorum, which will be useful, particularly under massive screening situations.
Introduction
Salmonellosis is a zoonotic disease, which can cause gastroenteritis, diarrhea, and systemic typhoid fever (McGhie et al., 2009). So far, 2,610 different Salmonella serovars have been defined and classified based on the 46 lipopolysaccharide (O) and 114 flagellar (H) antigens (Guibourdenche et al., 2010). Salmonella enterica serovar Gallinarum biovars Pullorum (S. Pullorum) and Gallinarum (S. Gallinarum) belong to biovars of serovar Gallinarum within serotype D, and cause diseases only in avian species including turkeys, chickens, and other birds (Shivaprasad, 2000).
Salmonella Gallinarum is the causative pathogen of fowl typhoid and results in variable morbidity and high mortality, which often leads to a severe septicemic disease occurring primarily in adult birds. S. Pullorum can cause pullorum disease, which is a serious systemic disease with high mortality, especially in young birds (Barrow and Freitas Neto, 2011). Although part of infected birds could get recovered from pullorum disease, and some adult birds may not present with any clinical disease symptoms, most birds develop a carrier state, and thus may become a repository for the pathogens, which may then be transmitted vertically to newborn hatchlings and horizontally to other birds (Berchieri et al., 2001; Shivaprasad and Barrow, 2008). Although these diseases have been wiped out from the commercial poultry industries in some developed countries, they are widespread in developing countries such as Asia, Africa, and Central and South America, and have caused enormous economic losses (Jones et al., 2001; Kang et al., 2011).
To date, the only reliable method for Salmonella serotyping is the traditional White–Kauffmann–Le Minor scheme, but it cannot distinguish closely related biovars, such as S. Gallinarum and S. Pullorum (World Organization for Animal Health (OIE), 2012). These two biovars share many similar characteristics, including the same lipopolysaccharide (O) antigens, the lack of flagella because of mutations occurred in flagellar-associated genes, and the same avian host they infect (Barrow and Freitas Neto, 2011). Thus, it is not possible to differentiate the two biovars only by serotyping. Although very similar antigenic features are shared by the two Salmonella biovars, some biochemical reactions such as glucose, maltose, and dulcitol fermentation, and ornithine decarboxylase activity, have been established and used to differentiate them (Crichton and Old, 1990). However, biochemical reactions are labor-intensive and take approximately 2–3 days to obtain results (Cheraghchi et al., 2014).
In recent years, polymerase chain reaction (PCR) has been rapidly developed and widely used for the detection of different pathogens because of its relatively simple procedures, as well as its potent specificity and sensitivity (Hoorfar, 2011). DNA-based molecular technologies have also been reported for use in the identification of Salmonella biovars Pullorum and Gallinarum (Kisiela et al., 2005; Kang et al., 2011). However, these assays to identify the two biovars are laborious, complicated, and expensive due to the requirement of additional reagents such as restriction enzymes, or the need of more than one single reaction.
In this study, a reliable and rapid multiplex PCR method were developed and validated to identify and distinguish S. Gallinarum and S. Pullorum simultaneously. The approach was based on three primer pairs designed to target specific regions of stn, I137_08605, and ratA, respectively. The multiplex PCR selectivity was evaluated among various Salmonella serovars and different non-Salmonella pathogens. Besides, natural-occurring Salmonella isolates from a commercial chicken farm were tested by the PCR assay. The newly established multiplex PCR method will be a useful tool to accurately identify and distinguish the two specific Salmonella biovars, and will be particularly beneficial in situations requiring massive screening.
Materials and Methods
Bacterial Strains
Bacterial strains used in this study are commercially available or were isolated previously after our routine monitoring, and included 124 strains which belong to 31 various Salmonella serovars and 42 strains of different non-Salmonella pathogens. All Salmonella serotypes were confirmed according to the White–Kauffmann–Le Minor scheme (Grimont and Weill, 2007), based on the rapid agglutination assays with the Salmonella diagnostic antisera (Tianrun Bio-Pharmaceutical, Ningbo, China). Ornithine decarboxylation and dulcitol fermentation tests were conducted to differentiate the biovars Gallinarum and Pullorum.
Bacterial Culture and Genomic DNA Preparation
The bacterial growth and genomic DNA preparation were conducted as described previously (Xiong et al., 2016). In brief, frozen stocks of the bacterial strains were recovered on Luria-Bertani (LB) agar (Oxoid, Basingstoke, Hampshire, United Kingdom) or Brain Heart Infusion (BHI) agar (Becton, Dickinson and Company, Sparks, MD, United States) at 37°C overnight. The colonies were transferred to LB broth or BHI broth, and cultured at 37°C with constant shaking at a speed of 180 rpm for 16 h. Bacterial genomic DNA was prepared by a commercial QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The DNA purity and concentration were evaluated by the spectrophotometer NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, United States).
Bioinformatics Analysis
To detect and differentiate S. Gallinarum and S. Pullorum based on a PCR assay, we exploited the basic local alignment search tool algorithm from the National Center for Biotechnology Information. The I137_08605 (GenBank accession no. CP006575.1 segment 1847407-1847696) and ratA ROD (GenBank accession no. AM933173.1 segment 2636413-2636983) genes were each used in searches of the non-redundant nucleotide collection (nr/nt) database. To ensure that all aligned target sequences in the database were displayed, we set the number of nucleotide sequences to the maximum value of 20,000. Two pairs of primers specific for the two genes were designed using the online software Primer3 (Untergasser et al., 2012), and the specific primer pairs for the targets are listed in Table 1.

TABLE 1. Multiplex PCR primers used for identification and discrimination of S. Pullorum and S. Gallinarum.
Multiplex PCR Procedure
The multiplex PCR assays were performed in a final volume of 25-μL including 200 μM deoxynucleoside triphosphate mix, 0.25 μL (1 U) of Taq DNA polymerase, 2.5 μL of 10× Taq DNA polymerase buffer (Vazyme, Nanjing, China), 40 nM of the ratA ROD F/R primers, 80 nM of the I137_08605 F/R primers, 80 nM of the stn F/R primers, and 100 ng of the bacterial genomic DNA. The stn gene encodes Salmonella enterotoxin and is specific for S. enterica (Prager et al., 1995). Therefore, stn could be used as the internal amplification control in the multiplex PCR method. The PCR amplification was conducted using a T100 Thermal Cycler (Eppendorf, Hamburg, Germany) with the following protocol: initial denaturation at 94°C for 5 min; 30 sequential cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 40 s; and a final step of 72°C for 10 min. The PCR fragments were separated using 1% agarose gels after electrophoresis, and stained with 1× GelRed Nucleic Acid Gel Stain (Biotium, Fremont, CA, United States). The amplified PCR products were visualized using UV light excitation, and images were digitalized using a GelDoc XR Gel Documentation System (Bio-Rad).
Selectivity and Detecting Limit of the Multiplex PCR Assay
Genomic DNA extracted from 124 strains of various Salmonella serovars and 42 strains of different non-Salmonella pathogens were used to evaluate the selectivity and compatibility of the three primer sets in the multiplex PCR assay.
The detecting limit of the multiplex PCR assay regarding the lower limit of DNA that could be detected was determined by testing serially diluted samples of genomic DNA from S. Gallinarum strain SG9 and S. Pullorum strain S06004. The genomic DNA of the two Salmonella biovars was 10-fold serially diluted from 67.4 ng/μL to 674 fg/μL in deionized water. All diluted samples were separately served as the template in the established multiplex PCR assay.
To determine the fewest number of cells that could be detected by the multiplex PCR, S. Gallinarum strain SG9 and S. Pullorum strain S06004 were cultured overnight. The bacteria were centrifuged and washed with phosphate buffered saline three times, and the plate counting assays were used to calculate the bacterial concentrations. The templates were prepared by 10-fold serially dilution to obtain different concentrations of bacteria ranging from 2 × 107 to 2 × 102 colony forming units (CFU)/mL. All bacterial dilutions were boiled in a water bath for 10 min, and centrifuged at 10,000 rpm for 5 min. The supernatant containing the DNA from the lysed bacterial cells was collected and 5 μL of each dilution was used as template in the multiplex PCR assay.
Chicken Farm Salmonella Isolates
Naturally contaminated samples were collected from feces and floors of a commercial chicken farm in Yangzhou, China according to the method ISO 6579-1:2017. The indigenous Salmonella strains were isolated as described (Cai et al., 2016; Zhou et al., 2017). In brief, all samples were cultured in buffered peptone water (50 mL; Difco, BD, Sparks, MD, United States) for pre-enrichment at 37°C for 24 h. The bacterial culture (0.1 mL) was subsequently inoculated into Rappaport-Vassiliadis R10 broth (10 mL; Difco, BD) and cultured at 42°C for 24 h. Then the bacteria broth was streaked on xylose lysine tergitol 4 agar plates (Difco, BD) and incubated continuously at 37°C for 20–24 h. The presumptive Salmonella colonies on all plates were individually collected and the biochemical tests were performed based on an API 20E assay system (BioMerieux, Marcy l’Etoile, France).
Multiplex PCR Detection of the Chicken Farm Salmonella Isolates
To evaluate the diagnostic sensitivity of the multiplex PCR method on natural-occurring Salmonella isolates, the genomic DNA of Salmonella isolates from the chicken farm was extracted by the boiling method and used as PCR templates. The developed multiplex PCR results were then compared with the conventional Salmonella serotyping and biochemical tests.
Traditional Salmonella Serotyping and Biochemical Testing
The serovars of the clinical Salmonella isolates were characterized using traditional Salmonella serotyping assays with diagnostic antisera for rapid agglutination following the White–Kauffmann–Le Minor instructions (Grimont and Weill, 2007). For differentiation of S. Gallinarum and S. Pullorum, dulcitol fermentation and ornithine decarboxylation tests were used to distinguish the two biovars.
Results
Bioinformatics Analysis and Primer Design
Bioinformatics analysis revealed that I137_08605 existed only in S. Pullorum and S. Gallinarum, and was not present in any other Salmonella serovars, or in non-Salmonella strains that have 100% sequence similarity across the two Salmonella biovars that were included in the database (Supplementary Figure S1). The ratA gene of S. Pullorum was 4776 bp, covering 85% of the length of ratA in S. Gallinarum and other serovars. The truncated sequence was due to variations in the region of difference (ROD) of ratA, which was not found in any S. Pullorum. This truncation could be exploited to differentiate S. Pullorum from S. Gallinarum (Supplementary Figure S2). Therefore, two pairs of oligonucleotide primers that distinguished the two Salmonella biovars were designed, specifically targeting I137_08605 and ratA ROD (Table 1). The primer sets in the multiplex PCR assay produced an amplicon of 290 bp specific to I137_08605 for biovar Pullorum, and two amplicons of 290 and 571 bp specific to I137_08605 and ratA ROD, respectively, for biovar Gallinarum (Figure 1).
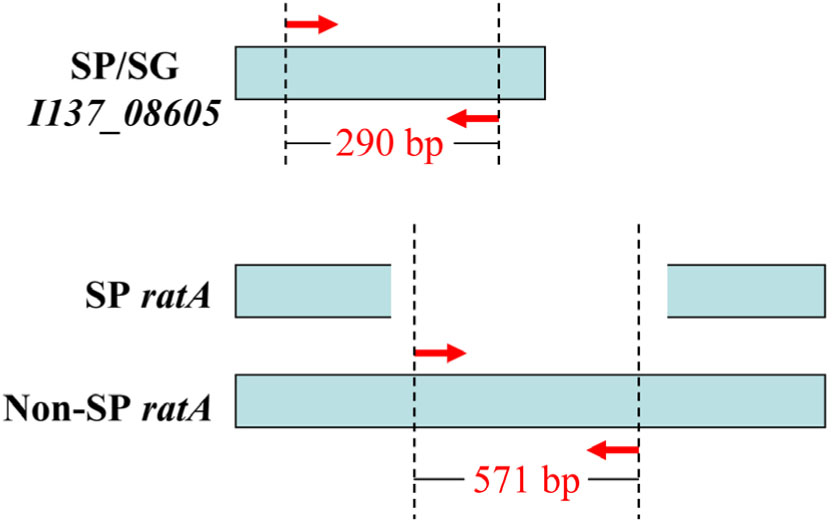
FIGURE 1. Primer design for the multiplex PCR method to distinguish Salmonella enterica serovar Gallinarum biovars Gallinarum (S. Gallinarum) and Pullorum (S. Pullorum) from other serovars. Gene I137_08605 exists only in the two Salmonella biovars, and gene ratA of S. Pullorum has a region of difference (ROD) compared with that of other serovars, which were exploited to design the primers. The red arrows indicate the positions of the designed primers.
Analytical Selectivity of the Multiplex PCR Assay
The multiplex PCR selectivity was evaluated by detecting the genomic DNA from 124 strains of various Salmonella serovars and 42 strains of different non-Salmonella pathogens. The multiplex PCR results showed that three specific bands corresponding to the amplification of stn, I137_08605, and ratA ROD from S. Gallinarum were generated, and only two specific bands corresponding to the amplification of stn and I137_08605 was generated from S. Pullorum. In contrast, only two amplifications corresponding to stn and ratA ROD were produced in a portion of the other 29 Salmonella serovars. No amplification was detected for any of the non-Salmonella strains, suggesting that the developed multiplex PCR method had an excellent selectivity with 100% inclusivity and exclusivity. The relative specificity and relative sensitivity to discriminate negative and positive samples were both 100% for the multiplex PCR method (Figure 2).
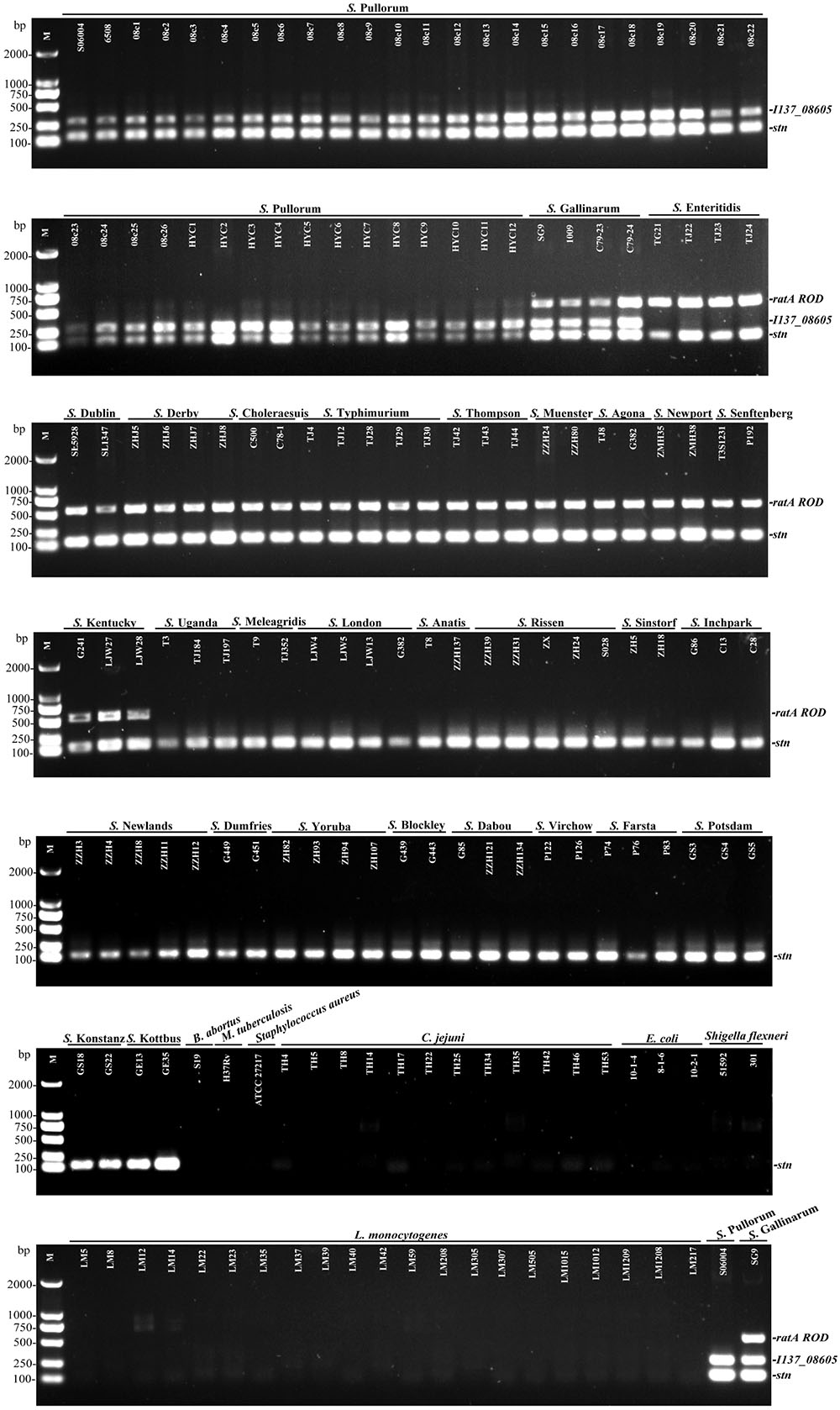
FIGURE 2. Selectivity of the multiplex PCR method for the identification of S. Pullorum and S. Gallinarum. The multiplex PCR assays, using genomic DNA from various Salmonella and non-Salmonella strains, were conducted using the designed primers targeting stn (131 bp), I137_08605 (290 bp), and ratA ROD (571 bp). The three specific PCR products could be amplified in S. Gallinarum, while only stn and I137_08605 genes could be amplified in S. Pullorum.
Analytical Detecting Limit of the Multiplex PCR Assay
The multiplex PCR sensitivity was evaluated by detecting serially diluted genomic DNA from S. Gallinarum and S. Pullorum. The results showed that stn, I137_08605, and ratA ROD could be amplified at the lowest concentration of 67.4 pg/μL, suggesting that at least 67.4 pg/μL of bacterial genomic DNA was required in order to identify and distinguish S. Gallinarum or S. Pullorum using this multiplex PCR method. In addition, the fewest number of S. Pullorum or S. Gallinarum cells the PCR method could detect was also determined. Based on the multiplex PCR assay on various concentrations of diluted Salmonella cells, we determined that the minimum number of bacteria able to be detected was 100 CFU (data not shown).
Application of the Multiplex PCR Method
To evaluate the sensitivity of the multiplex PCR, additional Salmonella isolates were used for the method. The isolates of unknown serovars were collected from naturally contaminated samples from a chicken farm and were prepared for use as a template in order to evaluate the diagnostic efficiency of this multiplex PCR method. The PCR results showed that 21 samples generated only two specific band corresponding to amplification of stn and I137_08605, suggesting that the 21 isolates were S. Pullorum. Two samples from the chicken farm generated the three specific bands consistent with stn, I137_08605, and ratA ROD amplification, suggesting that these two isolated strains were S. Gallinarum (Table 2).

TABLE 2. Salmonella strains isolated from one chicken farm to examine the application of the developed multiplex PCR method.
Traditional Serotyping and Biochemical Identification of the Salmonella Isolates
To validate the accuracy of the multiplex PCR assay, traditional Salmonella serotyping and biochemical assays were conducted on the Salmonella isolates of the chicken farm. The results showed that the 71 Salmonella isolates included 21 strains of S. Pullorum, two strains of S. Gallinarum, 13 strains of S. Indiana, and 35 strains of S. Enteritidis, which were completely concordant with the multiplex PCR results (Table 2).
Discussion
Salmonella Pullorum and S. Gallinarum belong to important bacterial pathogens and have resulted in enormous economic losses in poultry, particularly in the developing countries. S. Pullorum infection of young birds often leads to a high mortality. Infected adult birds show decreased egg laying, weight loss, dysentery, and most importantly persistent infection occurs in most recovered birds, which may lead to horizontal and vertical transmission (Barrow and Freitas Neto, 2011). S. Gallinarum can infect birds of any age and is highly pathogenic, causing fowl typhoid, which usually results in systemic infection, and may cause 40–80% mortality in the flock (Ribeiro et al., 2009). Although different diseases are caused by these two pathogens, they are genetically and phenotypically alike (Batista et al., 2015).
Definitive diagnosis and timely removal of infected birds are crucial to prevent the spread and control the prevalence of S. Gallinarum and S. Pullorum in poultry. Currently, the determination of Salmonella serotype is mainly based on the White–Kauffmann–Le Minor scheme by identifying the somatic (O) and flagellar (H) antigens, and some special biochemical tests (Grimont and Weill, 2007) according to ISO 6579-1:2017. However, it is difficult to differentiate between S. Gallinarum and S. Pullorum since they share the same 1, 9, and 12 O antigens (Christensen et al., 1993). Therefore, development of a rapid and reliable method for the accurate identification and discrimination of biovars Gallinarum and Pullorum would allow for earlier confirmation of the pathogens and subsequently a more effective elimination of the diseases from flocks.
Because Salmonella biovars Pullorum and Gallinarum belong to the same serovar, but different biovars, their identification and differentiation is primarily based on biochemical reactions. Dulcitol fermentation and ornithine decarboxylation are currently the most widely used methods, and are officially recognized. However, these traditional methods have inherited drawbacks, such as time-consumption, labor-intension, and costliness, especially when many samples must be analyzed in a short time. Thus, an affordable and rapid diagnosis method is urgently desired for the identification of S. Gallinarum and S. Pullorum.
Molecular assays have been shown to be highly specific and sensitive for identifying causative pathogens of infectious diseases, and for differentiating closely related bacteria (Hu et al., 2006). Many studies have shown that PCR-based detection of Salmonella serovars is more sensitive, easier, and faster than traditional microbiologic methods (Oliveira et al., 2002; Soria et al., 2012). Therefore, a novel multiplex PCR assay based on three specific genes, stn, I137_08605, and ratA ROD, was developed for the identification and differentiation of Salmonella serovar Gallinarum biovars Gallinarum and Pullorum. The unique gene I137_08605, present only in biovars Gallinarum and Pullorum, was a common feature shared by these biovars, but was not present in any other known Salmonella serovars or species. Sequence analysis of ratA ROD in serovar Gallinarum strains revealed a deletion in biovar Pullorum. The multiplex PCR developed in this study had 100% specificity, and the lower limit of DNA and the fewest number of bacteria that could be detected were 67.4 pg/μL and 100 CFU, respectively, which were comparable with our previous study (Xiong et al., 2017). Our results showed that the combined amplification of stn, I137_08605, and ratA ROD could be used to identify, as well as to distinguish, S. Gallinarum and S. Pullorum rapidly and reliably. This newly developed multiplex PCR assay may be applicable to epidemiological investigations and will be a potent diagnostic method in veterinary clinical laboratories.
Conclusion
In summary, we developed a simple, rapid, accurate, and cost-effective multiplex PCR assay to identify and discriminate S. Pullorum and S. Gallinarum from other serovars and non-Salmonella strains. The multiplex PCR method was based on the three genes stn, I137_08605, and ratA ROD, and it has excellent selectivity and potent sensitivity. The method was applied to detect a group of naturally occurring Salmonella isolates, thereby confirming its efficiency in the commercial farm. This highly accurate, biovar-specific, multiplex PCR system will be useful for the rapid differential diagnosis of biovars Pullorum and Gallinarum in veterinary laboratories. This assay could contribute to earlier confirmation of pathogens, and allow for more effective elimination of the diseases from flocks.
Author Contributions
ZP and XJ designed the experiments. DX and LS designed the experiments, performed the PCR assays, and analyzed the results. ZP, XJ, and DX wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program Special Project (2016YFD0501607), the National Natural Science Foundation of China (Nos. 31320103907 and 31730094), the Key Research and Development Program of Jiangsu Province (Modern Agriculture) Key Program (BE2017341), the Program for High-end Talents of Yangzhou University, the Yangzhou University Science and Technology Innovation Team, the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX18_2377), the Fund for Excellent Doctoral Dissertations from Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01718/full#supplementary-material
References
Barrow, P. A., and Freitas Neto, O. C. (2011). Pullorum disease and fowl typhoid–new thoughts on old diseases: a review. Avian Pathol. 40, 1–13. doi: 10.1080/03079457.2010.542575
Batista, D. F., Freitas Neto, O. C., Barrow, P. A., Oliveira, M. T., Almeida, A. M., Ferraudo, A. S., et al. (2015). Identification and characterization of regions of difference between the Salmonella Gallinarum biovar Gallinarum and the Salmonella Gallinarum biovar Pullorum genomes. Infect. Genet. Evol. 30, 74–81. doi: 10.1016/j.meegid.2014.12.007
Berchieri, A. Jr., Murphy, C. K., Marston, K., and Barrow, P. A. (2001). Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: effect of bacterial and host genetic background. Avian Pathol. 30, 221–231. doi: 10.1080/03079450120054631
Cai, Y., Tao, J., Jiao, Y., Fei, X., Zhou, L., Wang, Y., et al. (2016). Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 222, 56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020
Cheraghchi, N., Khaki, P., Moradi Bidhendi, S., and Sabokbar, A. (2014). Identification of isolated Salmonella enterica serotype Gallinarum biotype Pullorum and Gallinarum by PCR-RFLP. Jundishapur J. Microbiol. 7:e19135. doi: 10.5812/jjm.19135
Christensen, J. P., Olsen, J. E., and Bisgaard, M. (1993). Ribotypes of Salmonella enterica serovar Gallinarum biovars Gallinarum and Pullorum. Avian Pathol. 22, 725–738. doi: 10.1080/03079459308418960
Crichton, P. B., and Old, D. C. (1990). Salmonellae of serotypes Gallinarum and Pullorum grouped by biotyping and fimbrial-gene probing. J. Med. Microbiol. 32, 145–152. doi: 10.1099/00222615-32-3-145
Grimont, P. A. D., and Weill, F. X. (2007). Antigenic Formulae of the Salmonella Serovars, 9th Edn. Paris: Institut Pasteur.
Guibourdenche, M., Roggentin, P., Mikoleit, M., Fields, P. I., Bockemühl, J., Grimont, P. A., et al. (2010). Supplement 2003-2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161, 26–29. doi: 10.1016/j.resmic.2009.10.002
Hoorfar, J. (2011). Rapid detection, characterization, and enumeration of foodborne pathogens. APMIS Suppl. 119, 1–24. doi: 10.1111/j.1600-0463.2011.02767.x
Hu, H., Lan, R., and Reeves, P. R. (2006). Adaptation of multilocus sequencing for studying variation within a major clone: evolutionary relationships of Salmonella enterica serovar Typhimurium. Genetics 172, 743–750. doi: 10.1534/genetics.105.046466
Jones, M. A., Wigley, P., Page, K. L., Hulme, S. D., and Barrow, P. A. (2001). Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infect. Immun. 69, 5471–5476. doi: 10.1128/IAI.69.9.5471-5476.2001
Kang, M. S., Kwon, Y. K., Jung, B. Y., Kim, A., Lee, K. M., An, B. K., et al. (2011). Differential identification of Salmonella enterica subsp. enterica serovar Gallinarum biovars Gallinarum and Pullorum based on polymorphic regions of glgC and speC genes. Vet. Microbiol. 147, 181–185. doi: 10.1016/j.vetmic.2010.05.039
Kisiela, D., Kuczkowski, M., Kiczak, L., Wieliczko, A., and Ugorski, M. (2005). Differentiation of Salmonella Gallinarum biovar Gallinarum from Salmonella Gallinarum biovar Pullorum by PCR-RFLP of the fimH gene. J. Vet. Med. B Infect. Dis. Vet. Public Health 52, 214–218. doi: 10.1111/j.1439-0450.2005.00846.x
McGhie, E. J., Brawn, L. C., Hume, P. J., Humphreys, D., and Koronakis, V. (2009). Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12, 117–124. doi: 10.1016/j.mib.2008.12.001
Oliveira, S. D., Santos, L. R., Schuch, D. M., Silva, A. B., Salle, C. T., and Canal, C. W. (2002). Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 87, 25–35. doi: 10.1016/S0378-1135(02)00028-7
Prager, R., Fruth, A., and Tschäpe, H. (1995). Salmonella enterotoxin (stn) gene is prevalent among strains of Salmonella enterica, but not among Salmonella bongori and other Enterobacteriaceae. FEMS Immunol. Med. Microbiol. 12, 47–50. doi: 10.1111/j.1574-695X.1995.tb00173.x
Ribeiro, S. A., de Paiva, J. B., Zotesso, F., Lemos, M. V., and Berchieri Jánior, A. (2009). Molecular differentiation between Salmonella enterica subsp enterica serovar Pullorum and Salmonella enterica subsp enterica serovar Gallinarum. Braz. J. Microbiol. 40, 184–188. doi: 10.1590/S1517-838220090001000032
Shivaprasad, H. L. (2000). Fowl typhoid and pullorum disease. Rev. Sci. Tech. 19, 405–424. doi: 10.20506/rst.19.2.1222
Shivaprasad, H. L., and Barrow, P. A. (2008). “Pullorum disease and fowl typhoid,” in Disease of Poultry, 12th Edn, ed. Y. M. Saif (Ames, IA: Iowa State Press), 620–635.
Soria, M. C., Soria, M. A., and Bueno, D. J. (2012). Comparison of 2 culture methods and PCR assays for Salmonella detection in poultry feces. Poult. Sci. 91, 616–626. doi: 10.3382/ps.2011-01831
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
World Organization for Animal Health (OIE) (2012). “Fowl typhoid, and Pullorum disease,” in Manual of Diagnostic Tests, and Vaccines for Terrestrial Animals, 7th Edn, ed. OIE (Paris: OIE), 538–548.
Xiong, D., Song, L., Geng, S., Tao, J., An, S., Pan, Z., et al. (2016). One-step PCR detection of Salmonella Pullorum/Gallinarum using a novel target: the flagellar biosynthesis gene flhB. Front. Microbiol. 7:1863. doi: 10.3389/fmicb.2016.01863
Xiong, D., Song, L., Tao, J., Zheng, H., Zhou, Z., Geng, S., et al. (2017). An efficient multiplex PCR-based assay as a novel tool for accurate inter-serovar discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin. Front. Microbiol. 8:420. doi: 10.3389/fmicb.2017.00420
Keywords: Salmonella Pullorum, Salmonella Gallinarum, multiplex PCR, accurate discrimination, one-step diagnostic PCR
Citation: Xiong D, Song L, Pan Z and Jiao X (2018) Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay. Front. Microbiol. 9:1718. doi: 10.3389/fmicb.2018.01718
Received: 06 April 2018; Accepted: 10 July 2018;
Published: 31 July 2018.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Bradley D. Jones, University of Iowa, United StatesDario De Medici, Istituto Superiore di Sanità, Italy
Copyright © 2018 Xiong, Song, Pan and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiming Pan, zmpan@yzu.edu.cn Xinan Jiao, jiao@yzu.edu.cn
 Dan Xiong1,2,3,4
Dan Xiong1,2,3,4 Zhiming Pan
Zhiming Pan