- Institute of Analytical Chemistry and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering, Peking University, Beijing, China
Shigella flexneri is an important foodborne bacterial pathogen with infectious dose as low as 10–100 cells. SlyA, a transcriptional regulator of the MarR family, has been shown to regulate virulence in a closely related bacterial pathogen, Salmonella Typhimurium. However, the regulatory role of SlyA in S. flexneri is less understood. Here we applied unbiased proteomic profiling to define the SlyA regulon in S. flexneri. We found that the genetic ablation of slyA led to the alteration of 18 bacterial proteins among over 1400 identifications. Intriguingly, most down-regulated proteins (whose expression is SlyA-dependent) were associated with bacterial acid resistance such as the glutamate decarboxylation system. We further demonstrated that SlyA directly regulates the expression of GadA, a glutamate decarboxylase, by binding to the promotor region of its coding gene. Importantly, overexpression of GadA was able to rescue the survival defect of the ΔslyA mutant under acid stress. Therefore, our study highlights a major role of SlyA in controlling S. flexneri acid resistance and provides a molecular mechanism underlying such regulation as well.
Introduction
The bacterium Shigella flexneri (S. flexneri) is an intracellular pathogen that causes gut infection resulting in watery diarrhea or bacillary dysentery (Kotloff et al., 1999). It is estimated that there are over 160 million cases of shigellosis per year worldwide, leading to more than half a million deaths mostly associated with infants in developing countries (Kotloff et al., 1999). A major virulence determinant of S. flexneri is a large 230-kb plasmid that encodes the bacterial type III secretion system (T3SS) (Mattock and Blocker, 2017). Working as a molecular syringe, the T3SS is able to translocate an extensive repertoire of effector proteins into host cells, promoting initial bacterial internalization as well as intracellular replication. As a foodborne pathogen, S. flexneri infection of the gastrointestinal (GI) tract requires successful passage and survival through the highly acidic stomach. A rather striking feature of this bacterium is its extremely low infectious dose of as few as 10–100 bacterial cells. Therefore, bacterial acid resistance, in addition to its T3SS, constitutes another critical virulence determinant in Shigella spp.
Bacteria such as S. flexneri have evolved a plethora of strategies to cope with acid stress. A classical anti-acid mechanism of great importance is the amino acid (e.g., glutamate) decarboxylase-dependent systems (Jennison and Verma, 2007). In principle, protons are consumed by glutamate decarboxylation together with the release of CO2, thereby raising the cytoplasmic pH. GadA and GadB are the central enzymes in this process that catalyze the decarboxylating reaction (Bearson et al., 1997). Over the years, this important anti-acid mechanism has been well characterized (Hersh et al., 1996; De Biase et al., 1999; Castanie-Cornet and Foster, 2001; Foster, 2004); however, its transcriptional regulation still remains largely unclear in S. flexneri.
The MarR/SlyA family of transcription factors regulates a variety of bacterial cellular processes (George and Levy, 1983; Roper et al., 1993; Thomson et al., 1997; Srikumar et al., 1998; Galan et al., 2003). In Salmonella Typhimurium, for instance, SlyA has been shown to be an important regulator of bacterial virulence. The slyA deletion mutant exhibits a survival defect within macrophages as well as in a mouse model of infection (Libby et al., 1994; Buchmeier et al., 1997). Further transcriptome studies of the slyA deletion mutant in comparison to the wild-type strain revealed that it controls the expression of several known virulence genes (Navarre et al., 2005). In addition, many of the SlyA-controlled genes are also regulated by the PhoP-PhoQ two-component system (Navarre et al., 2005). In contrast, the regulatory role of SlyA in S. flexneri is less characterized thus far. In a pioneering study, Weatherspoon-Griffin and Wing (2016) recently reported the association of overexpressed SlyA with S. flexneri virulence. They also uncovered a survival defect of the slyA deletion mutant under acid stress, though the underlying mechanisms remain unclear (Weatherspoon-Griffin and Wing, 2016).
Here we performed unbiased proteomic profiling of a S. flexneri mutant lacking slyA in comparison to its parental strain. We found differential regulation of 18 proteins among over 1400 detected bacterial proteins. Notably, our data revealed SlyA-dependent expression of GadA and GadB (the glutamate decarboxylase), suggesting a direct link of this transcription factor to S. flexneri acid resistance. We provided further evidence that SlyA regulates the expression of Gad proteins by directly binding to the gene promoter regions. Importantly, S. flexneri mutants lacking slyA exhibited a severe survival defect under acid challenge and overproduction of the glutamate decarboxylase was able to restore, to a large extent, the bacterial anti-acid capacity of the slyA deletion mutant. Therefore, our study provides the first paradigm in bacteria that SlyA controls acid resistance by directly activating the glutamate decarboxylation system.
Materials and Methods
Bacterial Strains and Construction of Mutants
The S. flexneri serotype 2a 2457T (generously provided by Dr. Feng Shao from National Institute of Biological Sciences, Beijing, China) was used in this study, and the bacteria were routinely grown on trypticase soy agar plates with 1.5% agar and 30 μg/mL streptomycin at 37°C. A single colony picked from the plates was inoculated into trypticase soy broth (TSB) with 30 μg/mL streptomycin, and then the overnight culture was diluted 1:20 into 3 mL of TSB. The bacteria were harvested at indicated OD600 for different assays at 4°C. The slyA deletion mutant (ΔslyA) was constructed using the homologous recombination method as previously described (Liu et al., 2015). Successful deletion of a target gene was confirmed by both PCR analyses and sequencing. To construct the complementation strains harboring a plasmid-borne slyA or gadA gene in the ΔslyA background (ΔslyA+pSlyA or ΔslyA+pGadA), the slyA or gadA fragment was amplified, digested, and inserted into the pME6032 plasmid with an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter and a C-terminal 3 × FLAG tag. For β-galactosidase assays, the upstream region (1000 bp) of the gadA gene containing the promoter region was cloned and inserted into the pNN387 plasmid which contains a promoterless lacZ (Elledge and Davis, 1989). All primers and strains used in this study are listed in the Supplementary Table S1.
Sample Preparation, Protein Digestion, and Stable Isotope Dimethyl Labeling
To uncover potential SlyA-regulated proteins, we performed proteomic analyses of the ΔslyA strain together with the wild-type (WT) strain. In brief, bacterial proteins were digested and isotopically labeled prior to two-dimensional LC-MS/MS analyses. Bacterial subculture was grown to an OD600 of ∼0.9 and pelleted at 3000 ×g for 5 min. Bacterial pellets were washed with ice-cold PBS (pH = 7.5) and resuspended in 1 mL of 8 M urea with 50 mM NH4HCO3. Then bacterial cells were lysed by sonication and protein concentration was measured by using a BCA protein assay kit (Solarbio, China). After diluted to one fourth with 50 mM NH4HCO3, 1 mg of proteins from each sample was digested with trypsin (Promega, United States) overnight at an enzyme-to-protein ratio of 1:50 (w/w) at 37°C. Resulting tryptic peptides were desalted and labeled on C18 Sep-Pak cartridges (Welch, China). For desalting, the columns were activated with 1 mL of acetonitrile (ACN) containing 0.1% (vol/vol) formic acid (FA) and equilibrated with 1 mL of 0.1% FA. Then the resulting tryptic peptides were loaded onto the columns. After washing with 1 mL of 0.1% FA, the on-column isotopic labeling experiments were performed as described previously (Boersema et al., 2009). Peptides on the column were repeatedly treated for three times by 1 mL of the labeling solution containing 100 mM triethyl ammonium bicarbonate (TEAB), 30 mM sodium cyanoborohydride (NaBH3CN), 0.2% formaldehyde (CH2O) or deuterated formaldehyde (CD2O). Peptides from WT and ΔslyA strains were labeled with CH2O and CD2O, respectively. After labeling, the peptides were washed once with 0.1% FA and eluted with 70% ACN plus 0.1% FA. Then light- and heavy-labeled peptides were pooled and vacuum dried for further offline fractionation.
Offline High pH Reversed-Phase Fractionation and Nanoflow LC-MS/MS Analyses
Peptide samples were further fractionated using C18 Sep-Pak cartridges (Welch, China) under high pH conditions (10 mM NH4HCO3, pH 10). For peptide elution, the following percentages of ACN were used: 6, 9, 12, 15, 18, 21, 25, 30, and 35% (Lai et al., 2016). In total, nine fractions were collected and pooled into six samples (6+25%, 9+30%, 12+35%, 15%, 18%, and 21%). Samples were vacuum dried for subsequent LC-MS analyses. Nanoflow reversed-phase LC separation was carried out on an EASY-nLC 1200 system (Thermo Fisher Scientific). The capillary column (75 μm × 150 mm) with a laser-pulled electrospray tip (Model P-2000, Sutter Instruments) was home-packed with 4 μm, 100 Å Magic C18AQ silica-based particles (Michrom BioResources Inc., Auburn, CA, United States). The mobile phase was composed of Solvent A (97% H2O, 3% ACN, and 0.1% FA) and Solvent B (20% H2O, 80% ACN, and 0.1% FA). The LC separation was carried out at room temperature with the following gradient: Solvent B was started at 7% for 3 min, and then raised to 40% over 120 min; subsequently, Solvent B was rapidly increased to 90% in 2 min and maintained for 10 min before 100% Solvent A was used for column equilibration. Eluted peptides from the capillary column were electrosprayed directly into a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Fisher Scientific) for MS and MS/MS analyses in a data-dependent acquisition mode. One full MS scan (m/z 350–1200) was acquired and then MS/MS analyses were performed on the 10 most intense ions. The selected ions were fragmented by collision-induced dissociation (CID) in the ion trap with the following parameters: ≥+2 precursor ion charge, 2 Da precursor ion isolation window, and 35% normalized collision energy. Dynamic exclusion was set with repeat duration of 24 s and exclusion duration of 12 s.
Proteomic Data Processing
Raw MS files were processed by MaxQuant (version 1.5.4.1) and searched against the S. flexneri (strain 301/serotype 2a) protein database (4103 sequences, downloaded from UniProt). The precursor mass tolerance was set at 20 ppm and the fragment mass tolerance was set at 0.8 Da. Maximum missed cleavage was set to 2. Dimethyl (K, N-term) and dimethyl (D4K, D4N-term) were set as variable modifications for light (L)- and heavy (H)-labeled samples, respectively. Oxidation (M) was set as a variable modification. Both peptide and protein assignments were filtered to achieve a false discovery rate (FDR) <1%. Only proteins with at least two unique plus razor peptides were quantified. The protein group lists were further processed by using the Perseus software (version 1.5.4.1) for the calculation of logarithmic values (log2) of the H- and L-labeled protein intensity ratio and the p-values. Proteins with ratios (H/L) >2.0 or <0.5 and p-values <0.05 were considered as significant difference between the WT and ΔslyA strains.
Quantitative Real-Time PCR
Both the WT and ΔslyA strains were grown to an OD600 of ∼0.9 as described above. Total bacterial RNA from 1 mL of culture was extracted by using an EasyPure RNA Kit (TransGen Biotech, China) and then treated with DNase I. Reverse transcription of RNA was performed with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, China). RT-PCR analyses were carried out on an Applied Biosystems ViiATM 7 Real-Time PCR System by using UltraSYBR Mixture (Low ROX) (CWBIO, China). To quantitatively compare the levels of gadA, hdeB, slp, and SF2991 transcripts, the housekeeping 16S rRNA gene was used for normalization. The mRNA levels were determined using the comparative threshold cycle number (2-ΔΔCt) method (Livak and Schmitge, 2001).
β-Galactosidase Assays
Shigella cells were grown in TSB at 37°C to an OD600 of ∼1.0. Bacterial cells from 1.2 mL of culture were pelleted at 14,000 ×g for 2 min and then resuspended in 1.2 mL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4) plus 50 mM β-mercaptoethanol (freshly added). Subsequently, 30 μL of chloroform and 15 μL of 0.1% SDS were added and mixed upon vortexing. The assays were started by the addition of 240 μL of 4 mg/mL o-nitrophenyl-β-D-galactopyranoside (ONPG). Upon the observation of a faint yellow color, the reaction was quenched by the addition of 600 μL of 1 M Na2CO3 and the reaction time was recorded. Finally, samples were centrifuged at 14,000 ×g for 2 min, and the OD420 of the supernatant was recorded. Assay units were calculated as 1,000 × OD420/(OD600) (total reaction time).
Expression and Purification of Recombinant Proteins
His6-tagged SlyA was expressed in the E. coli strain BL21 (DE3) harboring the appropriate plasmid. Briefly, 15 mL of the overnight culture was added in 300 mL LB broth and grown until the OD600 reached 0.6–0.8. Upon the addition of IPTG to a final concentration of 0.2 mM, bacterial culture was incubated further in a shaker at 18°C for 16–18 h. Bacterial cells (from 300 mL of culture) were lysed in 30 mL of ice cold PBS buffer via sonication and cell lysates were clarified by centrifugation at 5,000 ×g for 10 min three times at 4°C. His-tagged proteins were captured with Ni-NTA Resin (GenScript) and washed serially with PBS and 20 mM imidazole. Finally, proteins were eluted with 300 mM imidazole. Purified proteins were further dialyzed overnight in 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% (vol/vol) glycerol, and 1 mM DTT with at least two buffer changes.
Electrophoretic Mobility Shift Assays (EMSAs)
The putative promoter sequence of gadA, a mutated version in which the SlyA-binding sequence (TTATCATGTTAA) was deleted and a DNA fragment of its coding sequence were amplified by PCR, purified with a Gel Extraction Kit (TransGen Biotech, China) and dissolved in water. Then purified SlyA proteins were incubated with 40 nM DNA fragments in 20 μL of binding buffer (10 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM EDTA, 0.1 mM DTT, 5% v/v glycerol, and 10 μg/mL BSA). Molar ratios between DNA fragment and SlyA were set at 1:0, 1:30, 1:35, 1:40, 1:45, and 1:50 (indicated as 0, 1.2, 1.4, 1.6, 1.8, and 2.0 μM SlyA, respectively, in Figure 3). The reaction mixtures were incubated at room temperature for 30 min and then loaded onto 8% native polyacrylamide gels. Electrophoresis was performed using 0.5 × TBE buffer (44.5 mM Tris, 44.5 mM boric acid, and 1 mM EDTA) in ice bath. The gel was stained with Gel Stain (Yeasen, China) and photographed by using a Tanon-1600 Gel Image System (Tanon, China).
Bacterial Acid Survival Assays
The overnight cultures of individual Shigella strains were diluted 1:20 into 3 mL of LB. When OD600 reached about 0.6, IPTG was added to a final concentration of 0.2 mM to induce the expression of plasmid-borne SlyA or GadA for 4 h. Then bacterial cells were diluted 1:50 in the acidified LB media (pH 2.5 adjusted with HCl). Viable cell counts were determined at 0, 1, and 2 h post acid challenge by serial dilution and plating on LB agar plates. At least three repetitions were performed for each experiment.
Results
Comparative Proteomic Profiling of S. flexneri Wild-Type and Its Isogenic ΔslyA Strains
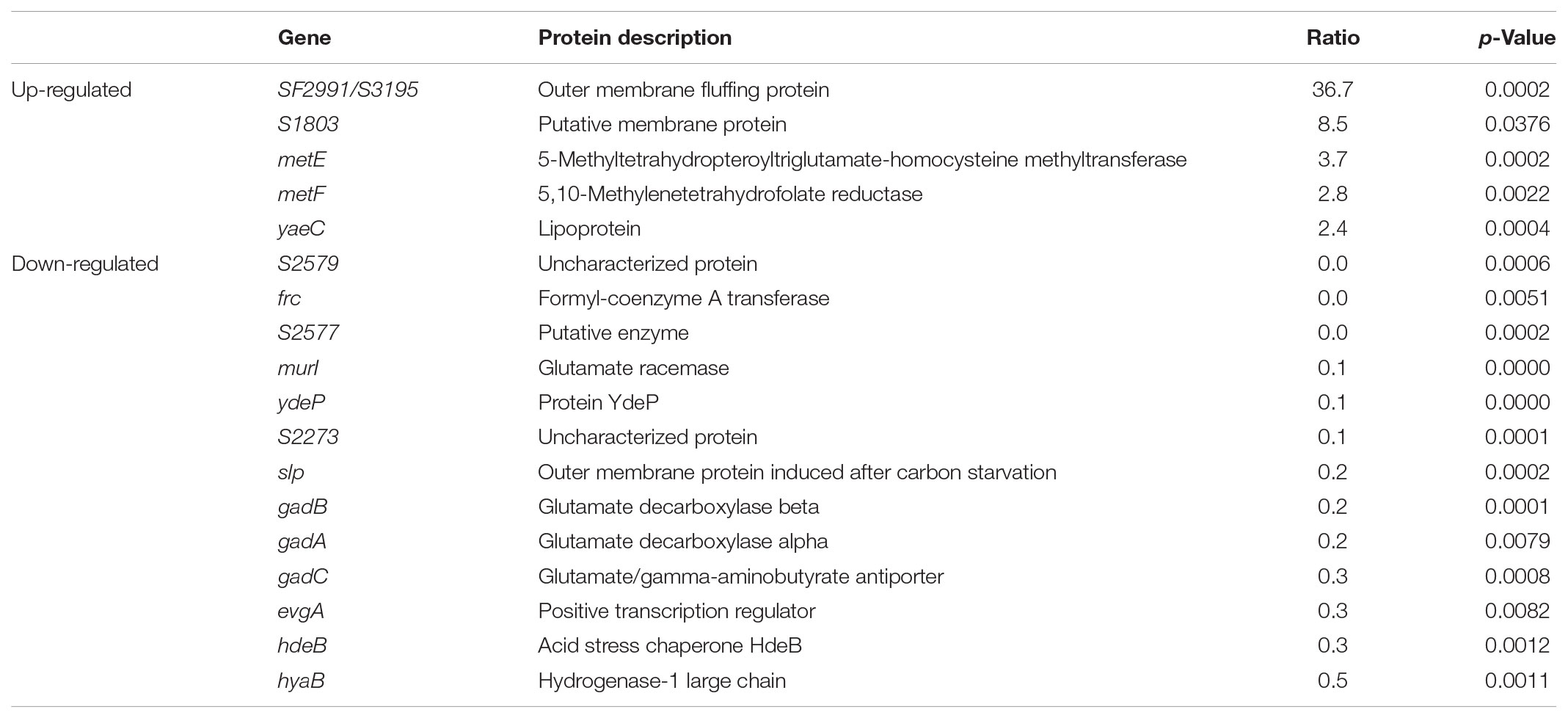
To study the SlyA regulon in S. flexneri, we carried out quantitative proteomic profiling of a bacterial strain lacking slyA (ΔslyA) and its parental wild-type (WT) strain. In total, we identified 1410 S. flexneri proteins from three biological replicates (i.e., six bacterial samples). A complete list of all identified bacterial proteins is provided in Supplementary Table S2. To get a global view of differentially expressed proteins, we plotted the ratio of protein abundance (ΔslyA/WT) as a function of calculated p-values (Figure 1). As seen in the protein-level volcano plot, the two proteomes are strikingly similar with vast majority of proteins lining up toward the center, indicating comparable expression levels of most proteins between two bacterial strains. By using the criteria described in the “Materials and Methods” section, 18 proteins were differentially regulated in the slyA-lacking strain, including 5 up-regulated and 13 down-regulated proteins (see Table 1). The most significant hit in the up-regulated proteins is SF2991/S3195, which is annotated as an outer membrane fluffing protein. The exact function of this protein has yet to be characterized. With respect to the down-regulated proteins, notably several proteins are well-known mediators of bacterial acid resistance such as GadA, GadB, GadC, and HdeB. The glutamate decarboxylation pathway represents arguably the most prominent example of bacterial anti-acid mechanisms. GadA and GadB are two glutamate decarboxylases catalyzing the proton-consuming reaction. MurI is a racemase catalyzing the interconversion of L-glutamate to D-glutamate, and therefore it is related to the GadAB-mediated bacterial anti-acid system.
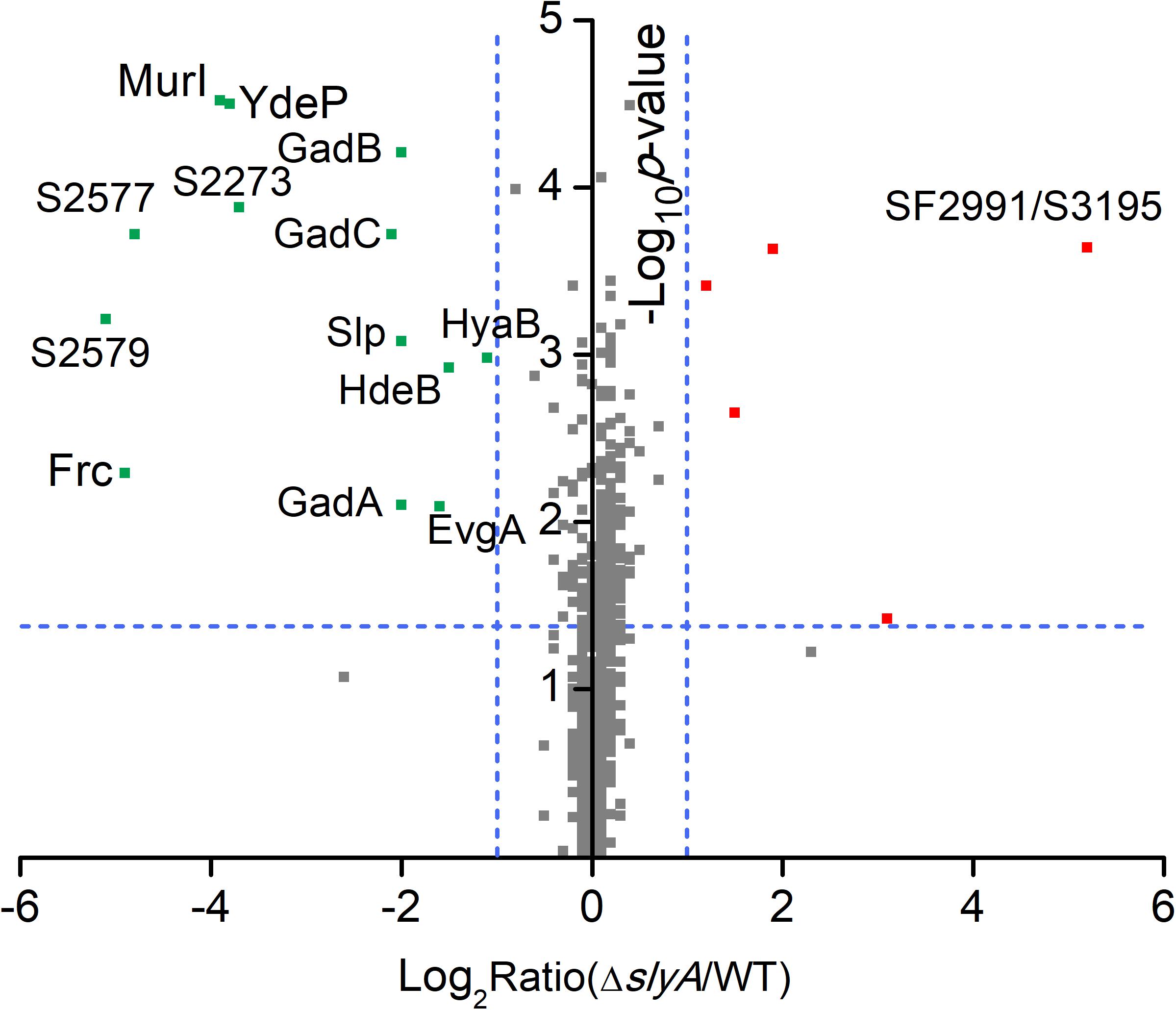
FIGURE 1. Quantitative proteomic data of Shigella WT and ΔslyA strains. A protein volcano plot was shown with the logarithmic values of the abundance ratios reported on the x-axis. The y-axis plots negative logarithmic p-values determined from the t-test on data from three biological replicates. Dotted lines denote two-fold (vertical) and p < 0.05 cutoff (horizontal). Proteins with homologous sequences may not be distinguished in LC-MS/MS experiments unless unique peptides were detected. As a result, the detection of SF2991 and S3195 is considered as a single protein identification (denoted as SF2991/S3195).
Furthermore, some of the other proteins that exhibited lower expression levels in the ΔslyA strain seem to be associated with bacterial acid resistance as well. For instance, YdeP was previously reported to confer acid resistance in E. coli (Masuda and Church, 2003). Interestingly, its regulator EvgA also appears in the list of those down-regulated proteins. In addition, the homologs of Frc and S2577 (i.e., YfdW and YfdU, respectively) were shown to play a role in E. coli acid resistance through a mechanism involving oxalate decarboxylation (Fontenot et al., 2013). Another protein, Slp, is an outer membrane protein that was depressed in the ΔslyA strain, consistent with a previous study of SlyA-regulated proteins in E. coli (Spory et al., 2002). Interestingly, we previously found that S. flexneri up-regulates Slp in response to acid stress (Yu et al., 2017). Taken together, these findings strongly suggest SlyA-dependent expression of many Shigella proteins associated with bacterial anti-acid mechanisms. Next we focused our attention on the characterization of the regulatory role of SlyA in mediating S. flexneri anti-acid capacities.
Transcriptional Regulation of S. flexneri Anti-acid Genes by SlyA
Proteomic observations of SlyA-dependent production of those anti-acid proteins prompted us to determine if SlyA, as a transcription factor, exerts its regulatory role on the transcript level. We carried out qRT-PCR measurements on some representative genes whose protein products were differentially expressed in the ΔslyA strain including gadA, hdeB, slp, and SF2991 (Figure 2A). Consistent with the down-regulation of their corresponding proteins, the mRNA levels of gadA, hdeB, and slp were profoundly lower in the slyA-deletion mutant than those in its parental strain. Additionally, the transcript of SF2991, a SlyA-repressed gene, was found to be substantially higher (∼60-fold) in the mutant strain. Unlike gadA and slp, the fold difference of hdeB and SF2991 transcript levels is markedly larger than that of their protein levels between the ΔslyA and WT strains, indicating potential mechanisms of post-transcriptional regulation.
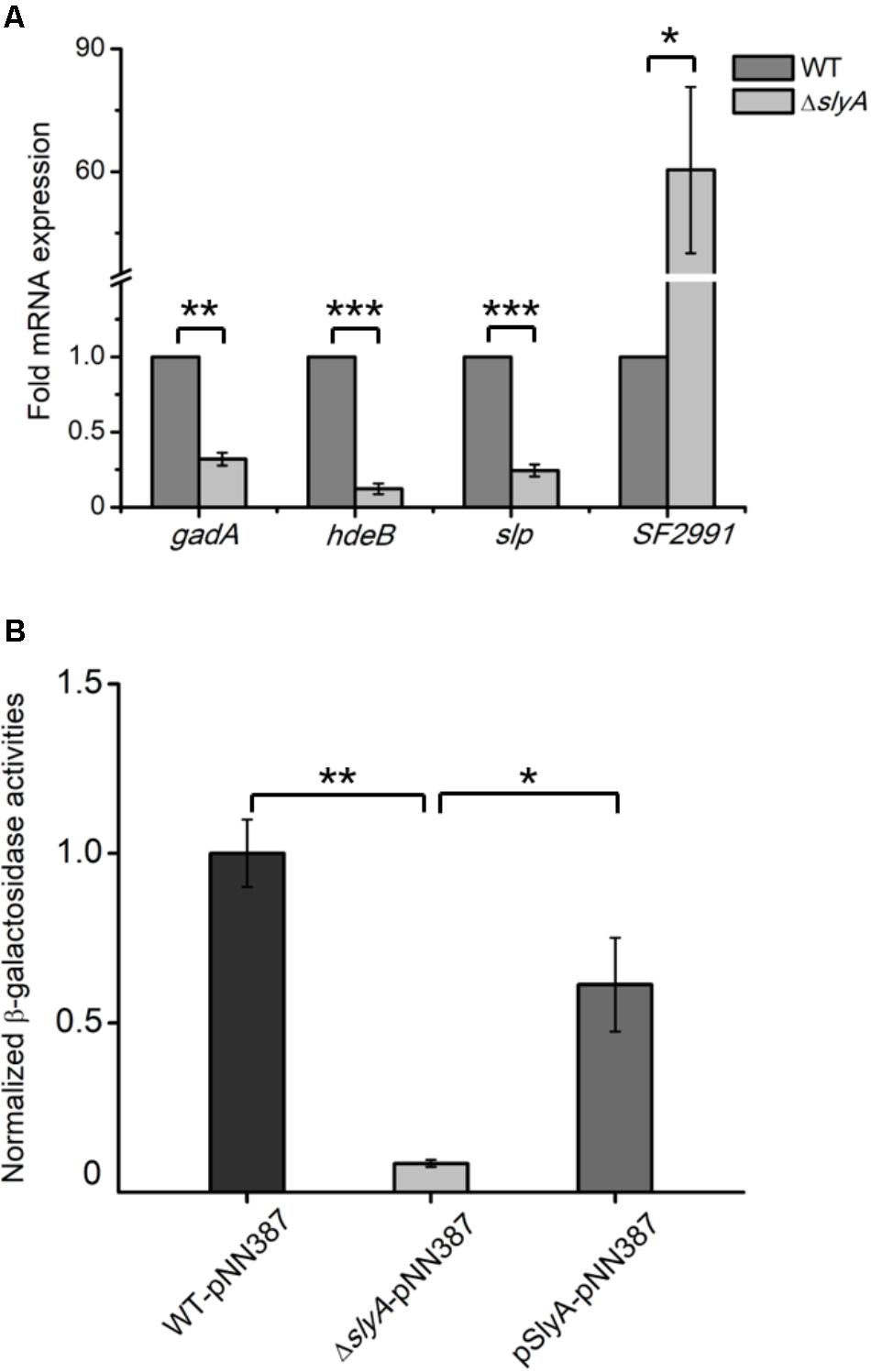
FIGURE 2. SlyA activates the expression of gadA at the transcriptional level. (A) qRT-PCR analyses of mRNA samples extracted from Shigella WT and ΔslyA strains. (B) Measurements of β-galactosidase activities in WT, ΔslyA, and ΔslyA+pSlyA strains harboring a plasmid encoding a promoterless lacZ fused with the gadA promoter. β-Galactosidase activities from three biological replicates are shown with values normalized to that of the WT strain. Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Given the prominent role of the glutamate decarboxylation system in acid resistance, next we further explored the transcriptional control of gadA by SlyA by utilizing a β-galactosidase-based reporter assay. We constructed a lacZ-expressing plasmid in which its promoter region was replaced by that of gadA and transformed this plasmid into Shigella strains of different genetic backgrounds (WT, ΔslyA, and ΔslyA+pSlyA). By measuring the enzymatic activities of expressed β-galactosidase in vitro, we could assess the promoter activity of the gadA gene in different strains. As shown in Figure 2B, β-galactosidase activity decreased substantially (by at least an order of magnitude) in the ΔslyA mutant relative to that in its parental strain, indicating that the promoter activity of gadA is strongly dependent on SlyA. Furthermore, ectopic expression of SlyA in the ΔslyA mutant largely restored the promoter activity to the wild-type level. Taken together, these results established that SlyA positively regulates the expression of gadA on the transcript level and likely other S. flexneri genes involved in acid resistance as well.
Direct Binding of S. flexneri SlyA to the Promoter Region of the gadA Gene
SlyA is a transcriptional regulator in the MarR family with a classical helix-turn-helix (HTH) DNA-binding domain (Ellison and Miller, 2006). Next we sought to determine if SlyA exerts a direct regulatory role in GadA expression by using electrophoretic mobility shift assays (EMSAs). Recombinantly purified SlyA proteins were incubated with various concentrations of DNA fragments spanning the gadA promoter region (the promoter region and putative SlyA-binding site are shown in Figure 3A). Electrophoretic experiments demonstrated that with increasing levels of SlyA a greater fraction of DNA fragments exhibited a mobility shift (Figure 3B). In contrast, when DNA fragments containing a partial sequence of gadA-coding region were used as a negative control, we did not observe any retardation in electrophoretic mobility at all concentrations of SlyA. Remarkably, deletion of the putative SlyA-binding site in the gadA promoter sequence completely abolished its binding to SlyA (Figure 3B). Together, these findings reveal that SlyA directly controls the expression of GadA by physically associating with the gene promoter region.
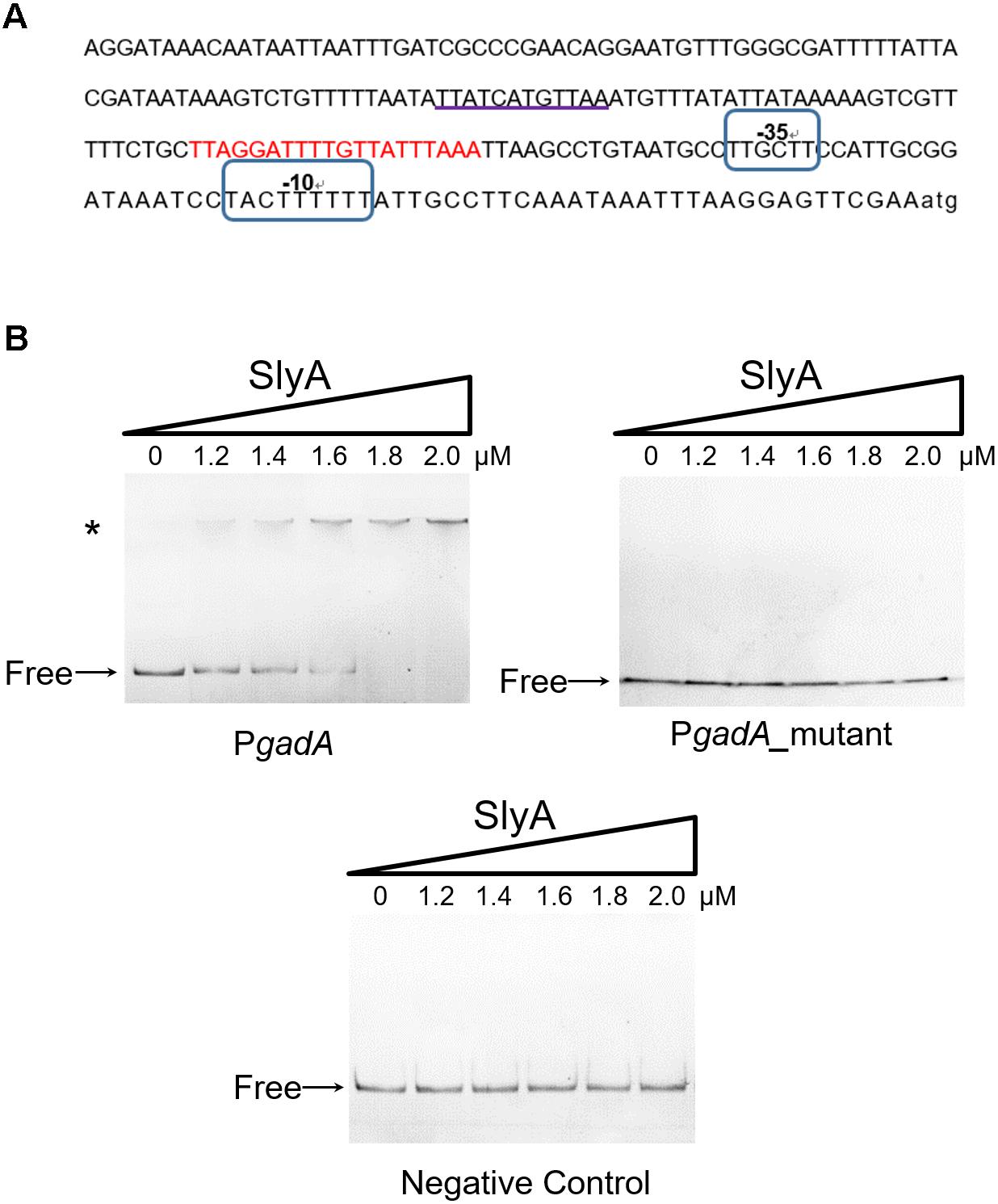
FIGURE 3. SlyA directly binds to the promoter region of the gadA gene. (A) Bioinformatics analysis of the gadA promoter region (PgadA). The sequence of the gadA promoter region of 224 bp upstream of the start codon is shown and the putative SlyA-binding site is underlined. Putative –10 and –35 elements are circled and the gad box is indicated in red. (B) EMSA experiments determined the direct binding of SlyA to PgadA. Different concentrations of purified SlyA (ranging from 0 to 2 μM) were incubated with PgadA, PgadA lacking the SlyA-binding site (PgadA_mutant) or negative control DNA prior to electrophoretic separation. “Free” indicates free DNA and the asterisk indicates DNA-protein complexes.
S. flexneri SlyA Mediates Acid Resistance by Regulating the Expression of Gad Proteins
Given its direct regulatory role in the glutamate decarboxylation system, we hypothesized that SlyA may modulate Shigella acid resistance. To test our hypothesis, we challenged the ΔslyA strain in highly acidic conditions (pH 2.5) for different durations (1 and 2 h) and then determined bacterial viability by CFU assays (Figure 4). Compared to its parental WT strain, upon 1 and 2 h of acid challenge the slyA-deletion mutant exhibited substantially lower survival percentages (>50-fold and 300-fold less, respectively). Furthermore, we were able to rescue such a survival defect by reintroducing a functional copy of slyA into the ΔslyA strain (ΔslyA+pSlyA). Next, we sought out to determine if the regulation of bacterial acid resistance by SlyA is largely accomplished by its control of the glutamate decarboxylation pathway (e.g., the expression of GadA). To explore this possibility, we further constructed a bacterial strain harboring a copy of plasmid-borne gadA in the ΔslyA background. Remarkably, ectopic expression of GadA can restore, to a large extent, bacterial acid resistance of the ΔslyA strain to the levels of the complementation strain (ΔslyA+pSlyA) as well as the WT bacteria. Nonetheless, we do note a minor difference in the rescuing ability of two complementation strains as suggested by the slightly lower (yet significant) survival percentage of the ΔslyA strain ectopically producing GadA than the SlyA-complemented mutant. Taken together, these data suggest Shigella SlyA plays a crucial role in bacterial acid resistance and importantly such regulation is mostly exerted through its control of the glutamate decarboxylation pathway.
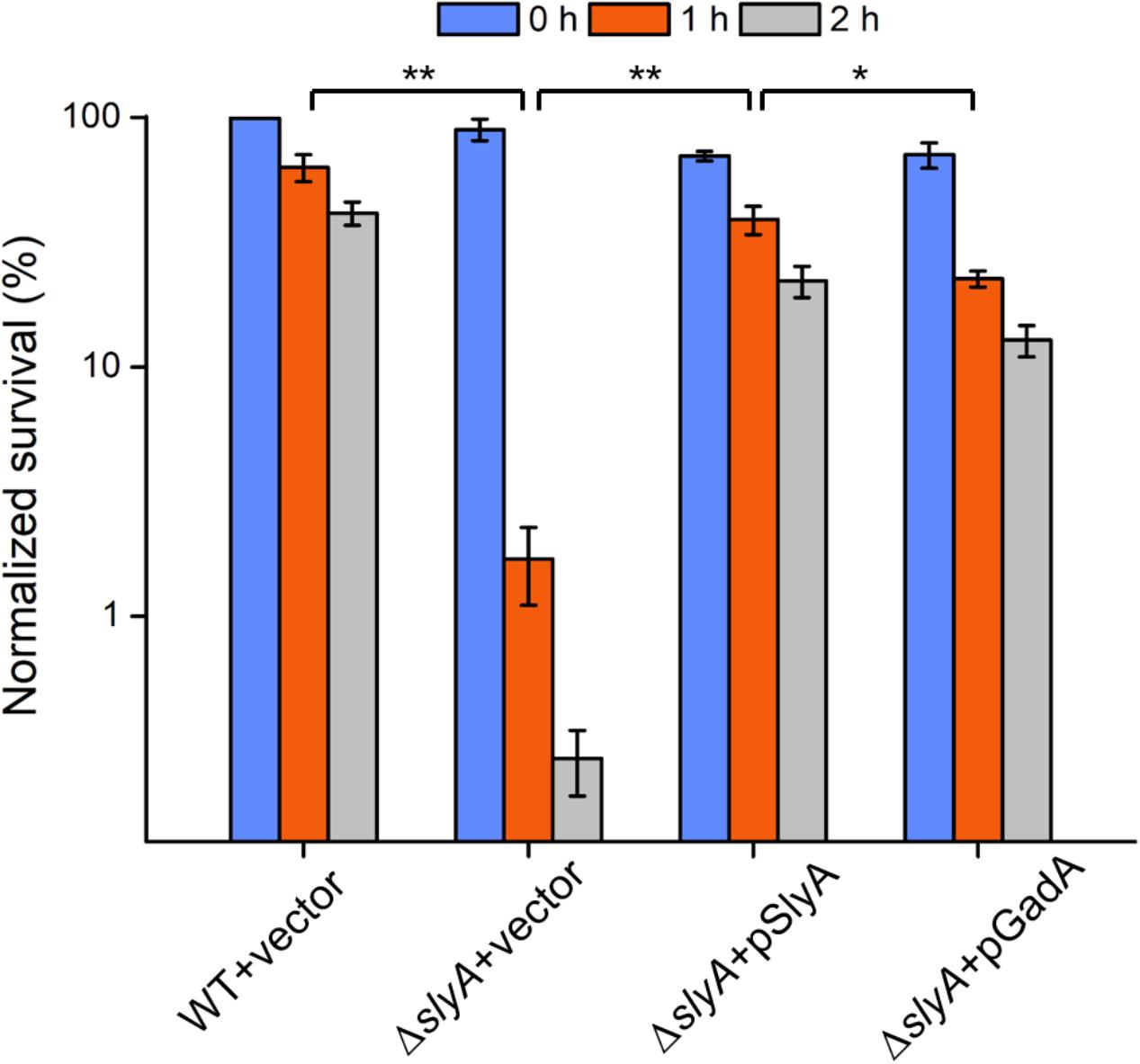
FIGURE 4. Acid survival assays of various Shigella strains. Survival percentages of Shigella wild-type and ΔslyA strains complemented with either empty vectors or SlyA/GadA-expressing plasmids were shown. Bacteria were incubated in acidic conditions for 0, 1, and 2 h and assayed for viability. Asterisks indicate significant differences (∗p < 0.05 and ∗∗p < 0.01).
Discussion
The MarR/SlyA family of transcriptional regulators has been identified in a variety of Gram-negative bacteria. In Salmonella Typhimurium, SlyA was reported to contribute to bacterial virulence by exerting a regulatory effect on SsrB (Navarre et al., 2005), a master regulator of Salmonella pathogenicity island 2 (SPI-2). Consistent with this notion, our previous work showed concurrent induction of S. Typhimurium SlyA and SPI-2-encoded virulence proteins during infection of epithelial cells (Liu et al., 2015, 2017). In addition to virulence, SlyA has been associated with other aspects of S. Typhimurium physiology including resistance to antimicrobial peptides (Navarre et al., 2005) and oxidative stress (Buchmeier et al., 1997). Unlike in S. Typhimurium, the regulatory role of SlyA in S. flexneri has been less defined. Previously, Weatherspoon-Griffin and Wing (2016) carried out the initial characterization of S. flexneri SlyA and identified a role in bacterial virulence as well. Our current study aims to examine the SlyA regulon in S. flexneri by using an unbiased proteomic approach. Quantitative profiling of a S. flexneri mutant lacking slyA in comparison to its parental strain led to the discovery of SlyA-dependent expression of many proteins associated with acid resistance such as GadA/B, HdeB, and Slp. Interestingly, the implication of SlyA in bacterial anti-acid defense was reported in Wing’s study, though the exact mechanism was not clarified (Weatherspoon-Griffin and Wing, 2016).
Among many bacterial anti-acid strategies, the glutamate decarboxylase-dependent pathway is one of the most effective systems for acid resistance. This system consists of three proteins, GadA, GadB, and GadC, all of which were down-regulated in the slyA deletion mutant. We further verified SlyA-dependent transcription of gadA by both mRNA measurements and β-galactosidase-based reporter assays. Importantly, we found SlyA directly regulates the expression of gadA by binding to its promoter region. After establishing the expression of GadA (and likely the entire Gad system) in a SlyA-dependent manner, we further examined the anti-acid capacities of the slyA-deletion mutant. Consistent with the previous report, the strain lacking slyA exhibited a severe survival defect upon acid challenge. Complementation with a SlyA-expressing plasmid was able to restore its anti-acid capacities to the level of the wild-type bacteria. If the glutamate decarboxylase-dependent pathway constitutes the major anti-acid system under the control of SlyA, we reasoned that the introduction of a GadA-expressing plasmid would rescue, at least in part, the survival defect of the slyA-deletion mutant under acid stress. Indeed, we found overproduction of GadA in the slyA-deletion strain largely restored its anti-acid capacities, though at slightly lower levels than the ΔslyA strain harboring a SlyA-producing plasmid as well as the WT bacteria. Such observations are in fact consistent with our proteomic data suggesting SlyA-dependent activation of multiple anti-acid pathways in addition to the Gad system. In other words, SlyA may regulate S. flexneri acid resistance through downstream proteins other than GadA.
Shigella flexneri gadB and gadC genes are located in a transcriptional unit different from that of gadA. In E. coli, a number of regulators have been identified to control the transcription of these gad genes, such as GadE (Ma et al., 2003a), GadW (Tucker et al., 2003), GadX (Tucker et al., 2003), PhoP (Zwir et al., 2005), YdeO (Ma et al., 2004), EvgA (Masuda and Church, 2003; Ma et al., 2004), RpoS (De Biase et al., 1999), and Crp (Ma et al., 2003b). Notably, GadE plays a central role in transcriptional regulation and activates the expression of gad genes by directly binding to the gad box (shown in Figure 3A), a 20-bp sequence located 63-bp upstream of the transcriptional start sites of gadA/B and gadC (Castanie-Cornet and Foster, 2001). Most of the other transcription factors above were found to regulate the expression of Gad proteins via the control of GadE. For instance, two AraC-like regulators, GadW and GadX, whose genes are located downstream of gadA, directly activate the transcription of gadE and hence the expression of gadA/B and gadC (Ma et al., 2002; Tramonti et al., 2002, 2003). However, under certain conditions, GadW and GadX can function as repressors by competitively binding to the gad box sequences (Castanie-Cornet and Foster, 2001; Ma et al., 2002; Tramonti et al., 2002). Thus, our discovery that S. flexneri SlyA regulates the transcription of gadA by directly binding to its promoter further expands the repertoire of regulators controlling the glutamate decarboxylase-dependent system. Previously, E. coli SlyA was also reported to regulate the expression of GadA and other proteins involved in acid resistance such as HdeA and HdeB (Spory et al., 2002). It is therefore tempting to speculate that the regulation of acid resistance by SlyA may be a conserved mechanism shared by many bacterial species.
Our proteomic data also revealed other SlyA-regulated proteins involved in acid resistance such as EvgA, YdeP, Frc, and S2577. EvgA is a member of the two-component regulatory system EvgS/EvgA, which activates the transcription of other regulators such as YdeO and GadE, thereby leading to increased expression of the Gad system (Ma et al., 2004). YdeP is a putative oxidoreductase under the control of EvgA. Intriguingly, overexpression of this enzyme in E. coli enhanced bacterial acid resistance (Masuda and Church, 2003). Frc and S2577 are homologous to the formyl-CoA transferase YfdW and the oxalyl-CoA decarboxylase YfdU in E. coli, respectively. Previously Fontenot et al. (2013) reported the contribution of these two enzymes to bacterial anti-acid capacities involving oxalate decarboxylation. Interestingly, the yfdXWUVE operon is also controlled by EvgA (Masuda and Church, 2002), and yfdX encodes a protein homologous to S2579, another down-regulated protein in our dataset. Given that EvgA regulates Frc, S2577, and S2579 in addition to the Gad system, it would be tempting to determine whether SlyA exerts a regulatory role in EvgA and thus indirectly influences the expression of the yfd operon. Furthermore, by using the gadA promoter as a positive control, the alignment of the SlyA-binding consensus (Stapleton et al., 2002) and the gadE gene sequence suggests direct binding of SlyA to the gadE promoter region (Figure 5). Considering the complicated regulatory network, we propose that SlyA might be a global transcriptional regulator of Shigella acid resistance (Figure 6), though experimental validation of SlyA-exerted regulatory roles in other known mediators of acid resistance would be highly desired in the future. Furthermore, given the direct binding of PhoP to the SlyA promoter in Salmonella Typhimurium (Shi et al., 2004), it is conceivable that SlyA is activated by other transcriptional factors such as PhoP rather than direct sensing of the environmental pH. Indeed, Shigella slyA promoter was also reported to respond to Mg2+ and be positively regulated by PhoP (Weatherspoon-Griffin and Wing, 2016).
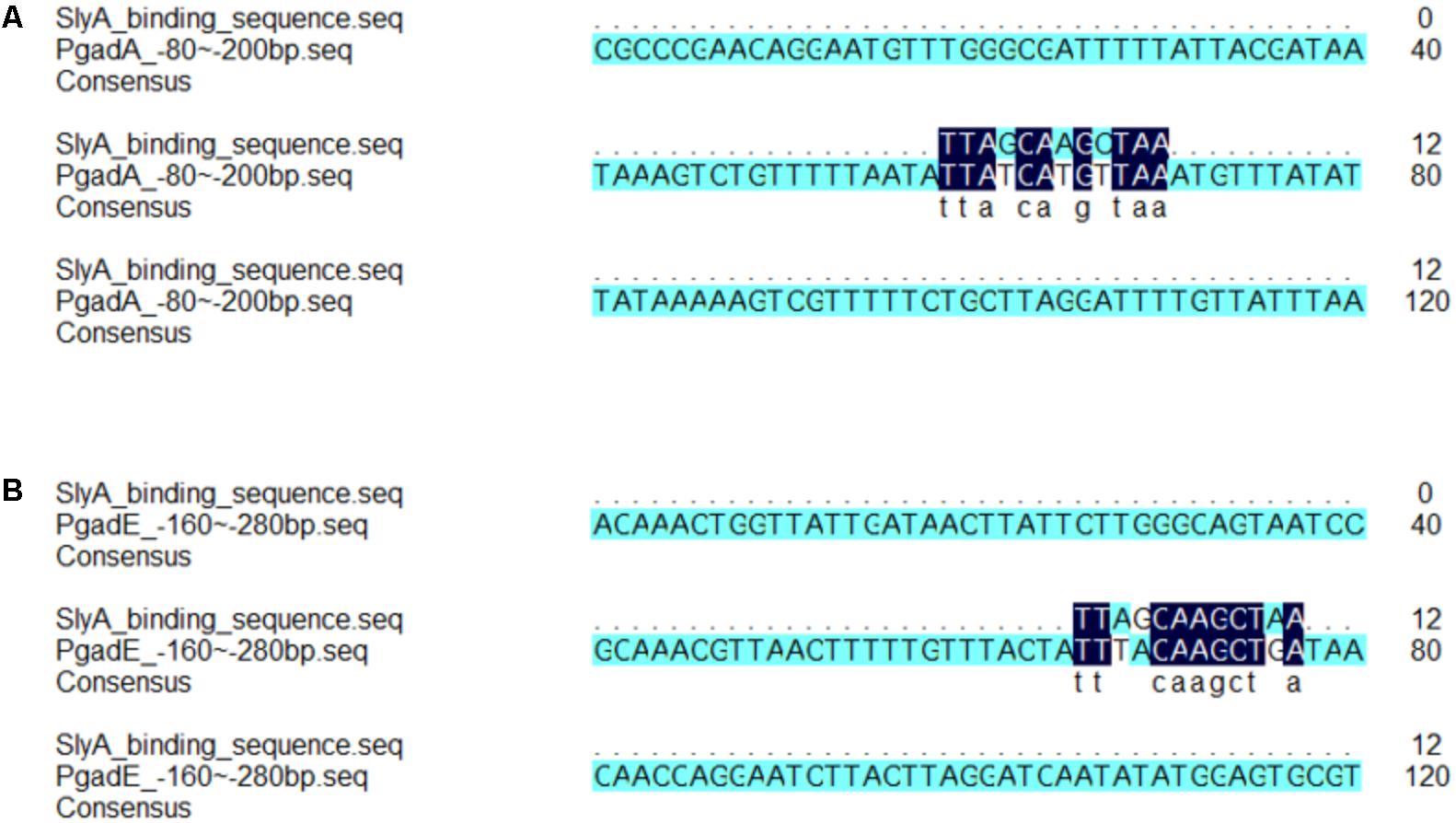
FIGURE 5. Alignment analyses revealed consensus SlyA-binding sequence in the promoter regions of the gadA and gadE genes. The alignment of consensus SlyA-binding sequence and fragments of the promoter of gadA (A) and gadE (B) was performed by the DNAMAN software.
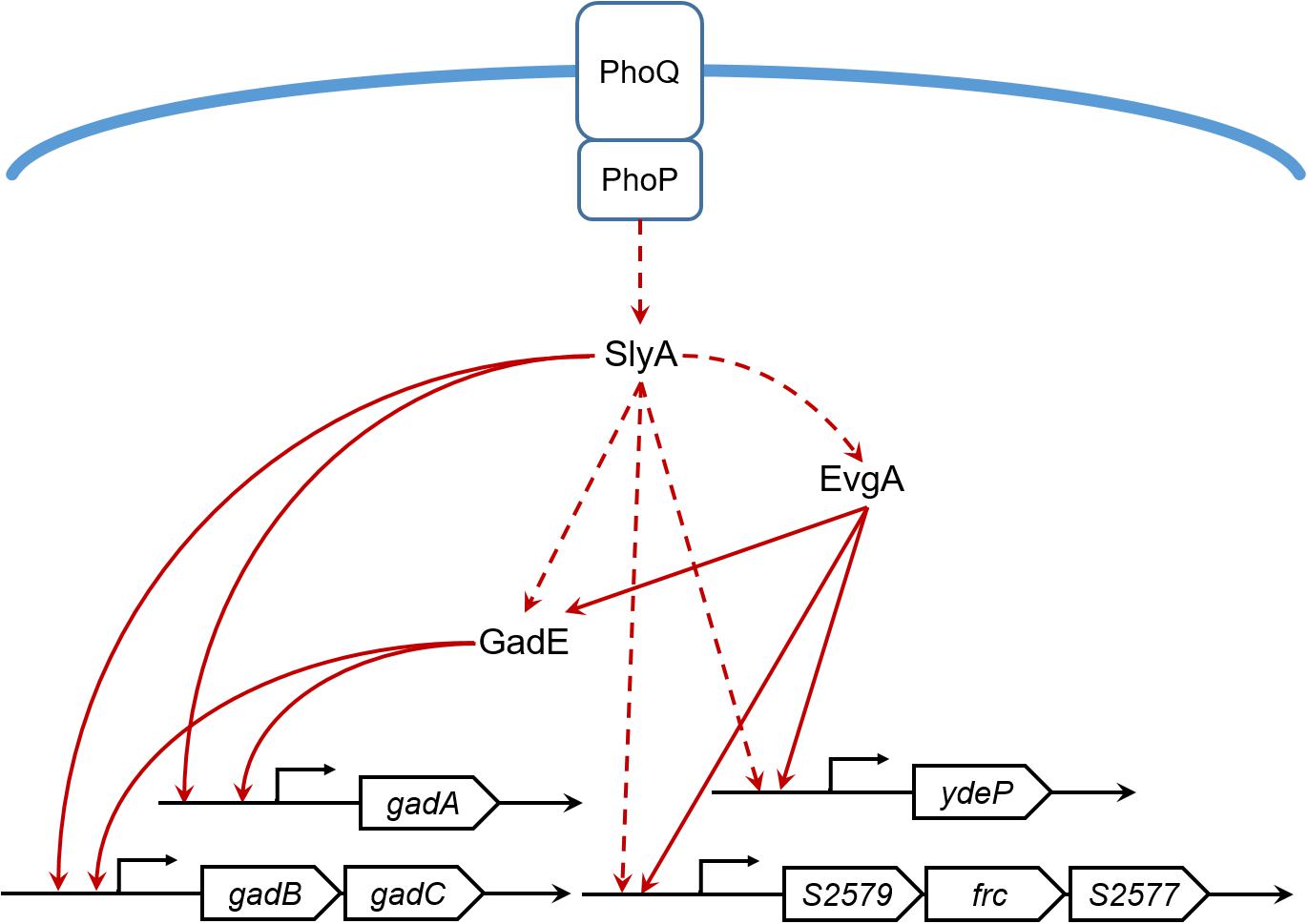
FIGURE 6. The SlyA regulatory network of S. flexneri acid resistance. Solid lines indicate direct or experimentally verified transcriptional regulation. Dashed lines indicate hypothetical regulation inferred from our proteomic results or literatures.
As a final note, we found insignificant difference in most T3SS-related proteins in the slyA deletion mutant (Supplementary Table S3), although Weatherspoon-Griffin and Wing (2016) suggested a potential role of SlyA in mediating S. flexneri virulence. In their previous work, ectopic expression of SlyA was found to increase the promoter activity of certain virulence genes and also rescue a virulence-associated phenotype. We reason that such observations may be resulted from the overproduction of SlyA from a high-copy-number plasmid, thereby differing to some extent from our proteomic measurements in which SlyA was expressed at physiological levels. Therefore, we propose that SlyA is likely to be a global transcriptional regulator of bacterial acid resistance in S. flexneri, which is in contrast to virulence regulation in S. Typhimurium.
Data Availability
The proteomics data reported in this paper have been deposited to the iProX database (URL: http://www.iprox.org/page/HMV006.html) under the accession number IPX0001250001.
Author Contributions
BZ and LR conducted most of the experiments with the assistance from MW, ZL, JJ, ZW, and SC. JF, BZ, LR, and XL wrote the manuscript with editorial inputs from all authors.
Funding
This work was financially supported by grants from the National Natural Science Foundation of China (21475005 and 21622501), Clinical Medicine Plus X-Young Scholars Project of Peking University, and the Thousand Young Talents Program of the Chinese government.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Liu laboratory for careful review of this manuscript. We also thank Dr. Guifang Jia for the use of the Applied Biosystems ViiATM 7 Real-Time PCR System and Dr. Aixin Yan for the kind gift of the plasmid pNN387.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02071/full#supplementary-material
TABLE S1 | All strains and primers used in this study.
TABLE S2 | All identified bacterial proteins.
TABLE S3 | Virulence proteins identified in the proteomic profiling.
References
Bearson, S., Bearson, B., and Foster, J. W. (1997). Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147, 173–180. doi: 10.1111/j.1574-6968.1997
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S., and Heck, A. J. (2009). Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494. doi: 10.1038/nprot.2009.21
Buchmeier, N., Bossie, S., Chen, C. Y., Fang, F. C., Guiney, D. G., and Libby, S. J. (1997). SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65, 3725–3730.
Castanie-Cornet, M. P., and Foster, J. W. (2001). Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147, 709–715. doi: 10.1099/00221287-147-3-709
De Biase, D., Tramonti, A., Bossa, F., and Visca, P. (1999). The response to stationary-phase stress conditions in Escherichia coli role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32, 1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x
Elledge, S. J., and Davis, R. W. (1989). Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3, 185–197. doi: 10.1101/gad.3.2.185
Ellison, D. W., and Miller, V. L. (2006). Regulation of virulence by members of the MarR SlyA family. Curr. Opin. Microbiol. 9, 153–159. doi: 10.1016/j.mib.2006.02.003
Fontenot, E. M., Ezelle, K. E., Gabreski, L. N., Giglio, E. R., McAfee, J. M., and Mills, A. C. et al. (2013). YfdW and YfdU are required for oxalate-induced acid tolerance in Escherichia coli K-12. J. Bacteriol. 195, 1446–1455. doi: 10.1128/JB.01936-1912.
Foster, J. W. (2004). Escherichia coli acid resistance tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. doi: 10.1038/nrmicro1021
Galan, B., Kolb, A., Sanz, J. M., Garcia, J. L., and Prieto, M. A. (2003). Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res. 31, 6598–6609. doi: 10.1093/nar/gkg851
George, A. M., and Levy, S. B. (1983). Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 155,541–548.
Hersh, B. M., Farooq, F. T., Barstad, D. N., Blankenhorn, D. L., and Slonczewski, J. L. (1996). A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178, 3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996
Jennison, A. V., and Verma, N. K. (2007). The acid-resistance pathways of Shigella flexneri 2457T. Microbiology 153, 2593–2602. doi: 10.1099/mic.0.2007/006718-0
Kotloff, K. L., Winickoff, J. P., Ivanoff, B., Clemens, J. D., Swerdlow, D. L., and Sansonetti, P. J. et al. (1999). Global burden of Shigella infections implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77, 651–666.
Lai, M., Liang, L., Chen, J., Qiu, N., Ge, S., and Ji, S. et al. (2016). Multidimensional proteomics reveals a role of UHRF2 in the regulation of epithelial-mesenchymal transition (EMT). Mol. Cell. Proteomics 15, 2263–2278. doi: 10.1074/mcp.M115.057448
Libby, S. J., Goebel, W., Ludwig, A., Buchmeier, N., Bowe, F., and Fang, F. C. et al. (1994). A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. U.S.A. 91, 489–493. doi: 10.1073/pnas.91.2.489
Liu, Y., Yu, K., Zhou, F., Ding, T., Yang, Y., and Hu, M. (2017). Quantitative proteomics charts the landscape of Salmonella carbon metabolism within host epithelial cells. J. Proteome Res. 16, 788–797. doi: 10.1021/acs.jproteome.6b00793
Liu, Y., Zhang, Q., Hu, M., Yu, K., Fu, J., and Zhou, F. et al. (2015). Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect. Immun. 83, 2897–2906. doi: 10.1128/IAI.02882-14
Livak, K. J., and Schmitge, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Z., Gong, S., Richard, H., Tucker, D. L., Conway, T., and Foster, J. W. (2003a). GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49, 1309–1320.
Ma, Z., Richard, H., and Foster, J. W. (2003b). pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 185,6852–6859.
Ma, Z., Masuda, N., and Foster, J. W. (2004). Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 186, 7378–7389. doi: 10.1128/JB.186.21.7378-7389.2004
Ma, Z., Richard, H., Tucker, D. L., Conway, T., and Foster, J. W. (2002). Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184, 7001–7012. doi: 10.1128/JB.184.24.7001-7012.2002
Masuda, N., and Church, G. M. (2002). Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184, 6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002
Masuda, N., and Church, G. M. (2003). Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48, 699–712. doi: 10.1046/j.1365-2958.2003.03477.x
Mattock, E., and Blocker, A. J. (2017). How do the virulence factors of Shigella work together to cause disease. Front. Cell. Infect. Microbiol. 7:64. doi: 10.3389/fcimb.2017.00064
Navarre, W. W., Halsey, T. A., Walthers, D., Frye, J., McClelland, M., and Potter, J. L. (2005). Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56, 492–508. doi: 10.1111/j.1365-2958.2005.04553.x
Roper, D. I., Fawcett, T., and Cooper, R. A. (1993). The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol. Gen. Genet. 237, 241–250. doi: 10.1007/BF00282806
Shi, Y., Latifi, T., Cromie, M. J., and Groisman, E. A. (2004). Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J. Biol. Chem. 279, 38618–38625. doi: 10.1074/jbc.M406149200
Spory, A., Bosserhoff, A., von Rhein, C., Goebel, W., and Ludwig, A. (2002). Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184, 3549–3559. doi: 10.1128/JB.184.13.3549-3559.2002
Srikumar, R., Kon, T., Gotoh, N., and Poole, K. (1998). Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexCMexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42, 65–71.
Stapleton, M. R., Norte, V. A., Read, R. C., and Green, J. (2002). Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277, 17630–17637. doi: 10.1074/jbc.M110178200
Thomson, N. R., Cox, A., Bycroft, B. W., Stewart, G. S., Williams, P., and Salmond, G. P. (1997). The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26, 531–544. doi: 10.1046/j.1365-2958.1997.5981976.x
Tramonti, A., De Canio, M., Bossa, F., and De Biase, D. (2003). Stability and oligomerization of recombinant GadX, a transcriptional activator of the Escherichia coli glutamate decarboxylase system. Biochim. Biophys. Acta. 1647, 376–380. doi: 10.1016/S1570-9639(03)00098-0
Tramonti, A., Visca, P., De Canio, M., Falconi, M., and De Biase, D. (2002). Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184, 2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002
Tucker, D. L., Tucker, N., Ma, Z., Foster, J. W., Miranda, R. L., and Cohen, P. S. et al. (2003). Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185, 3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003
Weatherspoon-Griffin, N., and Wing, H. J. (2016). Characterization of SlyA in Shigella flexneri identifies a novel role in virulence. Infect. Immun. 84, 1073–1082. doi: 10.1128/IAI.00806-15
Yu, K., Wang, Z., Zhou, F., Jiang, J., Liu, Y., and Hu, M. et al. (2017). Quantitative analysis of Shigella flexneri protein expression under acid stress. Proteomics 17:1600381. doi: 10.1002/pmic.201600381
Keywords: Shigella flexneri, SlyA, acid resistance, glutamate decarboxylation, GadA
Citation: Zhang B, Ran L, Wu M, Li Z, Jiang J, Wang Z, Cheng S, Fu J and Liu X (2018) Shigella flexneri Regulator SlyA Controls Bacterial Acid Resistance by Directly Activating the Glutamate Decarboxylation System. Front. Microbiol. 9:2071. doi: 10.3389/fmicb.2018.02071
Received: 14 June 2018; Accepted: 14 August 2018;
Published: 31 August 2018.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Laura R. Jarboe, Iowa State University, United StatesFeng Jiang, Chinese Academy of Medical Sciences, China
Yi-Cheng Sun, Institute of Pathogen Biology (CAMS), China
Copyright © 2018 Zhang, Ran, Wu, Li, Jiang, Wang, Cheng, Fu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Liu, eGlhb3l1bi5saXVAcGt1LmVkdS5jbg==
†These authors have contributed equally to this work
 Buyu Zhang
Buyu Zhang Longhao Ran†
Longhao Ran† Xiaoyun Liu
Xiaoyun Liu