- 1Key Laboratory of Applied Technology on Green-Eco-Healthy Animal Husbandry of Zhejiang Province, College of Animal Science and Technology, Zhejiang A&F University, Hangzhou, China
- 2Department of Animal Science, Wenzhou Vocational College of Science and Technology, Wenzhou, China
- 3Faculty of Health Sciences, University of Macau, Macau, China
- 4College of Life Science, Nanjing Agricultural University, Nanjing, China
Bacterial pathogens maintain disulfide bonds for protein stability and functions that are required for pathogenesis. Vibrio parahaemolyticus is a Gram-negative pathogen that causes food-borne gastroenteritis and is also an important opportunistic pathogen of aquatic animals. Two genes encoding the disulfide bond formation protein A, DsbA, are predicted to be encoded in the V. parahaemolyticus genome. DsbA plays an important role in Vibrio cholerae virulence but its role in V. parahaemolyticus is largely unknown. In this study, the activities and functions of the two V. parahaemolyticus DsbA proteins were characterized. The DsbAs affected virulence factor expression at the post-translational level. The protein levels of adhesion factor VpadF (VP1767) and the thermostable direct hemolysin (TDH) were significantly reduced in the dsbA deletion mutants. V. parahaemolyticus lacking dsbA also showed reduced attachment to Caco-2 cells, decreased β-hemolytic activity, and less toxicity to both zebrafish and HeLa cells. Our findings demonstrate that DsbAs contribute to V. parahaemolyticus pathogenesis.
Introduction
Vibrio parahaemolyticus is a Gram-negative halophilic bacterium that lives in estuarine, marine and coastal surroundings. It is one of the leading causes of human food borne gastroenteritis due to consumption of raw or under-cooked seafood (Yeung and Boor, 2004; Su and Liu, 2007; Zhang and Orth, 2013), and is also one of the major threats in aquaculture (Tey et al., 2015). V. parahaemolyticus can cause acute hepatopancreas necrosis disease (AHPND) in shrimp and is one of the major pathogens of cultured mud crabs (Xiao et al., 2017; Zhang et al., 2019).
Vibrio parahaemolyticus secretes virulence factors to establish successful infection in the host (Broberg et al., 2011; Letchumanan et al., 2014). Virulence factors that contain free thiol-groups (cysteine residues) require disulfide bonds for proper folding and function. Studies of oxidoreductases are therefore important for understanding bacterial pathogenesis and for developing novel therapeutics (Heras et al., 2009). Disulfide bond formation pathways in Gram-negative bacteria have been well studied in Escherichia coli (Kadokura and Beckwith, 2009; Depuydt et al., 2011). Disulfide bond formation protein A, DsbA, introduces disulfide bonds into proteins between consecutive cysteine residues as they pass through the inner membrane translocation system (Bardwell et al., 1991; Berkmen et al., 2005; Kadokura and Beckwith, 2009).
The Dsb redox system plays a pivotal role in the virulence of many pathogens (Braun et al., 2001; Heras et al., 2009; Qin et al., 2009; Ren et al., 2014; Mariano et al., 2018). Francisella tularensis requires the DsbA redox system to promote the proper folding of virulence proteins (Qin et al., 2009; Ren et al., 2014). Pseudomonas aeruginosa DsbA is required for the expression of elastase, exotoxin A, protease IV, and is also required for the formation of a functional type III secretion system (Braun et al., 2001; Ha et al., 2003). Serratia marcescens DsbA is required for virulence and for proper deployment of the T6SS (Mariano et al., 2018). We previously found that DsbA is indispensable to Vibrio cholerae pathogenesis. A dsbA deletion mutant of V. cholerae did not colonize the intestine because the expression of the master virulence gene regulator ToxT was abolished in the dsbA deletion mutant (Yang et al., 2013; Xue et al., 2016).
Here, we investigated the role of V. parahaemolyticus DsbA in pathogenesis. Two DsbA genes are predicted to be encoded in the V. parahaemolyticus genome. We analyzed the reductase activities and the redox potential of the purified DsbA proteins and characterized the virulence properties of single and double dsbA mutants. Deleting the dsbA genes affected V. parahaemolyticus virulence factor expression at the post-translational level.
Materials and Methods
Bacterial Strains, Plasmids, and Media
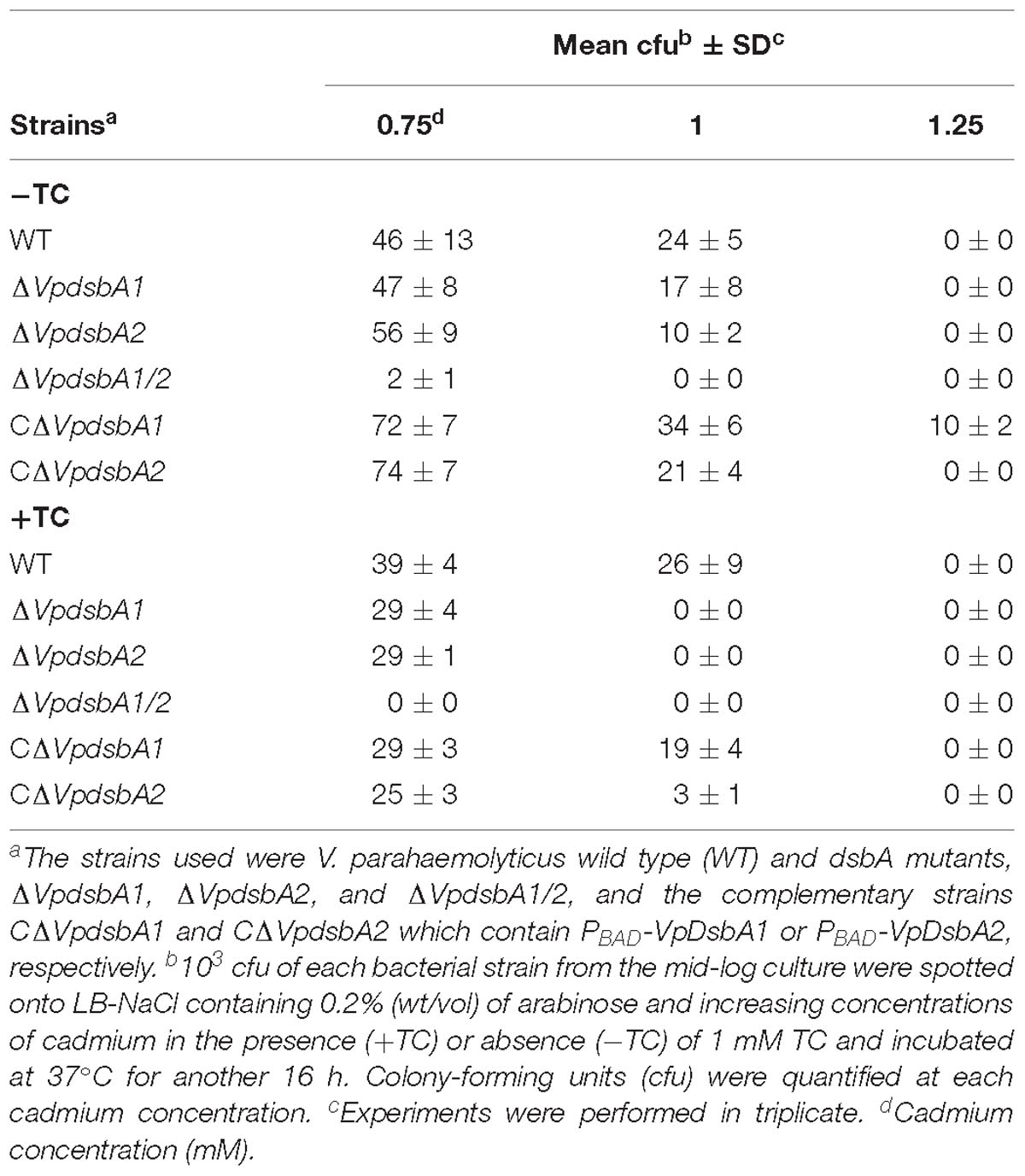
The bacterial strains, plasmids, and oligonucleotides used in this study are summarized in Tables 1, 2. All V. parahaemolyticus strains used in this study were derived from the HZ strain, a clinical isolate from the Zhejiang Provincial Center for Disease Control and Prevention, Zhejiang, China (Yu et al., 2015). Escherichia coli strains DH5α, BL21, and CC118λpir were used for general manipulation of plasmids, prokaryotic expression of proteins, and mobilization of plasmids into V. parahaemolyticus, respectively. The bacterial strains were grown at 37°C in Luria-Bertani (LB) broth with 1% NaCl (Sambrook et al., 1989) (for E. coli) or LB-NaCl (for V. parahaemolyticus) which is LB broth supplemented with 3% NaCl containing appropriate antibiotics. Plasmids for overexpressing VpDsbAs or VpDsbB or other proteins in V. parahaemolyticus were constructed by cloning the PCR-amplified coding regions into pBAD24Cm (Guzman et al., 1995) and introduced into V. parahaemolyticus by conjugation with the helper strain of CC118λpir (Yu et al., 2015). Plasmids for overexpressing VpDsbAs in E. coli were constructed by cloning the coding regions into pacyc177 which has been modified to obtain an arabinose operon (Chang and Cohen, 1978) and introduced into E. coli strain by electroporation (Sambrook et al., 1989).
Construction of the dsbA and dsbB Mutants
In-frame deletion strains used in this study were described in previous publications (Yu et al., 2015; Jiang et al., 2018). In-frame deletions of VpdsbA1 (vp3054), VpdsbA2 (vpa1271), VpdsbB (vp2073) or EcDsbA were constructed by cloning the regions flanking the target genes into suicide vector pDS132 (Philippe et al., 2004) containing a sacB counter selectable marker (Metcalf et al., 1996). The resulting plasmids were introduced into V. parahaemolyticus or E. coli by conjugation (Boyer, 1966; Yu et al., 2015) and deletion mutants were selected for double homologous recombination events (Kaniga et al., 1991).
Zebrafish Virulence Evaluation
Zebrafish virulence assays were designed based on a previous publication (Paranjpye et al., 2013). All animal experiments were carried out in strict accordance with the animal protocols that were approved by the Institutional Animal Care and Use Committee of Zhejiang A&F University (Permit Number: ZJAFU/IACUC_2011-10-25-02). Care and feeding of zebrafish followed established protocols1. Zebrafish were obtained from a commercial supplier between 5 and 6 months old and were raised at our animal facility at Zhejiang A&F University for at least 2 weeks before challenge experiments following previously published protocols (Paranjpye et al., 2013). For challenge experiments, zebrafish were injected intraperitoneally using a repeater dispenser (Hamilton #83700) and a 33 gauge needle (Hamilton 1750LTSN, 33/0.3759/PT4) following previously published protocols (Lefebvre et al., 2009). Ten zebrafish per group were inoculated with each strain at a concentration of 107 cfu 10 μl−1 of the inoculum and observed every 4 or 8 h for 48 h. Control fish injected with 10 μl PBS were included with each experiment. Experiments were repeated at least once. Tris-buffered tricaine at a concentration of 320 μg ml−1 was used to kill the fish on completion of the experiment. All aquaria and water were disinfected with 20% sodium hypochlorite after each experiment. Filters from air pumps were sterilized by autoclaving.
DsbA Cloning, Expression, and Purification
Vibrio parahaemolyticus VpdsbA1, VpdsbA2 or VPA1314 genes without the signal peptide sequence were amplified from the genomic DNA which was purified by QIAamp DNA Mini Kit (Qiagen) from bacterial culture. The PCR products without the signal peptide sequence were inserted into a modified pET-28a (Novagen, Inc.) vector encoding an N-terminal His6 tag. E. coli BL21(DE3)/pLysS (Karimova et al., 1998) cells were transformed with the plasmid containing the target gene and transformed cells were used for protein expression by autoinduction (Thompson et al., 1994). Proteins were expressed and purified on nickel columns according to the manufacturer’s instructions (Invitrogen).
Antibody Preparation
Ten milligrams of each protein, VpDsbA1, VpDsbA2, and VPA1314 (TDH) purified as described above were used as antigens, and then sent to GenScript Inc. for polyclonal antibodies preparation by immunizing rabbits. Antibody effectiveness was detected by Western blotting of each specific purified protein a week after the fourth immunization.
Insulin Reduction Assay
The protein disulfide reductase activity of VpDsbA was measured in vitro using the insulin-reduction assay in the presence of dithiothreitol (DTT) (Holmgren, 1979). Each DsbA protein (10 μM) was mixed with buffer consisting of 100 mM sodium phosphate, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.33 mM DTT. Insulin (170 μM) was added immediately before measurements were made. Insulin reduction by DTT was monitored spectrophotometrically at 650 nm.
Redox Potential Determination
VpDsbA redox potential assays were performed essentially as described by Walden et al. (2012). VpDsbA (2 μM) was incubated in fully degassed buffer consisting of 100 mM sodium phosphate, 1 mM EDTA pH 7.0 containing 1 mM oxidized glutathione (GSSG; Sigma–Aldrich, United States) and a range of reduced glutathione (GSH) concentrations (0.1–2.0 mM) for 24 h at room temperature. After incubation, the reactions were stopped with 10% trichloroacetic acid (TCA) and the precipitated protein pellets were collected by centrifugation at 16,000 × g for 30 min at 4°C. The pellets were washed with 100% ice-cold acetone and dissolved in a buffer consisting of 50 mM Tris-HCl pH 7.0, 1% sodium dodecyl sulfate (SDS), 10 mM 4-acetamide-4′-maleimidylstilbene-2,2′-disulfonate (AMS) to label the free thiols. Separation of the reduced and oxidized forms was performed on 12% SDS-polyacrylamide gels under denaturing conditions. Gels were stained with Coomassie Brilliant Blue and scanned. The relative intensity of the reduced and oxidized forms was analyzed using ImageJ2. The fraction of the reduced protein was plotted against the ratio [GSH]2/[GSSG]. The equilibrium constant Keq was calculated using the equation:
where R is the fraction of reduced protein at equilibrium. The standard redox potential was calculated using the Nernst equation,
where E0′GSH/GSSG is the standard potential of −240 mV (Gilbert, 1995), R is the universal gas constant 8.314 J K−1mol−1, T is the absolute temperature in Kelvin, n is the number of electrons transferred, F is the Faraday constant 9.648 × 104 C mol−1 and Keq is the equilibrium constant.
Real-Time Quantitative PCR (RT-qPCR) Analysis
Overnight cultures of V. parahaemolyticus WT or dsbA mutants were subcultured at a dilution of 1:100 in LB-NaCl medium and incubated without shaking at 37°C for 4 h. Total RNA was purified from bacterial cultures using TRIzol reagent (Invitrogen), DNase digestion and RNA reverse transcription was performed by using the PrimeScript RT reagent Kit with gDNA Eraser (TaKara). Quantitative real-time qPCR was performed in 20 μl reaction mixtures containing SYBR quantitative PCR mix (Toyobo) to measure the transcriptional levels of genes of interest using the Mx3000P PCR detection system (Agilent) with primers specific for tested genes. The 16s rRNA gene were used as internal controls in all reactions.
Spot Titers Cadmium Resistance
Cadmium resistance was performed to quantify the relative disulfide oxidase activity of the strains in vivo as described in Xue et al. (2016). Briefly, strains were grown overnight in LB and diluted 1:100 into fresh LB media with appropriate antibiotics. Strains were grown to mid-logarithmic phase at 37°C and serially diluted into phosphate buffer salt (PBS). A 5 μl aliquot of each dilution was plated onto LB plates with a cadmium gradient. Cells were grown at 37°C overnight. All spot titers were performed at least in triplicate.
Isothermal Titration Calorimetry
Affinity and thermodynamics of binding between taurocholate (TC) and VpDsbA proteins were assessed by isothermal titration calorimetry (ITC) using a VP-ITC instrument (MicroCalTM, GE Healthcare) according to the protocols as described in Xue et al. (2016). Briefly, the sample cell was loaded with 1.5 ml of purified protein at 200 μM concentration in PBS. The syringe was filled with TC in the same buffer as that was used to dilute the proteins at a concentration of 4 mM. Titrations were conducted at 25°C using 25 consecutive injections of 10 μl each delayed by 300 s with a stirring speed of 307 rpm. As a control for background noise, titration of TC into a solution containing the buffer only was performed. The association constant (Ka = 1/Kd), free energy (ΔG), and enthalpy change (ΔH) and entropy change (ΔS) were calculated by fitting the data to a single-site binding model using the MicroCal Origin software (Origin 7.0 SR4 version7.0552β). Parameters reported include the mean ± SD across three replicates. The calculated c-value for these measurements is 12.
Motility Assay in Soft Agar
Motility assay was performed essentially on soft LB (for E. coli strains) or LB-NaCl (for V. parahaemolyticus strains) agar (0.25%) as described (Paxman et al., 2009). E. coli or V. parahaemolyticus strains were grown overnight on LB or LB-NaCl agar and a single colony of each strain was inserted into soft agar by tooth pick and incubated at 37°C for 8–12 h. Motility was assessed by examining migration of bacteria through agar from the center toward the periphery of the colony. The diameter of the motility circle was analyzed using ImageJ (see text footnote 2).
VpDsbA Oxidized by VpDsbB Present in Membranes
VpDsbA oxidized by VpDsbB in vitro was performed as previously described (Xue et al., 2016). V. parahaemolyticus strain ΔdsbA harboring a plasmid encoding VpDsbB under the control of an arabinose inducible promoter was cultured at 37°C with 200 rpm shaking in LB-NaCl medium until OD600≈0.5. 0.2% of arabinose was added and bacteria were kept cultured in the same conditions for another 16 h. Membranes were prepared according to Kovach et al. (1995). Purified DsbA was reduced by incubation in 50 mM DTT for 10 min at 4°C. DTT was then removed by gel filtration on PD-10 Sephadex columns (GE). Reduced DsbA was stored at −80°C in the presence of 0.1 mM EDTA, pH 8.0. VpDsbB membrane suspension (1 mg/ml) was mixed with 10 mM ubiquinone 1 (UQ1) with or without different concentrations of TC as indicated in PBS. Reactions were started right after 2 mM of reduced VpDsbA was added. TCA (10%) was added at different time points to stop the reaction and protein was precipitated at 4°C overnight. Precipitated proteins were treated with AMS as described. Negative controls were the membrane of V. parahaemolyticus ΔdsbA/dsbB.
Cell Adhesion Assay
Caco-2 cells (Sigma-Aldrich Inc.) were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) at 37°C in 5% CO2. Adhesion assays were carried out as previously described (Liu and Chen, 2015). Briefly, Caco-2 cells were infected with freshly prepared V. parahaemolyticus cultures of tested strains at a multiplicity of infection (MOI) of 10. As a control, bacterial cells were added to the empty wells of the cell culture plates and incubated for the same time as the binding experiment to determine the final total number of V. parahaemolyticus for the experiment. After 1 h infection, cells were washed three times with PBS and then lysed with 1% Triton X-100 at 37°C for 10 min. The cell lysates and control bacteria were serially diluted and plated on LB-NaCl agar. Attachment rate was calculated by dividing bound bacteria to the total bacterial load.
Kanagawa Hemolytic (KH) Activity Assay
Kanagawa hemolytic (KH) hemolytic activity was determined as described in Chun et al. (1975). Briefly, Wagatsuma blood agar (WBA) (Chun et al., 1975) contained 10% washed rabbit erythrocytes was prepared freshly before each assay. Bacteria from freshly prepared V. parahaemolyticus cultures of test strains were pelleted by centrifugation and washed with PBS. ∼108 CFU in 10 μl bacterial suspension were dropped on WBA plates, and the result was observed after incubation for 24 h at 37°C. Well-defined, clear hemolysis around the bacterial growth was recorded and the hemolysis around area was analyzed using ImageJ (see text footnote 2).
Cytotoxicity Assay
The cytotoxic assays were performed as described previously (Hiyoshi et al., 2010). HeLa cells (Thermo Fisher Scientific Inc.) were seeded in 96-well plates. The overnight cultured strains were sub-cultured at the ratio 1:50 to the fresh LB-NaCl broth medium and grow at 37°C to OD600≈0.5. Bacteria were pelleted by centrifugation and washed with PBS, then suspended in two volumes of unsupplemented DMEM (DMEM without FBS). Before infection, HeLa cells were also washed with unsupplemented DMEM. Infection was performed at an MOI of 10. After infection, the release of lactate dehydrogenase (LDH) into the medium was quantified at each indicated time point with a CytoTox96 kit (Promega, Madison, WI, United States) used according to the manufacturer’s instructions.
Statistical Analysis
All data are presented as the mean ± SD of three determinations in each experimental condition. Statistical significance was determined using one-way ANOVA. P < 0.05 was considered statistically significant.
Results
V. parahaemolyticus Encodes Two dsbA Genes
VP3054 (NP_799433.1) and VPA1271 (NP_800781.1) are predicted to be DsbA homologs in V. parahaemolyticus. The amino acid sequences of VP3054 and VPA1271 were compared and thioredoxin-fold molecular features were observed (Figure 1A). Both VP3054 and VPA1271 show about 40% amino acid sequence identity to E. coli DsbA (EcDsbA), Klebsiella pneumonia DsbA (KpDsbA) or Salmonella enterica DsbA (SeDsbA), and the identities are even higher when compared to V. cholerae DsbA from (VcDsbA). VP3054 shows 79% identity to VcDsbA while VPA1271 is 59% identical (Figure 1A). Both VP3054 and VPA1271 encode a DsbA-like domain, including a conserved CXXC active-site motif (CPHC in this case) and a putative cisPro motif (Ren et al., 2009) (Figure 1A). Comparison of loop sequences on the catalytic face of these DsbA proteins revealed that both the two DsbAs from V. parahaemolyticus should belong to Class-Ia DsbA (McMahon et al., 2014; Totsika et al., 2018). To differentiate these two proteins, we named VP3054 as VpDsbA1 and VPA1271 as VpDsbA2.
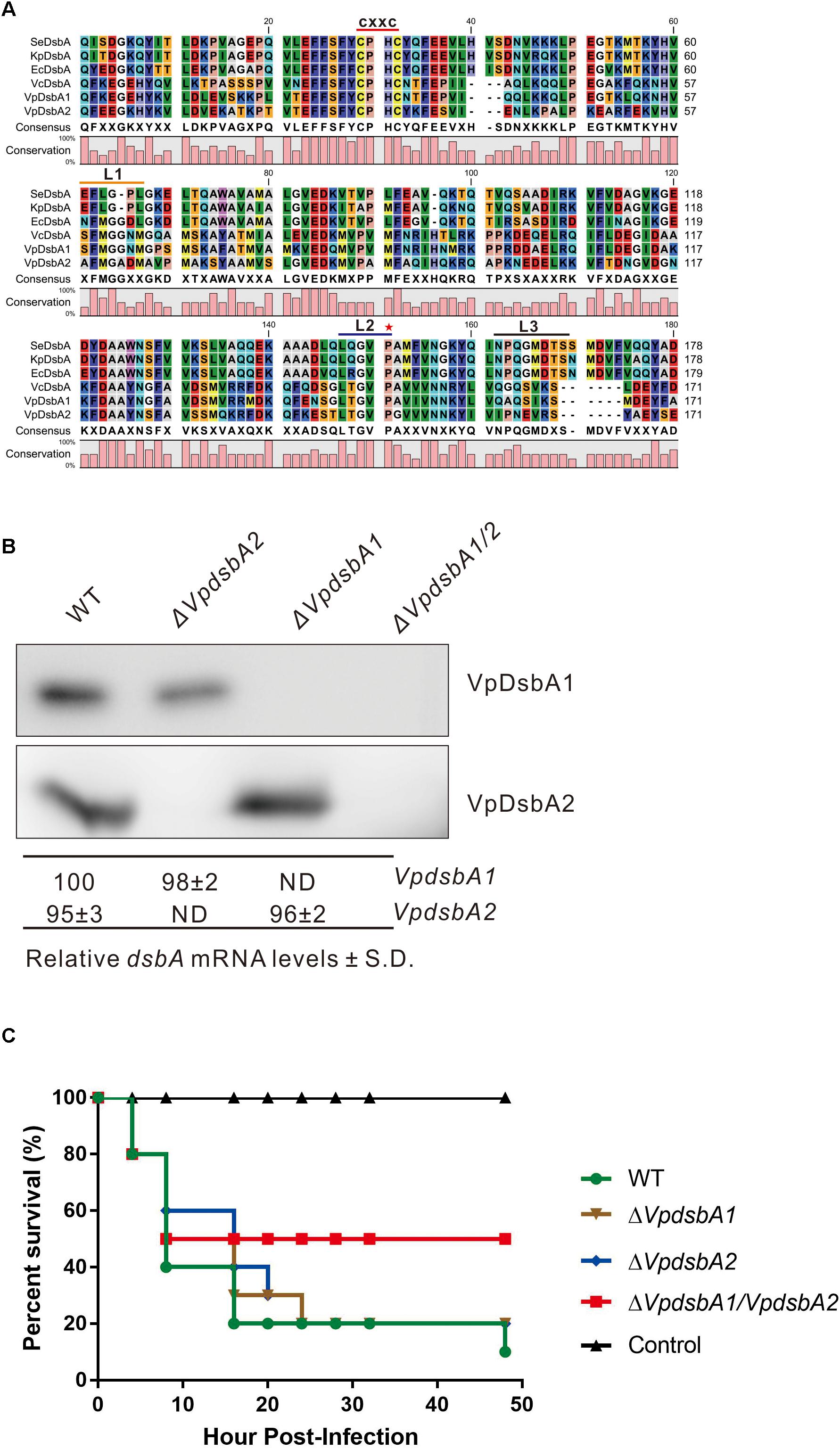
Figure 1. VpDsbAs modulate V. parahaemolyticus virulence in zebrafish. (A) Amino acid alignment of VpDsbA1, VpDsbA2, VcDsbA, and EcDsbA. The red line indicates the conserved CXXC motif. Three loop sequences of L1, L2, and L3 are indicated as an orange, blue, and black line, respectively, and a red star indicates the predicted cisPro. (B) Top, analysis of VpDsbA1 and VpDsbA2 expression level. V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 were grown in LB-NaCl until OD600≈0.8. Cell lysates (1 mg) were separated by SDS-PAGE and VpDsbA1 or VpDsbA2 was detected by the Western blot using antiserum specific for either VpDsbA1 or VpDsbA2. Blot shown is representative of at least three separate experiments. Bottom, analysis of VpdsbA1 and ΔVpdsbA2 mRNA levels by qRT-PCR. RNA was purified from freshly prepared cultures grown in LB-NaCl. The % dsbA mRNA levels ± standard deviation (SD) were normalized to 16S RNA for each strain and relative to WT (set to 100%). ND, none detected. (C) Survival curves (Kaplan–Meier) of zebrafish following intraperitoneal challenges of V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 strains.
We found that both VpdsbA1 and VpdsbA2 are expressed in V. parahaemolyticus (Figure 1B), and mutating one gene did not appreciably affect the expression of the other gene. In a zebrafish infection model, ΔVpdsbA1/2 had significantly reduced virulence while both ΔVpdsbA1 and ΔVpdsbA2 were as virulent as the wild type strain (Figure 1C). Each mutant grew similarly to the wild-type strain in broth culture (Supplementary Figure S1). These results suggest that both VpDsbA1 and VpDsbA2 are functional in V. parahaemolyticus and they may function redundantly, because a virulence defect was only seen in the double mutant.
Redox Properties of VpDsbA1 and VpDsbA2
To investigate the enzyme activity of VpDsbA1 and VpDsbA2, we performed a standard reductase activity assay using folded insulin as the substrate and DTT as the electron donor (Holmgren, 1979). Insulin comprises A and B chains which are linked by two disulfide bonds, and the reduction of the disulfide bonds leads to B-chain precipitation, which can be monitored as an increase in absorbance at 650 nm. B chain precipitation and a consequent increase in the OD650 was measurable after 22 min with EcDsbA (Figure 2A). B chain precipitation was observed after 15 min with VpDsbA1 and after 22 min with VpDsbA2. Insulin reduction was observed after 10 min with VcDsbA.
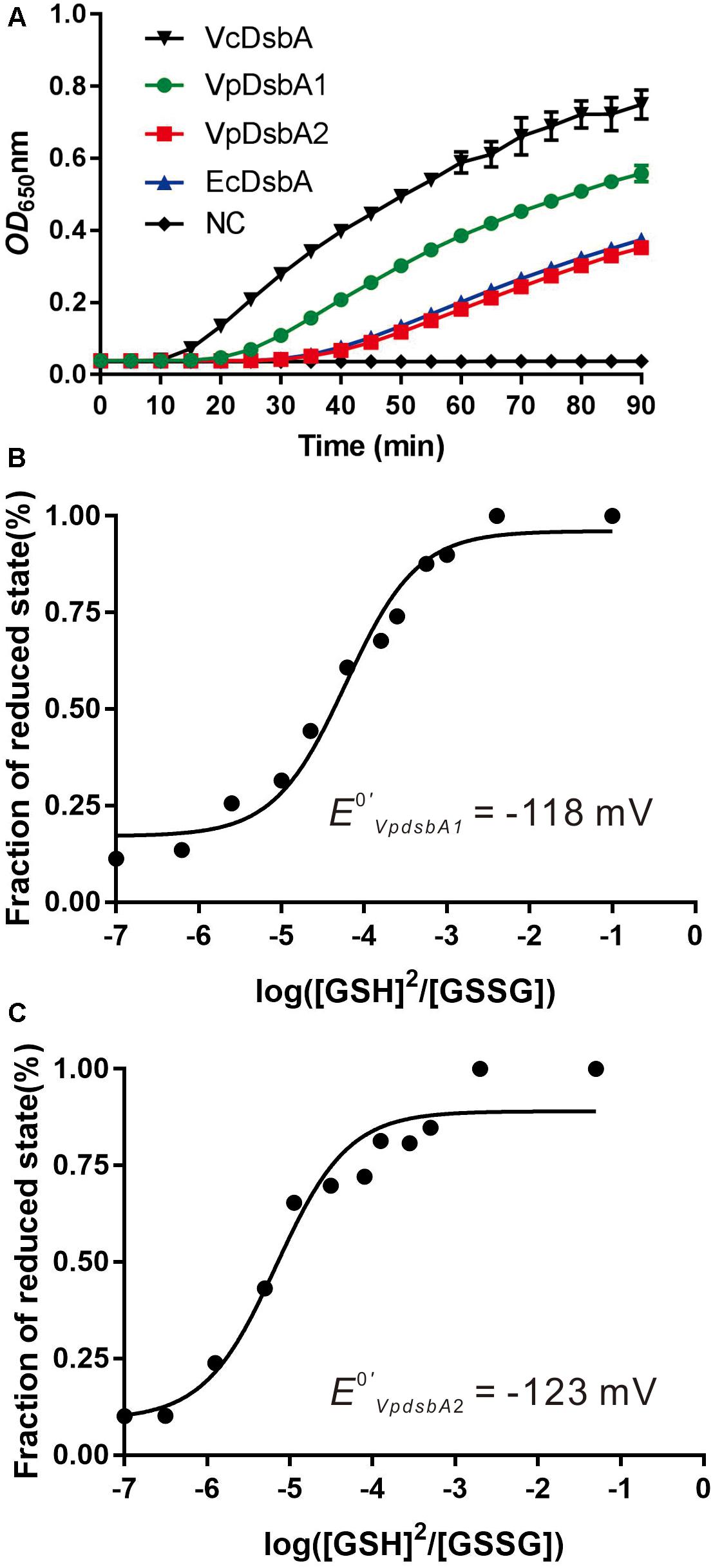
Figure 2. Reductase activity and redox potential of VpDsbAs. (A) Insulin (170 μM) and 0.33 mM DTT was incubated with 10 μM of each DsbA protein. The reduction of insulin was measured by monitoring the increase in absorbance at OD650. Reaction without reductase protein is used as negative control (NC). (B,C) Characterization of the redox potential of VpDsbA1 (B) and VpDsbA2 (C). Non-linear fit to the fraction of reduced VpDsbA at different ratios of reduced:oxidized glutathione. This fit was used to obtain the equilibrium constant Keq and the redox potential [calculated relative to the GSH/GSSG standard potential of –240 mV (Gilbert, 1995)]. The curve was fit to the averaged data from three replicate experiments.
To better characterize the redox properties of VpDsbA1 and VpDsbA2, we determined the standard redox potential of these two proteins relative to the redox potential of GSH/GSSG (−240 mV) (Gilbert, 1995). By calculating the fraction of reduced VpDsbA at different concentrations of [GSH]2/[GSSG], the equilibrium constants were determined (Figures 2B,C). The Keq for VpDsbA1 was 8.4+0.06 × 10−5 M and VpDsbA2 was 1.2+0.03 × 10−4 M. The corresponding redox potentials of VpDsbA1 and VpDsbA2 are −118 mV and −123 mV rat pH 7.0. Thus, VpDsbA1 is relatively oxidizing, with a redox-potential value similar to that of the thioredoxin-fold proteins, VcDsbA (E0′ = −116 mV) (Walden et al., 2012) and the redox-potential value of VpDsbA2 is similar to that of VcDsbA (E0′ = −122 mV) (Huber-Wunderlich and Glockshuber, 1998).
VpDsbAs Function as Oxidases in V. parahaemolyticus
It was previously reported that E. coli ΔdsbA showed cadmium sensitivity due to the high affinity of Cd2+ for protein free thiols (Vallee and Ulmer, 1972). We tested V. parahaemolyticus dsbA mutant cadmium sensitivity as an indicator of oxidase capacity. V. parahaemolyticus WT and mutants were grown in the presence of increasing concentrations of cadmium (0–1.25 mM). The wild type strain of V. parahaemolyticus grew on concentrations of up to 1 mM cadmium, but the ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 mutants were cadmium-sensitive (Figure 3A and Table 3). These results suggest that both VpDsbA1 and VpDsbA2 possess oxidase activity.
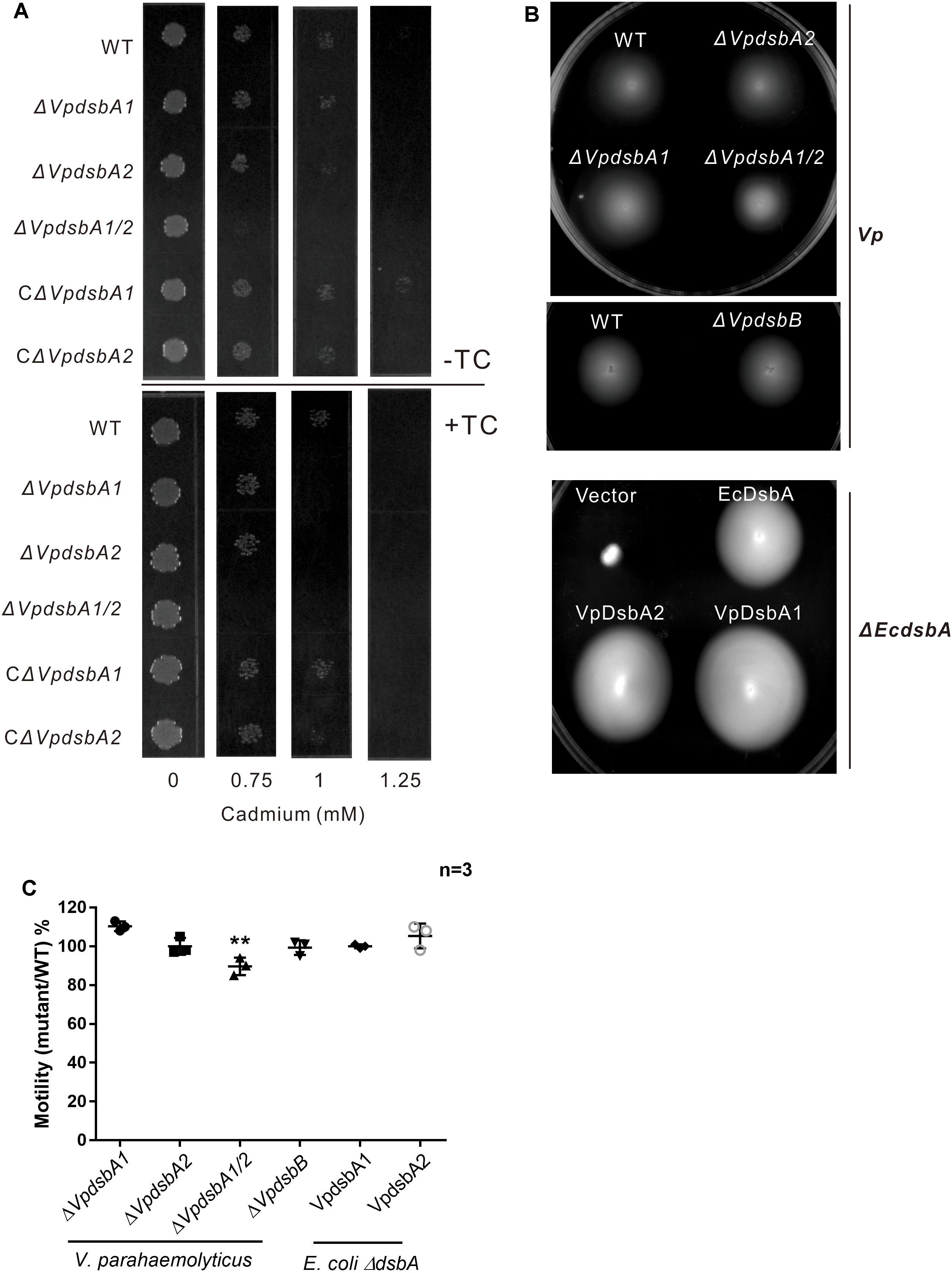
Figure 3. VpDsbA in vivo activity assay. (A) V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 strains were tested in cadmium sensitivity assays by spotting 103 cfu of each bacterial strain onto LB-NaCl containing increasing concentrations of cadmium (0–1.25 mM) in the presence or absence of 1 mM TC. (B) Motility assay of V. parahaemolyticus dsb mutants and E. coli ΔEcdsbA complemented with either EcdsbA, VpDsbA1, or VpDsbA2. (C) Quantification of the diameter of the motility circle of each strain shown in panel (B) was performed using ImageJ software. Graph represents percentages of each V. parahaemolyticus dsb mutant versus WT strain or E. coli ΔEcdsbA complemented with either VpDsbA1 or VpDsbA2 versus that with EcdsbA. Statistical analysis was calculated by one-way ANOVA. n = 3. ∗∗P < 0.001.
E. coli ΔdsbA or ΔdsbB were reported to be defective in motility because they fail to assemble flagella due to a lack of disulfide bonds in the P-ring protein (FlgI) (Dailey and Berg, 1993). However, we did not observe the same motility defect in V. parahaemolyticus dsb mutants (Figures 3B,C). To test if VpDsbA1 and VpDsbA2 can rescue this defect in E. coli ΔdsbA, we introduced VpdsbA1 or VpdsbA2 on plasmids with a low copy number replication origin under the control of the PBAD promoter in E. coli ΔdsbA. Both VpDsbA1 and VpDsbA2 were able to restore the motility of E. coli ΔdsbA (Figures 3B,C).
Bile Salts Repress VpDsbAs Activity
When V. parahaemolyticus enters into the human body through the consumption of contaminated water or uncooked food, a set of virulence determinants controlled by a regulatory network are produced in response to the chemical signals present in the small intestine where it normally causes disease (Rivera-Cancel and Orth, 2017). V. parahaemolyticus uses bile salts to regulate virulence factor expression (Gotoh et al., 2010; Li et al., 2016). Bile salts also induce virulence gene expression in V. cholerae by interfering with the redox reaction of DsbA proteins (Xue et al., 2016). We first investigated the interaction between the bile salt TC and VpDsbA1 and VpDsbA2 by using ITC. We found that both VpDsbA1 and VpDsbA2 can bind TC, with a KD of 131 ± 7 μM and 164 ± 11 μM, respectively (Figure 4A). We also tested TrxA from E. coli binding to TC by ITC, but EcTrxA and TC did not show any specific interaction (Supplementary Figure S2) which indicated that the binding of VpDsbA1 or VpDsbA2 with TC is a specific interaction event.
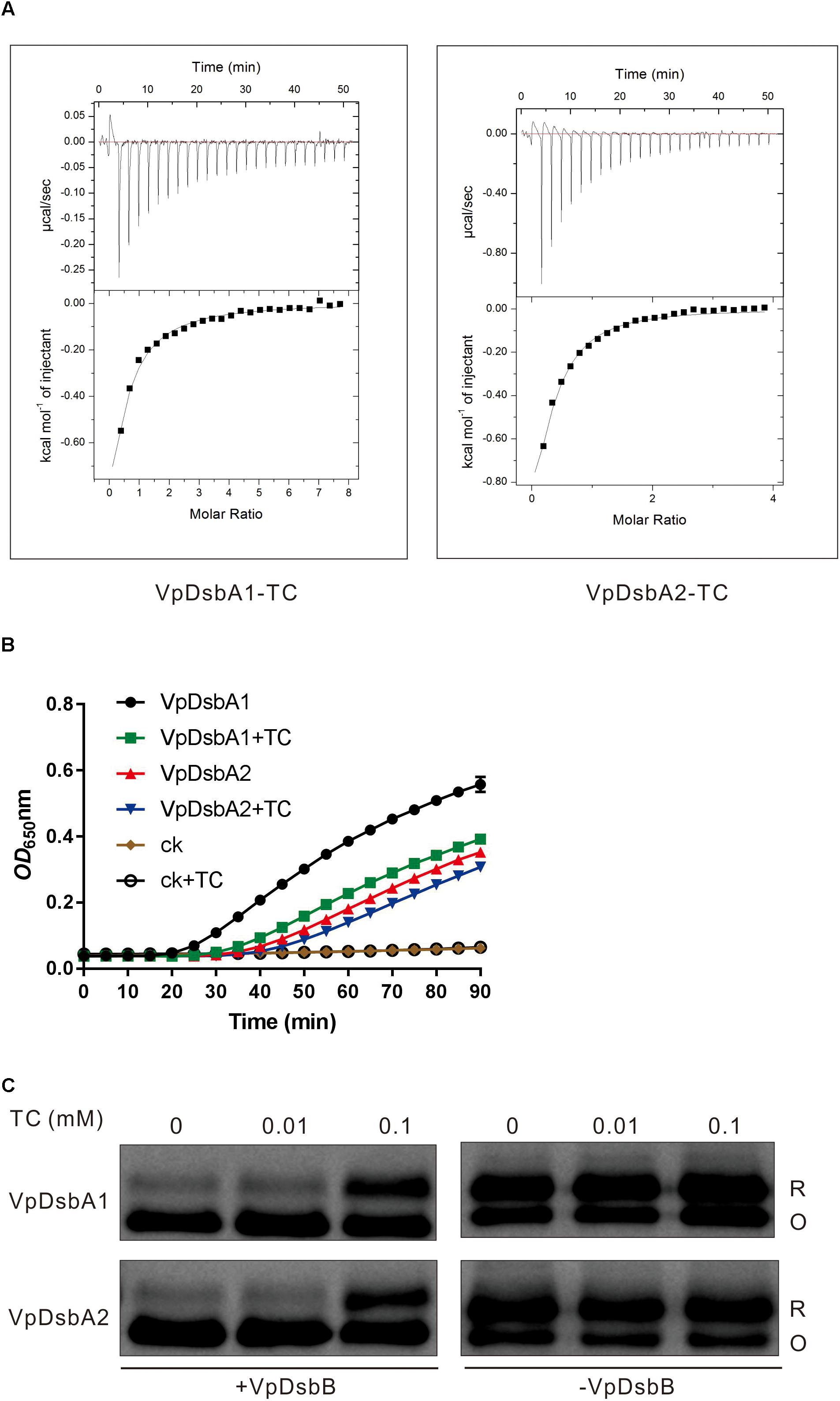
Figure 4. Bile salts inhibit VpDsbAs activity. (A) Bile salts interact with VpDsbA. ITC data titrating 4 mM of TC into 0.2 mM of tested proteins, VpDsbA1 (left) and VcDsbA2 (right). (B) Insulin reduction by VpDsbA1 or VpDsbA2 was inhibited in the presence of TC. Insulin (170 μM) and 0.33 mM DTT was incubated with 10 μM of VpDsbA, and 0.1 mM of TC were added at the beginning of reaction. The reduction of insulin was measured by monitoring the increase in absorbance at OD650. (C) Reduced VpDsbA oxidized by VpDsbB present in the membranes in vitro. Reduced VpDsbA (2 μM) was incubated with or without VpDsbB containing membranes and trapped with AMS after various incubation times. After incubation with VpDsbB membrane, VpDsbA (O) shifts to a lower molecular weight band. VpDsbA was detected using Western blot with an anti-VpDsbA antibody.
Insulin reduction mediated by either VpDsbA1 or VpDsbA2 was inhibited by TC (Figure 4B). The cadmium sensitivity assay showed that V. parahaemolyticus was more sensitive to cadmium in the presence, than in the absence of TC (Figure 3A and Table 3), indicating that TC might reduce the efficiency of disulfide bond formation catalyzed by VpDsbAs in the periplasm of V. parahaemolyticus. DsbA reductase activity correlates with DsbA reoxidation by DsbB (Ren et al., 2009). To test if VpDsbA activity inhibited by TC in vivo is because TC interferes with the reaction between VpDsbA and VpDsbB, we studied the oxidization of VpDsbA by VpDsbB in vitro. The oxidization of both VpDsbA1 and VpDsbA2 by the membrane protein VpDsbB was inhibited by TC in vitro (Figure 4C).
VpDsbAs Impact V. parahaemolyticus Pathogenesis
Vibrio parahaemolyticus has evolved several regulatory networks that control the production of a wide range of virulence factors which enables them to cause disease in the host, including the thermostable direct hemolysin (tdh) and TDH related hemolysin (trh), adhesin, and secreted effectors (Zhang and Orth, 2013; Liu and Chen, 2015). We found that V. parahaemolyticusΔVpdsbA1/2 exhibits reduced virulence in the zebrafish infection model (Figure 1C). To understand why, we first investigated the effect of VpDsbAs on host cell adhesion (Liu and Chen, 2015). Deleting either VpdsbA1 or VpdsbA2 decreased V. parahaemolyticus attachment to Caco-2 epithelial cells (Figure 5A). Several adhesion factors present at the surface of V. parahaemolyticus have been reported to be important in host cell binding, including MAM7 (Multivalent Adhesion Molecule 7) (Krachler et al., 2011), MSHA pilus (O’Boyle et al., 2013), PilA (Shime-Hattori et al., 2006), and VpadF (Liu and Chen, 2015). To determine whether VpDsbAs are involved in maintaining the stability of these adhesion factors, we quantified the abundance of MAM7 (VP1611), MshA (VP2697), PilA (VP2523), and VpadF (VP1767). The abundance of VpadF, but not MAM7 was significantly reduced in the ΔVpdsbA1/2 mutant but not in ΔVpdsbA1 or ΔVpdsbA2 single knock-out mutant (Figure 5B) which indicates that either one of these two VpDsbA proteins is necessary for VpadF stability. The VpDsbAs had no effect on the transcription levels of all these adhesion factors (Figure 5B and Supplementary Figure S3). These results indicate that the stability of VpadF depends on a functional DsbA protein present in the periplasm of V. parahaemolyticus which plays an important role in V. parahaemolyticus adhesion to epithelia cells.
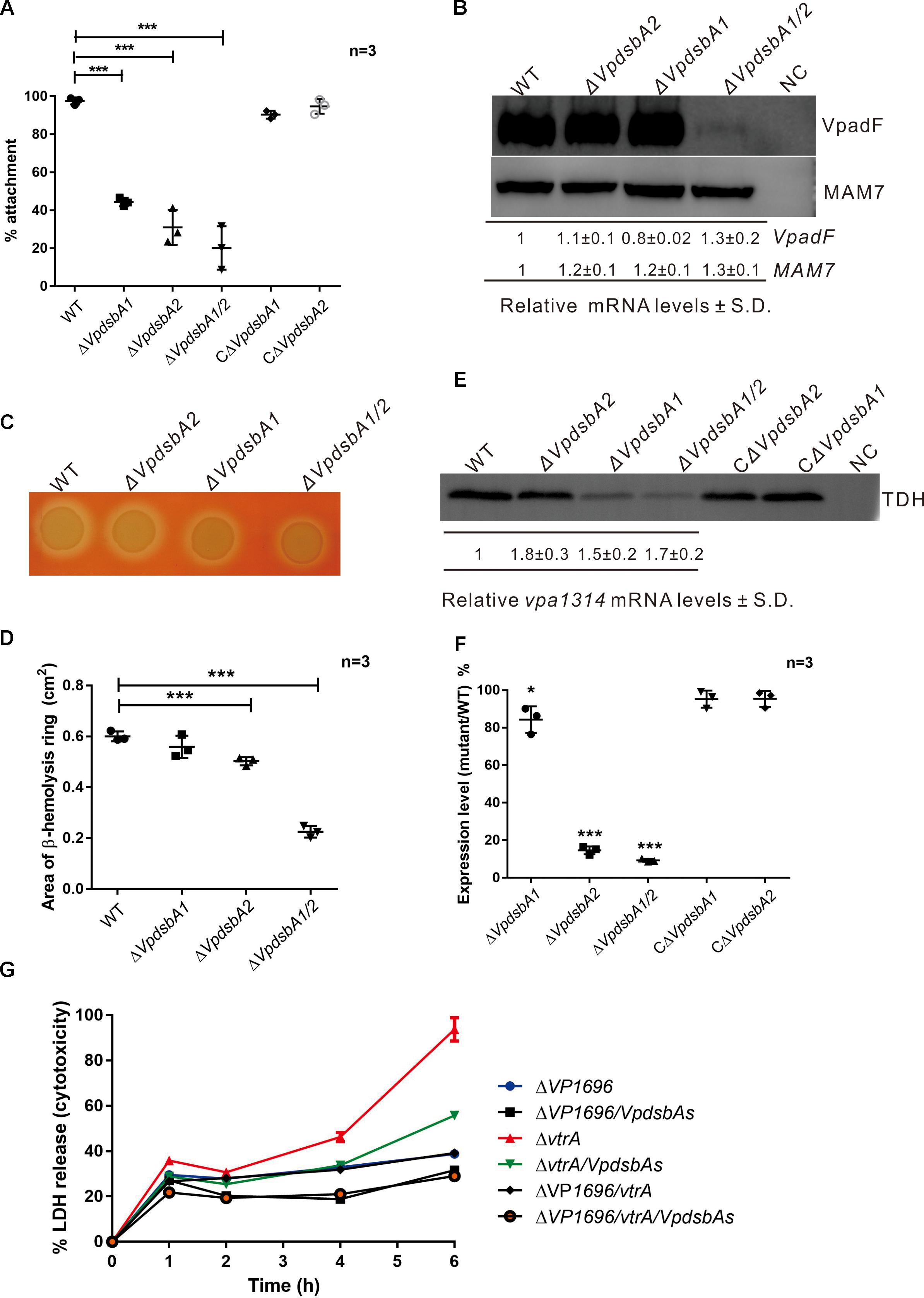
Figure 5. VpDsbAs is required in pathogenesis of V. parahaemolyticus. (A) Adhesion of V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 and the complement strains to Caco-2 cells. Statistical analysis was calculated by one-way ANOVA. n = 3. ∗∗∗P < 0.001. (B) Top, analysis of VpadF and MAM7 protein levels. V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 containing PBAD-VpadF-cFLAG or PBAD-MAM7-cFLAG were grown in LB-NaCl until OD600≈0.8. Cell lysates (1 mg) were separated by SDS-PAGE and VpadF or MAM7 was detected by the Western blot using anti-FLAG monoclonal antibody (Sigma-Aldrich, United States). Blot shown is representative of at least three separate experiments. Bottom, analysis of VpadF and MAM7 mRNA levels by qRT-PCR. RNA was purified from freshly prepared cultures grown in LB-NaCl. The relative mRNA levels ± SD were normalized to 16S RNA. (C) Hemolysis activity of V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 strains on Wagatsuma blood agar. (D) Quantification of area of the hemolysis circle of each strain shown in panel (B) was performed using ImageJ software. Statistical analysis was calculated by one-way ANOVA. n = 3. ∗∗∗P < 0.001. (E) Top, analysis of TDH protein level. V. parahaemolyticus WT, ΔVpdsbA1, ΔVpdsbA2, and ΔVpdsbA1/2 and the complement strains were kept growing in LB-NaCl containing 1 mM of taurodeoxycholate acid (TDCA) at 37°C until 10% (vol/vol) trichloroacetic acid (TCA) was added. Protein samples were suspended in a buffered solution containing 100 mM Tris-HCl pH 7.5 and 1% (wt/vol) SDS and 1 mg of each sample was separated by SDS-PAGE and TDH was detected by the Western blot using anti-TDH antibody. Blot shown is representative of at least three separate experiments. Bottom, analysis of vpa1314 mRNA levels by qRT-PCR. RNA was purified from freshly prepared cultures grown in LB-NaCl containing 1 mM of TDCA. The relative mRNA levels ± SD were normalized to 16S RNA compared with that of WT. (F) Quantification of band intensities from blot shown in panel (E) was performed using ImageJ software. Graph represents percentages of TDH compared with that of WT. Protein expression levels were normalized to that of WT. Data shown are averages of three independent experiments. Statistical analysis was calculated by one-way ANOVA. n = 3. ∗P < 0.05, ∗∗∗P < 0.001. (G) Cytotoxicity assay of V. parahaemolyticus mutant strains against HeLa cells by detecting the release of LDH into the medium at each indicated time. Parameters reported include the mean ± SD across three replicates.
Vibrio parahaemolyticus strains isolated from clinical samples are able to lyse human erythrocytes when plated on a high-salt media called Wagatsuma agar, a process termed the Kanagawa (KP) test (Nishibuchi and Kaper, 1995). To determine whether VpDsbAs contribute to the β-hemolytic activity of V. parahaemolyticus, we performed the KP test with WT and the dsbA mutants. We found that the β-hemolytic activity significantly decreased when both VpdsbA genes are deleted (Figures 5C,D). TDH is the major toxin that contributes to the β-hemolytic activity of V. parahaemolyticus (Honda et al., 1988; Nishibuchi et al., 1992). The mRNA level of TDH was not affected by VpDsbAs (Figures 5E,F). However, the amount of TDH protein decreased significantly in the ΔVpdsbA1 and ΔVpdsbA1/2 mutants (Figures 5E,F), suggesting that VpDsbA1 plays a major role in maintaining TDH stability.
Vibrio parahaemolyticus harbors two type III secretion systems (T3SS) encoded in chromosomes 1 (T3SS1) and 2 (T3SS2) (Makino et al., 2003). T3SS1 is responsible for cytotoxicity and T3SS2 is primarily involved in enterotoxicity, as well as in cytotoxic activity against some specific cell lines (Hiyoshi et al., 2010). To test whether VpDsbAs affect cytotoxicity of V. parahaemolyticus against HeLa cells, HeLa cell lysis was measured by monitoring the release of LDH after infection with V. parahaemolyticus tdhAs and T3SS deletion mutant strains in the presence or absence of VpDsbAs. VtrA is the master regulator of T3SS2 in V. parahaemolyticus, so T3SS2 will stop working with vtrA deletion (Kodama et al., 2010); and T3SS1 will lose function when the inner membrane protein VP1696 is deleted from the genome (Park et al., 2004). Consistent with previous reports (Park et al., 2004; Hiyoshi et al., 2010), T3SS1 of V. parahaemolyticus works dominantly in cytotoxicity of HeLa cells (Figure 5G). After 6 h infection, HeLa cells were nearly completely lysed by V. parahaemolyticus T3SS2 mutant strains containing a functional T3SS1, but strains without VpDsbAs showed ∼40% less lysis (Figure 5G). This suggests that VpDsbAs play an important role in epithelial cell infection through T3SS1. By assessing the transcription level and the secretion level of some major effectors and the activators of T3SS1, we found that VpDsbAs do not have a significant effect on the transcription of genes involved in T3SS1 activity (Supplementary Figure S4). However, the secretion efficiency of VPA0450 decreased in dsbA mutant compared with that in WT strain, while VP1683 was secreted more efficiently in the mutant strain under the conditions that we tested (Supplementary Figure S5). We did not figure out what’s the mechanism that VpDsbAs work differently in the secretion of these two effectors, yet, this strongly suggests that VpDsbAs affect T3SS1 functions at the post-translational level.
Discussion
DsbA participates in protein folding by introducing disulfide bonds into proteins secreted to the periplasm (Hu et al., 1997; Heras et al., 2009). In this study, we characterized the redox properties of two DsbA proteins from V. parahaemolyticus and investigated their essential role for several important virulence factors that affect V. parahaemolyticus pathogenesis.
Two DsbA genes, VpDsbA1 and VpDsbA2, are encoded on V. parahaemolyticus chromosomes 1 and 2, respectively. Both genes share high sequence similarity with that of VcDsbA and EcDsbA and contain the classical CXXC active-site motif and cisPro motif (Figure 1A), catalyzed insulin reduction in vitro (Figure 2A), and introduced disulfide bonds to the proteins secreted to the periplasm of V. parahaemolyticus in vivo (Figure 3A). Unlike E. coli or V. cholerae, the motility of which is mainly driven by one set of flagellar system, V. parahaemolyticus possesses dual flagellar systems, a single polar flagellum which propels the bacterium in liquid (swimming) and the lateral flagella which drive the bacterium move on the surface (swarming) (McCarter and Silverman, 1990; McCarter, 2004). V. parahaemolyticus dsb mutant strains were still motile (Figure 3B). We speculate that if the polar flagellum stops working, which might be the case in the dsb mutants, the lateral flagella will be activated and enable V. parahaemolyticus to remain motile (McCarter et al., 1988; McCarter and Silverman, 1989).
Like V. cholerae which activates virulence production by sensing bile salts (Yang et al., 2013; Xue et al., 2016), V. parahaemolyticus also hijacks bile salts as an intestinal signal to regulate virulence production (Gotoh et al., 2010). V. parahaemolyticus activates T3SS2 gene expression by sensing bile salts taurodeoxycholate acid (TDCA; Rivera-Cancel and Orth, 2017). Here, we found that both VpDsbA1 and VpDsbA2 can bind TC and TC repressed disulfide bond formation in the periplasm of V. parahaemolyticus by inhibiting the reoxidation of VpDsbA1 or VpDsbA2 by VpDsbB (Figure 4). Bile salts act as a stressor to bacteria that transit the intestinal tract, so it is reasonable to speculate that bile salts might regulate V. parahaemolyticus bile resistance by affecting the activity of the Dsb system.
This study also describes the essential role of the two DsbA proteins for several important virulence factors that affect V. parahaemolyticus pathogenesis. Both VpDsbA1 and VpDsbA2 are transcribed and expressed in V. parahaemolyticus under the conditions that we tested. V. parahaemolyticus dsbA mutants, especially the double mutant, showed defects in attachment to Caco-2 cells, β-hemolytic activity, and cytotoxicity against HeLa cells (Figure 5). All the mutants of ΔVpdsbA1,ΔVpdsbA2, and ΔVpdsbA1/2 showed defects in attachment to Caco-2 cells compared with that of the wild type strain (Figure 5A), however, the amount of adhesion factor VpadF decreased dramatically only in ΔVpdsbA1/2 mutant and either VpDsbA1 or VpDsbA2 protein alone was sufficient for VpadF stability (Figure 5B). All the other adhesion factors that we tested in this study are not affected by VpDsbA1 or VpDsbA2. This might indicate that some other unknown adhesion factors which are required VpDsbAs to fold correctly also contribute to the adhesion of V. parahaemolyticus to mammal cells. Compared to VpDsbA2, VpDsbA1 played a more important role in affecting the stability of virulence factor TDH (Figure 5D) in V. parahaemolyticus. However, TDH can be oxidized by either VpDsbA1 or VpDsbA2 in vitro (Supplementary Figure S6). So VpDsbA1 and VpDsbA2 might work differently in maintaining TDH protein stability in vivo.
Vibrio parahaemolyticus dsbA mutant also showed dramatically reduced cytotoxicity against HeLa cells in the T3SS2 knock-out background (containing functional T3SS1) (Figure 5E). Although the molecular mechanism by which VpDsbAs affect T3SS1 is not clear yet, it is reasonable to assume that the catalytic activity of VpDsbA to form a disulfide bond for the functional folding of various proteins is required for its virulence. V. parahaemolyticus T3SS1 is similar to the Ysc secretion system in Yersinia (Troisfontaines and Cornelis, 2005). Yersinia pestis DsbA is required for the formation of a ring-shaped structure in a type III secretion apparatus to secrete virulence effectors (Jackson and Plano, 1999). Like Yersinia, the DsbA proteins of V. parahaemolyticus might also be required for the formation of the type III secretion apparatus. VcDsbA from V. cholerae was reported to be indispensible for the pathogenesis of this organism. VcDsbA is required for the functional maturation of secreted virulence factors in V. cholerae (Peek and Taylor, 1992; Hu et al., 1997). VcDsbA is also essential for the homodimerization of TcpP to activate virulence gene expression in V. cholerae (Yang et al., 2013; Xue et al., 2016). VtrA, which is the master regulator of T3SS2 genes in V. parahaemolyticus, adopts the same topology and function as TcpP (Rivera-Cancel and Orth, 2017). The dsbA mutants did not show a defect in the cell invasion assay; on the contrary, the efficiency of invasion to HeLa cells was increased without VpDsbA (Supplementary Figure S7). We have checked the mRNA level of some T3SS2 relevant genes, vpa1321, vpa1327, vpa1332, vpa1346, vpa1348 and vpa1370 all of which have been reported to be important for T3SS2 function (Broberg et al., 2011; Kodama et al., 2015). We found that in the presence of TDCA, the mRNA level of all these tested genes in dsbA mutant did not show much difference from that of WT (Supplementary Figure S8). All these indicate that VpDsbAs might somehow affect T3SS2 function through an unknown mechanism.
Overall, this study describes the roles of DsbA in host cell adhesion, cytotoxicity, and β-hemolytic activity, which suggests that VpDsbAs could be a potential target for the development of antibacterial compounds to control V. parahaemolyticus infections.
Data Availability
This manuscript contains previously unpublished data. The name of the repository and accession number are not available.
Ethics Statement
All procedures performed in studies involving animals were in accordance with the ethical standards of the Institutional Animal Care and Use Committee of Zhejiang A&F University at which the studies were conducted (Permit Number: ZJAFU/IACUC_2011-10-25-02).
Author Contributions
C-qW, TZ, WZ, MS, FT, AY, and ML performed the research (the acquisition, analysis, or interpretation of the data). C-qW, TZ, WZ, and MY analyzed the data. MY designed the research and wrote the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31470244 and 31770151) and by the Science Development Foundation of Zhejiang A&F University (Grant No. 2013FR012) to MY, and also supported by Wenzhou Technology Innovation Platform Structure Foundation (Grant No. GC201701) to C-qW. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01103/full#supplementary-material
Footnotes
References
Bardwell, J. C., McGovern, K., and Beckwith, J. (1991). Identification of a protein required for disulfide bond formation in vivo. Cell 67, 581–589. doi: 10.1016/0092-8674(91)90532-4
Berkmen, M., Boyd, D., and Beckwith, J. (2005). The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J. Biol. Chem. 280, 11387–11394. doi: 10.1074/jbc.M411774200
Braun, P., Ockhuijsen, C., Eppens, E., Koster, M., Bitter, W., and Tommassen, J. (2001). Maturation of Pseudomonas aeruginosa elastase. formation of the disulfide bonds. J. Biol. Chem. 276, 26030–26035. doi: 10.1074/jbc.M007122200
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes. Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156.
Chun, D., Chung, J. K., Tak, R., and Seol, S. Y. (1975). Nature of the Kanagawa phenomenon of Vibrio parahaemolyticus. Infect. Immun. 12, 81–87.
Dailey, F. E., and Berg, H. C. (1993). Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 1043–1047. doi: 10.1073/pnas.90.3.1043
Depuydt, M., Messens, J., and Collet, J. F. (2011). How proteins form disulfide bonds. Antioxid. Redox Signal. 15, 49–66. doi: 10.1089/ars.2010.3575
Gilbert, H. F. (1995). Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 251, 8–28. doi: 10.1016/0076-6879(95)51107-5
Gotoh, K., Kodama, T., Hiyoshi, H., Izutsu, K., Park, K. S., Dryselius, R., et al. (2010). Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 5:e13365. doi: 10.1371/journal.pone.0013365
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Ha, U. H., Wang, Y., and Jin, S. (2003). DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 71, 1590–1595. doi: 10.1128/iai.71.3.1590-1595.2003
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/s0022-2836(83)80284-8
Heras, B., Shouldice, S. R., Totsika, M., Scanlon, M. J., Schembri, M. A., and Martin, J. L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. doi: 10.1038/nrmicro2087
Hiyoshi, H., Kodama, T., Iida, T., and Honda, T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780. doi: 10.1128/IAI.01051-1059
Holmgren, A. (1979). Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632.
Honda, T., Ni, Y. X., and Miwatani, T. (1988). Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56, 961–965.
Hu, S. H., Peek, J. A., Rattigan, E., Taylor, R. K., and Martin, J. L. (1997). Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J. Mol. Biol. 268, 137–146. doi: 10.1006/jmbi.1997.0940
Huber-Wunderlich, M., and Glockshuber, R. (1998). A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold. Des. 3, 161–171. doi: 10.1016/S1359-0278(98)00024-28
Jackson, M. W., and Plano, G. V. (1999). DsbA is required for stable expression of outer membrane protein yscc and for efficient yop secretion in Yersinia pestis. J. Bacteriol. 181, 5126–5130.
Jiang, N., Tang, L., Xie, R., Li, Z., Burkinshaw, B., Liang, X., et al. (2018). Vibrio parahaemolyticus RhsP represents a widespread group of pro-effectors for type VI secretion systems. Nat. Commun. 9:3899. doi: 10.1038/s41467-018-06201-6205
Kadokura, H., and Beckwith, J. (2009). Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138, 1164–1173. doi: 10.1016/j.cell.2009.07.030
Kaniga, K., Delor, I., and Cornelis, G. R. (1991). A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141. doi: 10.1016/0378-1119(91)90599-7
Karimova, G., Pidoux, J., Ullmann, A., and Ladant, D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756. doi: 10.1073/pnas.95.10.5752
Kodama, T., Gotoh, K., Hiyoshi, H., Morita, M., Izutsu, K., Akeda, Y., et al. (2010). Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 5:e8678. doi: 10.1371/journal.pone.0008678
Kodama, T., Hiyoshi, H., Okada, R., Matsuda, S., Gotoh, K., and Iida, T. (2015). Regulation of Vibrio parahaemolyticus T3SS2 gene expression and function of T3SS2 effectors that modulate actin cytoskeleton. Cell. Microbiol. 17, 183–190. doi: 10.1111/cmi.12408
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Krachler, A. M., Ham, H., and Orth, K. (2011). Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc. Natl. Acad. Sci. U.S.A. 108, 11614–11619. doi: 10.1073/pnas.1102360108
Lefebvre, K. A., Tilton, S. C., Bammler, T. K., Beyer, R. P., Srinouanprachan, S., Stapleton, P. L., et al. (2009). Gene expression profiles in zebrafish brain after acute exposure to domoic acid at symptomatic and asymptomatic doses. Toxicol. Sci. 107, 65–77. doi: 10.1093/toxsci/kfn207
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, P., Rivera-Cancel, G., Kinch, L. N., Salomon, D., Tomchick, D. R., Grishin, N. V., et al. (2016). Bile salt receptor complex activates a pathogenic type III secretion system. Elife 5:e15718. doi: 10.7554/eLife.15718
Liu, M., and Chen, S. (2015). A novel adhesive factor contributing to the virulence of Vibrio parahaemolyticus. Sci. Rep. 5:14449. doi: 10.1038/srep14449
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-12651
Mariano, G., Monlezun, L., and Coulthurst, S. J. (2018). Dual role for DsbA in attacking and targeted bacterial cells during type vi secretion system-mediated competition. Cell Rep. 22, 774–785. doi: 10.1016/j.celrep.2017.12.075
McCarter, L., Hilmen, M., and Silverman, M. (1988). Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54, 345–351. doi: 10.1016/0092-8674(88)90197-3
McCarter, L., and Silverman, M. (1989). Iron regulation of swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 171, 731–736. doi: 10.1128/jb.171.2.731-736.1989
McCarter, L., and Silverman, M. (1990). Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4, 1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x
McCarter, L. L. (2004). Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7, 18–29. doi: 10.1159/000077866
McMahon, R. M., Premkumar, L., and Martin, J. L. (2014). Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim. Biophys. Acta 1844, 1391–1401. doi: 10.1016/j.bbapap.2014.01.013
Metcalf, W. W., Jiang, W., Daniels, L. L., Kim, S. K., Haldimann, A., and Wanner, B. L. (1996). Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13. doi: 10.1006/plas.1996.0001
Nishibuchi, M., Fasano, A., Russell, R. G., and Kaper, J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60, 3539–3545.
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099.
O’Boyle, N., Houeix, B., Kilcoyne, M., Joshi, L., and Boyd, A. (2013). The MSHA pilus of Vibrio parahaemolyticus has lectin functionality and enables TTSS-mediated pathogenicity. Int. J. Med. Microbiol. 303, 563–573. doi: 10.1016/j.ijmm.2013.07.010
Paranjpye, R. N., Myers, M. S., Yount, E. C., and Thompson, J. L. (2013). Zebrafish as a model for Vibrio parahaemolyticus virulence. Microbiology 159(Pt 12), 2605–2615. doi: 10.1099/mic.0.067637-67630
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Paxman, J. J., Borg, N. A., Horne, J., Thompson, P. E., Chin, Y., Sharma, P., et al. (2009). The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J. Biol. Chem. 284, 17835–17845. doi: 10.1074/jbc.M109.011502
Peek, J. A., and Taylor, R. K. (1992). Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci U S A 89, 6210–6214. doi: 10.1073/pnas.89.13.6210
Philippe, N., Alcaraz, J. P., Coursange, E., Geiselmann, J., and Schneider, D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. doi: 10.1016/j.plasmid.2004.02.003
Qin, A., Scott, D. W., Thompson, J. A., and Mann, B. J. (2009). Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77, 152–161. doi: 10.1128/IAI.01113-1118
Ren, G., Champion, M. M., and Huntley, J. F. (2014). Identification of disulfide bond isomerase substrates reveals bacterial virulence factors. Mol. Microbiol. 94, 926–944. doi: 10.1111/mmi.12808
Ren, G., Stephan, D., Xu, Z., Zheng, Y., Tang, D., Harrison, R. S., et al. (2009). Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J. Biol. Chem. 284, 10150–10159. doi: 10.1074/jbc.M809509200
Rivera-Cancel, G., and Orth, K. (2017). Biochemical basis for activation of virulence genes by bile salts in Vibrio parahaemolyticus. Gut Microbes 8,366–373. doi: 10.1080/19490976.2017.1287655
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Shime-Hattori, A., Iida, T., Arita, M., Park, K. S., Kodama, T., and Honda, T. (2006). Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264, 89–97. doi: 10.1111/j.1574-6968.2006.00438.x
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Tey, Y. H., Jong, K. J., Fen, S. Y., and Wong, H. C. (2015). Genetic variation in Vibrio parahaemolyticus isolated from the aquacultural environments. Lett. Appl. Microbiol. 60, 321–327. doi: 10.1111/lam.12372
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Totsika, M., Vagenas, D., Paxman, J. J., Wang, G., Dhouib, R., Sharma, P., et al. (2018). Inhibition of diverse DsbA enzymes in multi-DsbA encoding pathogens. Antioxid. Redox Signal. 29, 653–666. doi: 10.1089/ars.2017.7104
Troisfontaines, P., and Cornelis, G. R. (2005). Type III secretion: more systems than you think. Physiology 20, 326–339. doi: 10.1152/physiol.00011.2005
Vallee, B. L., and Ulmer, D. D. (1972). Biochemical effects of mercury, cadmium, and lead. Annu. Rev. Biochem. 41, 91–128. doi: 10.1146/annurev.bi.41.070172.000515
Walden, P. M., Heras, B., Chen, K. E., Halili, M. A., Rimmer, K., Sharma, P., et al. (2012). The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production. Acta Crystallogr. D Biol. Crystallogr. 68(Pt 10), 1290–1302. doi: 10.1107/S0907444912026388
Xiao, J., Liu, L., Ke, Y., Li, X., Liu, Y., Pan, Y., et al. (2017). Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci. Rep. 7:42177. doi: 10.1038/srep42177
Xue, Y., Tu, F., Shi, M., Wu, C. Q., Ren, G., Wang, X., et al. (2016). Redox pathway sensing bile salts activates virulence gene expression in Vibrio cholerae. Mol. Microbiol. 102, 909–924. doi: 10.1111/mmi.13497
Yang, M., Liu, Z., Hughes, C., Stern, A. M., Wang, H., Zhong, Z., et al. (2013). Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. U.S.A. 110, 2348–2353. doi: 10.1073/pnas.1218039110
Yeung, P. S., and Boor, K. J. (2004). Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 1, 74–88. doi: 10.1089/153531404323143594
Yu, Y., Fang, L., Zhang, Y., Sheng, H., and Fang, W. (2015). VgrG2 of type VI secretion system 2 of Vibrio parahaemolyticus induces autophagy in macrophages. Front. Microbiol. 6:168. doi: 10.3389/fmicb.2015.00168
Zhang, L., and Orth, K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16, 70–77. doi: 10.1016/j.mib.2013.02.002
Keywords: Vibrio parahaemolyticus, DsbA, reductase, virulence, pathogenesis
Citation: Wu C-q, Zhang T, Zhang W, Shi M, Tu F, Yu A, Li M and Yang M (2019) Two DsbA Proteins Are Important for Vibrio parahaemolyticus Pathogenesis. Front. Microbiol. 10:1103. doi: 10.3389/fmicb.2019.01103
Received: 20 February 2019; Accepted: 30 April 2019;
Published: 16 May 2019.
Edited by:
Yuji Morita, Meiji Pharmaceutical University, JapanReviewed by:
Rheinallt Jones, Emory University, United StatesTakashi Uebanso, Tokushima University, Japan
Begoña Heras, La Trobe University, Australia
Copyright © 2019 Wu, Zhang, Zhang, Shi, Tu, Yu, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Menghua Yang, yangmh@zafu.edu.cn
†These authors have contributed equally to this work
 Chun-qin Wu
Chun-qin Wu Ting Zhang1†
Ting Zhang1† Wenwen Zhang
Wenwen Zhang Menghua Yang
Menghua Yang